Background: Myofibroblasts are responsible for excessive collagen expression during scarring and fibrosis.
Results: Myocardin-related transcription factor-A (MRTF-A) potentiates collagen overproduction through transcription complexes containing Sp1 and SRF on noncanonical DNA elements.
Conclusion: MRTF-A promotes myofibroblast differentiation and collagen production through diverse molecular mechanisms.
Significance: MRTF-A may be a molecular target in developing strategies to impede fibrosis.
Keywords: Collagen, Co-regulator Transcription, Fibrosis, Myofibroblast, Sp1, MRTF-A, Serum-response Factor (SRF)
Abstract
Pulmonary fibrosis is characterized by the excessive deposition of a collagen-rich extracellular matrix. The accumulation of collagen within the lung interstitium leads to impaired respiratory function. Furthermore, smooth muscle actin-positive myofibroblasts within the fibrotic lung contribute to disease progression. Because collagen and smooth muscle cell α-actin are coordinately expressed in the setting of fibrosis, the hypothesis was tested that specific transcriptional regulators of the myocardin family might also regulate collagen gene expression in myofibroblasts. Myocardin-related transcription factors (MRTFs), through their interaction with the serum-response factor (SRF) on CArG box regulatory elements (CC(A/T)6GG), are important regulators of myofibroblast differentiation. MRTF-A transactivated type I collagen gene reporters as much as 100-fold in lung myofibroblasts. Loss of functional MRTF-A using either a dominant negative MRTF-A isoform, shRNA targeting MRTF-A, or genetic deletion of MRTF-A in lung fibroblasts significantly disrupted type I collagen synthesis relative to controls. Analysis of the COL1A2 proximal promoter revealed a noncanonical CArG box (CCAAACTTGG), flanked by several Sp1 sites important for MRTF-A activation. Chromatin immunoprecipitation experiments confirmed the co-localization of MRTF-A, SRF, and Sp1 bound to the same region of the COL1A2 promoter. Mutagenesis of either the noncanonical CArG box or the Sp1 sites significantly disrupted MRTF-A activation of COL1A2. Together, our findings show that MRTF-A is an important regulator of collagen synthesis in lung fibroblasts and exhibits a dependence on both SRF and Sp1 function to enhance collagen expression.
Introduction
The interstitial lung disease, idiopathic pulmonary fibrosis, is characterized by progressive and irreversible scarring of lung tissue (1, 2). The pathological hallmarks in fibrotic lung tissue include excessive extracellular matrix (ECM)6 deposition, notably fibrillar collagen, and myofibroblast expansion generating fibroblastic foci (3, 4). Resident in these foci are myofibroblasts, which express smooth muscle cell α-actin (SMA), vimentin, and desmin but not smooth muscle myosin heavy chain (3–5). Through expression of collagen and contractile proteins, myofibroblasts contribute significantly to fibrotic disease progression (3).
The triple helical type I collagen molecule is the most abundant up-regulated protein in fibrosis (6). Collagen type I consists of two chains of α1(I) and one chain of α2(I) encoded by separate genes (COL1A1 and COL1A2). The transcriptional regulation of the COL1A2 gene provides an informative model of combinatorial interactions among promoter-bound proteins at specific DNA sites (7). Previous work has identified several key positive and negative regulators of collagen transcription in the setting of fibrotic disease. The Sp1 family of proteins activates collagen transcription through G/C-rich sites (8, 9), whereas the Ets domain family of proteins both activates and represses collagen gene expression in fibroblasts (10, 11). These transcriptional regulators respond to pro- and anti-fibrotic growth factors and cytokines, including transforming growth factor β (TGFβ), endothelin-1, and interferon-γ (IFNγ) (5, 12, 13). For example, transforming growth factor β (TGFβ) acts via SMAD proteins that bind to the COL1A2 proximal promoter regions (14). TGFβ acts on the COL1A1 distal promoter (15), and IFNγ-dependent repressive mechanisms modulate collagen type I expression via CIITA and RFX complexes at the proximal promoter near the transcription start site (16–18).
Although collagen production is one characteristic of the myofibroblastic phenotype, these cells also express the contractile isoform of actin, SMA. In fibroblasts and vascular smooth muscle cells, SMA is regulated at the transcriptional level by the serum-response factor (SRF), and co-activators of the myocardin family are essential regulators of SMA (19, 20). Myocardin, a potent nuclear transcriptional co-activator expressed specifically in the cardiac and smooth muscle lineage, is required for expression of smooth muscle-specific gene expression (16, 21). The myocardin-related transcription factors MRTF-A (also called MKL1/MAL/BSAC) and MRTF-B (also called MKL2) are ubiquitously expressed proteins that respond to changes in actin dynamics leading to nuclear accumulation (22). Once in the nucleus, MRTF-A drives transcription of cytoskeleton genes, including SMA (23). Myocardin family members interact with SRF as homo- or heterodimers and stimulate transcription via conserved CArG box DNA elements (24–26). Signals of stress, mechanical force, and migration converge on the activation of Rho GTPases resulting in polymerization of the actin cytoskeleton into stress fibers thereby permitting nuclear translocation of MRTFs, which links actin dynamics with gene transcription (27–32).
Because collagen and SMA are coordinately expressed in the setting of fibrosis, the hypothesis that specific transcriptional regulators of the myocardin family might also regulate collagen gene expression in myofibroblasts was tested. While our study was in progress, it was recently demonstrated that MRTF-A regulates collagen synthesis in the context of cardiomyocytes (33). The results from our current study provide a significant mechanistic advance demonstrating that MRTF-A regulates collagen expression in fibroblasts through several regulatory sites. Building on these findings, we determined that the MRTF-A transcription factor complexes recruited to the collagen gene include a novel MRTF-A interaction with Sp1 that enhances collagen expression. This collagen regulatory complex differs from the MRTF-A complexes on the SMA promoter. These studies identified a novel mechanism where MRTF-A can coordinate collagen gene expression in SRF-independent and -dependent mechanisms in lung fibroblasts.
EXPERIMENTAL PROCEDURES
Cell Culture
Rat lung fibroblasts (RFL6) and IMR90 fetal human fibroblasts (population doublings between 10 and 40) were grown in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% fetal bovine serum (Atlanta Biologicals and Hyclone) and 1% penicillin/streptomycin and incubated in 5% CO2 at 37 °C. For mithramycin A experiments, cells were plated at 2 × 104 cells/cm2 overnight and treated for 24 h with 50 nm mithramycin A (Sigma) dissolved in DMSO.
Primary Mouse Lung Fibroblast Isolation and Culture
Mice, 3–6 weeks old, were euthanized by CO2 asphyxiation and perfused with 10 ml of PBS. Dissected lungs were minced into ∼1-mm pieces and suspended in 10 ml of digestion buffer (1× PBS with calcium and magnesium, 500 units/ml dispase (BD Biosciences), 10 μg/ml collagenase A (Sigma), 10 μg/ml DNase I (Qiagen)) at 37 °C for 1 h and established in culture as described previously (34). Cells were cultured in DMEM with 10% FBS and used between passages 2 and 6. The Boston University School of Medicine Institutional Animal Care and Use Committee approved the animal experiments.
Plasmids and Cloning
The −351 COL1A2, −224 COL1A2, and −311 COL1A1 luciferase reporter vectors are described previously (16, 17). Site-directed mutants and deletion constructs were generated by PCR using Pfu polymerase followed by DpnI digestion. Specific mutants generated include the following: CArG1a, CCAAACTTGG→AAGGCCTTGG; CArG1b, TTCCAAACTTGG→CCGGGCTTGG; CArG2, CCAATTTAAG→CGGATCCAAG; Sp1a, CCCTCCCCC→CCCTCAAAC; Sp1b, CCTCCC→GAATCCC. Expression vectors for MRTF-A, MRTF-B, and myocardin are as described previously (35). The plasmids pcDNA3-SRF and pcDNA3-DN-SRF were used as described previously (36). Site-directed mutagenesis was used to mutate SRF amino acids RVKI in the DNA binding domain to LVAG, shown to eliminate SRF binding to CArG box elements as described previously (37).
Gene Reporter Assays
IMR90 and RFL6 fibroblasts were transfected using Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Roche Applied Science), and reporter activity was measured as described previously (17). Drosophila D.Mel-2 cells (D.Mel-2) were cultured and transfected with Cellfectin (Invitrogen) as described previously (36).
SDS-PAGE and Western Blotting
Equal amounts of total protein (7–10 μg) were diluted in SDS-PAGE sample buffer and boiled. Protein samples were run on 4–12% Novex SDS-polyacrylamide gels (Invitrogen) and prepared for Western blot according to standard procedures. Antibodies used include type I collagen (Southern Biotech, 1:300), collagen α1(I), (Rockland, 1:1000), GAPDH (Santa Cruz Biotechnology, 1:200), SMA (Sigma, 1:5000), MRTF-A (Santa Cruz Biotechnology, 1:250), Sp1 (Millipore, 1:5000), γ-tubulin (Sigma, 1:500), SRF (Santa Cruz Biotechnology, 1:200), and FLAG (Sigma, 1:500). Blots were imaged and quantified using a Kodak 440 one-dimensional instant imager.
Collagen Secretion, Western Blot Assays
For detection of collagen secreted into the media, cultures were washed twice with PBS and incubated in 0.4% FBS media supplemented with 250 μm sodium ascorbate for the indicated time. Media were removed and treated with 2 mm EDTA, and proteins were precipitated with chloroform/methanol protein precipitation and dissolved in 2× sample buffer (38). The cell layer was lysed for total protein quantification. To normalize for cell density, media samples were loaded proportionally to whole cell lysate concentration and run on 7.5% SDS-polyacrylamide gels.
Sircol Assay for Total Secreted Collagen
For quantification of total collagen in the media, cell cultures were grown to equivalent densities (∼50%) in the presence of 250 μm ascorbate. Cells were then washed with PBS and cultured in media containing 0.4% FBS and 250 μm ascorbate for 24 h. Media were removed, and cell layers were harvested for total protein. Equivalent amounts of media (200 μl) were incubated with 5× volume of Sircol Assay buffer (Biocolour, UK) for 30 min followed by centrifugation at 15,000 × g for 10 min to pellet precipitated collagen. Supernatant was discarded, and pellets were dissolved in 300 μl of 0.2 m KOH for 10 min. Sircol dye was quantified by reading the absorbance (A520), and absolute values were calculated using the linear regression of absorbance values derived from a standard curve of rat tail collagen and expressed as micrograms of total collagen per μg of total protein from the cell layer.
Inducible Adenovirus Expression System
Doxycycline-controlled adenoviral constructs for MRTF-A and dominant negative (DN) MRTF-A were generated by cloning the appropriate C-terminal FLAG-tagged cDNAs in the TRE-Shuttle2 vector (Clontech). After ligating the linearized vectors to the Adeno-X DNA, the ligation product was transformed into bacteria, and correct clones were transfected into AD293 cells (Stratagene). Recombinant adenovirus was expanded and purified with the Vivapure AdenoPack 100 (Vivascience) or Fast-Trap adenovirus purification kits (Millipore) according to the manufacturer's instructions. Concentrated adenovirus titer was calculated using optical particle units/ml.
Quantitative Real Time PCR
RNA was isolated from cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Generation of cDNA was achieved using 500 ng of total mRNA and Superscript III reverse transcriptase kit (Invitrogen). Quantification of mRNA was carried out on an ABI 7300 instrument using TaqMan and SYBR probes as described previously (16, 34). To detect mRNA transcripts that were not spliced, hnRNA TaqMan probes were designed to amplify the 1st exon/intron border of the COL1A1 and COL1A2 genes (see supplemental Table 1). This has been shown to quantify newly transcribed RNA with similar results to the nuclear run-on assay (39). Samples were treated with 1500 Kunitz units of DNase I to digest genomic DNA contamination. Controls were performed without reverse transcriptase to measure any residual DNA contamination.
shRNA Lentivirus
Lentivirus preparation and infection were carried out as described previously (16). Lentiviral Mission shRNA constructs (Sigma) targeting MRTF-A were used. Lentivirus genomic RNA was purified from virus stocks and quantified by quantitative PCR detection of the LV2 amplicon using the linear regression of a standard curve from lentivirus plasmid (40). Two thousand (2 × 103) particles/cell were used for all lentivirus infections.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP experiments were carried out using the ChIP-IT Express kit (Active Motif) according to the manufacturer's instructions and as described previously (41). Antibodies used in ChIP assays include SRF, Sp1, and MRTF-A (Santa Cruz Biotechnology). Precipitated DNA was measured using quantitative PCR (primers in supplemental Table 2) and quantified using linear regression of serial dilutions of IMR90 genomic DNA, and quantifiable IgG detection was subtracted from sample values.
Statistical Analysis
Statistical tests for significance of mean values were performed using analysis of variance employing Scheffe's post hoc procedure with SPSS 11.0 statistical software (p ≤ 0.05 was considered statistically significant).
RESULTS
Myocardin-related Transcription Factors Activate Type I Collagen Promoter
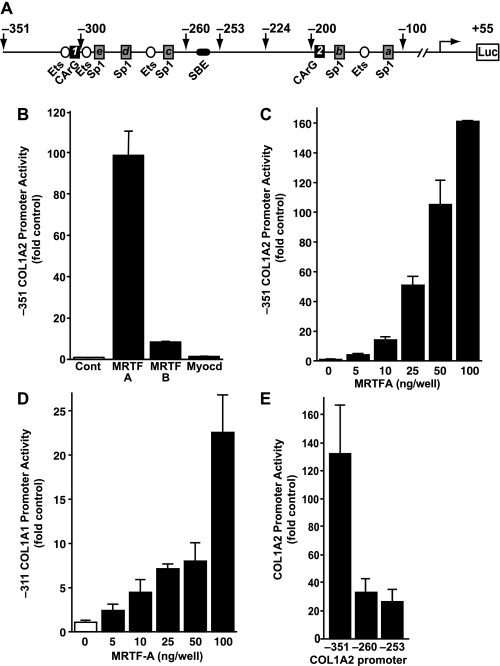
Myofibroblasts coordinately express ECM genes, including collagen and the contractile gene SMA (42). Because of the importance of SRF and its co-activators in regulating the contractile genes, the hypothesis that these factors could also regulate collagen expression was tested. The COL1A2 promoter has numerous positive transcription factor-binding sites, including Ets sites, Sp1 sites, and potential noncanonical CArG sequences (Fig. 1A). To study the effect of the MRTFs on collagen gene expression, rat lung fibroblasts (RFL6) were cultured to ∼50% confluence and transfected with equivalent amounts of MRTF-A, MRTF-B, or myocardin expression constructs in the presence of luciferase reporter plasmid containing −351/+55 of the COL1A2 proximal promoter. Relative to empty vector control, MRTF-A expression stimulated collagen promoter activity ∼100-fold (Fig. 1B). Although considerably less active than MRTF-A, MRTF-B stimulated the −351 gene reporter ∼10-fold, and myocardin did not stimulate collagen promoter activity (Fig. 1B). Collagen gene reporter activity was observed to significantly increase with as little as 10 ng of transfected MRTF-A plasmid (Fig. 1C, COL1A2, and Fig. 1D, COL1A1) and showed a dose-dependent response to increasing amounts of MRTF-A. In these assays, MRTF-A activated COL1A2 more than COL1A1.
FIGURE 1.
Myocardin-related transcription factors potently activate type I collagen promoters. A, type I collagen proximal promoter contains several evolutionarily conserved regulatory elements, including Sp1 (gray rectangles), Ets (white circles), and SMAD (SBE) (black oval). In addition, two potential SRF-binding CArG-like sequences were identified at −326 (CCAAACTTGG) and −193 (CCAATTTAAG) (black squares). B, rat lung fibroblasts (RFL6) were co-transfected with −351 COL1A2 promoter (100 ng/well) with the myocardin family expression constructs MRTF-A, MRTF-B, and myocardin (100 ng/well) with CMV-β to normalize for transfection efficiency. Normalized luciferase activity is expressed as fold of empty vector control. C, RFL6 cells were transfected as in A with increasing amounts of MRTF-A. D, RFL6 cells transfected with the −311 COL1A1 promoter and increasing concentrations of MRTF-A. E, RFL6 fibroblasts were transiently transfected with (100 ng) MRTF-A and a deletion set of COL1A2 luciferase reporter constructs. The activity of the various truncations was assessed relative to the wild type construct (−351/+55 bp). Results are representative of three independent experiments each performed in triplicate.
To identify the collagen promoter region mediating MRTF-A transactivation, 5′ truncation constructs of the −351 construct were generated that eliminated multiple Sp1 sites (−260) or both the Sp1 and SMAD elements (SBE) (−253). MRTF-A activation was decreased to about 30-fold of empty vector control in both the −260 and −253 deletion constructs (Fig. 1E) and was similar in magnitude to the COL1A1 promoter. These results indicate that potent MRTF-A-responsive elements exist both within the −351 to −260 enhancer-like region and in the proximal promoter region.
Knockdown of MRTF-A Decreases Collagen Synthesis
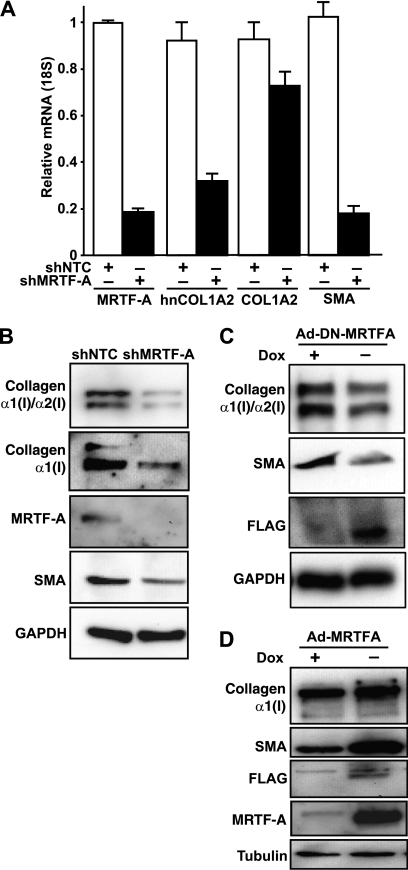
To determine whether MRTF-A is required for collagen mRNA expression and protein production, IMR90 myofibroblasts were infected with lentiviral shRNA for 48 h to reduce MRTF-A expression. Cells infected with shMRTF-A showed a >80% decrease in MRTF-A mRNA levels compared with control, confirming an efficient knockdown (Fig. 2A). Because type I collagens have long half-lives (43), we measured levels of newly synthesized hnCOL1A2 by quantitative PCR. Importantly, COL1A2 hnRNA was decreased significantly following the reduction of MRTF-A (>50%) compared with nontargeting control-treated cells (Fig. 2A). At this time point, steady-state COL1A2 mRNA was decreased by ∼20%. Furthermore, SMA transcript was decreased ∼80% in shMRTF-A-infected cells relative to controls. These results indicate that loss of MRTF-A can reduce both collagen and SMA mRNA levels.
FIGURE 2.
MRTF-A regulates synthesis of type I collagen in human lung fibroblasts. A, IMR90 lung fibroblasts transiently infected for 48 h with lentivirus encoding control shRNA (shNTC, white bars) or shRNA targeting MRTF-A (shMRTF-A, black bars). Message levels for MRTF-A, hnCOL1A2, COL1A2, and SMA were measured by qRT-PCR and normalized to those measured in shNTC-infected cells. B, Western blot analysis was performed on total cell lysates prepared from IMR90 cells infected with lentivirus. Lysates were analyzed for collagen type I (Southern Biotech), collagen α1(I) (Rockland), MRTF-A, and SMA and normalized to GAPDH. C, IMR90 cells were infected with doxycycline (Dox) controlled adenoviral vectors for dominant negative MRTF-A (Ad-DN-MRTF-A) as described under “Experimental Procedures” for 48 h. Cells were treated with (+) or without doxycycline (−) to control DN-MRTF-A expression and subjected to Western blot analysis for collagen type I (Southern Biotech), SMA, FLAG, and normalized to GAPDH. D, IMR90 cells were infected with doxycycline controlled adenoviral vectors for MRTF-A (Ad-MRTF-A) as described under “Experimental Procedures” for 48 h. Cells were treated with (+) or without doxycycline (−) to control MRTF-A expression and subjected to Western blot analysis for collagen α1(I) (Rockland), SMA, FLAG, MRTF-A, and normalized to tubulin. Results are representative of three independent experiments.
Consistent with the mRNA findings, cell extracts treated with shMRTF-A showed efficient knockdown of MRTF-A protein compared with shNTC samples (Fig. 2B). As anticipated, knockdown of MRTF-A resulted in a reduction in SMA protein levels. An antibody (Southern Biotech) was used to detect both type I collagen chains (Fig. 2B, top panel). To determine whether the α1(I) chain was increased, an antibody (Rockland) that specifically recognizes the pro-α1(I) chain, the mature α1(I) chain, and the heterotrimer of type I collagen was used (Fig. 2B, 2nd panel) (44). This antibody does not recognize the α2(I) chain and does not cross-react with other collagens. Importantly, compared with controls, knockdown of MRTF-A significantly reduced collagen protein levels 52% (p < 0.05) based on the Western blot quantification from three independent experiments.
Gain- and Loss-of-Function MRTF-A Alters Collagen Expression in Lung Fibroblasts
To investigate the effect of functional MRTF-A on collagen synthesis, the full-length MRTF-A cDNA and a C-terminally truncated cDNA lacking the transcriptional activation domain, which acts as a dominant negative isoform (DN-MRTF-A) (45), were cloned into a Tet-Off tetracycline-inducible adenovirus expression system. Both MRTF-A constructs were designed to contain a C-terminal FLAG tag. IMR90 cells were infected with Ad-DN-MRTF-A or Ad-MRTF-A constructs along with the rtTA adenovirus (Clontech) and were treated with or without 250 nm doxycycline to regulate MRTF-A expression. Immunostaining of cells for FLAG epitope showed nuclear localization in ∼75% of cells infected with MRTF-A and >90% of cells infected for DN-MRTF-A without doxycycline (data not shown). These experiments confirmed the efficacy of this infection procedure. Cells were analyzed for protein expression after 48 h. DN-MRTF-A expression (Fig. 2C) resulted in a 35% (p < 0.05, n = 3) decrease in collagen protein levels similar to the shRNA experiment. MRTF-A was overexpressed in cells in the absence of doxycycline compared with cells with doxycycline treatment as determined by anti-FLAG and total MRTF-A Western blotting (Fig. 2D). Collagen was increased as a result of ectopic MRTF-A expression (Fig. 2D) and resulted in enhanced SMA expression (Fig. 2D). Quantification of three separate experiments indicated that collagen expression increased an average of 40% with MRTF-A expression (p < 0.05). Taken together, these results indicate that MRTF-A regulates collagen expression in fibroblasts.
Sp1 Sites Are Involved in MRTF-A Transactivation of Collagen
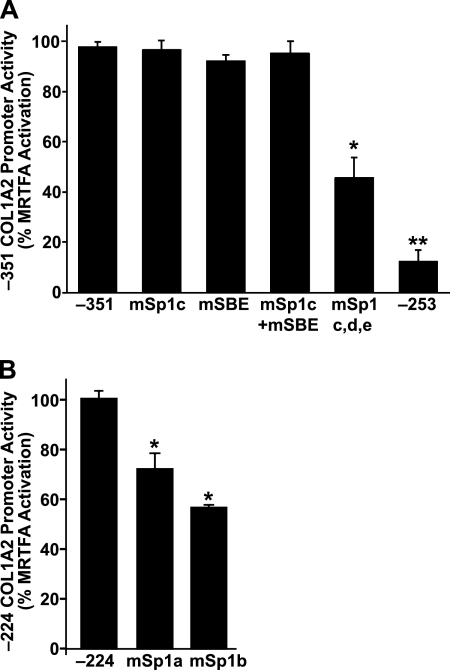
Because our studies indicated a role for both proximal and distal elements in MRTF-A-mediated collagen expression (Fig. 1), we next examined the function of Sp1 and SMAD binding sites using a series of −351 collagen promoter point mutation constructs (Fig. 1A, designated Sp1c, Sp1d, Sp1e, and SBE). Mutation of the Sp1c site alone, the SBE site alone, or in combination was ineffective at blunting the MRTF-A activity (Fig. 3A). Interestingly, mutation of all three Sp1 sites (Fig. 1A, mSp1c, -d, and -e) resulted in ∼60% decrease in activation by MRTF-A, whereas deletion of all these sites in the −253 region also resulted in a loss of much of the MRTF-A-stimulated collagen promoter activity (Fig. 3A).
FIGURE 3.
Sp1 sites contribute to MRTF-A transactivation of collagen. A, IMR90 fibroblasts (30,000/well) were transfected with MRTF-A (40 ng/well) and a series of COL1A2 promoter mutation and deletion constructs (100 ng/well) (described under “Experimental Procedures” and in Fig. 1A) (*, p < 0.05; **, p < 0.01). B, IMR90 cells were transiently transfected as in A with −224 COL1A2 wild type and Sp1 mutant constructs (described under “Experimental Procedures” and in Fig. 1A). Normalized luciferase activity is expressed as percent of the wild type −224/+55 reporter (*, p < 0.05). Results are averages of three independent experiments.
To evaluate the importance of the proximal promoter Sp1 sites, two additional mutants were generated. Although truncation of the promoter to −224 region resulted in reduction of MRTF-A activity compared with the full-length construct, MRTF-A was able to stimulate this promoter significantly relative to empty vector control (∼30-fold). Mutation of the two Sp1 sites (Sp1a and Sp1b, Fig. 1A) significantly impaired MRTF-A-dependent collagen promoter activity (Fig. 3B). Taken together, these data indicate that MRTF-A transactivation relies on functional Sp1-binding sites.
Collagen Promoter MRTF-A Transcription Factors Complexes
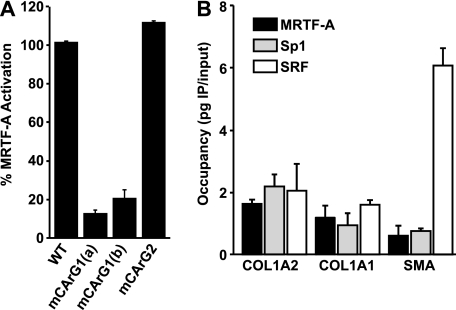
Although MRTF-A has not been shown to directly bind to DNA, several studies have demonstrated that it associates with SRF via CArG regulatory elements (45, 46). Our results above demonstrate that Sp1 is an important mediator of MRTF-A activation of collagen. While our work was in progress, a study by Small et al. (33) demonstrated that SRF can bind to the type I collagen promoter via a single CArG-like sequence (CArG1) at −326, which is adjacent to Sp1 sites (c, d, and e) (Fig. 1A, CArG1) (33), and contains a previously identified Ets site (11). Our sequence analysis identified a second CArG-like sequence (CArG2) within the COL1A2 promoter (CCAATTTAAG) at bp −193 (Fig. 1A). To test the potential function of these sequences, we mutated CArG1 to AAGGCCTTGG (CArG1a) or CCGGGCTTGG (CArG1b) and CArG2 to CGGATCCAAG in the context of the −351 COL1A2 promoter. Both of these mutations are predicted to abrogate SRF binding (47). However, the CArG1 mutation was predicted to also abrogate Ets binding as well as SRF. To rule out a contribution to MRTF-A activity from the Ets1 box element, CArG1b was generated and predicted to only disrupt SRF binding. Transfection results indicated that a substantial portion of the MRTF-A-dependent activation was lost with mutation of the CArG1 but not the CArG2 site (Fig. 4A). We observed that mutation of CArG1 inhibited MRTF-A-dependent gene reporter activation.
FIGURE 4.
MRTF-A occupies the collagen promoter with Sp1 and SRF. A, IMR90 cells were transiently transfected with MRTF-A (40 ng) together with 100 ng of −351 COL1A2 promoter constructs containing mutations of the CArG-like sequences (described under “Experimental Procedures”). Normalized luciferase activity is expressed relative to the −351 COL1A2 construct. B, IMR90 cells were seeded at a density of 2 × 104/cm2 and cultured for 16 h before cross-linking and chromatin isolation. Aliquots of 7 μg of total chromatin were used for each immunoprecipitation reaction. ChIP samples (n = 3) were detected by quantitative RT-PCR and quantified using the linear regression of a genomic DNA standard curve for each primer set. IgG amplification was subtracted from ChIP values and normalized relative to COL1A2. Results are averages of three independent experiments.
Based on the above results, ChIP was used to define the relative occupancy of Sp1, SRF, and MRTF-A transcription factors on the collagen promoters in IMR90 cells. Probes were designed to amplify regions centered directly on the CArG1 element and adjacent Sp1 sites of the COL1A2 and COL1A1, and as a positive control for binding, the canonical CArG element of the SMA promoter was examined. Antibody specificity was validated by amplification of a probe within COL1A2 exon 1, which was predicted to have no transcription factor-binding sites. Transcription factor occupancy on the probe sites was quantified and expressed as picograms of immunoprecipitated DNA per input and subtracted for nonspecific immunoprecipitation using IgG controls. Immunoprecipitation of MRTF-A revealed occupancy on both collagen and SMA gene promoters (Fig. 4B). Sp1 binding to the COL1A2 probe region was significantly higher compared with controls. Interestingly, ChIP of the SRF protein revealed an ∼3-fold greater occupancy on the SMA promoter compared with either of the collagen promoters (p < 0.01). These results reveal that Sp1, MRTF-A, and SRF can be detected at the collagen promoters.
Sp1 Is Important for MRTF-A Activation of Collagen
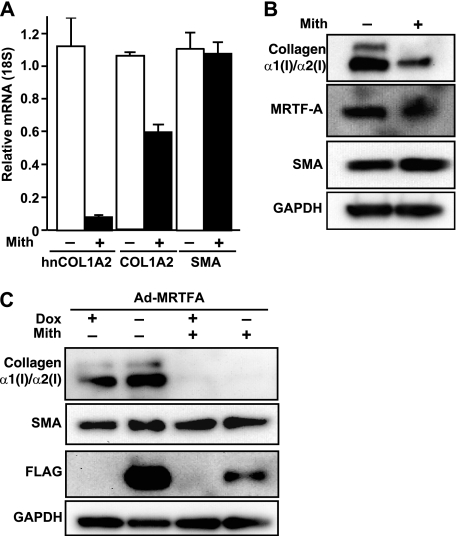
Based on the above ChIP results, we next wanted to determine the relative contribution of Sp1 to the transactivation potential of MRTF-A. To define the contributions of Sp1 to MRTF-A-dependent collagen expression, cells were treated with the antibiotic mithramycin A, which has been previously shown to inhibit collagen expression by selectively blocking Sp1 family binding to GC-rich regions (8, 48, 49). After IMR90 cells were treated with 50 nm mithramycin A for 24 h, total RNA was isolated, and the corresponding cDNA was quantified by quantitative PCR. Relative to vehicle (DMSO), mithramycin treatment resulted in a decrease in newly synthesized, heterogeneous, and steady-state COL1A2 mRNA levels (Fig. 5A). Importantly, SMA mRNA was unaffected by mithramycin A treatment. Similarly, protein analysis revealed that cells exposed to vehicle showed abundant production of type I collagen and SMA proteins (Fig. 5B). Treatment with mithramycin A strongly inhibited collagen protein expression but had no effect on SMA (Fig. 5B). To determine whether MRTF-A can activate collagen gene expression without Sp1 activity, cells were infected with MRTF-A adenovirus followed by treatment with mithramycin A. As expected, overexpression of MRTF-A increased collagen protein levels significantly compared with control (Fig. 5C). Mithramycin significantly decreased collagen protein expression, whereas ectopic MRTF-A was unable to rescue collagen synthesis in cells with inhibited Sp1 binding activity. Interestingly, SMA production was not altered by mithramycin A treatment. These data indicate an important mechanistic difference in MRTF-A activation of collagen versus SMA expression.
FIGURE 5.
Contribution of Sp1 sites to MRTF-A-dependent collagen expression. A, IMR90 cells were treated for 24 h with DMSO (control, white bars) or with 50 nm mithramycin A (Mith, black bars), which inhibits transcription factor binding to GC-rich DNA elements. Expression levels of hnCOL1A2, COL1A2, and SMA were measured by quantitative RT-PCR (n = 3). Results are expressed relative to control. B, Western blots were performed on cells treated as in A for collagen type I (Southern Biotech), MRTF-A, and SMA. Blots were normalized to GAPDH. C, IMR90 cells were infected with a doxycycline-controlled adenoviral MRTF-A vector and treated in the presence (+) or absence (−) of doxycycline (Dox) to control MRTF-A expression or mithramycin A to regulate transcription factor binding. Western blots were analyzed for collagen I, SMA, FLAG, and normalized to GAPDH. Data are representative of three independent experiments.
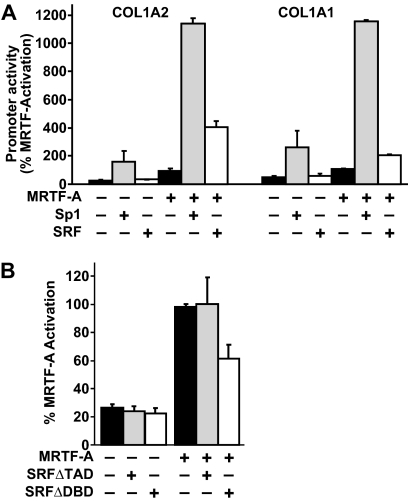
Our studies indicate that MRTF-A regulates collagen expression through both Sp1- and SRF-dependent mechanisms. We next investigated the potential synergy between MRTF-A and Sp1 or SRF. Because of the high levels of endogenous Sp1 in mammalian cells, reporter gene assays were performed in Drosophila melanogaster cells (D-Mel.2) (36, 50, 51). In this system, the transcriptional activation by MRTF-A in the absence of Sp1 proteins could be explored. Reporter constructs of the collagen type I promoters were analyzed for activity in the presence of equivalent amounts of MRTF-A and Sp1 or SRF expression constructs. Sp1, but not SRF, alone was able to induce collagen reporter activity significantly relative to MRTF-A activation (set at 100%, Fig. 6A). Co-transfection of MRTF-A and SRF leads to an increase in reporter activity (∼4-fold), whereas MRTF-A and Sp1 co-expression displayed a stronger synergistic activation (>10-fold) of both collagen reporter constructs (Fig. 6A). Taken together, these results show that Sp1 is important to potentiate maximum MRTF-A stimulation of collagen gene expression.
FIGURE 6.
MRTF-A-dependent collagen promoter activity is regulated by both SRF and Sp1. A, DMel.2 cells, which do not express Sp1 homologues, were transiently transfected with collagen COL1A2 and COL1A1 luciferase reporters in the presence of equivalent amounts of MRTF-A, Sp1, and SRF. Luciferase values are normalized to percent of MRTF-A activation. B, IMR90 cells were transfected with the −351 COL1A2 and MRTF-A along with two different SRF dominant negative expression vectors (SRFΔTAD and SRTFΔDBD). Luciferase activity is expressed as % of MRTF-A activation. Results are averages of three independent experiments.
Effect of Dominant Negative SRF on Collagen Promoter Activity
Because we observed SRF occupancy on the collagen promoter in ChIP assays (Fig. 4B) and a reduction of MRTF-A-dependent collagen in CArG site mutants (Fig. 4A), we sought to test whether dominant negative forms of SRF could alter collagen expression. To do this, two loss-of-function expression constructs of SRF were generated as follows: 1) SRFΔTAD, which bears a truncation of the transcriptional activation domain (36), and 2) SRFΔDBD, which substitutes the residues RVKI to LVAG in the DNA binding domain and renders it incapable of binding to CArG elements (37). Neither of these constructs altered basal collagen promoter activity (Fig. 6B). Relative to MRTF-A activation of collagen promoter alone, co-transfection of SRFΔTAD did not alter reporter activity. However, co-transfection of MRTF-A with the SRFΔDBD construct showed a decrease in gene reporter activation of both collagen promoter activities. These results indicate that collagen transcription is independent of the SRF transactivation domain but may require SRF binding for MRTF-A recruitment.
MRTF-A KO Lung Fibroblasts Have Aberrant Collagen and SMA Synthesis
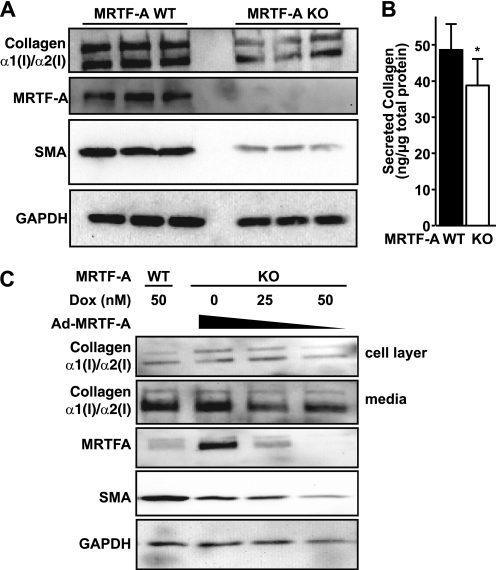
Two groups have disrupted the MRTF-A gene in mice and have observed changes in mammary gland myoepithelial cells (16, 52). However, nothing is currently known about the effect of MRTF-A loss in lung fibroblasts. To begin to address this question, we isolated lung cells from 3- to 5-week-old MRTF-A WT and MRTF-A KO littermates by standard techniques (34). Comparison of the myofibroblast phenotype between WT and KO lung cells by confocal immunofluorescence microscopy revealed high intracellular collagen staining and SMA stress fiber formation in control cells, whereas KO lung cells showed little collagen expression and low disorganized SMA expression (data not shown). In low passage lung fibroblasts, compared with MRTF-A WT controls, KO fibroblasts exhibited reduced levels of collagen expression and SMA expression (Fig. 7A). The Western analysis of cell lysates, performed three times in triplicate, demonstrate a 54% decrease (p < 0.05) of type I collagen. The reduced levels of intracellular collagen type I in the MRTF-A KO cells correlated with a reduction in all secreted collagen types (Fig. 7B).
FIGURE 7.
MRTF-A KO lung fibroblasts exhibit reduced collagen expression. A, low passage MRTF-A WT and MRTF-KO mouse lung fibroblasts (n = 3) were plated for 24 h and subjected to Western blot analysis for collagen type I (Southern Biotech), MRTF-A, and SMA. Blots were normalized to GAPDH. B, mouse lung fibroblasts plated at equal densities in media supplemented with 250 μm ascorbate for 48 h. Secreted collagen levels were determined by the Sircol assay and normalized to total protein levels in cell lysates. C, MRTF-A KO lung fibroblasts were infected with MRTF-A adenoviral vectors and treated with various doses of doxycycline to control MRTF-A expression along with 250 μm ascorbate for 48 h. Total cell layer proteins and secreted proteins were analyzed by Western blot for collagen I, MRTF-A, SMA. Blots were normalized to GAPDH. Results are representative of two independent experiments.
Ectopic Expression of MRTF-A Rescues Defective Myofibroblast Differentiation in KO Cells
To determine whether the defective myofibroblast phenotype in the MRTF-A KO cells could be restored, MRTF-A KO cells were infected with adenoviral MRTF-A as in Fig. 5C. Ectopic MRTF-A expression was regulated in KO cultures by modulation of doxycycline concentrations (Fig. 7C). Ectopic MRTF-A expression enhanced SMA expression in lung fibroblasts and served as a positive control for MRTF-A activity. Importantly, in two independent experiments, MRTF-A overexpression in the absence of doxycycline rescued both intracellular and secreted collagen protein expression in these cells. Loss of MRTF-A in lung cells results in decreased collagen and SMA expression. Taken together, these data indicate that knock-out of MRTF-A in lung fibroblasts decreases collagen synthesis and disrupts myofibroblast differentiation.
DISCUSSION
This study demonstrates an important role for the myocardin family activator, MRTF-A, in regulating collagen expression in myofibroblasts. We discovered that MRTF-A functions through both SRF and Sp1 complexes to regulate collagen promoter activity and production. Together with the known role for MRTF-A in SMA gene expression (19, 20, 45, 53), our results indicate that MRTF-A may serve to coordinate both contractile and ECM gene expression in the setting of fibrosis.
The pathological deposition of the collagen-rich ECM in fibrosis is mediated by the hyper-proliferative and synthetic properties of several cell types, including myofibroblasts within the fibrotic lesion. Critical to the progression of fibrosis is the activation of several signaling cascades (54). For example, activation of the Rho GTPase signaling node has long been implicated in the regulation of focal adhesion assembly and stress fiber formation (55), which coincides with myofibroblast differentiation. Rho-mediated activation of MRTF-A has emerged as an important transcriptional regulator of actin cytoskeleton remodeling, myogenic programming, and cardiac hypertrophy (53, 56, 57). MRTF-A, in particular, is a critical mediator in response to mechanical stress (30, 53), and previous studies have shown that collagen production responds to mechanical stress (58).
MRTF-A activated collagen transcription significantly greater than other members of the myocardin family (Fig. 1B). The COL1A2 basal promoter (−224 bp) was activated by MRTF-A to approximately the same amount as the COL1A1 promoter (−311 bp) (Fig. 1). However, the COL1A2 promoter contains an enhancer-like region (−221 to −345 bp) with overlapping binding sites for several transcription factors. This region confers additional activation potential to the COL1A2 promoter. Regulation of constitutive transcription occurs through four clusters of cis-acting elements that bind positive and negative trans-acting factors (7). The first region occurs around the methylation-sensitive CpG at +7 where repressor complexes of RFX/CIITA proteins down-regulate collagen transcription when the site is methylated or during IFNγ treatment (16–18). There is a canonical CCAAT motif around −80 where CBF-NFY complex activates transcription (59). There are two TCC-enriched regions that interact with Sp1 family and cKrox proteins located between −160 and −125 that both activate and repress depending on cell type (9, 59–61). Finally, the region around −300 contains multiple binding sites for Sp1 proteins and Ets family proteins as well as other transcription factors, including Ap1, SMAD, and C/EBP (9–11, 62, 63).
MRTF-A is sequestered in the cytoplasm through G-actin binding to its RPEL domain that blocks residues necessary for nuclear localization (64). Signaling events that promote actin polymerization, such as Rho GTPase activation, deplete G-actin pools and permit MRTF-A translocation into the nucleus (19). In addition, Rac1 induces SMA expression during epithelial to mesenchymal transition, and expression of DN-MRTF-A inhibited these changes (65). Future studies will investigate the signaling events that regulate MRTF-A nuclear localization and collagen gene activation.
In this study, we demonstrate that MRTF-A overexpression in human lung fibroblasts increases collagen and SMA protein levels, whereas overexpression of a dominant negative MRTF-A reduces collagen protein levels (Fig. 2C). Interestingly, knockdown of MRTF-A reduces both protein synthesis and heterogeneous nuclear levels of collagen RNA (Fig. 2, A and B). Because collagen and ECM gene products are regulated by many post-transcriptional mechanisms, including miRNA, post-transcriptional storage, and mRNA translatability, this observation may suggest that MRTF-A plays additional roles in collagen regulation downstream of transcription (66, 67). Further investigation of this finding is necessary to explore this possibility.
Because MRTF-A is a co-activator that interacts with DNA-binding proteins, detailed mutational analysis was performed on the COL1A2 promoter (Fig. 3). The loss of known Sp1 sites reduced MRTF-A activation of the collagen promoter, whereas the mutation of CCAAT region did not (data not shown). Collagen expression is activated by TGFβ through cooperation of Sp1 and SMAD through the Sp1c and SMAD sites (68). MRTF-A family members also interact with SMAD proteins (69, 70), although deletion or mutation of the SMAD-binding site did not reduce MRTF-A activation of the collagen promoter suggesting that the MRTF-A/SMAD interaction is not functional under these culture conditions. However, future studies with TGFβ stimulation may indicate additional roles for MRTF-A in myofibroblasts.
We also demonstrate that Sp1 functions synergistically with MRTF-A to activate collagen transcription (Fig. 6A). Mithramycin A is a potent inhibitor of Sp1 function (48). This effect is achieved by mithramycin intercalation into GC-rich DNA sequences, such as Sp1-binding sites. The inhibition of collagen expression using mithramycin has been well documented and attributed to Sp1 disruption (8). The observation that adenoviral expression of MRTF-A was unable to rescue mithramycin-induced inhibition of collagen synthesis (Fig. 5C) strongly suggests that MRTF-A relies on Sp1 to stimulate collagen expression.
Close examination of the COL1A2 promoter suggests that a noncanonical CArG box does exist that harbors a cytosine substitution within the A/T-rich core of the CArG box consensus sequence (33). Similar mutations to the SMA promoter have been shown to significantly diminish the binding capacity of the SRF-myocardin complex (71). Furthermore, noncanonical (i.e. degenerate) CArG boxes can dampen SMA expression by smooth muscle cells in response to injury (24). During injury to smooth muscle cells, there is an interaction of canonical CArG and GC-rich regions to repress smooth muscle-specific genes that is due to reduced myocardin/SRF binding (24). It has been postulated that such injury can promote the redifferentiation of SMCs into synthetic myofibroblasts that highly express collagen (72). Our data suggest that both noncanonical CArG (Fig. 4) and GC-rich regions (Fig. 3) are important for up-regulation of collagen through MRTF-A and Sp1 proteins. Although mutation of these CArG boxes reduced MRTF-A activation to 20–25% of control (Fig. 4A), truncated collagen promoter constructs that completely lack these elements (Fig. 1D) still exhibited significant MRTF-A activation of reporter gene expression. Together, these findings support a model where MRTF-A stimulates transcription via both CArG and GC-rich sites and may serve to coordinate expression within the overall myofibroblast program.
The occupancy of transcription factors complexed with MRTF-A on the collagen promoter was probed using quantitative ChIP assays. Quantitative detection of DNA bound to these transcription factors was measured using quantitative PCR (Fig. 4B). Positive binding was calculated by subtracting detectable levels of signal from IgG samples (i.e. nonspecific binding), thus providing a scale of transcription factor binding with zero representing no specific binding. Specificity of the immunoprecipitation reaction was verified by probe amplification within intron 1 of the COL1A2 promoter. In each immunoprecipitation sample, no amplification of this control region was detected. The chromatin immunoprecipitation data in this paper indicate that MRTF-A, SRF, and Sp1 are present at the collagen promoters in human lung fibroblasts in culture (Fig. 4B). These results indicate that binding of SRF on the collagen promoter is reduced relative to the SMA promoter. Two different mutations of the noncanonical CArG box with and without preservation of a neighboring Ets1-binding site (10, 11) significantly blunted, but did not eliminate, MRTF-A activation of collagen gene reporters (Fig. 4A). This implies that SRF binding to this noncanonical CArG site contributes to MRTF-A recruitment. To delineate its function, SRF was mutagenized to inactivate either its DNA binding or transactivation domain. Mutation of SRF DNA binding significantly disrupted MRTF-A transactivation and potentially indicates that the binding capacity of SRF, regardless of CArG box sequence homology, contributes to MRTF-A transactivation of collagen (Fig. 6B). Interestingly, however, truncation of the transcription activation domain of SRF had no effect on MRTF-A transactivation of the collagen promoter. C-terminal truncation of SRF decreases gene reporter activity by transgelin promoter (also called SM22) (36), which is a cytoskeletal gene that has been demonstrated to rely on myocardin/SRF activation via a conserved CArG box (21). Together, these studies identified functional differences of SRF between the collagen promoter and other MRTF/SRF-regulated genes.
Mice with a loss-of-function MRTF-A mutation lack mammary myoepithelial cells (52) and are resistant to cardiac hypertrophy (33, 53, 57). It is clear from this study that loss-of-function MRTF-A lung fibroblasts have decreased collagen protein synthesis as well as decreased cytoskeletal protein expression (Fig. 7). Sircol assays for total collagen also indicated that MRTF-A KO cells secreted less collagen, although the results were modest. This result likely reflects the fact that Sircol measures all secreted collagen types and not type I collagen specifically. Most importantly, expression of MRTF-A restored expression of collagen and SMA levels similar to those seen in wild type controls, thereby rescuing the phenotype. While our study was in progress, it was recently demonstrated that MRTF-A regulates collagen synthesis in cardiomyocytes (33). Our study builds on that finding by identifying important CArG-dependent and CArG-independent mechanisms of collagen expression in lung fibroblasts. First, we show that MRTF-A associates with Sp1 and SRF in the region of the CArG box elements on both the collagen and SMA promoter in lung fibroblasts. Second, we describe a novel mechanism by which Sp1 interacts with MRTF-A to transactivate the collagen promoter, where disruption of Sp1 family binding to DNA with mithramycin A strongly inhibits collagen while not altering SMA expression. Finally, mutation of the transcriptional activation domain within SRF is deleterious for transcriptional activation of SMA, although no effect was observed on collagen promoter activity. These results suggest a differential function for SRF and Sp1 in mediating activation of the two genes.
Our working model is that MRTF-A activates collagen transcription through Sp1 family and SRF through noncanonical CArG sites within the proximal promoter of COL1A2. This mechanism appears to differ relative to other SRF-dependent genes such as SMA, which are dependent on myocardin as well as MRTF interaction with full-length SRF to achieve efficient activation. The Sp1 family member KLF4 has been demonstrated to inhibit MRTF-A activation of SMA, suggesting that additional transcription factor components may be necessary to fine-tune the specificity of these two transcriptional activation complexes (73).
In conclusion, we show that MRTF-A is an important activator of collagen synthesis. These results show active collagen transcription and protein synthesis correlate positively with increased MRTF-A activity. Loss of MRTF-A in knockdown and knock-out models shows an inhibition of collagen and SMA expression, suggesting a disruption of myofibroblast function. Our findings that lung fibroblasts from MRTF-A null mice have reduced collagen production indicate that MRTF-A may have an essential regulatory function in fibrotic diseases. Future studies will address the hypothesis that MRTF-A coordinates ECM and myofibroblast gene expression in the fibrotic lung.
Supplementary Material
Acknowledgments
We greatly appreciate the gift of the MRTF-A heterozygous mice from Dr. Eric Olson (University of Texas Southwestern Medical Center). We thank Anupma Agarwal and William Monis for technical assistance in some of the preliminary experiments.
This work was supported, in whole or in part, by National Institutes of Health Grants HL68094 (to B. D. S.) and HL078869 (to M. D. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- ECM
- extracellular matrix
- SMA
- α-smooth muscle actin
- MRTF-A
- myocardin related transcription factor
- SRF
- serum response factor
- DN
- dominant negative
- hn
- heterogeneous
- SBE
- SMAD element.
REFERENCES
- 1. Wilson M. S., Wynn T. A. (2009) Mucosal Immunol. 2, 103–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wynn T. A. (2008) J. Pathol. 214, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuhn C., McDonald J. A. (1991) Am. J. Pathol. 138, 1257–1265 [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang K., Rekhter M. D., Gordon D., Phan S. H. (1994) Am. J. Pathol. 145, 114–125 [PMC free article] [PubMed] [Google Scholar]
- 5. Adler K. B., Low R. B., Leslie K. O., Mitchell J., Evans J. N. (1989) Lab. Invest. 60, 473–485 [PubMed] [Google Scholar]
- 6. Selman M., Montaño M., Ramos C., Chapela R. (1986) Thorax 41, 355–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramirez F., Tanaka S., Bou-Gharios G. (2006) Matrix Biol. 25, 365–372 [DOI] [PubMed] [Google Scholar]
- 8. Nehls M. C., Brenner D. A., Gruss H. J., Dierbach H., Mertelsmann R., Herrmann F. (1993) J. Clin. Invest. 92, 2916–2921 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Tamaki T., Ohnishi K., Hartl C., LeRoy E. C., Trojanowska M. (1995) J. Biol. Chem. 270, 4299–4304 [DOI] [PubMed] [Google Scholar]
- 10. Czuwara-Ladykowska J., Sementchenko V. I., Watson D. K., Trojanowska M. (2002) J. Biol. Chem. 277, 20399–20408 [DOI] [PubMed] [Google Scholar]
- 11. Czuwara-Ladykowska J., Shirasaki F., Jackers P., Watson D. K., Trojanowska M. (2001) J. Biol. Chem. 276, 20839–20848 [DOI] [PubMed] [Google Scholar]
- 12. Zhang K., Flanders K. C., Phan S. H. (1995) Am. J. Pathol. 147, 352–361 [PMC free article] [PubMed] [Google Scholar]
- 13. Evans J. N., Kelley J., Low R. B., Adler K. B. (1982) Am. Rev. Respir. Dis. 125, 89–94 [DOI] [PubMed] [Google Scholar]
- 14. Chen S. J., Yuan W., Mori Y., Levenson A., Trojanowska M., Varga J. (1999) J. Invest. Dermatol. 112, 49–57 [DOI] [PubMed] [Google Scholar]
- 15. Ritzenthaler J. D., Goldstein R. H., Fine A., Lichtler A., Rowe D. W., Smith B. D. (1991) Biochem. J. 280, 157–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sengupta P., Xu Y., Wang L., Widom R., Smith B. D. (2005) J. Biol. Chem. 280, 21004–21014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sengupta P. K., Fargo J., Smith B. D. (2002) J. Biol. Chem. 277, 24926–24937 [DOI] [PubMed] [Google Scholar]
- 18. Xu Y., Wang L., Butticè G., Sengupta P. K., Smith B. D. (2004) J. Biol. Chem. 279, 41319–41332 [DOI] [PubMed] [Google Scholar]
- 19. Morita T., Mayanagi T., Sobue K. (2007) Exp. Cell Res. 313, 3432–3445 [DOI] [PubMed] [Google Scholar]
- 20. Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., Treisman R. (2009) Nat. Cell Biol. 11, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 22. Pawłowski R., Rajakylä E. K., Vartiainen M. K., Treisman R. (2010) EMBO J. 29, 3448–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olson E. N., Nordheim A. (2010) Nat. Rev. Mol. Cell Biol. 11, 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hendrix J. A., Wamhoff B. R., McDonald O. G., Sinha S., Yoshida T., Owens G. K. (2005) J. Clin. Invest. 115, 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pipes G. C., Creemers E. E., Olson E. N. (2006) Genes Dev. 20, 1545–1556 [DOI] [PubMed] [Google Scholar]
- 26. Miano J. M. (2003) J. Mol. Cell. Cardiol. 35, 577–593 [DOI] [PubMed] [Google Scholar]
- 27. Miralles F., Posern G., Zaromytidou A. I., Treisman R. (2003) Cell 113, 329–342 [DOI] [PubMed] [Google Scholar]
- 28. Kuwahara K., Barrientos T., Pipes G. C., Li S., Olson E. N. (2005) Mol. Cell. Biol. 25, 3173–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mack C. P., Hinson J. S. (2005) J. Thromb. Haemost. 3, 1976–1984 [DOI] [PubMed] [Google Scholar]
- 30. Zhao X. H., Laschinger C., Arora P., Szászi K., Kapus A., McCulloch C. A. (2007) J. Cell Sci. 120, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 31. Du H., Wang X., Wu J., Qian Q. (2010) Hypertens. Res. 33, 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan M. W., Chaudary F., Lee W., Copeland J. W., McCulloch C. A. (2010) J. Biol. Chem. 285, 9273–9281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Small E. M., Thatcher J. E., Sutherland L. B., Kinoshita H., Gerard R. D., Richardson J. A., Dimaio J. M., Sadek H., Kuwahara K., Olson E. N. (2010) Circ. Res. 107, 294–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu Y., McDonald J., Perloff E., Butticè G., Schreiber B. M., Smith B. D. (2007) Mol. Immunol. 44, 1709–1721 [DOI] [PubMed] [Google Scholar]
- 35. Chen C. H., Wu M. L., Lee Y. C., Layne M. D., Yet S. F. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Layne M. D., Yet S. F., Maemura K., Hsieh C. M., Liu X., Ith B., Lee M. E., Perrella M. A. (2002) Circ. Res. 90, 728–736 [DOI] [PubMed] [Google Scholar]
- 37. Poser S., Impey S., Trinh K., Xia Z., Storm D. R. (2000) EMBO J. 19, 4955–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wessel D., Flügge U. I. (1984) Anal. Biochem. 138, 141–143 [DOI] [PubMed] [Google Scholar]
- 39. Elferink C. J., Reiners J. J., Jr. (1996) BioTechniques 20, 470–477 [DOI] [PubMed] [Google Scholar]
- 40. Sastry L., Johnson T., Hobson M. J., Smucker B., Cornetta K. (2002) Gene Ther. 9, 1155–1162 [DOI] [PubMed] [Google Scholar]
- 41. Sun Y., Boyd K., Xu W., Ma J., Jackson C. W., Fu A., Shillingford J. M., Robinson G. W., Hennighausen L., Hitzler J. K., Ma Z., Morris S. W. (2006) Mol. Cell. Biol. 26, 5809–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stefanovic B., Brenner D. A. (2003) J. Biol. Chem. 278, 927–933 [DOI] [PubMed] [Google Scholar]
- 43. Stefanovic B., Hellerbrand C., Holcik M., Briendl M., Aliebhaber S., Brenner D. A. (1997) Mol. Cell. Biol. 17, 5201–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stefanovic B., Schnabl B., Brenner D. A. (2002) J. Biol. Chem. 277, 18229–18237 [DOI] [PubMed] [Google Scholar]
- 45. Cen B., Selvaraj A., Burgess R. C., Hitzler J. K., Ma Z., Morris S. W., Prywes R. (2003) Mol. Cell. Biol. 23, 6597–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee S. M., Vasishtha M., Prywes R. (2010) J. Biol. Chem. 285, 22036–22049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shore P., Sharrocks A. D. (1995) Eur. J. Biochem. 229, 1–13 [DOI] [PubMed] [Google Scholar]
- 48. Blume S. W., Snyder R. C., Ray R., Thomas S., Koller C. A., Miller D. M. (1991) J. Clin. Invest. 88, 1613–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sleiman S. F., Langley B. C., Basso M., Berlin J., Xia L., Payappilly J. B., Kharel M. K., Guo H., Marsh J. L., Thompson L. M., Mahishi L., Ahuja P., MacLellan W. R., Geschwind D. H., Coppola G., Rohr J., Ratan R. R. (2011) J. Neurosci. 31, 6858–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Courey A. J., Tjian R. (1988) Cell 55, 887–898 [DOI] [PubMed] [Google Scholar]
- 51. Chin M. T., Pellacani A., Wang H., Lin S. S., Jain M. K., Perrella M. A., Lee M. E. (1998) J. Biol. Chem. 273, 9755–9760 [DOI] [PubMed] [Google Scholar]
- 52. Li S., Chang S., Qi X., Richardson J. A., Olson E. N. (2006) Mol. Cell. Biol. 26, 5797–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kuwahara K., Kinoshita H., Kuwabara Y., Nakagawa Y., Usami S., Minami T., Yamada Y., Fujiwara M., Nakao K. (2010) Mol. Cell. Biol. 30, 4134–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jordan J. D., Landau E. M., Iyengar R. (2000) Cell 103, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ridley A. J., Hall A. (1992) Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 56. Fan L., Sebe A., Péterfi Z., Masszi A., Thirone A. C., Rotstein O. D., Nakano H., McCulloch C. A., Szászi K., Mucsi I., Kapus A. (2007) Mol. Biol. Cell 18, 1083–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liao X. H., Wang N., Liu Q. X., Qin T., Cao B., Cao D. S., Zhang T. C. (2011) IUBMB Life 63, 54–61 [DOI] [PubMed] [Google Scholar]
- 58. Chiquet M., Matthisson M., Koch M., Tannheimer M., Chiquet-Ehrismann R. (1996) Biochem. Cell Biol. 74, 737–744 [DOI] [PubMed] [Google Scholar]
- 59. Maity S. N., Golumbek P. T., Karsenty G., de Crombrugghe B. (1988) Science 241, 582–585 [DOI] [PubMed] [Google Scholar]
- 60. Karsenty G., de Crombrugghe B. (1990) J. Biol. Chem. 265, 9934–9942 [PubMed] [Google Scholar]
- 61. Galéra P., Park R. W., Ducy P., Mattéi M. G., Karsenty G. (1996) J. Biol. Chem. 271, 21331–21339 [DOI] [PubMed] [Google Scholar]
- 62. Chung K. Y., Agarwal A., Uitto J., Mauviel A. (1996) J. Biol. Chem. 271, 3272–3278 [DOI] [PubMed] [Google Scholar]
- 63. Greenwel P., Tanaka S., Penkov D., Zhang W., Olive M., Moll J., Vinson C., Di Liberto M., Ramirez F. (2000) Mol. Cell. Biol. 20, 912–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hinson J. S., Medlin M. D., Lockman K., Taylor J. M., Mack C. P. (2007) Am. J. Physiol. Heart Circ. Physiol. 292, H1170–H1180 [DOI] [PubMed] [Google Scholar]
- 65. Sebe A., Masszi A., Zulys M., Yeung T., Speight P., Rotstein O. D., Nakano H., Mucsi I., Szászi K., Kapus A. (2008) FEBS Lett. 582, 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lindquist J. N., Marzluff W. F., Stefanovic B. (2000) Am. J. Physiol. Gastrointest. Liver Physiol. 279, G471–G476 [DOI] [PubMed] [Google Scholar]
- 67. Cushing L., Kuang P. P., Qian J., Shao F., Wu J., Little F., Thannickal V. J., Cardoso W. V., Lü J. (2011) Am. J. Respir. Cell Mol. Biol. 45, 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang W., Ou J., Inagaki Y., Greenwel P., Ramirez F. (2000) J. Biol. Chem. 275, 39237–39245 [DOI] [PubMed] [Google Scholar]
- 69. Iwasaki K., Hayashi K., Fujioka T., Sobue K. (2008) J. Biol. Chem. 283, 21230–21241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Morita T., Mayanagi T., Sobue K. (2007) J. Cell Biol. 179, 1027–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leung S., Miyamoto N. G. (1989) Nucleic Acids Res. 17, 1177–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 73. Davis-Dusenbery B. N., Chan M. C., Reno K. E., Weisman A. S., Layne M. D., Lagna G., Hata A. (2011) J. Biol. Chem. 286, 28097–28110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.