Background: MicroRNA processing is a tightly controlled multistep process involving accessory proteins.
Results: Expression of microRNAs 15a and 16 is controlled by nucleolin expression and localization.
Conclusion: Nucleolin facilitates the biogenesis of microRNAs 15a and 16 through direct interaction with the microprocessor complex.
Significance: Nucleolin represents a novel component of the microRNA processing pathway.
Keywords: Bcl-2, MicroRNA, Nucleolus, Ribosomal RNA (rRNA), RNA Processing, Nucleolin, Biogenesis, Microprocessor Complex
Abstract
MicroRNAs (miRNA) are endogenous, short, non-coding RNA that undergo a multistep biogenesis before generating the functional, mature sequence. The core components of the microprocessor complex, consisting of Drosha and DGCR8, are both necessary and sufficient for this process, although accessory proteins have been found that modulate the biogenesis of a subset of miRNA. Curiously, many of the proteins involved in miRNA biogenesis are also needed for ribosomal RNA processing. Here we show that nucleolin, another protein critical for rRNA processing, is involved in the biogenesis of microRNA 15a/16 (miR-15a/16), specifically at the primary to precursor stage of processing. Through overexpression and knockdown studies, we show that miR-15a/16 levels are directly correlated to nucleolin expression. Furthermore, we found that cellular localization is critical for the proper functioning of nucleolin in this pathway and that nucleolin directly interacts with DGCR8 and Drosha in the nucleus. Nucleolin can bind to the primary miRNA both directly and specifically. Finally, we show that in the absence of nucleolin, cell extracts are unable to process miR-15a/16 in vitro and that this can be rescued by the addition of nucleolin. Our findings offer a new protein component in the microRNA biogenesis pathway and lend insight into miRNA dysregulation in certain cancers.
Introduction
MicroRNA (miRNA)3 are short ∼21-nucleotide single-stranded non-coding RNA that affect gene expression by inhibiting translation or degrading mRNA targets by binding to their 3′-untranslated region (3′-UTR) (1). Transcripts from miRNA-encoding genes generate primary miRNA (pri-miRNA) that vary in size from one to tens of kilobases and contain a 5′ cap and a poly(A) tail. These pri-miRNA are processed in the microprocessor complex composed of Drosha and DGCR8 into a 60–90-nucleotide pre-miRNA (2). After being exported to the cytoplasm by Exportin-5 and Ran-GTP, pre-miRNA are cleaved by Dicer, which transfers the double-stranded RNA to the Argonaut complex to generate the mature miRNA (3–5).
The miRNA biogenesis pathway is tightly regulated with numerous other proteins transiently associating with the individual complexes to either stimulate or inhibit processing. For example, transient interaction of p53, p68, and p72 with Drosha increases the processing of a subset of miRNA (6, 7). Negative regulators of miRNA processing include the estrogen receptor, NF45-NF90 complexes, and Lin28 (8–10). The AU-rich binding protein KH-type splicing regulatory protein (KSRP) can interact with both the microprocessor and the Dicer complexes and affect their function (11, 12). The effect of these regulators on miRNA processing can be very specific, as is the case of heterogeneous nuclear ribonucleoprotein A1, which facilitates the specific processing of miR-18a from the polycistronic miRNA miR-17-92 and Lin28, which blocks the biogenesis of let-7 family miRNA (10, 13). Although KH-type splicing regulatory protein and Lin28 are the only two regulatory proteins known to affect Dicer, most of the transient effectors characterized thus far interact exclusively with the microprocessor complex, suggesting that this may be a critical step in the regulation of miRNA expression.
An interesting parallel exists between miRNA biogenesis and ribosomal RNA (rRNA) biogenesis with a number of proteins having important roles in both pathways. Drosha was originally identified for its role in rRNA biogenesis, cleaving the 48 S pre-rRNA into the 12 S rRNA intermediate (14). Both p68 and p72 are responsible for cleavage of the 12 S rRNA to generate the mature 5.8 S rRNA species (15, 16). Additionally, Dicer has recently been found to associate with the chromatin encoding rRNA (17). Depletion of either Dicer or Ago2 results in an accumulation of 5.8 S rRNA species, indicating that they are necessary for processing (18). Recently, it was found that Drosha co-localizes with nucleolin in the nucleolus to increase the biogenesis of a mouse long non-coding RNA mrhl, which is a 2.8-kb RNA that gets cleaved into an 80-nucleotide RNA that can be further processed into a 21-nucleotide RNA by Dicer (19). Strikingly, this pathway, like that of rRNA biogenesis, is very similar to the miRNA biogenesis pathway with regard to proteins and RNA substrates.
Nucleolin has long been known as a protein critical for rRNA biogenesis (20). Evidence suggests that it may also have a role as an accessory protein in miRNA biogenesis. Nucleolin is predominantly a nucleolus-localized protein; however, in a number of different cancers, nucleolin is found largely in the cytoplasm (21–24). In the cytoplasm, nucleolin functions to stabilize the mRNA of bcl-2, thereby inhibiting apoptosis (21). When cytoplasmic levels of nucleolin are decreased upon treatment with all-trans-retinoic acid, the levels of miR-15a and miR-16 increase in both acute promyelocytic leukemia cell lines and patients with acute promyelocytic leukemia treated with all-trans-retinoic acid (25, 26). miR-15a and -16 have been shown to target bcl-2 mRNA and are greatly decreased in chronic lymphocytic leukemia, where nucleolin is predominantly localized to the cytoplasm (21, 27).
Therefore, we sought out to investigate the role of nucleolin in controlling the expression of miR-15a and miR-16. We determined how overall expression of nucleolin and its cellular localization impacts miRNA expression. Additionally, we characterized the interaction of nucleolin with components of the miRNA biogenesis pathway and with miRNA directly.
EXPERIMENTAL PROCEDURES
Cell Culture
All cell lines were purchased from ATCC (Manassas, VA). HEK293 and MCF-7 cells were grown in DMEM with 10% FBS. MOLM-13 cells were grow in RPMI with 10% heat-inactivated FBS.
Antibodies, Drugs, and Oligonucleotides
Antibodies to nucleolin (clone 4E2, MBL International), FLAG M2 (Sigma-Aldrich), HA (Roche Applied Science, clone 3F10), Drosha (Cell Signaling, D28B1), GAPDH (Santa Cruz Biotechnology, SC-32233), PARP (Santa Cruz Biotechnology, SC-1019), and β-actin (Sigma-Aldrich, A2228) were obtained from the suppliers indicated. Parthenolide was purchased from Sigma-Aldrich. Oligonucleotides used in this study were obtained from Sigma-Aldrich and are listed in supplemental Table 1.
Plasmids and siRNA Transfection
The pCMV2-FLAG-NCL was generously donated by Paula Bates (University of Louisville) and previously described (28). The tandem affinity purification-tagged DGCR8 expression vector was generated by the Tuschl laboratory and was obtained through Addgene (Plasmid ID: 10921) (29). Cells stably transfected with the pCMV2-FLAG-NCL plasmid were selected for 3 weeks with 800 μg/ml G418 and maintained with 200 μg/ml G418. Cells transfected with pFLAG/HA-DGCR8 were selected in 10 μg/ml puromycin for 1 week and maintained with 1 μg/ml. Primary miRNA for in vitro processing assays were reverse-transcribed from cDNA using the SuperScript III first strand cDNA synthesis kit (Invitrogen), and PCR products included ∼200 nucleotides of the flanking sequences. The gel-purified products were inserted into the pGEM-T-easy vector (Promega) by TA cloning, and inserts were verified by sequencing. Small interfering RNA were purchased from Dharmacon and Sigma-Aldrich and routinely achieved >70% knockdown efficiency. Transfections were carried out using Lipofectamine 2000 with 40 nm siRNA for 48–72 h before knockdown experiments were carried out.
RNA Extraction and qRT-PCR
RNA was extracted using TRIzol according to the manufacturer's instructions. RNA was reverse-transcribed and quantified using the TaqMan miRNA assay kit (Applied Biosystems) according to the manufacturer's instructions.
Northern Blotting
10 μg of RNA was run on a 15% PAGE-urea gel and transferred to BrightStar-Plus positively charged nylon membranes (Ambion, Inc.) by semidry transfer. The membrane was cross-linked using a Stratalinker 1800 (Stratagene) at 0.12 J/cm2 followed by baking for 30 min at 80 °C. Membranes were prehybridized in ULTRAhyb-Oligo (Ambion) for 30 min at 42 °C before the addition of 106 cpm of 32P-labeled synthetic LNA probes (Exiqon) and incubated overnight at 42 °C. Membranes were washed two times for 30 min at 37 °C in 2× SSC and 0.5% SDS before being exposed on a phosphorimaging screen overnight.
Cellular Fractionation
Cells were trypsinized and washed three times with PBS. Three packed cell volumes of cytoplasmic extraction buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, pH 8.0, 0.1 mm EGTA, 1 mm DTT) were used to resuspend the pellet. After 15 min on ice, Nonidet P-40 was added to a final concentration of 0.3%, gently mixed, and centrifuged at 4 °C for 1 min at 10,000 × g. The supernatant was saved as the cytoplasmic fraction. The pellet was washed once with 2 volumes of cytoplasmic extraction buffer. The pellet was resuspended in 1 volume of nuclear extract buffer (20 mm Tris, pH 7.9, 400 mm NaCl, 0.2 mm EDTA, pH 8.0), incubated for 30 min on ice, and centrifuged for 5 min at 10,000 × g. The supernatant was saved as the nuclear extract, and the pellet was discarded.
Immunoprecipitation
HEK293 cells stably expressing either FLAG/HA-DGCR8 or FLAG-NCL were collected, washed with PBS, and lysed in Nonidet P-40 whole cell extraction buffer (20 mm HEPES, pH 8.0, 150 mm NaCl, 0.1% Nonidet P-40, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride (PMSF), and 1 μg/ml each of leupeptin, pepstatin, and aprotinin). 2.5 μg of antibody was added to the 500-μl immunoprecipitation overnight at 4 °C. RNase A (20 μg/ml) treatments were overnight at 4 °C. 50 μl of protein A/G-agarose (Santa Cruz Biotechnology) was added to the lysates for 2 h at 4 °C. Beads were washed three times in lysis buffer (20 mm HEPES, pH 8.0, 150 mm NaCl, 0.1% Nonidet P-40) and boiled in 60 μl of 2× Laemmli sample buffer. 20 μl of the boiled beads and 10% of input and supernatant were resolved by SDS-PAGE and Western blotted with the described antibodies.
RNA Immunoprecipitation
RNA immunoprecipitation was carried out as described previously with slight modifications (30). HEK293 cells were trypsinized, washed twice with PBS, and resuspended in 10 ml of PBS. Cells were cross-linked with 1% formaldehyde for 10 min followed by quenching with 0.25 m glycine for 10 min at room temperature and washed twice with PBS. The pellet was resuspended in buffer A (5 mm PIPES (pH 8.0), 85 mm KCl, 0.5% Nonidet P-40) and incubated on ice for 10 min. Nuclei were collected by spinning at 2500 × g for 5 min at 4 °C. The pellet was washed once with buffer A without Nonidet P-40. The nuclear pellet was resuspended in 500 μl of buffer B (1% SDS, 10 mm EDTA, 50 mm Tris (pH 8.1)) and sonicated. Insoluble material was pelleted at 14,000 × g for 10 min at 4 °C. The supernatant was diluted into immunoprecipitation buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris (pH 8.1), 167 mm NaCl) and incubated with 5 μg of FLAG antibody overnight at 4 °C. Beads were washed once with low salt wash (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris (pH 8.1), 150 mm NaCl), high salt wash (same as low salt but with 500 mm NaCl), and LiCl wash (0.25 m LiCl 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris (pH 8.1)) and twice with Tris-EDTA buffer. Immunoprecipitated complexes were eluted from the beads by treatment with 100 μl of elution buffer (50 mm Tris-HCl (pH 7.5), 10 mm EDTA, 1% SDS) at 65 °C for 10 min. Samples were reverse cross-linked by incubation for 2 h at 42 °C and 6 h at 65 °C in 0.5× elution buffer plus 0.5 mg/ml proteinase K. RNA was extracted with TRIzol and reverse-transcribed in a 20-μl final volume using the high capacity cDNA kit (Applied Biosystems) with random primers. Following reverse transcription, 5 μl of sample was analyzed by PCR with 500 nm primer concentration, and the reaction was allowed to proceed for 30–35 cycles before 10 μl of product was analyzed on a 2% agarose gel.
In Vitro miRNA Processing Assay
The processing assay was carried out as described previously (31). Labeled primary miRNA were generated using the MAXIscript kit (Ambion) according to the manufacturer's instructions. Briefly, 1 μg of linearized plasmid was labeled with 32P-labeled UTP. RNA products were gel-purified and eluted in 0.5 m NH4OAc, 1 mm EDTA, 2% SDS overnight at 37 °C followed by ethanol precipitation. All in vitro transcribed probes were made fresh for each experiment. HEK293 cells were treated with siRNA for 48 h before being collected. Cells were washed in PBS, resuspended in in vitro processing lysis buffer (50 mm Tris (pH 8.0), 100 mm KCl, 0.2 mm EDTA), and sonicated. Insoluble material was pelleted by spinning at 14,000 rpm for 10 min at 4 °C. 15 μl of cell extract was mixed with 3 μl of 64 mm MgCl2, 3 μl of labeled pri-miRNA (∼3 × 104 cpm), and 1 unit/μl SUPERase-in (Ambion), and the final volume was brought to 30 μl with diethyl pyrocarbonate-treated water. For rescue experiments, 5 or 10 μl of beads from a FLAG-nucleolin immunoprecipitation was added to the reaction. The reaction was incubated for 90 min at 37 °C before the RNA was extracted with TRIzol LS (Invitrogen). Purified RNA was run on a 12.5% denaturing PAGE and exposed to a phosphorimaging device overnight.
RESULTS
Expression of miR-15a and miR-16 Correlates with Nucleolin Expression
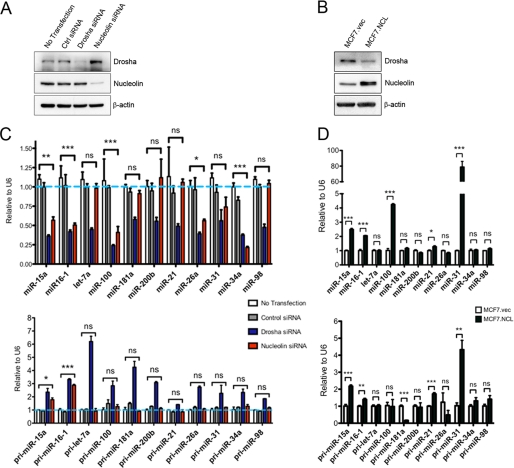
To determine whether nucleolin expression impacts the levels of miRNA, nucleolin was knocked down with siRNA, and cancer-associated mature miRNA was assessed by qRT-PCR. As a positive control, Drosha was also knocked down (Fig. 1A). As expected, all mature miRNA were decreased in Drosha knockdown to 25–50% of control siRNA cells (Fig. 1C). In nucleolin knockdown cells, five miRNA were significantly reduced. Because none of the miRNA tested were increased upon loss of nucleolin, we investigated whether nucleolin directly controlled their expression through processing. Primary miRNA levels were analyzed because loss of proteins involved in processing typically causes an increase in primary miRNA, whereas mature miRNA decrease (2, 15). All primary miRNA increased in Drosha knockdown, whereas only two primary miRNA, miR-15a and miR-16, increased in the absence of nucleolin. This suggests that nucleolin may directly control the expression of these two miRNA, whereas the other miRNA decreased in the absence of nucleolin may be controlled through indirect mechanisms. To determine whether the opposite held true when nucleolin is increased, MCF-7 cells were stably transfected with a CMV-driven expression vector containing FLAG-tagged nucleolin, and miRNA expression was determined by real-time PCR. We were able to achieve an ∼2-fold increase in nucleolin expression (Fig. 1B). Upon overexpression of nucleolin, the levels of mature miR-15a and miR-16 increased ∼2.5-fold (Fig. 1D). Two other miRNA, miR-100 and miR-31, increased significantly with miR-31 increasing nearly 80-fold. This increase may be transcriptionally regulated because the pri-miRNA for miR-31 significantly increased in nucleolin-overexpressing cells. pri-miRNA for miR-15 and miR-16 also increase, however; this may be indirectly regulated through c-Myc. Nucleolin can inhibit c-Myc through formation of a G-quadruplex in the promoter of c-myc, and c-Myc can transcriptionally inhibit miR-15a and miR-16 (32, 33). An interesting observation was that nucleolin expression inversely correlates to Drosha expression (Fig. 1, A and C). It is possible that this relationship is the way cells compensate for altered ribosome biogenesis as this is a common function between the two. We speculate that upon loss of nucleolin, the cell tries to compensate by increasing Drosha, and whenever nucleolin is plentiful, there is less of a need for Drosha in ribosome biogenesis. Altogether, these data are consistent with the hypothesis that nucleolin plays an ancillary role in the biogenesis of miR-15a and miR-16.
FIGURE 1.
Nucleolin expression correlates with miR-15a and miR-16 expression. A, expression of Drosha and nucleolin 72 h following siRNA treatment. Ctrl siRNA, control siRNA. B, expression of stably expressed FLAG-tagged nucleolin in MCF-7 cells. vec, vector. C, real-time PCR analysis of mature miRNA (top) and primary miRNA (bottom) in cells with Drosha or nucleolin knocked down. Results are normalized to U6 small nucleolar RNA expression. D, real-time PCR analysis of mature miRNA (top) and primary miRNA (bottom) in cells overexpressing nucleolin. Error bars = S.E. *, p < 0.1, **, p < 0.01, ***, p < 0.001. ns, not significant.
Induced Cytoplasmic Nucleolin Inhibits Processing of Primary miRNA
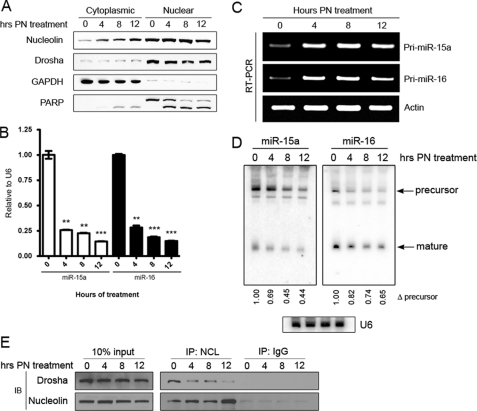
Clinical data from chronic lymphocytic leukemia patients show that nucleolin is localized primarily in the cytoplasm, which leads to increased bcl-2 mRNA levels (21). Because miR-15a and miR-16 are known to target bcl-2 mRNA, we sought to determine whether cytoplasmic nucleolin could lead to decreased levels of miR-15a and miR-16. We found that when MOLM-13 acute myelogenous leukemia cells are treated with parthenolide, a natural product small molecule that affects multiple signal transduction pathways, it induces a dramatic increase in cytoplasmic nucleolin levels (Fig. 2A). Analysis of mature miRNA by qRT-PCR revealed a significant decrease in both miR-15a and miR-16 concurrent with cytoplasmic localization of nucleolin (Fig. 2B). To determine whether treatment with parthenolide decreased the transcription of these miRNA, the pri-miRNA were amplified by PCR. Astonishingly, we found not a decrease but rather an increase in the pri-miRNA precursor of miR-15a and miR-16 (Fig. 2C). Given this finding, we analyzed the precursor species by Northern blot and found that levels of the pre-miR-15 and -16 precursors decreased similarly to that of the mature miRNA following parthenolide treatment (Fig. 2D). These data are similar to that observed when either Drosha or DGCR8 are knocked down, resulting in a decrease of mature species and an increase in the primary precursors (21). Although it was clear that nucleolin increased in cytoplasmic localization, it appeared that nucleolin did not decrease in nuclear expression despite the miRNA expression indicating this. Nucleolin normally resides in the nucleolus and is not evenly dispersed throughout the nucleus. Upon activation of p53, nucleolin leaves the nucleolus (39). We previously reported that parthenolide potently activates p53 (34). It has been reported that Drosha co-localizes with nucleolin in the nucleolus, and so we hypothesized that the interaction between nucleolin and Drosha is decreased upon relocalization of nucleolin out of the nucleolus. To determine this, MOLM13 cells were treated with parthenolide, and nucleolin was immunoprecipitated from nuclear extracts. We found that nucleolin interacts with Drosha and that this interaction is disrupted upon treatment with parthenolide (Fig. 2E). From these data, we concluded that nucleolin facilitates the processing of miR-15a and miR-16 likely through the microprocessor complex, which is localized exclusively in the nucleus. Upon cellular stress, nucleolin alters its localization and is spatially separated from the microprocessor complex, which is responsible for the cleavage of primary miRNA to generate the precursor. We suspect that in the absence of nucleolin, the microprocessor can less effectively cleave the primary species, resulting in the observed buildup of the pri-miRNA for miR-15a and miR-16. Conversely, when the acute promyelocytic leukemia HL-60 cell line was treated with all-trans-retinoic acid, nucleolin was observed to move from the cytoplasm to the nucleus (supplemental Fig. 1A). The increased nuclear localization of nucleolin was accompanied by significant increases in mature miR-15a/16 (supplemental Fig. 1B). The primary miRNA were not increased in these cells following treatment (supplemental Fig. 1C), indicating post-transcriptional regulation.
FIGURE 2.
Decreased association of nucleolin with the microprocessor complex affects miRNA processing. A, MOLM-13 cells were treated with 10 μm PN for the times indicated. The efficiency of cellular fractionation was determined using GAPDH and PARP. Cleaved PARP is visible in the cytoplasm after 8 h of treatment. B, expression of mature miRNA following treatment with PN for the times indicated as determined by qRT-PCR and normalized to U6 small nucleolar RNA expression. Error bars = S.E. **, p < 0.01, ***, p < 0.001. C, PCR of primary miRNA following treatment with PN with actin as a loading control. D, Northern blot analysis of RNA from MOLM13 cells treated with PN. Quantitation of precursor species is indicated below each lane and is normalized to U6. E, Western blot (IB) of nucleolin immunoprecipitation (IP) from nuclear extracts to interrogate the interaction of nucleolin with Drosha in the nucleus following PN treatment.
Knockdown of Nucleolin Ablates Processing of Primary miRNA
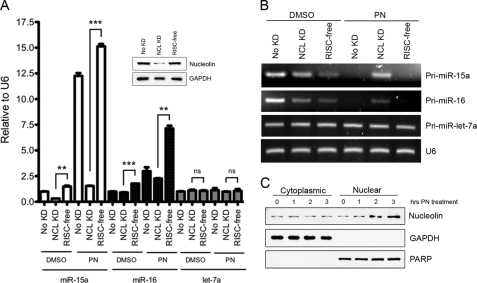
To further validate that nucleolin is involved in the processing of primary miRNA, nucleus-localized nucleolin was induced in MCF-7 cells. This was achieved through treatment of MCF-7 cells with parthenolide. Interestingly, in MCF-7 cells, parthenolide induced nucleus-localized nucleolin, whereas in MOLM-13 acute myelogenous leukemia cells, parthenolide increased cytoplasmic levels of nucleolin (Figs. 2A and 3C). This differential effect was independent of the capacity of parthenolide as a specific HDAC1 inhibitor, which is the same in both cell lines (34). Upon analyzing the miRNA expression in parthenolide-treated and DMSO-treated control cells, there was a substantial (>12-fold) increase in mature miR-15a in parthenolide-treated cells. miR-16 expression also significantly increased in parthenolide-treated cells, albeit to a lesser extent (∼2.5-fold). This could be due to the fact that the steady-state levels of mature miR-16 are over 50-fold higher under normal conditions than miR-15a4 and may not be as easily increased as miR-15a. To rule out that all miRNA are altered upon parthenolide (PN) treatment, we also analyzed let-7a expression, which was not altered following changes in nucleolin expression (Fig. 1, B and D). let-7a mature miRNA remain unchanged. To determine whether the large increases observed were truly due to nucleolin and not the effects of parthenolide, nucleolin or RISC-free control siRNA were transfected into MCF-7 cells, and 72 h later, treated with parthenolide. Incredibly, when nucleolin is dramatically knocked down (Fig. 3A, inset), the increase in miR-15a is almost completely ablated as the resulting miRNA levels were close to that of the DMSO-treated cells, whereas the RISC-free control remained comparable with untransfected cells. Similarly, miR-16 in nucleolin knockdown cells decreased significantly from the RISC-free control group following parthenolide treatment (Fig. 3A).
FIGURE 3.
Primary miRNA processing is inhibited in the absence of nucleolin. A, MCF-7 cells were treated with 25 μm PN or DMSO for 3 h before RNA extraction and qRT-PCR of mature miRNA. Nucleolin knockdown (NCL KD) and RISC-free siRNA control cells (No KD) were transfected 72 h prior to parthenolide treatment. **, p < 0.01, ***, p < 0.001. ns, not significant. B, PCR amplification of primary miRNA from RNA extracted in A. C, cellular fractionation of MCF-7 cells following treatment with PN demonstrating the increased nuclear nucleolin levels at the 3-h time point used in A and B.
We next sought to determine how the primary miRNA precursors of miR-15a and miR-16 were affected by the induced nuclear localization of nucleolin. From the data shown in Fig. 2, we concluded that cytoplasmic nucleolin inhibited the processing of pri-miRNA species as determined by their increase. Upon analyzing the pri-miRNA following parthenolide treatment in either the untransfected or the RISC-free control groups, we observed an absence of pri-miR-15a and pri-miR-16. These data indicate that in the presence of increased nuclear nucleolin, the primary miRNA are efficiently processed into precursor and ultimately mature miRNA, resulting in exhaustion of the pool of primary species in the cell. However, after knocking down nucleolin, a significant portion of the primary species persists, comparable with the levels observed in the DMSO-treated controls (Fig. 3B). The levels of pri-let-7a, like mature let-7a, were unaltered in all conditions. From these data, we conclude that it is likely that it is not off-target effects from parthenolide that induce the biogenesis of pri-miR-15a/16 but rather the increased nuclear localization of nucleolin. In the absence of nucleolin, the microprocessor complex appears unable to efficiently up-regulate the processing of the pri-miRNA, resulting in no increase of mature miR-15a and miR-16 species.
Nucleolin Interacts with Components of the Microprocessor Independent of RNA
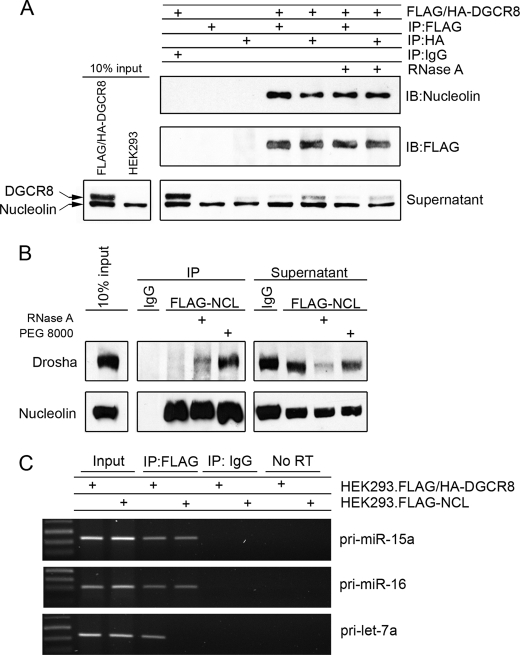
The data generated from the altered localization of nucleolin demonstrate that nucleolin is only able to exert an effect on miR-15a/16 expression while in the nucleus and that it is the primary miRNA that are affected, indicating that nucleolin may interact with the microprocessor complex. To determine whether it affects processing by directly binding to the proteins in the microprocessor complex composed of Drosha and DGCR8, an immunoprecipitation experiment was conducted. HEK293 cells stably expressing tandem affinity purification FLAG and HA tags at the N terminus of DGCR8 were used. Lysates were immunoprecipitated with either FLAG or HA antibody to bring down DGCR8, and Western blot was used to determine whether nucleolin was present in the pulldown. HEK293 cells not transfected with the expression plasmid were used as a control for nonspecific binding of FLAG or HA. Nucleolin was not present in the IgG isotype control pulldown or in the lysates not expressing DGCR8 with FLAG or HA but strongly came down with both FLAG and HA in FLAG/HA-DGCR8 expressing cells (Fig. 4A). Nucleolin is a well known RNA-binding protein, and to rule out the possibility that nucleolin came down as a result of tethering to a common RNA, the lysates were exhaustively treated with RNase A. The interaction between DGCR8 and nucleolin persisted even after complete digestion with RNase A, indicating a protein-protein interaction.
FIGURE 4.
Nucleolin interacts with the microprocessor complex and primary miRNA. A, immunoprecipitation (IP) of DGCR8 from whole cell extracts of HEK293 expressing FLAG/HA-DGCR8. IgG isotype control in DGCR8-expressing cells and FLAG or HA in cells not expressing tagged DGCR8 served as controls for nonspecific interactions. RNase A indicates incubation with 20 μg/ml overnight. Supernatant represents 10% of total supernatant. IB, immunoblot. B, immunoprecipitation of FLAG-nucleolin from HEK293 cells whole cell extracts and immunoblotting for Drosha. PEG 8000 indicates that polyethylene glycol (molecular weight 8000) was added to the wash buffer to a final concentration of 5% (w/v). C, HEK293 cells expressing either FLAG-nucleolin or FLAG/HA-DGCR8 were immunoprecipitated with either IgG or FLAG, and the bound primary miRNA was amplified by PCR. No RT indicates purified RNA from immunoprecipitates that was not reverse-transcribed before PCR analysis. Input represents 0.1% of total RNA.
Nucleolin has been shown to co-localize with Drosha to affect the processing of a long non-coding RNA mrhl in the nucleolus (19). However, in that study, a direct interaction was never investigated. FLAG-tagged nucleolin was immunoprecipitated in HEK293 lysates and probed for Drosha binding. No interaction was found when FLAG-nucleolin alone was used in the immunoprecipitation (Fig. 4B). When lysates were treated with RNase A, a weak band appeared, which could be the result of freeing Drosha from other complexes dependent on RNA. We consistently found that Drosha was decreased in the supernatant and yet failed to appear in the immunoprecipitation. We surmised that it was in fact binding to nucleolin but was lost during the washes due to a weak interaction or unstable complex. To overcome this issue, 5% polyethylene glycol with molecular mass 8000 Da was used in the wash buffer to reduce the void space during the washes. The addition of PEG to the wash buffer reduced the loss of Drosha and revealed that Drosha also interacts with nucleolin.
Nucleolin Binds to Primary miR-15a and miR-16
Nucleolin is a well known RNA-binding protein. It has no known catalytic function in RNA cleavage, but it necessary for rRNA cleavage (20). The proposed mechanism is that nucleolin holds the RNA in proper conformation to be cleaved by other components of rRNA biogenesis. Furthermore, nucleolin has been shown to co-localize with Drosha to affect the biogenesis of a long non-coding RNA but cannot by itself cleave the RNA (19). We determined that nucleolin binds to the components of the microprocessor but sought to determine whether it also binds to the primary miRNA, perhaps to stabilize its conformation for cleavage. An RNA immunoprecipitation experiment was conducted with HEK293 cells overexpressing either nucleolin or DGCR8, as a positive control. After cross-linking and sonication, nucleolin or DGCR8 was immunoprecipitated with FLAG antibody or IgG control, and the presence of primary miR-15a, miR-16, or let-7a was determined by PCR. DGCR8 bound to pri-miR-15a and -16, as expected (Fig. 4C). When primary miR-15a and miR-16 were amplified from nucleolin immunoprecipitates, we found that nucleolin bound them to a level equal to that of DGCR8, indicating that nearly the entire pool of pri-miR-15a and -16 in the cell that is bound by DGCR8 also contains nucleolin. To determine whether binding of nucleolin is specific, we also amplified pri-let-7a from both pools of extracted RNA and found that it was only bound by DGCR8 but not by nucleolin (Fig. 4C). The background contamination from genomic DNA was minimal as PCR amplification of extracted RNA without reverse transcription (No RT) revealed no amplification of primary miRNA (Fig. 4C). It also appears that a large pool of primary miR-15a and miR-16 exists in the cell that is poised to be processed but is not actively engaged with DGCR8 or nucleolin because the bound RNA was less than 0.1% of the total pool. This seems plausible because both miRNA are implicated in controlling the apoptotic response of a cell in addition to controlling the cell cycle, both of which require a rapid response to stimuli best provided from an inactive pool of unprocessed miRNA.
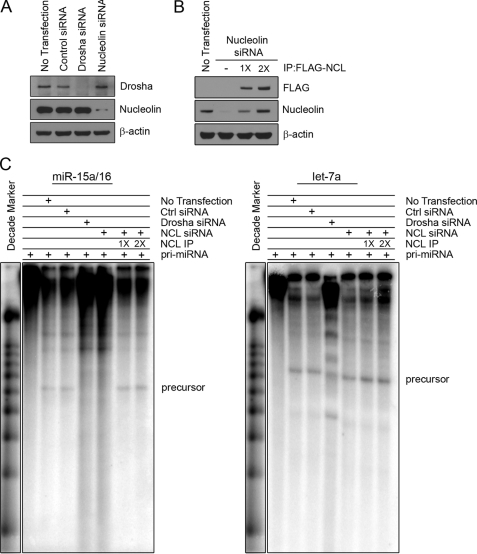
Nucleolin Affects Processing of Primary miRNA in Vitro
To confirm that nucleolin can indeed facilitate the processing of primary miRNA, we conducted an in vitro microRNA processing assay. Whole cell extracts were generated from HEK293 cells with nucleolin knocked down or Drosha knocked down as a positive control for defective processing (Fig. 5A). In extracts with nucleolin knocked down, nucleolin was reconstituted by the addition of immunoprecipitated FLAG-tagged nucleolin (Fig. 5B). Both control extracts (no transfection and control siRNA) were able to process the radiolabeled pri-miRNA to generate the precursor species for miR-15a/16 and let-7a (Fig. 5C). When Drosha was knocked down, the extracts were unable to generate the predicted precursors for both miRNA. Extracts lacking nucleolin were unable to process miR-15a/16 but could still generate the precursor for let-7a. The addition of immunoprecipitated nucleolin could rescue the processing in nucleolin-deficient cells, indicating that it can directly affect miRNA processing. There was no effect on let-7a processing with nucleolin rescue. Taken together, these data indicate that nucleolin directly and specifically affects the processing of primary miR-15a and miR-16.
FIGURE 5.
Nucleolin directly affects processing of primary miR-15a and miR-16. A, HEK293 cells treated with either Drosha or nucleolin siRNA. B, reconstituted nucleolin in nucleolin knockdown extracts. FLAG-nucleolin was immunoprecipitated (IP) from HEK293 stable cells and mixed at either 1× or 2× concentrations. 2× reconstitution restores nucleolin to levels equivalent in untransfected extracts. C, in vitro processing of primary miRNA. Gel-purified in vitro transcribed RNA was incubated with 15 μl of cell extracts from A for 90 min at 37 °C and analyzed on a 12.5% denaturing gel. Precursors of the predicted size are indicated. Ctrl siRNA, control siRNA.
DISCUSSION
In this study, we demonstrate that nucleolin affects the expression of miR-15a/16. We also found that cellular localization of nucleolin is important for its function and concluded that this is most likely because of its interaction with the microprocessor complex in the nucleus. When nucleolin is knocked down or is altered in its localization, the processing of primary miRNA is significantly decreased, indicating that it is necessary for proper expression of miR-15a/16. In addition, we found that nucleolin can bind directly to the primary miRNA species for miR-15a/16 and facilitate their processing in vitro. Taken together, these data suggest that nucleolin acts as an accessory protein to the microprocessor complex to facilitate miRNA biogenesis. As is the case with other accessory proteins involved in miRNA biogenesis, nucleolin does not globally regulate miRNA processing like Drosha or DGCR8.
We initially identified a correlation between nucleolin localization and miR-15a/16 expression before exploring how nucleolin may affect their expression. Nucleolin has been characterized as an AU-rich binding protein that stabilizes the 3′-UTR of bcl-2 mRNA upon cytoplasmic localization (21). The result of this is increased bcl-2 protein expression and inhibition of apoptosis, one of the hallmarks of cancer. miR-15a/16 have been well characterized as negative regulators of bcl-2 mRNA, and decreased expression of these miRNA is noted in numerous cancers. In some instances, decreased miR-15a/16 expression is due to chromosomal deletions; however, this only accounts for about half the patients with increased bcl-2, indicating that other mechanisms must be present (27). Our data indicate that increased cytoplasmic localization of nucleolin in cancer may be partially responsible for reduced miR-15a/16 expression. We propose that nucleolin plays a critical role in the balance between miR-15a/16 and bcl-2 mRNA to control the induction of apoptosis. Under normal conditions, nucleolin is localized in the nucleus of cells, specifically in the nucleolus, where it associates with the microprocessor complex. This results in an increase in expression of mature miR-15a/16, which then down-regulates bcl-2 mRNA. After induction of cellular stress, nucleolin leaves the nucleus to stabilize the 3′-UTR of bcl-2 while simultaneously decreasing the expression of miR-15a/16, thereby further stabilizing bcl-2 mRNA.
A number of proteins involved in miRNA processing are also essential for ribosomal RNA biogenesis including Drosha, one of the core proteins in the microprocessor complex (14). It is tempting to speculate that miRNA processing evolved from the existing machinery required for rRNA. Indeed a number of miRNA have been shown to localize to the nucleolus, a region long believed to be exclusive to rRNA biogenesis (35, 36). Moreover, nucleolin co-localizes with Drosha in the nucleolus to facilitate the processing of mrhl, a long non-coding RNA in mice. The transcript of this long non-coding RNA is 2.8 kb, which gets cleaved by the cooperative actions of nucleolin and Drosha to an 80-nucleotide precursor reminiscent of pre-miRNA (19). This product can be further processed by Dicer into a 22-nucleotide species, analogous to miRNA. DGCR8 localizes primarily to the nucleolus, and the dsRNA binding domains appear to be necessary for this because a truncation mutant lacking these domains is retained in the nucleus but fails to enter the nucleolus (37). Similarly, if the RNA recognition motifs of nucleolin are deleted, it fails to enter the nucleolus (38). Nucleolar retention of nucleolin requires a minimum of two of the four RNA recognition motif domains to be present. It is possible that cytoplasmic localization of nucleolin does not account for the disruption of miRNA processing we observed in Fig. 2 but instead is the result of nucleolin leaving the nucleolus because of the induction of stress on the cell, which has been previously reported (39).
The domain structure of nucleolin includes an acidic N-terminal region followed by a nuclear localization sequence, four RNA recognition motifs, and an arginine-glycine-rich repeat domain with putative RNA helicase activity (40). The four RNA recognition motifs bind to rRNA in a stem-loop conformation, stabilizing it for cleavage (41). Although the four RNA recognition motif domains are sufficient for RNA binding, the acidic N-terminal domain is required for cleavage of the precursor rRNA in vitro, indicating that it may be necessary for interactions with other proteins in the complex (20). Our data demonstrate that nucleolin interacts with DGCR8 and Drosha in an RNA-independent manner and yet also binds to the pri-miRNA. Based on this, we propose that although nucleolin may not possess any RNase III domains characteristic of the core proteins in the miRNA biogenesis pathway (4, 42), it instead facilitates cleavage of the primary miRNA by maintaining proper conformation of the pri-miRNA while simultaneously positioning Drosha and DGCR8 for binding and cleavage.
Supplementary Material
Acknowledgments
We are grateful to Paula Bates and Thomas Tuschl for making plasmids available.
This work was supported by the Robert A. Welch Foundation Grant R-1199 (to M. W. V. D.) and Cancer Prevention Research Institute of Texas Grant RP100726 (to D. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Fig. 1.
B. Pickering, unpublished observation.
- miRNA
- microRNA(s)
- pri-miRNA
- primary microRNA
- miR
- microRNA
- AML
- acute myelogenous leukemia
- NCL
- nucleolin
- PN
- parthenolide
- PARP
- poly(ADP-ribose) polymerase-2
- RISC
- RNA-induced silencing complex
- DMSO
- dimethyl sulfoxide
- qRT-PCR
- quantitative RT-PCR
- pre-miRNA
- precursor miRNA.
REFERENCES
- 1. Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 2. Gregory R. I., Yan K. P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. (2004) Nature 432, 235–240 [DOI] [PubMed] [Google Scholar]
- 3. Lund E, Güttinger S., Calado A., Dahlberg J. E., Kutay U. (2004) Science 303, 95–98 [DOI] [PubMed] [Google Scholar]
- 4. Provost P., Dishart D., Doucet J., Frendewey D., Samuelsson B., Rådmark O. (2002) EMBO J. 21, 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. (2004) Mol. Cell 15, 185–197 [DOI] [PubMed] [Google Scholar]
- 6. Suzuki H. I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. (2009) Nature 460, 529–533 [DOI] [PubMed] [Google Scholar]
- 7. Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008) Nature 454, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamagata K., Fujiyama S., Ito S., Ueda T., Murata T., Naitou M., Takeyama K., Minami Y., O'Malley B. W., Kato S. (2009) Mol. Cell 36, 340–347 [DOI] [PubMed] [Google Scholar]
- 9. Sakamoto S., Aoki K., Higuchi T., Todaka H., Morisawa K., Tamaki N., Hatano E., Fukushima A., Taniguchi T., Agata Y. (2009) Mol. Cell. Biol. 29, 3754–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Viswanathan S. R., Daley G. Q., Gregory R. I. (2008) Science 320, 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trabucchi M., Briata P., Garcia-Mayoral M., Haase A. D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M. G. (2009) Nature 459, 1010–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang X., Wan G., Berger F. G., He X., Lu X. (2011) Mol. Cell 41, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guil S., Cáceres J. F. (2007) Nat. Struct. Mol. Biol. 14, 591–596 [DOI] [PubMed] [Google Scholar]
- 14. Wu H., Xu H., Miraglia L. J., Crooke S. T. (2000) J. Biol. Chem. 275, 36957–36965 [DOI] [PubMed] [Google Scholar]
- 15. Fukuda T., Yamagata K., Fujiyama S., Matsumoto T., Koshida I., Yoshimura K., Mihara M., Naitou M., Endoh H., Nakamura T., Akimoto C., Yamamoto Y., Katagiri T., Foulds C., Takezawa S., Kitagawa H., Takeyama K., O'Malley B. W., Kato S. (2007) Nat. Cell Biol. 9, 604–611 [DOI] [PubMed] [Google Scholar]
- 16. Jalal C., Uhlmann-Schiffler H., Stahl H. (2007) Nucleic Acids Res. 35, 3590–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sinkkonen L., Hugenschmidt T., Filipowicz W., Svoboda P. (2010) PLoS One 5, e12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang X. H., Crooke S. T. (2011) Nucleic Acids Res. 39, 4875–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ganesan G., Rao S. M. (2008) RNA 14, 1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ginisty H., Amalric F., Bouvet P. (1998) EMBO J. 17, 1476–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Otake Y., Soundararajan S., Sengupta T.K., Kio E. A., Smith J. C., Pineda-Roman M., Stuart R. K., Spicer E. K., Fernandes D. J. (2007) Blood 109, 3069–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mi Y., Thomas S. D., Xu X., Casson L. K., Miller D. M., Bates P. J. (2003) J. Biol. Chem. 278, 8572–8579 [DOI] [PubMed] [Google Scholar]
- 23. Joo E. J., Yang H., Park Y., Park N. Y., Toida T., Linhardt R. J., Kim Y. S. (2010) J. Cell. Biochem. 110, 1272–1278 [DOI] [PubMed] [Google Scholar]
- 24. Mourmouras V., Cevenini G., Cosci E., Epistolato M. C., Biagioli M., Barbagli L., Luzi P., Mannucci S., Miracco C. (2009) J. Cutan. Pathol. 36, 637–646 [DOI] [PubMed] [Google Scholar]
- 25. Otake Y., Sengupta T. K., Bandyopadhyay S., Spicer E. K., Fernandes D. J. (2005) Mol. Pharmacol. 67, 319–326 [DOI] [PubMed] [Google Scholar]
- 26. Garzon R., Pichiorri F., Palumbo T., Visentini M., Aqeilan R., Cimmino A., Wang H., Sun H., Volinia S., Alder H., Calin G. A., Liu C. G., Andreeff M., Croce C. M. (2007) Oncogene. 26, 4148–4157 [DOI] [PubMed] [Google Scholar]
- 27. Cimmino A., Calin G. A., Fabbri M., Iorio M. V., Ferracin M., Shimizu M., Wojcik S. E., Aqeilan R. I., Zupo S., Dono M., Rassenti L., Alder H., Volinia S., Liu C. G., Kipps T. J., Negrini M., Croce C. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13944–13949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Teng Y., Girvan A. C., Casson L. K., Pierce W. M., Jr., Qian M., Thomas S. D., Bates P. J. (2007) Cancer Res. 67, 10491–10500 [DOI] [PubMed] [Google Scholar]
- 29. Landthaler M., Yalcin A., Tuschl T. (2004) Curr. Biol. 14, 2162–2167 [DOI] [PubMed] [Google Scholar]
- 30. Gilbert S. L., Pehrson J. R., Sharp P. A. (2000) J. Biol. Chem. 275, 36491–36494 [DOI] [PubMed] [Google Scholar]
- 31. Lee Y., Kim V. N. (2007) Methods Enzymol. 427, 89–106 [DOI] [PubMed] [Google Scholar]
- 32. González V., Hurley L. H. (2010) Biochemistry 49, 9706–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chang T. C., Yu D., Lee Y. S., Wentzel E. A., Arking D. E., West K. M., Dang C. V., Thomas-Tikhonenko A., Mendell J. T. (2008) Nat. Genet. 40, 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gopal Y. N., Arora T. S., Van Dyke M. W. (2007) Chem. Biol. 14, 813–823 [DOI] [PubMed] [Google Scholar]
- 35. Politz J. C., Hogan E. M., Pederson T. (2009) RNA 15, 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Politz J. C., Zhang F., Pederson T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18957–18962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shiohama A., Sasaki T., Noda S., Minoshima S., Shimizu N. (2007) Exp. Cell Res. 313, 4196–4207 [DOI] [PubMed] [Google Scholar]
- 38. Créancier L., Prats H., Zanibellato C., Amalric F., Bugler B. (1993) Mol. Biol. Cell 4, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Daniely Y., Dimitrova D. D., Borowiec J. A. (2002) Mol. Cell. Biol. 22, 6014–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ginisty H., Sicard H., Roger B., Bouvet P. (1999) J. Cell Sci. 112, 761–772 [DOI] [PubMed] [Google Scholar]
- 41. Allain F. H, Bouvet P., Dieckmann T., Feigon J. (2000) EMBO J. 19, 6870–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Rådmark O., Kim S., Kim V. N. (2003) Nature 425, 415–419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.