Abstract
Ubiquitin-conjugating enzymes (E2s) have a dominant role in determining which of the seven lysine residues of ubiquitin is used for polyubiquitination. Here we show that tethering of a substrate to an E2 enzyme in the absence of an E3 ubiquitin ligase is sufficient to promote its ubiquitination, whereas the type of the ubiquitin conjugates and the identity of the target lysine on the substrate are promiscuous. In contrast, when an E3 enzyme is introduced, a clear decision between mono- and polyubiquitination is made, and the conjugation type as well as the identity of the target lysine residue on the substrate becomes highly specific. These features of the E3 can be further regulated by auxiliary factors as exemplified by MDMX (Murine Double Minute X). In fact, we show that this interactor reconfigures MDM2-dependent ubiquitination of p53. Based on several model systems, we propose that although interaction with an E2 is sufficient to promote substrate ubiquitination the E3 molds the reaction into a specific, physiologically relevant protein modification.
Keywords: BRCA-BRCB, E3 Ubiquitin Ligase, Mass Spectrometry (MS), p53, Ubiquitin, Ubiquitin-conjugating Enzyme (Ubc), HERP, MDM2, gp78
Introduction
Targeting of most substrates to the 26 S proteasome requires covalent marking with polyubiquitin chains. Protein ubiquitination is a multistep process accomplished by the concerted action of three enzymes. The reaction begins with the ubiquitin-activating enzyme (E1), which initially adenylates the C-terminal glycine of ubiquitin and then forms a thioester bond between the activated glycine residue and a cysteine residue in its active site. Subsequently, a ubiquitin-conjugating enzyme (E2) acquires the activated ubiquitin through a trans-thioesterification reaction. Finally, a ubiquitin-protein ligase (E3) recruits a target protein and guides the transfer of the activated ubiquitin from the E2 to the substrate (1–3). Ubiquitin transfer from the E2 enzyme to the substrate is catalyzed directly by really interesting new gene (RING)3 finger-containing E3s or indirectly when a homologous to E6-AP C terminus (HECT) domain E3 is mediating the transfer (4).
Several forms of ubiquitination have been identified (5). Monoubiquitination or multiple monoubiquitinations are referred to as the conjugation of single or multiple ubiquitin moieties to distinct lysine residues on the substrate. These forms of ubiquitination were implicated in various cellular pathways, which include endocytosis and sorting of proteins to different cellular compartments (6, 7), as well as in several cases of proteasomal activity, such as the processing of the p105 precursor of the transcription regulator NF-κB (8). However, polyubiquitination is the most common post-translational modification of proteins destined for degradation (9).
In polyubiquitination assembly, ubiquitin conjugation was originally thought to be repeated in a cyclic manner whereby in each step a new moiety of ubiquitin is linked to one of the lysine residues of the previously conjugated ubiquitin. However, in view of recent findings, several alternative mechanisms have been proposed (10). Li et al. (11) demonstrated in a reconstituted cell free system that a preformed polyubiquitin chain is initially assembled on the active site cysteine of the E2 (E2G2) presumably by the action of an additional E2 acting in trans. Once assembled, an E3 enzyme (gp78) catalyzes the transfer of the polyubiquitin module to a lysine residue of the target substrate (the C terminus of homocysteine-induced endoplasmic reticulum protein (HERP), a known substrate of this E2/E3 enzyme pair) (11).
Because ubiquitin harbors seven lysine residues (Lys-6, Lys-11, Lys-27, Lys-29, Lys-33, Lys-48, and Lys-63), polyubiquitin chains can be theoretically formed on any one of them. Accordingly, seven different topologies of polyubiquitination can be generated (excluding mixed or multibranched topologies) (12). The current view holds that proteasomal degradation is mediated mainly by polyubiquitination, utilizing lysine 48 as the conjugated residue (13); however, chains based on other lysines have also been implicated in targeting proteins to the proteasome (14). Moreover, global in vivo analysis of the specific lysine residues engaged in polyubiquitination indicated that all seven lysine residues of ubiquitin participate in ubiquitin polymerization (15). However, it is unclear how lysine specificity in ubiquitin conjugation is being conferred by the ubiquitination system.
We have previously demonstrated in vitro that tethering of a substrate directly to any E2 by fusion of a ubiquitin moiety to the C terminus of the target protein is sufficient for its polyubiquitination (16). This tethering is due to direct interaction between the E2 and the substrate and occurs exclusively via the ubiquitin domain. Using this approach, we found that the capacity to polymerize ubiquitin via a specific or distinct subset of its seven lysines is an intrinsic property of the E2 enzymes (16). Because many E2 enzymes were found to hold broad lysine specificity, they potentially have the ability to assemble several types of chains on a target substrate simultaneously. However, de facto substrate ubiquitination is highly specific in terms of both the type of ubiquitin conjugates formed (lysine specificity) and the precise lysine that serves to anchor the polyubiquitin moiety on the target protein. This suggests that additional elements must direct the E2 to catalyze a specific configuration of ubiquitination. Obviously, the candidate for molding the E2 repertoire toward a more restricted set of activities is the E3 ubiquitin ligase. To our knowledge, these features of the E3 have not been systematically documented. Therefore, we set out to determine the contribution of E3 to the ubiquitination in the selection of mono- versus polyubiquitination, providing preference toward the usage of a single lysine in ubiquitin polymerization, and the targeting of the initial ubiquitin conjugation to specific lysine residue(s) on the substrate.
Our studies led us to conclude that the E3 component in addition to determining substrate specificity constrains the E2 wide conjugation potential and directs the attachment of polyubiquitin to the desired lysine residue on the target substrate. These capacities of the E3 enzymes could be reconfigured by auxiliary factors, such as MDMX.
EXPERIMENTAL PROCEDURES
Cloning of His-RFP Derivatives: RFP-20AA/40AA/80AA-Ub
The cherry RFP was fused to an N-terminal His tag and a C-terminal wild type ubiquitin in pRSET plasmid as described before (abbreviated RFP-Ub) (16). We initially amplified 60, 120, and 240 bp starting from the N terminus of β-casein while inserting NdeI sites on both ends of the fragment. To avoid potential ubiquitinations of the linker, we used a derivative of β-casein in which we replaced all of the lysine residues by arginines as a template. Following PCR amplification, the DNA fragments were purified, digested by NdeI, and ligated in-frame between the RFP and Ub domains.
RFP-Substrate Derivatives
The different RFP-substrate chimeras were constructed by fusing the amplified substrates to the C terminus of the cherry RFP in pRSET. The BRCA-associated RING domain-breast cancer (BARD-BRCA) fusion (XhoI/HindIII) and HERP (HindIII/HindIII) were cloned with the indicated restriction sites as described before (11, 17). The wild type, full-length p53 (XhoI/HindIII) was subcloned from a cDNA library using the indicated restriction sites. We used a PCR-based restriction-free cloning method to generate the RFP-substrate-Ub.
Purification of His-RFP-bound Derivatives
The desired plasmid was transformed into Escherichia coli (BL21 DE3). The bacteria were grown overnight in 50 ml of LB medium complemented with 100 μg of ampicillin/ml. This starter served to inoculate 2 liters of culture that were grown until an optical density of 0.6 at 600 nm was reached. Protein expression was induced by isopropyl 1-thio-β-d-galactopyranoside addition (0.5 mm final concentration) and further growth for 3 h. The bacteria were harvested by centrifugation; resuspended in 20 mm Hepes, pH 7.5, 20 mm imidazole; and lysed by sonication. The lysate was cleared by centrifugation (14,000 rpm for 30 min at 4 °C), and the supernatant was loaded on a homemade Ni-NTA column. Following absorption, the column was washed with 20 mm Hepes, pH 7.5, 1 m NaCl, 1% Tween 20; and the bound proteins were eluted with 0.5 m imidazole in PBS. The pure protein was dialyzed overnight against PBS and stored at −80 °C.
Cloning and Purification of E3 Ligases gp78c, Murine Double Minute 2 (MDM2), and MDMX
gp78c, MDM2, and MDMX were subcloned from a cDNA library by PCR using BamHI/SmaI restriction sites into the pGEX vector in-frame to a GST tag. The constructs in pGEX were transformed into E. coli (BL21 DE3). The bacteria were grown overnight in 50 ml of LB medium complemented with 100 μg of ampicillin/ml. This starter served to inoculate 2 liters of culture that were grown until an optical density of 0.6 at 600 nm was reached. Protein expression was induced by isopropyl 1-thio-β-d-galactopyranoside addition (0.5 mm final concentration) with further growth for 3 h. The bacteria were harvested by centrifugation, resuspended in PBS, and lysed by sonication. The lysate was cleared by centrifugation (14,000 rpm for 30 min at 4 °C), and the supernatant was loaded on a homemade glutathione column. Following absorption, the column was washed with PBS containing 1% Triton X-100, and the bound proteins were eluted with 20 mm free glutathione in PBS. The pure protein was dialyzed overnight against PBS and stored at −80 °C.
Polyubiquitination of His-RFP-bound Derivatives
His-RFP derivatives were conjugated to Ni-NTA beads to saturation. The RFP-conjugated beads were washed twice with 20 mm Hepes, pH 7.5; twice with 1 m NaCl, 1% Tween 20; and two more times with 20 mm Hepes, pH 7.5. 15 μl (200 μg) of the saturated beads were used for each polyubiquitination reaction in the presence of 30 nm human recombinant E1 (Boston Biochem), 0.5 μm indicated E2 enzyme, 1 μm indicated E3, and 10 μm wild type ubiquitin. The reactions were incubated for 4 h at 37 °C on a rotating platform (950 rpm). RFP derivatives were separated from the bulk solution by centrifugation and washed twice with 20 mm Hepes, pH 7.5; twice with 1 m NaCl, 1% Tween 20; and two times with 20 mm Hepes, pH 7.5. The beads were resuspended and boiled in sample buffer, separated by SDS-PAGE, and analyzed by Western blot with anti-ubiquitin (Covance).
LC-Tandem Mass Spectrometry (MS/MS) Analysis of Ubiquitination Reactions
The MS analysis of the ubiquitination reactions was performed by LC-MS/MS using HCTplus and OrbitrapXL instruments.
In Solution Trypsinization Followed by LC-MS/MS Using HCTplus Analysis
Proteins were reduced in 100 mm ammonium bicarbonate solution, pH 7.8 (Sigma) with 10 mm DTT (Sigma) for 30 min and subsequently alkylated with 50 mm chloroacetamide (Sigma). Digestion of proteins was carried out by the addition of 20 ng/μl trypsin (Promega) solution and incubation for 18 h at 37 °C as described previously (18).
For the analysis of digested protein material, liquid chromatography was performed using an Ultimate 3000 nano-HPLC system (Dionex) controlled by HystarTM (Bruker Daltonics) and DCMSLink 2.0 software. Samples were concentrated on a trapping column (LC Packings, 300-μm inner diameter × 5 mm) at a flow rate of 25 μl/min. For the separation with a C18 PepMap column (75-μm inner diameter, 15 cm; LC Packings), a flow rate of 300 nl/min was used as generated by a cap flow splitter cartridge (1/297). Peptides were eluted by the application of a 30-min linear gradient: solvent A (98% H2O, 2% acetonitrile, 0.1% formic acid), 0–45% solvent B (98% acetonitrile, 2% water, 0.1% formic acid). LC was interfaced directly with an ion trap mass spectrometer (HCTplus, Bruker Daltonics) utilizing 15-μm-inner diameter distal coated SilicaTips (New Objective) and nano-electrospray ionization mode. Raw LC-MS/MS data were processed, and Mascot-compatible files were created using DataAnalysisTM 3.4 software (Bruker Daltonics). Searches were performed using Mascot software (version 2.2) (19) and the Swiss-Prot Database with the following parameters: 2+ and 3+ ions; peptide tolerance, 1.5 Da; 13C = 1; fragment tolerance, 1.2 Da; missed cleavages, 3; instrument type, ESI-TRAP. In cases where ubiquitination was examined, the Gly-Gly tag on lysine residues (+114.1 Da) was included as a variable modification as described (20).
In Gel Separation and Trypsinization Followed by LC MS-MS Using OrbitrapXL Analysis
Samples were excised from the Imperial Blue (Pierce)-stained gel, reduced (4 mm DTT), modified with 16 mm iodoacetamide, and treated with trypsin or chymotrypsin (modified trypsin; Promega) at a 1:10 enzyme-to-substrate ratio. The resulting tryptic peptides were resolved by reversed phase chromatography on 0.075 × 200-mm fused silica capillaries (J&W) packed with Reprosil reversed phase material (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The peptides were eluted with linear 65-min gradients of 5–45% acetonitrile, 0.1% formic acid followed by 15 min of 95% acetonitrile, 0.1% formic acid in water. All through the run the flow rate was 0.25 μl/min. Mass spectrometry was performed by an ion trap mass spectrometer (OrbitrapXL, Thermo) in a positive mode using a repetitive full MS scan followed by collision-induced dissociation of the seven most dominant ions selected from the first MS scan. Validation of the results was done similarly using high collision dissociation. The mass spectrometry data were analyzed using Sequest 3.31 software (J. Eng and J. Yates, University of Washington and Finnigan, San Jose, CA) searching against the human part of the non-redundant NCBI Database and versus specific sequences of the constructs. An identification threshold of <5-ppm accuracy and Xcorr values of above 2.0 for doubly charged peptides and 2.5 for triply charged peptides were used. In addition, the identification of the ubiquitinated peptides was assessed visually by a trained operator. Ubiquitination was identified as a mass addition of 114 Da to the relevant peptide (the GG from the C terminus of ubiquitin attached to a lysine residue).
RESULTS
Tethering of E2 to Substrate Is Required and Sufficient to Promote Ubiquitination
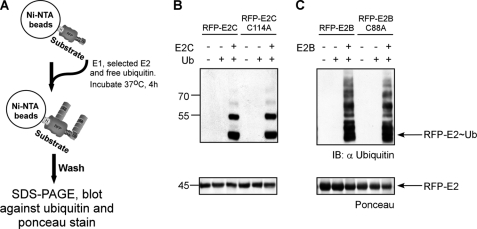
In a previous study, we demonstrated that fusing ubiquitin to RFP (termed RFP-Ub; supplemental Fig. S1) converts the RFP into a substrate for ubiquitination in vitro (16). We hypothesized that the ubiquitin recruits the E2 enzyme by binding to its active site cysteine through the action of an E1 enzyme (in analogy to wild type ubiquitin). This recruitment only occurs if the ubiquitin C-terminal glycine is intact. Mutating glycine 76 to tryptophan (G76W ubiquitin) abrogated the ability of the ubiquitin domain in the RFP-Ub fusion to recruit an E2 and promote E3-independent ubiquitination (16). Our working hypothesis is that once an E2 is recruited to the ubiquitin domain an additional E2 monomer catalyzes the polyubiquitination of the RFP-ubiquitin fusion in a manner reminiscent of the assembly of polyubiquitin on E2G2 prior to its transfer to the target substrate en bloc by the E3 (gp78) (11). Alternatively, ubiquitin molecules are transferred to the target in an alternating fusion by the two E2s engaged in the complex. In support, we have demonstrated the propensity of most E2 enzymes to spontaneously form dimers (16). Moreover, we have shown that a cross-linked dimer is recognized and charged with ubiquitin by an E1 enzyme (16). To directly test the ability of an E2 enzyme to associate and function in a dimeric state, we generated two chimeric proteins in which E2C and E2B were genetically fused to RFP (termed RFP-E2C and RFP-E2B, respectively). In addition, we constructed similar fusions with an active site mutant version of these E2s (RFP-E2C-C144A and RFP-E2B-C88A). We chose these two E2s as it was shown that E2C promotes monoubiquitination (21), whereas E2B holds the capacity to catalyze polyubiquitination (22). The rationale was to examine whether fusing an E2 to a substrate would be sufficient to promote ubiquitination by recruiting an untagged E2 as predicated by our model. Initially, Ni-NTA beads were saturated with either the wild type or active site mutant His-RFP-E2. Non-tagged wild type E2 was added into the polyubiquitination reaction mixture containing ubiquitin and E1 (see Fig. 1A for a schematic flowchart of the reaction). As shown in Fig. 1, B and C, both the wild type- and the mutant-bound RFP-E2s promote ubiquitination in an untagged E2-dependent manner. These results demonstrate that homodimerization of E2s is sufficient to promote autoubiquitination and that ubiquitination occurs in a mechanism that requires the recruitment of an additional E2 molecule (e.g. RFP linked to an active site mutant E2 was ubiquitinated as well as RFP linked to an active E2). We conclude that the ubiquitin domain in the RFP-Ub fusion recruits the E2 to the substrate by forming a thioester bond with the E2 active site cysteine. An additional E2 molecule associates with the bound E2 and provides an activated ubiquitin molecule to be conjugated. In summary, these findings demonstrate that stable association of an E2 with a substrate is required and sufficient to promote its ubiquitination.
FIGURE 1.
Attachment of E2 to substrate is sufficient to promote its ubiquitination. A, a schematic representation of a typical ubiquitination assay performed throughout this study. Initially, Ni-NTA beads were saturated with a His-tagged substrate. Next, E1, the selected E2, and ubiquitin were added and incubated for 4 h at 37 °C with constant shaking. Following the reaction, beads were washed thoroughly, boiled in sample buffer, and Western blotted against ubiquitin as well as Ponceau-stained. H, His6 tag; TEV, tobacco etch virus recognition site (in blue). B, RFP-E2C and its cognate active site mutant RFP-E2C-C114A were monoubiquitinated in the presence of untagged E2C. Similar reactions were performed with RFP-E2B and its active site mutant RFP-E2B-C88A and resulted in unbound E2B-dependent polyubiquitination (C). Protein input was detected by Ponceau staining. IB, immunoblot.
While examining RFP-Ub ubiquitination (16), we found that all the human E2 ubiquitin-conjugating enzymes were able to polyubiquitinate the ubiquitin domain of the RFP-Ub. However, only six of the E2s (E2B, E2C, E2D2, E2D3, E2D4, and E2T) conjugated ubiquitin to a lysine residue on the RFP itself. Notably, most ubiquitinations were on the anchored substrate as Western blot analysis of the supernatant revealed no free ubiquitin chains (16).
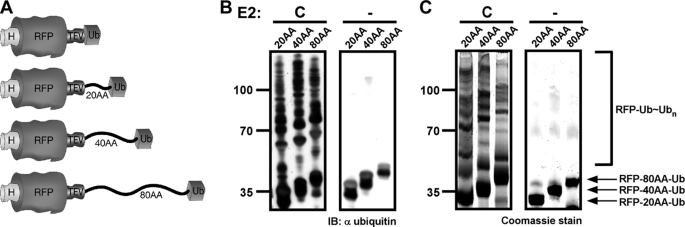
To test whether manipulating the distance between the RFP (target protein) and the ubiquitin (recruitment domain) moieties affects the efficiency and character of substrate ubiquitination, we inserted linkers of extended lengths, 20, 40 and 80 amino acids, derived from the loosely folded β-casein N terminus (termed RFP-20AA-Ub, RFP-40AA-Ub, and RFP-80AA-Ub, respectively) (Fig. 2A). The three modified RFP-Ub variants served as substrates in in vitro ubiquitination assays in the presence of the six E2s described above. Briefly, Ni-NTA beads were saturated with one of the RFP-Ub derivatives and used in ubiquitination reactions in the presence of E1, ubiquitin, and the indicated E2 enzyme (the schematic reaction is summarized in Fig. 1A). Once the reactions were completed, the polyubiquitinated RFP-Ub-bound beads were separated from the bulk solution, thoroughly washed by centrifugation, and boiled with sample buffer. 10% of the mixture was analyzed by Western blot with anti-ubiquitin antibody, whereas the rest was separated by SDS-PAGE, Coomassie-stained, and analyzed by MS/MS. A typical ubiquitination assay (performed in the presence of E2C) using the three extended linker RFP-Ub derivatives is presented in Fig. 2, B and C. Western blot analysis (Fig. 2B) clearly shows that ubiquitin was efficiently conjugated to all three variants irrespective of the linker length in an E2-dependent manner. A Coomassie-stained SDS gel of the same reactions (Fig. 2C) was analyzed by MS/MS to determine the type of ubiquitin conjugates and target lysines on the substrate. The ubiquitination reactions performed with the remaining five E2s are presented in supplemental Fig. S2. The MS/MS analysis (summarized in supplemental Table S1) demonstrates that ubiquitination was promoted by all six E2s, which conjugated ubiquitin to lysine residues on both the RFP and ubiquitin domains of the three RFP-Ub variants (see raw MS data in supplemental Fig. S10). Notably, the character of the ubiquitination was similar to that found when RFP-Ub lacking a linker was used (16). Although all of the detected RFP-ubiquitin conjugates were linked to lysine residues found on loops or edges of secondary structures (for illustration, see supplemental Fig. S3), we could not detect a common pattern or rule for the target ubiquitination site. Finally, MS/MS analysis of the conjugates confirmed that manipulating the linker size between the RFP and the Ub domains did not alter the lysine preference in the polyubiquitin chains assembled by the E2 enzymes examined. In fact, the type of detected conjugates was in agreement with that previously assigned for these E2s (16).
FIGURE 2.
Ubiquitination properties are not affected by length of linker tethering E2 and substrate. A, the E2 recruitment domain (Ub) and RFP were linked directly or through a stretch of 20, 40, and 80 amino acids as illustrated in the schematic. B, Western blot analysis of polyubiquitination assays performed in the presence of E2C showing effective ubiquitination of the target substrate (RFP) that is independent of the linker length. C, Coomassie stain of scaled up reactions shown in B. The stained ubiquitination products were used for mass spectrometry analysis (supplemental Table S1). Similar reactions performed with additional E2 enzyme are depicted in supplemental Fig. S2. H, His6 tag; TEV, tobacco etch virus recognition site; IB, immunoblot.
To rule out the possibility that the linker itself acts as a cryptic site in recruiting the E2 enzyme in a nonspecific manner, we generated an RFP derivative that contains the 80-AA linker but not the C-terminal Ub domain (termed RFP-80AA). This substrate was used in a ubiquitination assay in the presence of various E2s but did not undergo ubiquitination, demonstrating that the E2 enzymes are indeed recruited through the Ub domain to the RFP-Ub fusion proteins (supplemental Fig. S4). Taken together, these findings demonstrate that recruiting the E2 to a substrate is required and sufficient to promote its ubiquitination. Nevertheless, direct attachment of an E2 enzyme to a substrate resulted in the generation of multiple types of conjugates (broad lysine specificity) as well as attachment of the ubiquitin to multiple lysine residues on the substrate.
Recruitment of E2 to Substrate by E3 Results in Lysine-specific Conjugate Assembly
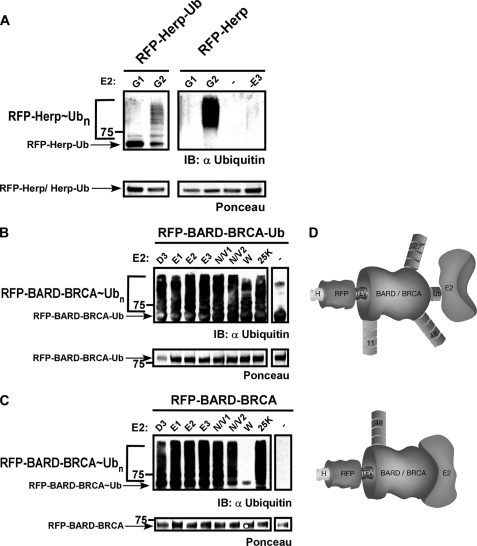
To directly evaluate the possibility that E3s may confer specificity onto the ubiquitination reaction in both the type of polyubiquitination and the site of conjugation, we examined the ubiquitination of HERP by its cognate E3 gp78 in association with E2G2 (11, 23). This ubiquitination was shown to be highly specific and dependent on the specific E2/E3 combination (11). Initially, we generated two chimeras, RFP-HERPc and RFP-HERPc-Ub, by cloning the cytosolic domain of the human HERP gene downstream of the His-tagged RFP. A tobacco etch virus (TEV) protease cleavage site was inserted between the RFP and the substrate. In the RFP-HERPc-Ub chimera, a ubiquitin was genetically fused in-frame to the C-terminal substrate domain in analogy to the RFP-Ub (supplemental Fig. S1). As an E3, we used the gp78c variant, which has improved solubility and activity equivalent to wild type gp78 (11). The RFP domain in the chimera allowed us to test whether ubiquitin attachment is strictly linked to the substrate itself or alternatively would “spill over” to the RFP domain when the substrate is recruited directly to the E2 or by an E3 enzyme.
Following expression, Ni-NTA beads were saturated with RFP-HERPc or RFP-HERPc-Ub and used in ubiquitination reactions in the presence of E2G2 and gp78c. As for all described ubiquitination reactions, we carefully optimized the concentrations of the substrate and E2 and E3 enzymes in the assay by a double titration experiment. A representative analysis is depicted in supplemental Fig. S5. As a negative control, we used E2G1, which is highly homologous to E2G2 (both orthologs of the yeast Ubc7) but does not interact with gp78c. Following ubiquitination, 10% of each reaction was separated side by side on a single SDS-polyacrylamide gel and analyzed by Western blotting with anti-ubiquitin as well as stained with Ponceau to verify equal protein input. To confirm ubiquitination, the rest of the mixture was separated by SDS-PAGE, Coomassie-stained, and analyzed by MS/MS. As seen in Fig. 3A (left panel), the ubiquitination profile of RFP-HERPc-Ub indicates that although both E2G1 and E2G2 were recruited via the Ub domain only E2G2 promoted HERPc ubiquitination in a gp78c-dependent manner. Although the analysis of RFP-HERPc lacking the C-terminal ubiquitin fusion seems similar to RFP-HERPc-Ub (Fig. 3A, right panel), the presence of the ubiquitin domain impeded the ubiquitination by E2G2. Thus, the recruitment of the substrate by the E3 supersedes a direct interaction of the E2 with the ubiquitin domain. Presumably in this case, the interaction between the E3 and the substrate prevents access by E2 to the ubiquitin domain. This demonstrates the ability of the E3 to recruit a specific E2 (E2G2 over E2G1) and hold the E2s as a dimer in the unique configuration needed to promote substrate ubiquitination. MS analysis of these reactions indicated that only Lys-48 chains were formed in the presence of E2G2 and gp78c independently of the HERPc variant used. Together, these findings demonstrate that HERPc Lys-48 ubiquitination strictly relies on the presence of the E3 ligase and its productive interaction with the E2 (see raw MS data in supplemental Fig. S11).
FIGURE 3.
Recruitment of E2 to substrate by E3 results in specific ubiquitination. Chimeras of RFP and the indicated substrate (RFP-substrate) as well as C-terminal fused ubiquitin versions of these substrates (RFP-substrate-Ub) were subjected to ubiquitination in the presence of the indicated E2 enzymes. A, left panel, RFP-HERPc-Ub was only polyubiquitinated by E2G2 in the presence of gp78c. Right panel, ubiquitination of RFP-HERPc promoted by E2G2/gp78c was markedly more efficient then when tethering of E2G2 to RFP-HERPc was promoted through the ubiquitin domain. Protein input was detected by Ponceau staining (lower panel). B, RFP-BARD-BRCA-Ub was polyubiquitinated by all the indicated E2s. C, BARD-BRCA acted in self-ubiquitination; monoubiquitination was catalyzed in the presence of E2W, whereas polyubiquitination occurred when associated with the remaining E2s. Protein input was detected by Ponceau staining (lower panel). D, schematic illustrating BARD-BRCA polyubiquitination upon tethering of the E2 either via a ubiquitin domain (upper panel) or directly by the E3 (lower panel). H, His6 tag; TEV, tobacco etch virus recognition site; 11/27/48, polyubiquitin chains types; IB, immunoblot.
Next, we used the heterodimeric E3 BARD-BRCA as it was shown to act with a variety of E2s in generating different autoubiquitinations. In analogy to HERPc, we used two derivatives of the BARD-BRCA: one containing an N-terminal RFP fusion (termed RFP-BARD-BRCA) and the other also containing a C-terminal ubiquitin domain (termed RFP-BARD-BRCA-Ub) (supplemental Fig. S1). Ni-NTA beads were saturated with the two purified BARD-BRCA derivatives and used in ubiquitination assays as described above. We used the E2 enzymes E2D3, E2E1, E2E2, E2E3, E2N-E2V1, E2N-E2V2, E2W, and E2-25K with BARD-BRCA because they were shown to act with this E3 (17). As seen in Fig. 3, B and C, and Table 1, ubiquitin was attached to RFP-BARD-BRCA-Ub and RFP-BARD-BRCA in the presence of all examined E2s with three major distinctions. 1) As previously reported (17), E2W promoted the attachment of only a single ubiquitin moiety (monoubiquitination) onto RFP-BARD-BRCA. In contrast, when RFP-BARD-BRCA-Ub was used, polyubiquitination was efficiently catalyzed by all the E2s including E2W. 2) The MS/MS results as depicted in Table 1 demonstrate that six different types of ubiquitin-ubiquitin conjugates were formed when RFP-BARD-BRCA-Ub was used in the reaction (see raw MS data in supplemental Fig. S6). In contrast, when the E2 enzyme was recruited by the BARD-BRCA domain (RFP-BARD-BRCA), only single specificity linkages (Lys-48 or Lys-63) were detected depending on the E2 used. 3) Ubiquitin was conjugated to multiple lysine residues of RFP-BARD-BRCA-Ub, whereas ubiquitination was confined to a single lysine residue on RFP-BARD-BRCA depending on the E2 enzyme used (Table 1). Notably, the lysine-specific chains generated by an E2 when it was recruited via the E3 were always within the repertoire of that E2 when BARD-BRCA-Ub was used. This was as if the E2 predetermines the set of possible lysine residues in ubiquitin from which the E3 could select (16). Together, these findings demonstrate that upon tethering of the E2 to the substrate via the ubiquitin domain multiple types of ubiquitin conjugates are formed. In contrast, recruitment of an E2 enzyme to a substrate by the E3 enzyme results in a specific conjugate that is confined to a single lysine residue on the target protein. The ubiquitination pattern of both BARD-BRCA substrates is schematically illustrated in Fig. 3D. See MS data in supplemental Fig. S6 and substrate sequence in supplemental Fig. S13.
TABLE 1.
Recruitment of E2 to substrate by E3 results in lysine-specific polyubiquitin modifications
Mass spectrometry analysis of the ubiquitination reactions performed on RFP-BARD-BRCA and RFP-BARD-BRCA-Ub chimeras presented in Fig. 3 is shown. ND, no modifications detected.
| E2 | Substrate |
|||
|---|---|---|---|---|
| RFP-BARD-BRCA-Ub |
RFP-BARD-BRCA |
|||
| Ub | Substrate | Ub | Substrate | |
| UBE2D3 | Lys-11a, Lys-27a, Lys-48a, Lys-63a | Lys-200a (RFP), Lys-338a (BARD) | Lys-48a | ND |
| UBE2E1 | Lys-11a, Lys-48a | Lys-191a (RFP) | Lys-48a | ND |
| UBE2E2 | Lys-11a, Lys-27a, Lys-48a, Lys-63a | Lys-191a (RFP), Lys-200a (RFP), Lys-338a (BARD) | Lys-48a | Lys-191a (RFP) |
| UBE2E3 | Lys-11a, Lys-27a, Lys-48a | Lys-191a (RFP), Lys-200a (RFP), Lys-338a (BARD) | Lys-48a | ND |
| UBE2N-UBE2V1 | Lys-6b, Lys-11b, Lys-29b, Lys-48a, Lys-63a | Lys-56b (BRCA), Lys-65b (BRCA) | Lys-63a | Lys-174b (RFP) |
| UBE2W | Polyubiquitin | ND | Monoubiquitin | ND |
| E2-25K | Lys-11a, Lys-48a | Lys-56b (BRCA), Lys-46b (BARD) | Lys-48a | ND |
a Ion trap data.
b Orbitrap data.
E3 Ubiquitin Ligase MDM2 Guides Ubiquitination of p53
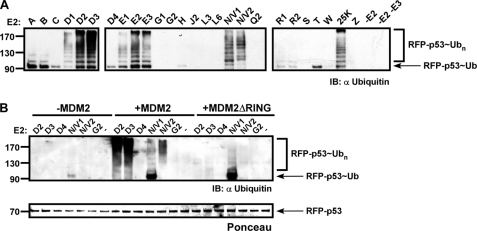
To further study the role of E3 ligases in directing substrate ubiquitination, we extended our study to the tumor suppressor gene p53. The analysis focused on one of its established physiological E3 ligases, MDM2 (24–26). p53 has two distinct ubiquitination regions, the C terminus (residues 301–393) and DNA binding domain (DBD) (residues 100–300), which are decorated with different ubiquitin conjugates by a variety of E3 enzymes (27–30). These ubiquitination sites contribute to both its nuclear export and the regulation of its cytoplasmic levels (31). In addition, auxiliary modulators, such as MDMX (MDM4), were suggested to affect the ubiquitination profile of p53 (28, 32). Initially, we cloned the human p53 gene and generated two derivatives, RFP-p53 and RFP-p53-Ub, as described above for BARD-BRCA and HERPc (supplemental Fig. S1). Following expression, Ni-NTA beads were saturated with either substrate and used in ubiquitination assays performed in the presence and absence of the E3 enzyme MDM2. The precise E2 enzymes that act with MDM2 in promoting p53 ubiquitination in vivo have not as yet been determined; therefore, we performed in vitro ubiquitination assays with all the human E2 conjugating enzymes. As seen in Fig. 4A, E2D2, E2D3, E2E2, E2E3, E2N-E2V1, E2N-E2V2, and E2-25K all catalyzed RFP-p53 ubiquitination in an MDM2-dependent manner. To examine whether the observed ubiquitinations require the RING domain of MDM2, similar reactions were performed using a mutant form of MDM2 lacking the RING domain (termed MDM2ΔRING) (33). Equal protein loading was verified by Ponceau staining of the membrane. Notably, using the MDM2ΔRING mutant abolished all but short E2N-E2V1 conjugation (Fig. 4B).
FIGURE 4.
Only a subset of E2 enzymes ubiquitinate RFP-p53 in MDM2-dependent manner. Beads saturated with RFP-p53 were incubated with all the human E2 enzymes in the presence of MDM2 (A). Similar reactions were also performed with the indicated E2s in the presence of MDM2ΔRING (B). Following the reactions, beads were washed thoroughly, boiled with sample buffer, and analyzed by Western blot with anti-ubiquitin as well as Ponceau-stained. IB, immunoblot.
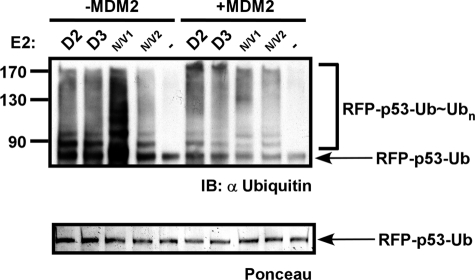
Subsequently, we analyzed whether the ubiquitin domain of RFP-p53-Ub would compensate for the requirement for MDM2 by directly recruiting the E2 enzyme to the substrate. To this end, we performed ubiquitination assays on RFP-p53-Ub with the indicated E2s in the presence or absence of MDM2 as described above (Fig. 1A). The findings presented in Fig. 5 demonstrate that in the absence of MDM2 RFP-p53-Ub ubiquitination was efficiently catalyzed. This ubiquitination was comparable or moderately reduced when MDM2 was present. We conclude that p53 ubiquitination is mediated by MDM2 in a RING-dependent manner by several E2 enzymes. However, the requirement of MDM2 can be abolished by direct recruitment of an E2 enzyme via a C-terminal ubiquitin domain (RFP-p53-Ub). Moreover, the presence of MDM2 did not overrule the ubiquitination catalyzed by tethering of p53 to the E2 enzyme via a ubiquitin domain.
FIGURE 5.
RFP-p53-Ub is ubiquitinated in MDM2-independent manner. The RFP-p53-Ub chimera in which a ubiquitin domain was fused to the C terminus of RFP-p53 was used in ubiquitination reactions with the indicated E2s in the absence (left panel) or presence (right panel) of MDM2. Reactions were boiled in sample buffer, separated by SDS-PAGE, and Western blotted with anti-ubiquitin as well as Ponceau-stained. IB, immunoblot.
The MS/MS analysis of selected ubiquitination reactions performed on the two p53 variants with the indicated E2/E3 combination is summarized in Table 2 (see MS data in supplemental Fig. S7 and substrate sequence in supplemental Fig. S13). These results demonstrate that MDM2 restricts RFP-p53 ubiquitination as compared with analogous E3-independent reactions performed on RFP-p53-Ub. For example, we detected Lys-11 and Lys-48 ubiquitin conjugates on p53 when E2D2 (Ubch5B) was used in an MDM2-dependent ubiquitination. In contrast, the same E2 enzyme catalyzed the conjugation of Lys-6, Lys-11, Lys-48, Lys-27, and Lys-29 polyubiquitin when RFP-p53-Ub was used. Moreover, E2D2 p53 ubiquitination by MDM2 led to ubiquitin conjugation at the DBD of p53 (Lys-101 and Lys-120), whereas in a similar reaction with RFP-p53-Ub, the DBD domain of p53 was not linked to ubiquitin but rather to the C-terminal domain (Lys-351) and the RFP (on multiple lysine residues). These findings further support the observation that the capacities of the E3 (MDM2) include specification of the target region of ubiquitination.
TABLE 2.
MDM2/MDMX ubiquitination of RFP-p53 and RFP-p53-Ub results in different ubiquitination pattern
MS analysis of the ubiquitination sites on RFP-p53 (left side) and RFP-p53-Ub (right side) upon ubiquitination in the presence and absence (−) of MDM2/MDMX is shown. ND, no modification detected.
| E2 | RFP-p53 |
RFP-p53-Ub |
||
|---|---|---|---|---|
| MDM2 | MDM2 + MDMX | − | MDM2 | |
| E2D2 | Lys-101a, Lys-120a (p53) | Lys-319a, Lys-320a (p53); Lys-11a, Lys-48a (Ub) | Lys-164b,c (RFP); Lys-6b,c, Lys-11b, Lys-48, Lys-27b, Lys-29b (Ub) | Lys-80b,c (RFP); Lys-351b,c (p53); Lys-6b,c, Lys-11b, Lys-48b, Lys-63b (Ub) |
| E2D3 | Lys-11b, Lys-48b (61) (Ub) | Lys-80b (RFP); Lys-6b, Lys-11b, Lys-48b (Ub) | Lys-174a (RFP); Lys-11a, Lys-48a (Ub) | |
| E2N-E2V1 | Lys-63b (63) | Lys-80b,c (RFP); Lys-142b,c (MDM2); Lys-11b,c, Lys-63b (Ub) | Lys-319b, Lys-320b (p53); Lys-63a (Ub) | Lys-63a (Ub) |
a Ion trap data.
b Orbitrap data.
c Non-tryptic peptide.
Finally, our findings are also supported by semiquantitative MS/MS data analysis in which we used the integrated peak areas corresponding to the specific peptides representing unique ubiquitin-ubiquitin linkage in different ubiquitination reactions. A representative analysis presented in supplemental Fig. S8 shows that although in three different substrate/E2/MDMX combinations Lys-11 and Lys-48 linkages were detected the ratios of these conjugates were altered. These results support our notion that the combination of different enzymes in the reaction and their recruitment to the substrate affects the final ubiquitination pattern of the substrate.
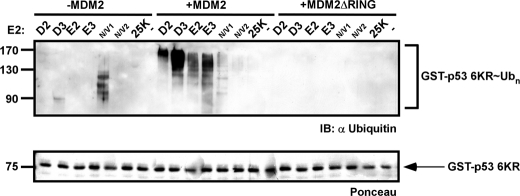
The C-terminal lysines of p53 are the predominant sites for MDM2-mediated ubiquitination (34). Although in vitro data demonstrate the importance of the six C-terminal lysines of p53 for MDM2-mediated ubiquitination, knock-in studies in which the corresponding lysines were mutated to arginines (p53–6KR knock-in mice) did not dramatically alter p53 protein levels (35, 36). Furthermore, the half-life of an analogous p53–7KR mutant (a mouse p53 construct with the seven C-terminal lysines mutated) showed no difference as compared with wild type p53 in embryonic fibroblasts and thymocytes derived from p53–7KR knock-in mice (36). Together, these two studies support the notions that the C-terminal lysines are not exclusively essential for efficient p53 degradation in vivo and that additional lysine ubiquitinations may have a role in p53 stability. Possibly, these lysine residues are located at the N terminus (DBD) of p53 as in vitro findings suggested (37). To test the function of MDM2 in imposing the selection of the target lysine residue, we mutated the six ubiquitination target lysine residues in the C-terminal tail of GST-p53 to arginines (termed p53–6KR) (34, 38). We then used p53–6KR in ubiquitination reactions as described above. As shown in Fig. 6, MDM2-dependent ubiquitination assays performed with p53–6KR (in the presence of various E2 enzymes) demonstrated that ubiquitination in this scenario may be directed to regions other than the C terminus (such as the DBD) as the p53–6KR mutant was still ubiquitinated by various E2s in an MDM2 RING-dependent manner.
FIGURE 6.
RFP-p53 6KR mutant is still ubiquitinated by MDM2. A p53 mutant in which six lysine residues at the C-terminal domain were mutated to arginine (termed p53–6KR) was used in ubiquitination reactions with the indicated E2s in the absence (left side) or presence of MDM2 (middle) or MDM2ΔRING (right side). Protein input was detected by Ponceau staining (lower panel). IB, immunoblot.
MDMX Alters Conjugation Type and Target Lysine Catalyzed by MDM2
Previous studies have demonstrated that MDM2 has a major role in the regulation of p53 stability during the stress response. However, in many cases, this function entails additional cofactors that act in concert with MDM2. One such effecter is MDMX, which is structurally related to MDM2, although its RING domain does not possess an E3 ligase activity (39–41). Presumably, in analogy to BRAD-BRCA, MDMX modulates the activity of MDM2 through heterodimerization of the conserved C-terminal RING domains of both proteins (42). However, in certain cases, MDMX also promotes the E3 ligase activity of MDM2 and thus probably the destruction of p53 (43–45). To directly test how MDMX affects the ubiquitination capacities of MDM2, we compared MDM2-dependent p53 ubiquitination patterns in the presence and absence of MDMX (supplemental Fig. S9). The MS analysis of MDMX-dependent reactions (Table 2) revealed that the manner by which the E2 is recruited to the substrate alters the ubiquitination outcome. Accordingly, the ubiquitination pattern of p53 changes when the E2 is recruited via MDM2 as compared with MDM2/MDMX. Notably, when the E2 was recruited to RFP-p53 via MDM2, the outcome was ubiquitination positioned in the DBD of p53 (lysines 101 and 120). However, the presence of MDMX resulted in ubiquitination of the p53 C-terminal region (lysines 319 and 320). The recruitment of the E2 by the ubiquitin domain (RFP-p53-Ub) led to the same relocation of ubiquitination from the DBD to the C terminus of p53 (Table 2). Unexpectedly, the chain topologies that were conjugated to RFP-p53 in the presence of both MDM2 and MDMX were outside the typical conjugation capacity predicted for the E2 enzymes used. For example, E2D3 catalyzed Lys-48 ubiquitination of p53 in the presence of MDM2. However, when both MDM2 and MDMX were present in the reaction, Lys-6, Lys-11, and Lys-48 chains were assembled. Surprisingly, the E2N-E2V1 complex, which typically produces Lys-63 chains (46), attached both Lys-11 and Lys-63 onto p53 in the presence of MDM2/MDMX (Table 2 and supplemental Fig. S12). Although these results were obtained in an in vitro setup and must be confirmed in vivo, these findings indicate that MDMX may affect the type of conjugates catalyzed by MDM2.
We hypothesize that the heterodimeric configuration of the MDM2/MDMX imposes a conformation that favors alternative types of polyubiquitin chains, such as Lys-6, to be catalyzed by the E2. Notably, this conjugate type was not detected in any of our other p53 ubiquitination assays or any other assay involving these E2s. Taken together, we conclude that MDM2/MDMX may act in a manner similar to the BARD-BRCA heterodimer, conferring the specificity in the E2 recruitment while imposing a conformation that dictates the conjugation type as well as the target lysine residue (47).
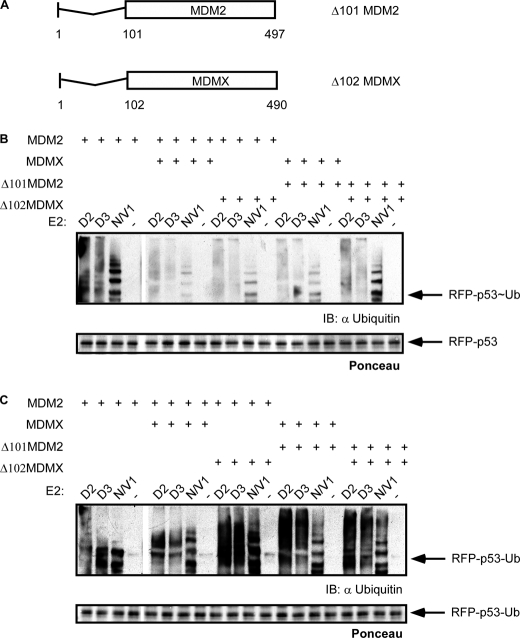
Previous studies proposed that both MDM2 and MDMX contain a p53 binding domain at their N termini (48). To test the significance of these domains in MDM2/MDMX p53-dependent ubiquitination, we prepared MDM2 and MDMX mutants, which lack their N-terminal p53 binding region (Fig. 7A). These mutants were used to catalyze RFP-p53 (Fig. 7B) and RFP-p53-Ub (Fig. 7C) ubiquitination in the presence of the indicated E2s as described above. The results of the ubiquitination analysis indicate that although different combinations of N-terminal MDM2/MDMX mutants failed to promote RFP-p53 ubiquitination RFP-p53-Ub was still efficiently ubiquitinated. These findings suggest that heterodimer formation is more favorable than either of the homodimers and that the N-terminal deletion acts as a dominant inhibitor with a lower capacity to catalyze p53 ubiquitination.
FIGURE 7.
Deletion of MDM2 and MDMX p53 binding domains differentially affects RFP-p53 and RFP-p53-Ub ubiquitination. A, Δ101MDM2 and Δ102MDMX mutants were generated by deletion of the first 101 and 102 amino acids, respectively. Δ101MDM2 and Δ102MDMX were used in ubiquitination reactions with the indicated E2 enzymes in different combinations with wild type MDM2 and MDMX on RFP-p53 (B) and RFP-p53-Ub (C). Reactions were boiled in sample buffer, separated by SDS-PAGE, and Western blotted with anti-ubiquitin as well as Ponceau-stained. IB, immunoblot.
In summary, the p53/MDM2/MDMX model system further supports the notion that E3 ligases control the conjugation type (lysine specificity) and define the target region on the substrate to be ubiquitinated. Importantly, these findings demonstrate a potential role of MDMX in determining the configuration of p53 ubiquitination.
DISCUSSION
Tethering E2 to Substrate Is Both Required and Sufficient for Its Ubiquitination
We demonstrated with the use of RFP-Ub that direct recruitment of an E2 enzyme to a substrate is both necessary and sufficient to promote its ubiquitination. However, in this scenario, the resulting ubiquitination was robust and not physiologically relevant. As shown in Fig. 2, supplemental Fig. S2, and supplemental Table S1, the ubiquitination of the RFP-Ub was nonspecific. These findings emphasize the importance of E3 enzymes as specificity factors and also provide an explanation for the great number of distinct E3 enzymes and their complex regulation in eukaryotic cells. We hypothesize that ubiquitination by direct tethering of the E2 to a substrate (for example via ubiquitin-like or ubiquitin-associated domains) if actually occurring in a cellular context would probably have a limited physiological significance.
In a simplistic manner, ubiquitin conjugation can be regarded as a protein-protein interaction between an E2 and a target substrate. Typically, this association is mediated by an E3. Direct recruitment of a substrate to the E2 via the attachment of a ubiquitin moiety (Fig. 2) allows the uncoupling of the E3 scaffold function of liaising both E2 and substrate from its additional role in guiding the specificity of the ubiquitination reaction. Our findings demonstrate that the ubiquitination character of a substrate directly associated with an E2 enzyme was unaffected by the length or amino acid sequence of the linker between the E2 and that substrate. Thus, converting the ubiquitin transfer from the E2 to the substrate into an intramolecular reaction by either linking the E2 to the substrate directly or via an E3 enzyme is the main criterion for efficient ubiquitination. In such a scenario, the highly activated bond between the ubiquitin and the active site cysteine of the E2 is predisposed to an attack by an ϵ-NH2 group of a lysine residue on the substrate. In support, upon tethering of the E2 and the substrate, the ubiquitination efficiency was unaffected by the linker properties. In this configuration, the intimate interaction between the ubiquitin and the E2 was not constrained by the E3 enzyme; thus, the ubiquitin was attached to the substrate on multiple lysine residues, which were all located on loops and edges of secondary structures, as suggested previously (49). The type of detected polyubiquitin conjugate reflected the full spectrum of conjugation capacities (lysine preference) of the E2 enzyme used (16) (Fig. 2 and supplemental Table S1) and was unaffected by the linker length.
E3 Ligase Narrows Capacity of Coupled E2 toward Single Lysine Preference
We reconstructed several ubiquitination model systems to investigate the role of each component in determining the ubiquitination profile of a substrate. The underlining rationale guiding these assays was to catalyze E3-dependent and -independent polyubiquitination reactions of a given substrate in parallel. E3-independent ubiquitinations were performed by recruiting the E2 to the substrate via a C-terminal ubiquitin domain on the substrate (supplemental Fig. S1). The ubiquitination products were compared with classical ubiquitination reactions in which an established cognate E2/E3 pair of specific substrates was used. The E2G2/gp78c/HERPc model system is an example for synchronized ubiquitination in which polyubiquitin is initially assembled on the E2 enzyme to then be transferred and linked to the substrate en bloc by the specific E3 enzyme. Thus, in this mode of action, the lysine specificity in the polyubiquitin is exclusively determined by the E2 prior to or independently of the engagement with the E3 enzyme (11, 23) (Fig. 3A). In other cases, such as BARD-BRCA and p53/MDM2 where the E2 could catalyze several types of polyubiquitin chains, recruitment via an interaction with the E3 restricted this specificity to a single ubiquitin conjugate type (Tables 1 and 2). In contrast, multiple types of conjugates were detected when the E2 enzymes were directly recruited to the substrate by the fused ubiquitin domain. This finding was independent of the substrate, E2, or the E3 enzyme used as similar results were obtained in non-related experimental systems. We conclude that the E3 enzyme has a major role in determining the type of ubiquitin conjugate that will be attached to a specific substrate by narrowing the inherent lysine spectrum of the cognate E2 enzyme.
E3 Enzyme Directs Ubiquitin Attachment to Specific Sites on Substrate
The mass spectrometry analysis suggests that direct recruitment of an E2 to a substrate results in multiple ubiquitination sites on that substrate. In contrast, recruitment of the E2 enzyme to a substrate by a dedicated E3 enzyme results in ubiquitination of specific sites (Tables 1 and 2). This is demonstrated by a smaller number of ubiquitination target sites on both BARD-BRCA and p53 when the E3 ligase rather than a fused ubiquitin domain mediated the reaction. In fact, in the case of p53, the entire region of ubiquitination was changed by the addition of the E3 MDM2 (Table 2). Whereas the DBD was targeted in the MDM2-dependent ubiquitination, recruitment of the E2 through the ubiquitin domain resulted in p53 C-terminal ubiquitination. These results are in agreement with past analysis of p53 ubiquitination showing that p53 is ubiquitinated in both regions (61–63). C-terminal ubiquitination by MDM2 was found to be crucial for p53 nuclear export and degradation involved in keeping homeostatic low levels of p53 (34, 36, 50). In support, expression of p53-Ub, which was used to mimic C terminus-ubiquitinated p53 in cells, was localized to the cytoplasm in MDM2-null cells (51). However, mutations of the C-terminal lysine residues of p53 were shown to have little effect on the stability of p53 (34, 36). Moreover, it has been found that MDM2-dependent DBD ubiquitination is essential for the proper nuclear export of p53 (31). We suggest that ubiquitination in both sites occurs in vivo in a cooperative or redundant manner. However, in the absence of the C-terminal lysine residues and perhaps under other cellular conditions possibly by a different combination of enzymes, MDM2 promotes ubiquitination of the p53 DBD to promote nuclear export of p53. Our results with both wild type and p53–6KR support this possibility (Fig. 7 and Table 2).
Role of E3 in Selecting Mono- versus Polyubiquitination
Another level of complexity is the decision between attaching a monoubiquitin versus the assembly of a polyubiquitin chain onto a substrate. To distinguish between the contribution of the E2 and the E3 enzymes in this selection, we took advantage of earlier studies, which illustrated that E2W catalyzes self-monoubiquitination of BRAD-BRCA (17). In contrast, when we used BRAD-BRCA fused to ubiquitin (supplemental Fig. S1), we detected polyubiquitination on the BRAD-BRCA in the presence of E2W presumably due to the direct recruitment of the E2 enzyme by the ubiquitin domain (Fig. 3, B and C, and Table 1). Based on these findings, we conclude that the capacity to catalyze mono- versus polyubiquitination is a result of constraints that are enforced by the manner in which the E3 binds the E2 and the substrate. Monoubiquitination can be imposed by preventing the “ping-pong” assembly of polyubiquitin on the E2 itself as an intermediate by a dual core E2 complex (11) or by establishing a non-processive mode of ubiquitination (52). It is also possible that polyubiquitination is promoted by the binding of an E2 dimer or a polyubiquitin-charged E2, whereas monoubiquitination is enforced by recruitment of a single molecule of E2 to the E3-substrate complex.
Therefore, beyond the highly regulated recruitment of substrates to an E2 enzyme, the E3 ligase also controls three additional aspects of the ubiquitination reaction: selecting the target lysine for ubiquitin attachment, determining the conjugation type (lysine specificity), and deciding between mono- and polyubiquitination. We hypothesize that the intimate contacts between the E2 and substrate conferred by the E3 as well as the positioning of the ubiquitin to be conjugated in respect to the attacking lysine residue facilitate these functions of the E3. In support, it has been previously illustrated that dictating the lysine preference of ubiquitin conjugates may be related to the manner in which the attacking ubiquitin interacts non-covalently with the E2. For example, analysis of structural motifs in certain E2 enzymes (E2-25K (53), E2D3 (E25C) (54), and UBC13-UEV1a (E2N-E2V1) (55, 56) shows that these E2s contain a region that binds to the attacking ubiquitin and affects the resulting conjugation type (57, 58). We suggest that the E3 fine tunes the non-covalent docking of the attacking ubiquitin to the E2, thus catalyzing only a specific conjugation type from the a priori capacities of that E2. This model is also sustained by earlier reports whereby a specific binding orientation of the attacking ubiquitin was shown to be favored by the tertiary structure of the E2s (such as E2-25K, E2S, E2N-E2V1/2, and E2G), providing the rationale for the single conjugation capacity of these E2s (53–56, 59).
It is noteworthy that in the current study we focused on the role of RING E3s in controlling substrate ubiquitination by pairing with specific E2s. The RING E3 family comprises more than 500 members and forms the majority of known cellular E3 enzymes. We did not address the manner by which HECT domain E3s direct ubiquitination. HECT E3s are composed of about 30 members and act in an alternative mechanism. These E3s contain an active site cysteine that acquires the ubiquitin from the E2 prior to its attachment to the substrate. This form of action endows the E3 with a broader control of the ubiquitination process. Indeed, several HECT E3s were shown to determine the specific type of the catalyzed polyubiquitin chain (50).
Auxiliary Factors May Alter Character of Ubiquitin Conjugation
We used the MDMX as an additional factor that may affect p53 ubiquitination in conjugation with MDM2. As mentioned above, MDMX shares structural homology with MDM2 but lacks an independent E3 ligase activity. MDMX is also missing the nuclear localization and nuclear export sequence signals and thus is thought to travel between the cytoplasm and the nucleus via its RING domain heterodimerization with MDM2. Both MDM2 and MDMX bind the p53 transactivation domain via their N terminus and were shown to affect both the activity and subcellular localization of p53. However, the exact role of MDMX in p53 ubiquitination has not as yet been elucidated. Our results demonstrate that the addition of MDMX to MDM2-dependent ubiquitination of p53 reduces the level of ubiquitination (Fig. 7 and supplemental Fig. S9). In addition, the presence of MDMX in the reaction altered the ubiquitination profile of p53 in terms of lysine specificity as well as the identity of the modified target lysine residues (Table 2). Whereas p53 ubiquitination by MDM2 was conjugated to the DBD, the presence of MDMX relocated the ubiquitination to the C terminus domain. Moreover, the presence of MDMX altered the polyubiquitin chain type attached to the p53. We hypothesize that MDMX reconfigures the MDM2-E2 complex into a conformation that promotes the catalysis of conjugation types (lysine specificity), which deviate from the typical conjugation range displayed by the E2 used in the reaction (16).
MDMX function may be analogous to that of BARD in the context of BRCA. Although the BARD-BRCA complex discriminates in selecting E2 partners, the E2 enzymes only bind to the BRCA domain (47). This notion is supported by previous evidence that the MDM2-MDMX heterodimer is more stable than the homodimers of either MDM2 or MDMX (60). In agreement, we found that deleting the N-terminal p53 binding domain of one of the partners (MDM2 or MDMX) leads to inactivation of the complex (Fig. 7). We propose that this type of interaction with E3 auxiliary factors marks a general mode of action that adds another layer of specificity to the function of the E3 enzymes.
Supplementary Material
This work was supported by the Israel Science Foundation (ISF), the Minerva Foundation (Germany), the German-Israeli Foundation for Scientific Research and Development (GIF), and a special donation from Rolando Uziel.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S13 and Table S1.
- RING
- really interesting new gene
- HECT
- homologous to E6-AP C terminus
- HERP
- homocysteine-induced endoplasmic reticulum protein
- RFP
- red fluorescent protein
- Ub
- ubiquitin
- BARD-BRCA
- BRCA-associated RING domain-breast cancer
- Ni-NTA
- nickel-nitrilotriacetic acid
- AA
- amino acid
- MDM2
- murine double minute 2
- DBD
- DNA binding domain.
REFERENCES
- 1. Busch H., Goldknopf I. L. (1981) Mol. Cell. Biochem. 40, 173–187 [DOI] [PubMed] [Google Scholar]
- 2. Hershko A., Ciechanover A., Rose I. A. (1981) J. Biol. Chem. 256, 1525–1528 [PubMed] [Google Scholar]
- 3. Hershko A., Heller H., Elias S., Ciechanover A. (1983) J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 4.(2005) Essays Biochem. 41, 15–30 [DOI] [PubMed] [Google Scholar]
- 5. Hicke L., (2001) Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 6. Hicke L., Dunn R. (2003) Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 7. Haglund K., Di Fiore P. P., Dikic I. (2003) Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 8. Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. (2009) Mol. Cell 33, 496–504 [DOI] [PubMed] [Google Scholar]
- 9. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 10. Hochstrasser M. (2006) Cell 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 11. Li W., Tu D., Brunger A. T., Ye Y. (2007) Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 12. Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. (2006) Mol. Cell 24, 701–711 [DOI] [PubMed] [Google Scholar]
- 13. Pickart C. M. (2000) Trends Biochem. Sci. 25, 544–548 [DOI] [PubMed] [Google Scholar]
- 14. Saeki Y., Kudo T., Sone T., Kikuchi Y., Yokosawa H., Toh-e A., Tanaka K. (2009) EMBO J. 28, 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 16. David Y., Ziv T., Admon A., Navon A. (2010) J. Biol. Chem. 285, 8595–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Christensen D. E., Brzovic P. S., Klevit R. E. (2007) Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 18. Kinter M., Sherman N. E. (2000) Protein Sequencing and Identification Using Tandem Mass Spectrometry, pp. 147–164, John Wiley and Sons, Inc., New York [Google Scholar]
- 19. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 20. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 21. Rape M., Kirschner M. W. (2004) Nature 432, 588–595 [DOI] [PubMed] [Google Scholar]
- 22. Haas A., Reback P. M., Pratt G., Rechsteiner M. (1990) J. Biol. Chem. 265, 21664–21669 [PubMed] [Google Scholar]
- 23. Das R., Mariano J., Tsai Y. C., Kalathur R. C., Kostova Z., Li J., Tarasov S. G., McFeeters R. L., Altieri A. S., Ji X., Byrd R. A., Weissman A. M. (2009) Mol. Cell 34, 674–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 25. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 26. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 27. Esser C., Scheffner M., Höhfeld J. (2005) J. Biol. Chem. 280, 27443–27448 [DOI] [PubMed] [Google Scholar]
- 28. Wade M., Wang Y. V., Wahl G. M. (2010) Trends Cell Biol. 20, 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G. D., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. (2004) Nature 429, 86–92 [DOI] [PubMed] [Google Scholar]
- 30. Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J. M., Lozano G., Hakem R., Benchimol S. (2003) Cell 112, 779–791 [DOI] [PubMed] [Google Scholar]
- 31. Nie L., Sasaki M., Maki C. G. (2007) J. Biol. Chem. 282, 14616–14625 [DOI] [PubMed] [Google Scholar]
- 32. Shvarts A., Bazuine M., Dekker P., Ramos Y. F., Steegenga W. T., Merckx G., van Ham R. C., van der Houven van Oordt W., van der Eb A. J., Jochemsen A. G. (1997) Genomics 43, 34–42 [DOI] [PubMed] [Google Scholar]
- 33. Minsky N., Oren M. (2004) Mol. Cell 16, 631–639 [DOI] [PubMed] [Google Scholar]
- 34. Lohrum M. A., Woods D. B., Ludwig R. L., Bálint E., Vousden K. H. (2001) Mol. Cell. Biol. 21, 8521–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng L., Lin T., Uranishi H., Gu W., Xu Y. (2005) Mol. Cell. Biol. 25, 5389–5395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krummel K. A., Lee C. J., Toledo F., Wahl G. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan W. M., Mak M. C., Fung T. K., Lau A., Siu W. Y., Poon R. Y. (2006) Mol. Cancer Res. 4, 15–25 [DOI] [PubMed] [Google Scholar]
- 38. Gu J., Nie L., Wiederschain D., Yuan Z. M. (2001) Mol. Cell. Biol. 21, 8533–8546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M.(2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 40. Linares L. K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharp D. A., Kratowicz S. A., Sank M. J., George D. L. (1999) J. Biol. Chem. 274, 38189–38196 [DOI] [PubMed] [Google Scholar]
- 42. Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 43. Poyurovsky M. V., Priest C., Kentsis A., Borden K. L., Pan Z. Q., Pavletich N., Prives C. (2007) EMBO J. 26, 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uldrijan S., Pannekoek W. J., Vousden K. H. (2007) EMBO J. 26, 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Francoz S., Froment P., Bogaerts S., De Clercq S., Maetens M., Doumont G., Bellefroid E., Marine J. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hofmann R. M., Pickart C. M. (1999) Cell 96, 645–653 [DOI] [PubMed] [Google Scholar]
- 47. Brzovic P. S., Keeffe J. R., Nishikawa H., Miyamoto K., Fox D., 3rd, Fukuda M., Ohta T., Klevit R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shvarts A., Steegenga W. T., Riteco N., van Laar T., Dekker P., Bazuine M., van Ham R. C., van der Houven van Oordt W., Hateboer G., van der Eb A. J., Jochemsen A. G. (1996) EMBO J. 15, 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- 49. Catic A., Collins C., Church G. M., Ploegh H. L. (2004) Bioinformatics 20, 3302–3307 [DOI] [PubMed] [Google Scholar]
- 50. Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 51. Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 52. Deffenbaugh A. E., Scaglione K. M., Zhang L., Moore J. M., Buranda T., Sklar L. A., Skowyra D. (2003) Cell 114, 611–622 [DOI] [PubMed] [Google Scholar]
- 53. Merkley N., Shaw G. S. (2004) J. Biol. Chem. 279, 47139–47147 [DOI] [PubMed] [Google Scholar]
- 54. Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. (2006) Mol. Cell 21, 873–880 [DOI] [PubMed] [Google Scholar]
- 55. VanDemark A. P., Hofmann R. M., Tsui C., Pickart C. M., Wolberger C. (2001) Cell 105, 711–720 [DOI] [PubMed] [Google Scholar]
- 56. McKenna S., Moraes T., Pastushok L., Ptak C., Xiao W., Spyracopoulos L., Ellison M. J. (2003) J. Biol. Chem. 278, 13151–13158 [DOI] [PubMed] [Google Scholar]
- 57. Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 58. Lewis M. J., Saltibus L. F., Hau D. D., Xiao W., Spyracopoulos L. (2006) J. Biomol. NMR 34, 89–100 [DOI] [PubMed] [Google Scholar]
- 59. Brzovic P. S., Klevit R. E. (2006) Cell Cycle 5, 2867–2873 [DOI] [PubMed] [Google Scholar]
- 60. Tanimura S., Ohtsuka S., Mitsui K., Shirouzu K., Yoshimura A., Ohtsubo M. (1999) FEBS Lett. 447, 5–9 [DOI] [PubMed] [Google Scholar]
- 61. Inoue T., Geyer R. K., Howard D., Yu Z. K., Maki C. G. (2001) J. Biol. Chem. 276, 45255–45260 [DOI] [PubMed] [Google Scholar]
- 62. Rodriguez M. S., Desterro J. M., Lain S., Lane D. P., Hay R. T. (2000) Mol. Cell. Biol. 20, 8458–8467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Laine A., Topisirovic I., Zhai D., Reed J. C., Borden K. L., Ronai Z. (2006) Mol. Cell. Biol. 26, 8901–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.