Abstract
Galectin-9 expression in endothelial cells can be induced in response to inflammation. However, the mechanism of its expression remains unclear. In this study, we found that interferon-γ (IFN-γ) induced galectin-9 expression in human endothelial cells in a time-dependent manner, which coincided with the activation of histone deacetylase (HDAC). When endothelial cells were treated with the HDAC3 inhibitor, apicidin, or shRNA-HDAC3 knockdown, IFN-γ-induced galectin-9 expression was abolished. Overexpression of HDAC3 induced the interaction between phosphoinositol 3-kinase (PI3K) and IFN response factor 3 (IRF3), leading to IRF3 phosphorylation, nuclear translocation, and galectin-9 expression. HDAC3 functioned as a scaffold protein for PI3K/IRF3 interaction. In addition to galectin-9 expression, IFN-γ also induced galectin-9 location onto plasma membrane, which was HDAC3-independent. Importantly, HDAC3 was essential for the constitutive transcription of PI3K and IRF3, which might be responsible for the basal level of galectin-9 expression. The phosphorylation of IRF3 was essential for galectin-9 expression. This study provides new evidence that HDAC3 regulates galectin-9 expression in endothelial cells via interaction with PI3K-IRF3 signal pathway.
Keywords: Histone Deacetylase, Inflammation, Interferon, Phosphatidylinositol 3-Kinase, Signal Transduction
Introduction
Galectins are a family of lectins classified into three groups (prototype, tandem-repeat, and chimera-type galectins) according to their conserved carbohydrate recognition domains and their ability to bind to β-galactosides (1). To date, 15 galectin proteins have been identified in mammals displaying wide tissue distribution whereas some have higher tissue specificity. They can function intracellularly and extracellularly. Accumulating evidence implicates galectins as immunoregulatory mediators in diverse physiological and pathological processes such as immune and inflammatory responses, tumor immunity, wound repair, and atherosclerosis (2, 3).
Galectin-9 belongs to the tandem-repeat galectins that contain two carbohydrate recognition domains, mainly functioning as a chemoattractant of eosinophils (4, 5) and immunomodulation in a physiological and pathological setting (6, 7). Galectin-9 exerts its immunosuppressive roles through its receptor, T cell immunoglobin domain and mucin domain 3 (Tim3),2 which can increase the survival of transplants (6, 8–11). Galectin-9 may also induce proinflammatory cytokines in monocytes and T cells in a Tim3-independent manner (12, 13). Galectin-9-deficient mice show enhanced susceptibility to arthritic induction with increased CD4+ TIM3+ cells (14) and were prone to increased mortality and died within 72 h of LPS induction (15). In endothelial cells, galectin-9 can be induced by interferon-γ (IFN-γ) (16, 17) and double-stranded RNA (18), which may play a role in inflammatory response by regulating interactions between the vascular wall and eosinophils. However, the mechanism of how galectin-9 is expressed in endothelial cells is unknown.
Histone deacetylase 3 (HDAC3) belongs to class I HDAC family (19). Distinct from other family members which are nuclear-exclusive, HDAC3 can shuttle in and out of the nucleus (20). Disruption of the HDAC3 gene in mouse germ line causes early embryonic lethality (21). In endothelial cells, HDAC3 can regulate agonist-induced tissue factor expression (22), eNOS expression (23) and activity (24) as well as VCAM-1 expression (25). In our laboratory, we have shown that HDAC3 is essential for stem/progenitor cell differentiation toward endothelial lineage (26, 27) and plays a prosurvival role in mature endothelial cells (28). This suggests that HDAC3 could be critical in the maintenance of endothelial integrity. It would be of interest to investigate whether HDAC3 has the ability to regulate galectin-9 expression in endothelial cells. In this study, we demonstrated that HDAC3 modulates the constitutive expression of galectin-9 in endothelial cells. We also show that HDAC3 is crucial for IFN-γ-induced galectin-9 expression via interaction with phosphoinositide 3-kinase (PI3K) and IFN response factor 3 (IRF3).
EXPERIMENTAL PROCEDURES
Materials
Cell culture media and serum were purchased from Invitrogen. Cell culture supplements were purchased from Sigma. Antibodies against GAPDH, H2B, and H4 were purchased from Santa Cruz Biotechnology, whereas antibodies against anti-FLAG, tubulin, and HDAC3 were purchased from Sigma. Antibodies against galectin-9, p85α, p-IRF3 (Ser-386), and IRF3 were purchased from Abcam. All secondary antibodies were from Dakocytomation. LY294002 was purchased from Merck Bioscience predissolved in dimethyl sulfoxide. Apicidin was purchased from Enzo Life Science and dissolved in dimethyl sulfoxide. IFN-γ was purchased from Sigma and dissolved in PBS.
Cell Culture and Treatment
Human umbilical vein endothelial cells (HUVECs) were purchased from Promocell and cultured on collagen I-coated flasks in M199 medium supplemented with 1 ng/ml β-EC growth factor, 3 μg/ml EC growth supplement from bovine neural tissue, 10 units/ml heparin, 1.25 μg/ml thymidine, 10% fetal bovine serum (FBS), 100 units/ml penicillin and streptomycin in humidified incubator supplemented with 5% CO2. The cells were split every 3 days at a ratio of 1:4. Cells up to passage 10 were used in this study. IFN-γ (10 ng/ml) treatment was carried out for the time indicated or for 16 h. Inhibitors, 5 μm LY294002 or 200 nm apicidin, were added 1 h prior to IFN-γ treatment.
Quantitative RT-PCR
Relative gene expression was determined by quantitative real time PCR, using 2 ng of cDNA (relative to RNA amount) for each sample with the SYBR Green Master Mix in a 25-μl reaction. Ct values were measured using ABI PRISM 7000 Sequence Detector (Applied Biosystems). The 18 S ribosomal RNA served as the endogenous control to normalize the amounts of RNA in each sample. For each sample, PCR was performed in triplicate in a MicroAmp Optical 96-well reaction plate (Applied Biosystems). The gene was considered undetectable beyond 35 cycles. The primer sets used for this study are as follows: 18 S forward, 5′-cccagtaagtgcgggtcataa-3′ and reverse, 5′-ccgagggcctcactaaacc-3′; GAPDH forward, 5′-acagtcagccgcatcttctt-3′ and reverse, 5′-tggaagatggtgatgggatg-3′; HDAC3 forward, 5′-ttccagccggttatcaacca-3′ and reverse, 5′-gttaaagcagcccaatcgatca-3′; galectin-9 forward, 5′-ctttcatcaccaccattctg3′ and reverse, 5′-atgtggaacctctgagcactg-3′; p85α forward, 5′-acgacagcctgcaccagcac-3′ and reverse, 5′-accgctcttggtctggcac-3′; IRF3 forward, 5′-caactggagggcgtggcctg-3′ and reverse, 5′-ttgcggttgagggcagagcg-3′.
Cellular Fractions Isolation
Cell membrane proteins were labeled with biotin and isolated using a commercially available kit (Cell Surface Protein Isolation kit; Pierce) according to the manufacturer's protocol. Cytosolic and nuclear fractions were isolated by the standard centrifuge procedure. Briefly, 150 μl of cytosolic buffer (10 mm HEPES, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 0.05% Nonidet P-40, and a mixture of protease and phosphatase inhibitors (Roche Applied Science), pH 7.9) was added to suspend the cell pellet obtained from a T-75-treated flask. The cell suspension was left on ice for 10 min and subsequently centrifuged at 4 °C at 3,000 rpm for 10 min. The supernatant, which contains the cytosolic fraction, was aliquoted. The remaining cell pellet was resuspended in 50 μl of lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100 plus a mixture of protease and phosphatase inhibitors) and sonicated for 9 s on a Radson Sonifier at the lowest power setting and left on ice for 30 min. The cell lysate was then centrifuged at 4 °C at 14,000 rpm for 20 min. This nuclear fraction was aliquoted. Both cytosolic and nuclear fractions were measured by the Bradford method, and 25 μg of protein was applied to Western blot analysis.
Immunofluorescent Staining
The immunofluorescent staining of galectin-9 was performed using standard procedures. Briefly, HUVECs were seeded at 2.5 × 104cells/chamber on a chamber slide (BD Biosciences) and treated as indicated. The cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 in PBS, and blocked with 10% normal swine serum. Incubation with galectin-9 primary antibody (1:100 in 10% normal swine serum) was carried out at 4 °C overnight and subsequently washed three times with PBS. Swine anti-rabbit FITC-conjugated (1:50 in 10% normal swine serum) secondary antibody was applied for 45 min at room temperature and washed with PBS. Cells were counterstained with DAPI (1:1,000 in PBS) for 2 min at room temperature. Cells were mounted with fluorescent mounting media (Dako), and images were taken with the Axio Imager.M2 microscope and AxioVision Digital Imaging System (Carl Zeiss Ltd.) and processed with Adobe Photoshop software.
Immunoprecipitation and Western Blotting
Ad-FLAG-HDAC3 infected or uninfected HUVECs were treated with IFN-γ treatment (10 ng/ml) for 4 h in the absence or presence of apicidin. The cells were lysed in 400 μl/T75 flask IP-A buffer (25 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, pH 8.0, 1%Triton X-100 plus protease inhibitors) on ice for 45 min. The protein concentration was measured by the Bradford method. The cell lysate (1 mg) was mixed with 3 volumes of IP-B buffer (IP-A buffer without Triton X-100, plus protease inhibitors). The cell lysate was precleared with 2 μg of mouse, rabbit, or goat normal IgG and 10 μl of protein G-agarose beads for 1 h and then incubated with 2 μg of rabbit anti-HDAC3 or goat anti-IRF3 and 10 μl of protein G-agarose beads or directly with 10 μl of anti-FLAG-agarose beads, followed by Western blotting. 50 μg (5%) of lysate was included as input control. For Western blotting, cells were lysed in IP-A buffer. 50 μg of lysate was applied to SDS-PAGE and transferred to Hybond PVDF membrane (Amersham Biosciences), followed by standard Western blotting procedure. The bound primary antibodies were detected by the use of HRP-conjugated secondary antibody and the ECL detection system (Amersham Biosciences). The band density was semiquantified by Adobe Photoshop Elements software using the histogram tool.
Plasmid Transient Transfection
The plasmid pcDNA3-IRF3 (29) was purchased from Addgene (Addgene plasmid 22860). Mutated pcDNA3IRF3S386A was created by PCR-based mutagenesis with a primer set of 5′-gctggcaccccctacccgggccatttc-3′ and 5′-gctctggagaatactgtggacctgcac-3′ and verified by DNA sequencing. The expression vector pShuttle2-FLAG-HDAC3, pcDNA3-IRF3, or pcDNA3-IRF3S386A was transfected into HUVECs (2 μg of plasmid/2 × 106 cells) by electroporation using the HUVEC nucleofection kit (Lonza) according to the manufacturer's recommendation. pShuttle2-FLAG or pcDNA3 empty vector was included as mock control. The cells were harvested 48 h later.
Adenoviral Gene Transfer and shRNA Lentiviral Particle Transduction
Ad-FLAG-HDAC3 adenovirus and HDAC3 shRNA lentivirus were amplified in HEK293 and 293T cells, respectively, as described previously (28). For adenoviral DNA transfer, HUVECs were seeded on collagen I-coated plates 24 h prior to infection and incubated with Ad-FLAG-HDAC3 at a 10 multiplicity of infection (m.o.i.) for 6 h. Subsequently, fresh medium was added, and the cells were cultured or treated for the time indicated. For shRNA lentiviral transduction assays, HUVECs were seeded on collagen I-coated plates 24 h prior to transduction and incubated with HDAC3 shRNA lentivirus (HD3sh) or nontarget shRNA lentivirus (NTsh) at 1 × 107 units/1 × 106cells in the presence of 10 μg/ml Polybrene for 24 h. Subsequently, the medium was refreshed with complete growth medium and the cells treated with IFN-γ 48 h after transduction. The shRNA lentivirus-infected cells were assessed 72 h after transduction.
Statistical Analysis
Data expressed as the mean ± S.E. were analyzed with a two-tailed Student's t test for two-groups or pair-wise comparisons. A value of p < 0.05 was considered significant.
RESULTS
HDAC3 Is Necessary for IFN-γ-induced Galectin-9 Expression in Endothelial Cells
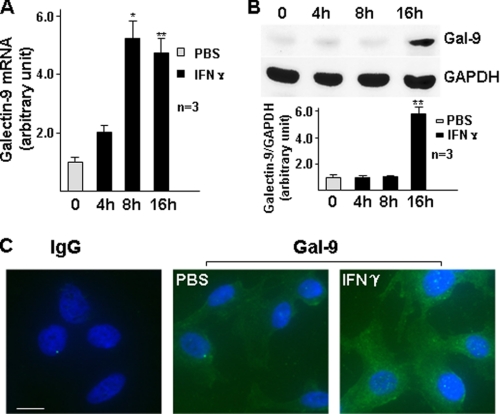
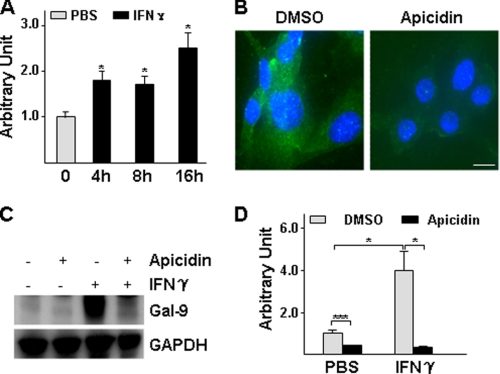
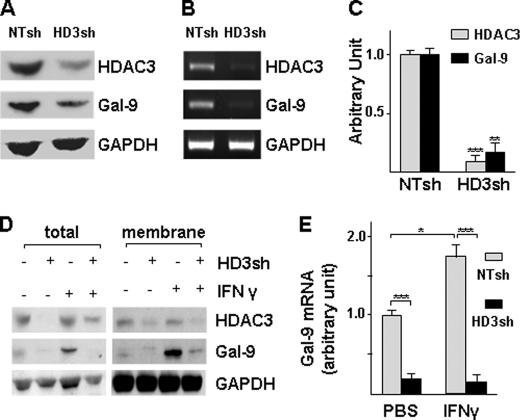
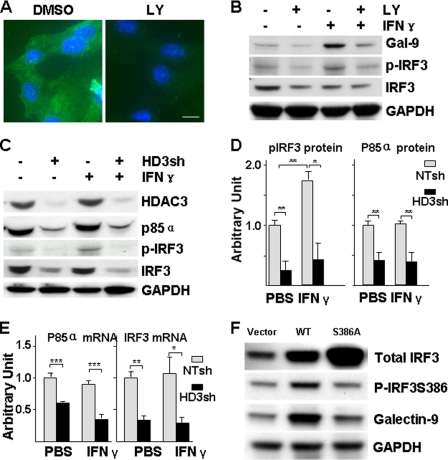
To explore the mechanism of galectin-9 expression, we first assessed the basal level and responsive expression of galectin-9 in HUVECs under IFN-γ treatment in our culture system. Galectin-9 was expressed at low level in HUVECs and significantly up-regulated by IFN-γ at mRNA (Fig. 1A) and protein (Fig. 1, B and C) levels. Because HDAC3 participates in IFN-γ downstream signaling, we wondered whether HDAC3 is involved in IFN-γ-induced galectin-9 expression in HUVECs. To test this, we assessed whether IFN-γ up-regulated HDAC activity. Indeed, IFN-γ activated HDAC in a time-dependent manner (Fig. 2A), whereas HDAC1 and HDAC3 protein levels were not affected (data not shown). The inhibitor experiments showed that both the general HDAC inhibitor, trichostatin A (data not shown), and HDAC3-specific inhibitor, apicidin (30) (Fig. 2, B–D) abolished IFN-γ-induced galectin-9 expression at mRNA (Fig. 2D) and protein (Fig. 2, B and C) levels. Further experiments with shRNA lentivirus-mediated gene knockdown assay demonstrated that knockdown of HDAC3 not only decreased the basal level of galectin-9 expression (Fig. 3, A–C) but also abolished the IFN-γ-induced galectin-9 expression (Fig. 3, D and E). Importantly, a cell plasma membrane fraction assay revealed that IFN-γ induced galectin-9 translocation to plasma membrane whereas the location of HDAC3 on the membrane remained unchanged (Fig. 3D). These results suggest that HDAC3 is essential for the basal level and IFN-γ-induced galectin-9 expression in HUVECs.
FIGURE 1.
IFN-γ induces galectin-9 expression in HUVECs at mRNA and protein levels. A and B, HUVECs were treated with 10 ng/ml IFN-γ for the time indicated, followed by quantitative RT-PCR analysis of galectin-9 mRNA (A) and Western blot analysis of galectin-9 protein (B, lower panel represents the relative galectin-9 protein level to GAPDH). *, p < 0.05; **, p < 0.01. C, HUVECs were treated with 10 ng/ml IFN-γ for 16 h, followed by immunofluorescent staining with galectin-9 antibody (green). DAPI was included to counterstain the nucleus (blue). PBS was used as vehicle control. IgG was included as primary antibody control of the staining. Scale bar, 5 μm. Note that the fluorescence intensity in IFN-γ-treated cells is significantly higher than untreated cells. Data presented are representative of mean ± S.E. (error bars) of three independent experiments.
FIGURE 2.
HDAC3 activity is essential for galectin-9 expression. A, IFN-γ increases HDAC activity. HUVECs were treated with 10 ng/ml IFN-γ for the time indicated, followed by HDAC activity assay. Arbitrary unit was defined as A420 nm/μg of protein with untreated set as 1.0. *, p < 0.05. B–D, apicidin decreased the basal level and IFN-γ-induced galectin-9 expression. HUVECs were pretreated with 200 nmol/liter apicidin for 1 h, then treated with 10 ng/ml IFN-γ for 16 h in the presence of apicidin, followed by immunofluorescent staining (B: green, galectin-9; blue, DAPI; Scale bar, 5 μm), Western blot assay (C) and quantitative RT-PCR analysis (D) are shown. *, p < 0.05; ***, p < 0.0001. Data presented are representative of mean ± S.E. (error bars) of three independent experiments. DMSO, dimethyl sulfoxide.
FIGURE 3.
Knockdown of HDAC3 decreases basal level and IFN-γ-induced galectin-9 expression. A–C, knockdown of HDAC3 decreased basal level of galectin-9 expression. HUVECs were infected with nontarget (NTsh) or HDAC3 (HD3sh) shRNA lentiviruses at 1 × 107 units/1 × 106 cells for 72 h, followed by Western blotting (A), routine RT-PCR(B), and quantitative RT-PCR (C) assays. An arbitrary unit was defined as the ratio of target gene mRNA to 18 S RNA with that of NTsh set as 1.0. **, p < 0.01; ***, p < 0.0001. D and E, knockdown of HDAC3 abolishes IFN-γ-induced galectin-9 expression. HUVECs were infected with NTsh or HD3sh RNA at 1 × 107 units/1 × 106 cells for 48 h, then treated with 10 ng/ml IFN-γ for 16 h, followed by membrane fraction isolation and RNA extraction. HDAC3 and galectin-9 protein levels were detected in total cell lysate and membrane fractions by Western blotting (D), and mRNA was detected with quantitative RT-PCR (E). *, p < 0.05; ***, p < 0.0001. Data presented are representative of mean ± S.E. (error bars) of three independent experiments.
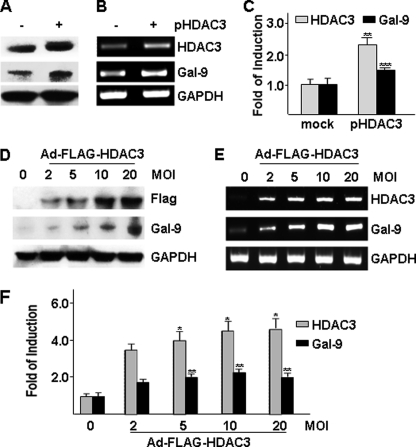
Overexpression of HDAC3 Up-regulates Galectin-9 Expression in HUVECs
To explore whether up-regulation of HDAC3 protein level could affect galectin-9 expression, HUVECs were transfected with pShuttle2-HDAC3 plasmid (Fig. 4, A–C, pHDAC3) or infected with Ad-FLAG-HDAC3 virus (Fig. 4, D–F). pShuttle2-HDAC3 plasmid transfection significantly up-regulated galectin-9 protein level (Fig. 4A) due to mRNA increase (Fig. 4, B and C). Similarly, Ad-DAC3 gene transfer up-regulated galectin-9 protein (Fig. 4D) and mRNA (Fig. 4, E and F) levels in a dose-dependent manner. These results suggest that elevated HDAC3 is sufficient to up-regulate galectin-9 expression in HUVECs.
FIGURE 4.
Overexpression of HDAC3 increases galectin-9 expression. A–C, HUVECs were transfected with pShuttle2-FLAG-HDAC3 plasmid (pHDAC3, 2 μg/2 × 106cells) via electroporation, followed by Western blotting (A), routine RT-PCR (B), and real time RT-PCR (C) assays 48 h after transfection. pShuttle2-FLAG vector was included as mock control. **, p < 0.01; ***, p < 0.0001. D–F, HUVECs were infected with Ad-FLAG-HDAC3 virus at the m.o.i. indicated, followed by Western blotting (D), routine RT-PCR (E), and real time RT-PCR (F) assays 48 h after infection. Ad-null virus was included as negative control and to compensate the m.o.i. *, p < 0.05; **, p < 0.01. -Fold of induction was defined as the ratio of target gene mRNA to 18 S RNA with that of control group set as 1.0. Data presented are representative of mean ± S.E. (error bars) of three independent experiments.
HDAC3 Regulates PI3K and IRF3 Expression in HUVECs
It has been reported that the PI3K and IRF3 signal pathway is involved in double-stranded RNA-induced galectin-9 expression in vascular endothelial cells (18). We wondered whether a similar pathway was involved in IFN-γ-mediated galectin-9 expression in HUVECs. To explore this hypothesis, HUVECs were pretreated with PI3K inhibitor LY294002 prior to IFN-γ treatment. Immunofluorescent staining showed that LY294002 significantly reduced the galectin-9 protein level (Fig. 5A). Western blotting confirmed that LY294002 reduced the basal level and IFN-γ induced IRF3 phosphorylation at serine residue 386 (Fig. 5B). These results suggest that PI3K-mediated IRF3 phosphorylation at Ser-386 is responsible for the basal and IFN-γ-induced galectin-9 expression in HUVECs. To investigate further the involvement of HDAC3 in this signal pathway, HDAC3 shRNA lentivirus-mediated knockdown assays were performed. Surprisingly, HDAC3 knockdown dramatically decreased PI3K (p85α) and IRF3 protein levels together with IRF3 phosphorylation at a basal level and in response to IFN-γ treatment (Fig. 5, C and D). Quantitative RT-PCR analysis demonstrated that the decrease in protein levels was due to decreased mRNA levels (Fig. 5E). To explore whether IRF3 phosphorylation was necessary for galectin-9 expression, mutated IRF3 was created by substituting Ser-386 with Ala-386. Transient transfection with wild-type IRF3 showed elevated phosphorylation and galectin-9 expression, in contrast to IRF3S386A, although there was robust expression of mutant IRF3 (Fig. 5F). These results suggest that HDAC3 plays a crucial role in regulating galectin-9 expression in endothelial cells through modulating PI3K and IRF3 transcription and that IRF3 phosphorylation is essential for galectin-9 expression.
FIGURE 5.
HDAC3 is essential for IFN-γ-induced PI3K-IRF3 activation and galectin-9 expression. A and B, PI3K is crucial for IFN-γ-induced galectin-9 expression and IRF3 activation. HUVECs were pretreated with PI3K inhibitor LY294002 (5 μmol/liter) for 1 h, then treated with 10 ng/ml IFN-γ for 16 h in the presence of LY294002, followed by immunofluorescent staining (A: green, galectin-9; blue, DAPI; Scale bar, 5 μm) and Western blot assay (B). The same amount of dimethyl sulfoxide was included as vehicle control. p-IRF3 indicates phosphorylation of IRF3 at Ser-386. C–E, HDAC3 is crucial for basal level expression of PI3K and IRF3 and IFN-γ-induced IRF3 activation. HUVECs were infected with NTsh or HD3sh RNA for 48 h, then treated with 10 ng/ml IFN-γ for 16 h, followed by Western blot analysis (C, representative image; D, quantitative analysis) and quantitative RT-PCR assays (E). *, p < 0.05; **, p < 0.01. F, IRF3 phosphorylation is essential for galectin-9 expression. HUVECs were transfected with pcDNA3 (vector), pcDNA3-IRF3 (WT), or pcDNA3-IRF3S386A (S386A) plasmids for 48 h, followed by Western blot analysis of IRF3 phosphorylation and galectin-9 expression. Data presented are representative or average ± S.E. (error bars) of three independent experiments.
HDAC3 Is Essential for IRF3 Nuclear Translocation
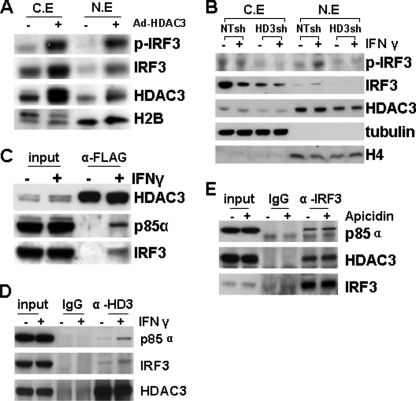
Further experiments were performed to assess the effect of HDAC3 on IRF3 subcellular location. As shown in Fig. 6A, overexpression of HDAC3 via adenoviral gene transfer increased IRF3 nuclear translocation and its phosphorylation. However, when HDAC3 was knocked down by shRNA lentivirus, IRF3 expression, phosphorylation, and nuclear location were significantly reduced (Fig. 6B). Most importantly, IFN-γ-induced IRF3 phosphorylation and nuclear translocation were totally ablated (Fig. 6B). These results suggest that HDAC3 is essential for IFN-γ-induced IRF3 nuclear translocation. Data presented are representative or the average of three independent experiments.
FIGURE 6.
IFN-γ enhances the HDAC3-p85α-IRF3 complex formation. A, overexpression of HDAC3 induced IRF3 nuclear translocation. HUVECs were infected with 10 m.o.i. Ad-HDAC3 virus for 48 h, followed by cellular fraction assay. Ad-null virus was included as control. C.E, cytosolic extract; N.E, nuclear extract. B, HDAC3 knockdown ablated IFN-γ-induced IRF3 nuclear translocation. HUVECs were infected with NTsh or HD3sh for 72 h, and then treated with 10 ng/ml IFN-γ for 4 h, followed by cellular fraction assay. Anti-tubulin and histone H4 antibodies were included to identify cytosol and nuclear fractions. C, IFN-γ induced the formation of the HDAC3-p85α-IRF3 complex. HUVECs were infected with Ad-FLAG-HDAC3 virus at 10 m.o.i. and 48 h later treated with 10 ng/ml IFN-γ for 4 h, followed by immunoprecipitation assay with anti-FLAG antibody. 50 μg of lysate was included as input. D, IFN-γ enhanced endogenous HDAC3 association with p85α-IRF3. HUVECs were treated with 10 ng/ml IFN-γ for 4 h, followed by immunoprecipitation assay with anti-HDAC3 antibody. E, HDAC3 activity is unnecessary for HDAC3-p85α-IRF3 complex formation. HUVECs were treated with 10 ng/ml IFN-γ for 4 h in the presence or absence of apicidin, followed by immunoprecipitation with anti-IRF3 antibody. Dimethyl sulfoxide was included as vehicle control.
HDAC3 Forms a Complex with PI3K and IRF3 in Response to IFN-γ
HDAC3 is reported to interact with nonhistone proteins, such as Akt1 and MEF2 (28, 31, 32). To investigate whether HDAC3 physically interacts with PI3K and IRF3 in addition to regulating their expression, Ad-HDAC3-infected or uninfected HUVECs were treated with IFN-γ followed by immunoprecipitation assay with anti-FLAG (for exogenous HDAC3) or anti-HDAC3 (endogenous). No association of exogenous HDAC3 was detected in untreated cells. However, IFN-γ treatment induced HDAC3 association with PI3K and IRF3 (Fig. 6B). Surprisingly, when we used anti-HDAC3 antibody to pull down endogenous HDAC3, the association of HDAC3 with PI3K and IRF3 could be detected at low levels in untreated cells, which was significantly enhanced by IFN-γ treatment (Fig. 6C). Because HDAC3 is a deacetylase, we wondered whether the deacetylase activity is necessary for this interaction. To test this hypothesis, immunoprecipitation was performed with anti-IRF3 antibody under IFN-γ treatment in the absence or presence of HDAC3 inhibitor, apicidin. As shown in Fig. 6D, the presence of apicidin did not affect the interaction of HDAC3, IRF3, and PI3K. These results suggest that HDAC3 may be directly involved in PI3K-IRF3 signal transduction acting as a scaffold protein independent of deacetylase activity.
DISCUSSION
Through interaction with β-galactosides containing proteins, galectin-9 may function in cell-to-cell communication involved in inflammatory response and tumor metastasis. In the present study, we demonstrated that HDAC3 is required for the constitutive galectin-9 expression. In addition, HDAC3 is activated by IFN-γ and forms a complex with PI3K and IRF3, which increases PI3K-mediated IRF3 phosphorylation, leading to up-regulation of galectin-9 transcription. This is the first time that HDAC3 has been shown to play a role in regulating the expression of immune modulator molecules such as galectin-9 in vascular endothelial cells.
Galectin-9 can exist extracellularly, on the plasma membrane, and intracellularly. Different locations may determine its different functions. In extracellular matrix, galectin-9 may function as chemoattractant of eosinophils (33). On the cellular membrane, galectin-9 may mediate cell-to-cell communication via its specific receptor, Tim3, or other β-galactosides containing proteins (34, 35).
Intracellularly, galectin-9 may interact with β-galactosides containing signal transducer, leading to gene transcription regulation, such as cytokine production in monocytes and T cells (12, 13). In endothelial cells, the expression of galectin-9 can be controlled by HDAC3. Inhibition of HDAC3 activity by apicidin or knockdown of HDAC3 protein level decreases the galectin-9 expression. In this study, we found that IFN-γ exerted dual effects on galectin-9: induction of expression and membrane location. The former is HDAC3-dependent, as apicidin and HDAC3 shRNA abolish IFN-γ-induced galectin-9 expression at mRNA and protein levels. However, the latter is HDAC3-independent. In HDAC3 shRNA lentivirus-infected cells, although the total protein level of galectin-9 is significantly decreased, IFN-γ still increases the galectin-9 location on membrane compared with untreated cells. Galectin-9 on endothelial membrane increases eosinophil adhesion (16). These findings suggest that HDAC3 may function in inflammatory responses through regulating galectin-9 expression, which in turn affects cell-to-cell interaction.
Our previous study indicated that disturbed flow induced HDAC3 translocation to plasma membrane in endothelial cells (28). On plasma membrane, HDAC3 can associate with c-Src and is phosphorylated by c-Src, which is essential for HDAC3 activation (36, 37). In this study, IFN-γ may activate HDAC3 in another unknown way, because membrane-associated HDAC3 remains unchanged under IFN-γ treatment. Activation of HDAC3 is crucial for IFN-γ-induced galectin-9 expression. HDAC3-specific inhibition with apicidin can abolish the IFN-γ-induced galectin-9 expression.
HDAC3 may regulate galectin-9 expression in two ways. On the one hand, HDAC3 may function as a scaffold protein for PI3K and IRF3 interaction, which is independent of HDAC3 activity, as the presence of apicidin does not affect PI3K and IRF3 interaction. This HDAC3-PI3K-IRF3 complex may be responsible for the constitutive expression of galectin-9 in untreated cells. On the other hand, activated HDAC3 activity may promote IRF3 phosphorylation and its access to gene promoter. Further experiments will be needed to address this issue. Furthermore, it has been reported that the N-terminal region of HDAC3 is responsible for its protein/protein interaction. In this study, immunoprecipitation with anti-FLAG antibody that detects exogenous HDAC3 binding with PI3K and IRF3 was in contrast to that detected with endogenous HDAC3 interactions. Immunoprecipitation with anti-HDAC3 antibody that detects endogenous HDAC3 shows association of HDAC3 with PI3K and IRF3 in untreated cells. This basal level of interaction is not seen with exogenous immunoprecipitation of HDAC3. Exogenous HDAC3 is expressed as a fusion protein, in which a FLAG tag is fused into the N-terminal of HDAC3. Anti-FLAG antibody will affect the access of associated proteins to the N-terminal region of HDAC3. The anti-HDAC3 recognizes an epitope in the far C terminus. Thus, these results also imply that the N-terminal domain of HDAC3 may mediate its association with PI3K and IRF3.
IRF3 dimerization and transcriptional activity are regulated by phosphorylation (for review see Ref. 38), one of which is the phosphorylation site at Ser-386. IRF3 Ser-386 phosphorylation is mediated by PI3K in response to virus infection (39). In our study, IFN-γ induces IRF3 Ser-386 phosphorylation in a PI3K- and HDAC3-dependent manner. Both PI3K inhibitor and HDAC3 knockdown abolish IFN-γ induced IRF3 Ser-386 phosphorylation. The Ser-386 phosphorylation is imperative for the IRF3-mediated galectin-9 expression, as S386A mutant IRF3 losses its regulatory function on galectin-9 expression. Importantly, HDAC3 is essential for the constitutive expression of PI3K and IRF3. Knockdown of HDAC3 decreases PI3K-IRF3 protein and mRNA levels, implying that HDAC3 positively regulates PI3K-IRF3 transcription. This finding complements the observation that HDAC3 knockdown decreased Akt phosphorylation, substrate of PI3K (28). HDAC3 may regulate PI3K and IRF3 transcription through either a direct or an indirect manner. In the direct manner, HDAC3 may function through its protein/protein interaction or through its deacetylase activity for the activation/inactivation of transcriptional factors responsible for PI3K and IRF3 transcription. For example, Ozaki and colleagues demonstrate that the E2F1 transcription factor can be regulated by its acetylation status (40). Increasing the acetylation of E2F1 by trichostatin A treatment enhanced its transcriptional activity on its target genes. Surprisingly, Xu et al. have shown that IRF3 transcription can be negatively regulated by the E2F1 transcription factor through binding on the human IRF3 promoter region (41). Taken together, HDAC3 knockdown or inhibition of HDAC3 activity may promote E2F1 negative activity toward IRF3 transcription, reducing IRF3 transcription. In the indirect manner, HDAC3 may function as a standard deacetylase co-repressor negatively regulating the transcriptional expression of a repressor. HDAC3 deficiency will increase the expression of the repressor, which in turn down-regulates PI3K and IRF3 transcription. However, the detailed mechanism needs further investigations. Considering the fact that PI3K is involved in multiple signaling pathways, it implicates HDAC3 in multiple cellular processes including endothelial/inflammatory cell interaction.
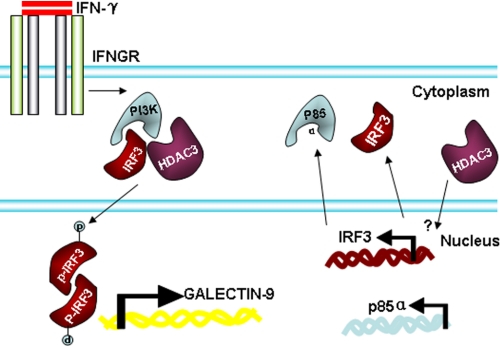
In summary, HDAC3 regulates the basal level of PI3K and IRF3 transcription, which may be responsible for the constitutive expression of galectin-9 in endothelial cells. Under IFN-γ treatment, HDAC3 is activated and forms a complex with PI3K and IRF3, leading to PI3K-mediated IRF3 phosphorylation and galectin-9 expression (Fig. 7). This study provides new evidence of HDAC3 involvement in not only modulating IRF3 and galectin-9 expression but also in other signal pathways possibly via regulating PI3K transcription in endothelial cells.
FIGURE 7.
Schematic illustration of IFN-γ-mediated galectin-9 expression in HUVECs. In HUVECs, HDAC3 is involved in the constitutive expression of IRF3 and the p85α subunit of PI3K. Under IFN-γ treatment, HDAC3, PI3K, and IRF3 form a complex, leading to the phosphorylation of IRF3 at serine 386, which in turn activates galectin-9 expression.
This work was supported by grants from British Heart Foundation and Oak Foundation.
- Tim3
- T cell immunoglobin domain and mucin domain 3
- HDAC
- histone deacetylase
- HD3sh
- HDAC3 shRNA lentivirus
- HUVEC
- human umbilical vein endothelial cell
- IRF
- IFN response factor
- m.o.i.
- multiplicity of infection
- NTsh
- nontarget shRNA lentivirus.
REFERENCES
- 1. Leffler H., Carlsson S., Hedlund M., Qian Y., Poirier F. (2004) Glycoconj. J. 19, 433–440 [DOI] [PubMed] [Google Scholar]
- 2. Norling L. V., Perretti M., Cooper D. (2009) J. Endocrinol. 201, 169–184 [DOI] [PubMed] [Google Scholar]
- 3. Yang R. Y., Rabinovich G. A., Liu F. T. (2008) Expert Rev. Mol. Med. 10, e17. [DOI] [PubMed] [Google Scholar]
- 4. Asakura H., Kashio Y., Nakamura K., Seki M., Dai S., Shirato Y., Abedin M. J., Yoshida N., Nishi N., Imaizumi T., Saita N., Toyama Y., Takashima H., Nakamura T., Ohkawa M., Hirashima M. (2002) J. Immunol. 169, 5912–5918 [DOI] [PubMed] [Google Scholar]
- 5. Matsumoto R., Matsumoto H., Seki M., Hata M., Asano Y., Kanegasaki S., Stevens R. L., Hirashima M. (1998) J. Biol. Chem. 273, 16976–16984 [DOI] [PubMed] [Google Scholar]
- 6. Zhu C., Anderson A. C., Schubart A., Xiong H., Imitola J., Khoury S. J., Zheng X. X., Strom T. B., Kuchroo V. K. (2005) Nat. Immunol. 6, 1245–1252 [DOI] [PubMed] [Google Scholar]
- 7. Arikawa T., Saita N., Oomizu S., Ueno M., Matsukawa A., Katoh S., Kojima K., Nagahara K., Miyake M., Yamauchi A., Kohrogi H., Hirashima M. (2010) Eur. J. Immunol. 40, 548–558 [DOI] [PubMed] [Google Scholar]
- 8. Wada J., Ota K., Kumar A., Wallner E. I., Kanwar Y. S. (1997) J. Clin. Invest. 99, 2452–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He W., Fang Z., Wang F., Wu K., Xu Y., Zhou H., Du D., Gao Y., Zhang W. N., Niki T., Hirashima M., Yuan J., Chen Z. K. (2009) Transplantation 88, 782–790 [DOI] [PubMed] [Google Scholar]
- 10. Wang F., Wan L., Zhang C., Zheng X., Li J., Chen Z. K. (2009) Immunobiology 214, 342–349 [DOI] [PubMed] [Google Scholar]
- 11. Wang F., He W., Zhou H., Yuan J., Wu K., Xu L., Chen Z. K. (2007) Cell. Immunol. 250, 68–74 [DOI] [PubMed] [Google Scholar]
- 12. Su E. W., Bi S., Kane L. P. (2011) Glycobiology 21, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Matsuura A., Tsukada J., Mizobe T., Higashi T., Mouri F., Tanikawa R., Yamauchi A., Hirashima M., Tanaka Y. (2009) Genes Cells 14, 511–521 [DOI] [PubMed] [Google Scholar]
- 14. Seki M., Oomizu S., Sakata K. M., Sakata A., Arikawa T., Watanabe K., Ito K., Takeshita K., Niki T., Saita N., Nishi N., Yamauchi A., Katoh S., Matsukawa A., Kuchroo V., Hirashima M. (2008) Clin. Immunol. 127, 78–88 [DOI] [PubMed] [Google Scholar]
- 15. Tsuboi Y., Abe H., Nakagawa R., Oomizu S., Watanabe K., Nishi N., Nakamura T., Yamauchi A., Hirashima M. (2007) Clin. Immunol. 124, 221–233 [DOI] [PubMed] [Google Scholar]
- 16. Imaizumi T., Kumagai M., Sasaki N., Kurotaki H., Mori F., Seki M., Nishi N., Fujimoto K., Tanji K., Shibata T., Tamo W., Matsumiya T., Yoshida H., Cui X. F., Takanashi S., Hanada K., Okumura K., Yagihashi S., Wakabayashi K., Nakamura T., Hirashima M., Satoh K. (2002) J. Leukocyte Biol. 72, 486–491 [PubMed] [Google Scholar]
- 17. Imaizumi T., Kumagai M., Nishi N., Hirashima M., Hatakeyama M., Tamo W., Yoshida H., Nakamura T., Okumura K., Satoh K. (2003) Int. Arch. Allergy Immunol. 131, 57–61 [DOI] [PubMed] [Google Scholar]
- 18. Imaizumi T., Yoshida H., Nishi N., Sashinami H., Nakamura T., Hirashima M., Ohyama C., Itoh K., Satoh K. (2007) Glycobiology 17, 12C–15C [DOI] [PubMed] [Google Scholar]
- 19. Gray S. G., Ekström T. J. (2001) Exp. Cell Res. 262, 75–83 [DOI] [PubMed] [Google Scholar]
- 20. Yang W. M., Tsai S. C., Wen Y. D., Fejer G., Seto E. (2002) J. Biol. Chem. 277, 9447–9454 [DOI] [PubMed] [Google Scholar]
- 21. Montgomery R. L., Potthoff M. J., Haberland M., Qi X., Matsuzaki S., Humphries K. M., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) J. Clin. Invest. 118, 3588–3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang J., Mahmud S. A., Bitterman P. B., Huo Y., Slungaard A. (2007) J. Biol. Chem. 282, 28408–28418 [DOI] [PubMed] [Google Scholar]
- 23. Zhang M. X., Zhang C., Shen Y. H., Wang J., Li X. N., Chen L., Zhang Y., Coselli J. S., Wang X. L. (2008) Mol. Biol. Cell 19, 3997–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jung S. B., Kim C. S., Naqvi A., Yamamori T., Mattagajasingh I., Hoffman T. A., Cole M. P., Kumar A., Dericco J. S., Jeon B. H., Irani K. (2010) Circ. Res. 107, 877–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inoue K., Kobayashi M., Yano K., Miura M., Izumi A., Mataki C., Doi T., Hamakubo T., Reid P. C., Hume D. A., Yoshida M., Aird W. C., Kodama T., Minami T. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 2652–2659 [DOI] [PubMed] [Google Scholar]
- 26. Zeng L., Xiao Q., Margariti A., Zhang Z., Zampetaki A., Patel S., Capogrossi M. C., Hu Y., Xu Q. (2006) J. Cell Biol. 174, 1059–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao Q., Zeng L., Zhang Z., Margariti A., Ali Z. A., Channon K. M., Xu Q., Hu Y. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 2244–2251 [DOI] [PubMed] [Google Scholar]
- 28. Zampetaki A., Zeng L., Margariti A., Xiao Q., Li H., Zhang Z., Pepe A. E., Wang G., Habi O., deFalco E., Cockerill G., Mason J. C., Hu Y., Xu Q. (2010) Circulation 121, 132–142 [DOI] [PubMed] [Google Scholar]
- 29. Kirshner J. R., Karpova A. Y., Kops M., Howley P. M. (2005) J. Virol. 79, 9320–9324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Strobl J. S., Cassell M., Mitchell S. M., Reilly C. M., Lindsay D. S. (2007) J. Parasitol. 93, 694–700 [DOI] [PubMed] [Google Scholar]
- 31. Fischle W., Dequiedt F., Fillion M., Hendzel M. J., Voelter W., Verdin E. (2001) J. Biol. Chem. 276, 35826–35835 [DOI] [PubMed] [Google Scholar]
- 32. Grégoire S., Xiao L., Nie J., Zhang X., Xu M., Li J., Wong J., Seto E., Yang X. J. (2007) Mol. Cell. Biol. 27, 1280–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chagan-Yasutan H., Shiratori B., Siddiqi U. R., Saitoh H., Ashino Y., Arikawa T., Hirashima M., Hattori T. (2010) Clin. Mol. Allergy 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jayaraman P., Sada-Ovalle I., Beladi S., Anderson A. C., Dardalhon V., Hotta C., Kuchroo V. K., Behar S. M. (2010) J. Exp. Med. 207, 2343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Irie A., Yamauchi A., Kontani K., Kihara M., Liu D., Shirato Y., Seki M., Nishi N., Nakamura T., Yokomise H., Hirashima M. (2005) Clin. Cancer Res. 11, 2962–2968 [DOI] [PubMed] [Google Scholar]
- 36. Matteucci E., Ridolfi E., Maroni P., Bendinelli P., Desiderio M. A. (2007) Mol. Cancer Res. 5, 833–845 [DOI] [PubMed] [Google Scholar]
- 37. Longworth M. S., Laimins L. A. (2006) Oncogene 25, 4495–4500 [DOI] [PubMed] [Google Scholar]
- 38. Savitsky D., Tamura T., Yanai H., Taniguchi T. (2010) Cancer Immunol. Immunother. 59, 489–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao Y., Rivieccio M. A., Lutz S., Scemes E., Brosnan C. F. (2006) Glia 54, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ozaki T., Okoshi R., Sang M., Kubo N., Nakagawara A. (2009) Biochem. Biophys. Res. Commun. 386, 207–211 [DOI] [PubMed] [Google Scholar]
- 41. Xu H. G., Ren W., Lu C., Zhou G. P. (2010) Mol. Biol. Rep. 37, 3073–3080 [DOI] [PubMed] [Google Scholar]