Background: Salivary histatin 5 (Hst 5) kills the fungal pathogen Candida albicans upon its internalization.
Results: Hst 5 requires C. albicans polyamine transporters Dur3 and Dur31 for its uptake and toxicity.
Conclusion: C. albicans Dur polyamine transporters recognize and internalize cationic peptides competitively with spermidine.
Significance: Modification of Hsts based upon polyamine structure may improve targeting and fungicidal activity.
Keywords: Antimicrobial Peptides, Fungi, Peptide Transport, Polyamines, Protein Translocation, Candida albicans, Dur, Histatin 5, Polyamine Transporters
Abstract
Histatin 5 (Hst 5) is a salivary gland-secreted cationic peptide with potent fungicidal activity against Candida albicans. Hst 5 kills fungal cells following intracellular translocation, although its selective transport mechanism is unknown. C. albicans cells grown in the presence of polyamines were resistant to Hst 5 due to reduced intracellular uptake, suggesting that this cationic peptide may enter candidal cells through native yeast polyamine transporters. Based upon homology to known Saccharomyces cerevisiae polyamine permeases, we identified six C. albicans Dur polyamine transporter family members and propose a new nomenclature. Gene deletion mutants were constructed for C. albicans polyamine transporters Dur3, Dur31, Dur33, Dur34, and were tested for Hst 5 sensitivity and uptake of spermidine. We found spermidine uptake and Hst 5 mediated killing were decreased significantly in Δdur3, Δdur31, and Δdur3/Δdur31 strains; whereas a DUR3 overexpression strain increased Hst 5 sensitivity and higher spermidine uptake. Treatment of cells with a spermidine synthase inhibitor increased spermidine uptake and Hst 5 killing, whereas protonophores and cold treatment reduced spermidine uptake. Inhibition assays showed that Hst 5 is a competitive analog of spermidine for uptake into C. albicans cells, and that Hst 5 Ki values were increased by 80-fold in Δdur3/Δdur31 cells. Thus, Dur3p and Dur31p are preferential spermidine transporters used by Hst 5 for its entry into candidal cells. Understanding of polyamine transporter-mediated internalization of Hst 5 provides new insights into the uptake mechanism for C. albicans toxicity, and further suggests design for targeted fungal therapeutic agents.
Introduction
Human saliva is the source of most innate host defense proteins within the oral cavity. Salivary proteins play a key role in modulating the oral flora and preventing overgrowth of Candida albicans. Among over 120 distinct antimicrobial salivary proteins, salivary histatins are the primary source of direct fungicidal activity (1). Histatins (Hsts)2 are rich in basic amino acids including histidine, arginine, and lysine and have a net cationic charge. Hst 5 (a 24-amino acid protein) has the most potent fungicidal activity among the major secreted histatins (2), whereas smaller proteolytic fragments (processed by enzymatic cleavage in saliva) of only 12 amino acid residues retain full antifungal activity (3, 4). Although Hsts were named because of high numbers of histidine residues, other amino acids including lysines (Lys5 and Lys13) rather than histidines have key importance for fungicidal activity (3). Thus the small size and a net basic charge including selected lysine residues are required for Hsts antifungal activity.
Many cationic antimicrobial peptides have the capacity to form amphipathic structures (5). These peptides generally exert their activity through spontaneous insertion into microbial membranes by forming pores, and disrupting cell structure and integrity (6, 7). However, Hst 5 does not act as a pore forming protein as it has no defined secondary structure in aqueous solutions such as saliva (8, 9). Substitution of functionally nonessential residues of Hst 5 with proline to disrupt any secondary structure resulted in unaltered candidacidal activity (10), showing that the degree of the amphipathic molecular moment is not involved in its fungicidal activity. Furthermore, synthetic variants of Hst 5 with increased (dhvar4) and decreased (dhvar5) amphipathic features both killed C. albicans (11). Comparison with amphipathic peptides that translocated across and disrupted liposome membranes showed that Hst 5 did not interact with model membranes (12). Thus, unlike many antimicrobial peptides, Hst 5 does not disrupt membranes by pore formation nor does it rely on amphipathicity for its biological function.
Histatin fungicidal activity is a distinctive multistep mechanism requiring binding to Candidal cell wall proteins, followed by translocation to intracellular compartments (13). Although the exact intracellular target of Hst 5 is not defined, lethality is a result of overall induction of osmotic stress in C. albicans cells as a result of release of intracellular ions and small nucleotides including ATP (14–17). Indeed, Hst 5 can kill C. albicans when expressed from a chromosomally encoded human Hst 5 gene in the absence of externally applied Hsts (18). Histatin binding with the cell wall and its intracellular translocation are two independent events, although a threshold level of cell wall binding is a requirement for subsequent uptake (17). The independence of these two processes is illustrated by the complete blockade of intracellular transport of a modified Hst 5 fragment (substituting two key lysine residues), although high levels of cell wall binding were retained (4). Thus fungal cell wall binding in itself does not result in uptake of histatins, instead a second independent process is required for its passage into the cytosol.
Intracellular translocation of histatins is inhibited by low temperatures (19), or pretreatment of cells with proton ionophores including carbonyl cyanide m-chlorophenylhydrazone (CCCP) and dinitrophenol or energy metabolism inhibitors such as sodium azide (14). Although it has been suggested that azide blocks Hst 5 internalization by cell membrane rigidification due to ATP depletion (20), two other energy-dependent mechanisms for intracellular transport are possible including endocytosis and active transport of histatin through a cellular transporter. Indeed, Hst 5 is internalized by endocytotic transport to the vacuole (21), although substantial levels of cytosolic Hst 5 are found prior to endocytotic uptake (17). Vacuolar sequestration of Hst 5 does not reduce its toxicity (17), most likely because high levels of cytosolic peptide are attained before endocytosis occurs. Collectively these data show that rapid translocation of Hst 5 across the fungal cell membrane is required for toxicity, and that this process involves cellular metabolic energy and a proton motive force (as well as ambient temperature), all consistent with an active transport mechanism.
Various membrane-bound transport proteins belonging to ABC (ATP binding cassette) or major facilitator superfamily families have been well described for both C. albicans and Saccharomyces cerevisiae (22–24). These transporters are involved in the uptake of amino acids, peptides, sugar molecules, as well as efflux of azole- and polyene-based antifungal drugs (25–27). Typically active transporters have a relatively narrow substrate specificity, thus we hypothesized that polyamine transporters (such as putrescine, spermidine, and spermine) that catalyze the uptake of polycationic molecules may also translocate Hsts as polycationic substrates. Polyamines are essential organic cations required for protein and nucleic acid synthesis and therefore cell growth. Putrescine is a diamine precursor of the larger molecules spermidine and spermine that are synthesized by the aminopropyl transferases spermidine and spermine synthases, respectively (28). Despite structural variations, putrescine, spermidine, and spermine all carry a net positive charge (pKa values of 9–10) similar to Hst 5.
Fungal cells tightly regulate their intracellular concentration of polyamines by balancing biosynthesis and degradation with cellular polyamine transport (both uptake and efflux). Because membrane transporters are major regulators of polyamine homeostasis, they have been studied as drug targets in bacteria (29, 30) and more recently in S. cerevisiae (31). Four plasma membrane-localized polyamine transporters in S. cerevisiae have been characterized: Gap1, Agp2, Dur3, and Sam3 (32–34). The amino acid permeases Gap1 and Agp2 catalyze the uptake of smaller polyamines (primarily putrescine and to a lesser degree spermidine) as well as amino acids. Both Gap1 and Agp2 preferentially transport amino acids rather than polyamines when both are present in the media. In contrast, Dur3 and Sam3 are polyamine-preferential transporters that catalyze the uptake of polyamines, although they may also transport urea (Dur3), or S-adenosylmethionine, glutamic acid, and lysine (SAM3) (34). Polyamine uptake is repressed by high intracellular levels of polyamines at a transcriptional level as illustrated by the reduction in DUR3 and SAM3 mRNA in cells cultured in the presence of polyamines. A decrease in putrescine uptake in S. cerevisiae cells was comparable with the reduction in mRNA levels of DUR3 and SAM3, thus showing the tight regulation of polyamine uptake proteins Dur3 and Sam3 with polyamine concentrations in S. cerevisiae (34). Therefore, it was expected that C. albicans cells grown in the presence of high concentrations of polyamines would have reduced uptake of polyamine substrates including Hst 5. We report here that C. albicans has reduced intracellular transport of Hst 5 upon growth in medium rich in spermidine, implicating polyamine transporters in uptake of this peptide.
The identity of polyamine transporter proteins is relatively well defined in S. cerevisiae, however, virtually nothing is known about these proteins or their regulation of polyamine transport in C. albicans. Thus, the aims of the present study were to identify polyamine transporters in C. albicans, with the goal of gaining insight into the mode of Hst 5 intracellular translocation by cells, as we hypothesized that Hst 5 is a polycationic molecule recognized as a substrate for polyamine transporters. Indeed, we identified a novel Dur family of transporter proteins in C. albicans that catalyze spermidine uptake, with Dur3 and Dur31 proteins being the primary carriers for intracellular translocation of Hst 5. The study provides novel understanding of how Hst 5 enters C. albicans cells to exert its fungicidal activity, and is invaluable information for designing new therapeutic agents with improved cellular uptake.
EXPERIMENTAL PROCEDURES
Strains, Media, and Reagents
The strains generated by this study and their genotypes are listed in Table 1. The ura3− auxotrophic C. albicans CAF4-2 strain (35) was the parental strain for DUR gene deletion mutants. Yeast carbon base (Sigma) was used for induction of FLP-mediated excision of the URA3 flipper cassette. Yeast nitrogen base (Difco) without uridine and supplemented with 2% glucose was used for selection of the URA3+ transformants. Cells were maintained in yeast extract/peptone/dextrose (YPD; Difco) medium with the addition of uridine when required and stored as −78 °C stock. For routine work, the strains were maintained on YPD agar plates and recultured monthly from −78 °C stock. Cells were routinely manipulated at 28 °C to maintain the cells in blastospore morphology. Escherichia coli DH5α competent cells (Invitrogen) were used as a host for plasmids and were grown in Luria-Bertani (LB) medium (Difco). Solid media were made with 1.8% Bacto-agar (Difco). All oligonucleotides used in strain constructions were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Restriction endonucleases and GeneRuler DNA markers were purchased from Fermentas. The Fast-Link DNA ligation kit was purchased from Roche Diagnostics. Plasmid DNA was isolated from E. coli by the alkaline lysis method, employing a QIAprep Spin miniprep kit from Qiagen. Purification of restriction digest mixtures, PCR mixtures, and DNA fragment isolation from agarose gels were done by using QIAquick PCR purification and QIAquick gel extraction kits, respectively, both from Qiagen. Genomic DNA of C. albicans strain was isolated by PUREGENE DNA isolation kit from Qiagen. Ampicillin, fluoroorotic acid, and concanavalin A were purchased from Sigma at the highest purity available. Polyamines (putrescine dihydrochloride, spermidine trihydrochloride, and spermine tetrahydrochloride) were obtained from Sigma. Unlabeled Hst 5 (DSHAKRHHGYKRKFHEKHHSHRGY) and N terminus FITC-labeled Hst 5 were synthesized by GeneMed Synthesis, Inc. (San Antonio, TX).
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Genotype | Phenotype | Reference |
|---|---|---|---|---|
| CAF4-2 | CAF2-1 | Δura3::imm434/ura3::imm434 | URA− | 35 |
| CAI4 | CAF2-1 | Δura3::imm434/Δura3::imm434 | URA− | 35 |
| Δdur3 | CAF4-2 | Δura3::imm434/Δura3::imm434, Δdur3::FRT | URA− | This study |
| Δdur31 | CAF4-2 | Δura3::imm434/Δura3::imm434, Δdur31::FRT | URA− | This study |
| Δdur33 | CAF4-2 | Δura3::imm434/Δura3::imm434, Δdur33::FRT | URA− | This study |
| Δdur34 | CAF4-2 | Δura3::imm434/Δura3::imm434, Δdur34::FRT | URA− | This study |
| Δdur3/dur31 | Δdur3 | Δura3::imm434/Δura3::imm434, Δdur3::FRT/Δdur31::FRT | URA− | This study |
| dur3/DUR3 | Δdur3 | Δura3::imm434/Δura3::imm434, dur3/DUR3::FRT/RP10::DUR3ORF | URA+ | This study |
| DUR3 O/E | CAF4-2 | Δura3::imm434/Δura3::imm434, DUR3/DUR3:FRT RP10::DUR3ORF | URA− | This study |
Construction of DUR Gene Knock-out Plasmids
The URA3-flipper technique (36) was used to replace the first allele of each DUR gene. The pSFUC2 plasmid (kindly provided by Dr. J. Morschhauser) containing two FRT sites, a SAP2 promoter-driven FLP gene and a URA3 marker, was used as the starting vector. The 5′-flanking sequence from DUR3, DUR31, DUR33, and DUR34 genes was subcloned and ligated with the KpnI- and XhoI-digested plasmid. The constructed plasmids containing the 5′-flanking region of DUR genes were confirmed by PCR. Fragments including 3′-flanking sequence, spanning the untranslated region of DUR genes, were digested by BglII and SacI and ligated into the plasmid containing 5′-flanking region of the respective DUR gene. Insertion of DUR genes into the vector was confirmed by restriction digestion and PCR. For deletion of the second allele of the DUR genes, the pDBU3 plasmid (∼5.1 kb), containing 2.37 kb length of the URA3 open reading frame, was used as a starting vector. The strategy used was as described above. In addition to the 5′- and 3′-second target sequences (STR) of the DUR gene (5′ STR and 3′ STR), a second 3′-target region (3′ STR-2) amplified from the sequence immediately downstream of the first 3′-STR was added. The PCR product of 3′ STR-2 was introduced into the vector between the 5′ STR and URA3 marker. This additional flanking region served as the recognition site for fluoroorotic acid-induced intra-chromosomal recombination. The final gene disruption constructs were used to target the second allele of DUR genes.
Disruption of DUR Genes in CAF4-2 Strain
Gene deletion strains were constructed for DUR genes to characterize CaDUR gene products in Hst 5 induced killing. Because of the diploid genome of C. albicans, a combination of two genetic strategies was employed to sequentially remove both alleles of DUR genes. To inactivate the first allele of DUR genes, C. albicans strain CAF4-2 was first transformed with linearized pUFC plasmid containing flanking regions of the DUR gene using the Frozen-EZ Yeast Transformation II KitTM (Zymo Research, CA). Transformed cells were spread onto uracil-deficient agar plates. Candida genomic DNA was isolated, and integration of the gene disruption cassette was confirmed by PCR with control primers. Selected transformants were grown on yeast carbon base/BSA medium for induction of FLP-mediated excision of the UFC (36). Randomly selected ura− colonies were analyzed using PCR. The heterozygous strain of DUR genes was further transformed with a linearized fragment of pURA to obtain homozygous DUR gene knock-out mutant strains. Successful replacement of the second allele of the DUR genes was confirmed by PCR. Selected colonies were spread onto yeast nitrogen base-agar plates containing fluoroorotic acid (1 mg/ml) and uridine (50 μg/ml) to induce intra-chromosomal recombination and eliminate colonies containing the URA3 gene. For additional deletion of DUR genes, these mutants were transformed with another series of cassettes. To construct a strain overexpressing the Dur3 protein (DUR3 O/E), one copy of DUR3 was placed at the RPS10 locus in WT cells for its constitutive expression. Details of gene deletion strains are listed in Table 1.
Phylogenetic Analysis
Amino acid sequences of Dur proteins were obtained from the Candida genome data base and used for sequence alignment by ClustalX version 2.0.11 (37). These profiles were used to calculate a phylogenetic tree with use of the neighbor joining algorithm (38). Graphical representation and editing of the phylogenetic tree was performed using PhyloDraw version 0.82 (39). Transmembrane domains were predicted for Dur proteins and visual models were constructed using TMHMM version 2.0 (40) and TMRPres2D version 0.93 (41). The conserved domains within the predicted amino acid sequences were identified using the PFAM data base (42).
Synthesis of BODIPY-Spermidine Conjugate
The amine-reactive BODIPY® 630/650 succinimidyl ester (Invitrogen) containing an additional 7-atom aminohexanoyl spacer (“X”) was used to label spermidine. Spermidine was acylated using BODIPY 630/650-X with stirring for 2 h at 4 °C in dimethyl sulfoxide, while keeping it protected from light. BODIPY-X-spermidine (Spd) was purified using Sephadex G-25 column chromatography with dimethyl sulfoxide/sterile-distilled water (1:3). Pooled fractions of purified BODIPY-Spd were assayed for protein content and stored at −80 °C.
Spermidine Uptake and Hst 5 Competition Assays
Wild-type (CAF4-2) and mutant strains were inoculated in YPD broth supplemented with uridine, and incubated at 30 °C with shaking overnight. Cells were subcultured and grown to mid-log phase, washed twice in 10 mm sodium phosphate buffer (NaPB, pH 7.4), and the cell number was adjusted to 1 × 106 cells/50 μl of NaPB. Cells were used immediately in assays performed on 96-well, clear bottom, black plates and handled in subdued lighting. Upon optimization, 100 μm BODIPY-Spd was used in uptake and competition assays, as this concentration allowed for saturation of polyamine transporters and consistent competition with 15.5 and 31 μm Hst 5. A standard curve of fluorescence intensities for known amounts of BODIPY-Spd was determined with a linear range (R2 = 0.9899) between 3.5 and 10 nmol of BODIPY-Spd. For all assays, 1 × 106 cells were added to each well of the assay plate as appropriate, with a total reaction volume of 100 μl/well. BODIPY-Spd was added to the cells immediately before recording fluorescent counts at 30 °C using the Bio-Tek multifunction plate reader and Gen5 software provided by the University at Buffalo School of Medicine and Biomedical Sciences Confocal Microscopy and Flow Cytometry Core Facility. Readings were taken at 1-min intervals from 0 to 15 min. The cellular uptake of BODIPY-Spd resulted in a decrease in fluorescence, which was normalized by subtraction from the fluorescent count values from the control wells containing only BODIPY-Spd as well as from the readings at time 0. For Hst 5 competition assays, 100 μm BODIPY-Spd was added 1.5 min after the addition of 15.5 μm Hst 5 to the cells that allowed Hst 5 to access the transporters before high affinity saturation with spermidine. To determine the Km, Vmax, and Ki values, uptake as well as competition assays were performed at different concentrations of BODIPY-Spd and Hst 5. Statistical analysis and determination of kinetic parameters were performed using GraphPad Prism version 5.0 for Windows (GraphPad Software, San Diego, CA).
Time Lapse Confocal Microscopy for Hst 5 Binding and Uptake
C. albicans CAF4-2, Δdur3, and DUR3 O/E cells were treated with FITC-labeled Hst 5 (F-Hst 5) to observe relative binding and uptake of salivary Hst 5. Overnight grown cells (A600 ∼ 0.8–1.0) were diluted to obtain 106 cells/ml of NaPB. The chambered coverglass (Lab-TekII) was coated with 100 μg/ml of concanavalin A for 30 min and washed twice with water. Cells (1 × 106) were fixed on concanavalin A-coated slides for 20 min at room temperature. The plates were then washed twice with NaPB followed by addition of 31 μm FITC-Hst 5 and images were captured under Zeiss LSM 510 Meta Confocal Microscope (Carl Zeiss, Germany) using a Plan Apochromat ×63/1.4 (oil) objective. The average fluorescence intensity was calculated using ImageJ software. A similar procedure was used for competition assays with CAF4-2 and Δdur3 cells where 2 mm spermidine was added in test samples in addition to 31 μm FITC-Hst 5. Confocal images of cells were captured and compared to determine the relative binding and uptake of Hst 5.
Candidacidal Assays of Hst 5
For candidacidal assays, single colonies of C. albicans wild-type and mutant cells were inoculated in YPD media to A600 of 1.6–1.8. For polyamine growth assays, cells were grown in the YPD media supplemented with different concentrations of polyamines (putrescine, spermidine, and spermine). For selective polyamine biosynthesis inhibition, 5 mm cyclohexylamine (CHA) was used as spermidine synthesis inhibitor. Overnight grown cells were diluted to A600 = 0.3–0.4 and were incubated at 28 °C with shaking at 250 rpm until A600 of ∼1.0 was attained. We did not observe any growth inhibition in the presence of CHA. Cells were washed twice with 10 mm NaPB and cells (1 × 106) were mixed with different concentrations of the Hst 5. Following incubation at 28 °C with shaking (250 rpm) for 30 min, cell suspensions were diluted in 10 mm NaPB, and aliquots of 500 cells were spread onto YPD agar plates and incubated for 48 h to visualize surviving colonies. All killing assays were performed in triplicate and repeated at least twice. Percent cell killing was calculated as 1 − (numbers of colonies from suspensions with Hst 5/numbers of colonies from control suspensions) × 100. Statistical analysis was performed using GraphPad Prism 5 software.
Reverse Transcription-PCR
For RT-PCR analysis, total RNA was isolated using the RNeasy Mini-Kit (Qiagen) and cDNA was synthesized using 1 μg of total RNA and oligo(dT) primers and Moloney murine leukemia virus reverse transcriptase (Retroscript, Ambion, Austin, TX). Using 2 μl of cDNA synthesized, PCR was performed using GoTaq® Hot Start polymerase (Promega Corp., WI). The sequence of primers were: 5′-GACCAATGACTGCTGCTGAA-3′ and 5′-GCCAGTTTTGACGTTTGGAT-3′ for DUR3; 5′-GATCATCTGTGCTGCTGGAA-3′and 5′-AGCAGCTGAAGCCAATGT-3′ for DUR31; 5′-CGATGGAAGTTTGAGGCAATA-3′ and 5′-CTCTCGGCCAAGGCTTATACT-3′ for 18S RNA. The PCR products were separated on 1.2% agarose gels and visualized by ethidium bromide staining.
RESULTS
Identification of C. albicans Dur Transporter Family and Nomenclature
To identify C. albicans polyamine transporter proteins, we screened the Candida genome data base for orthologs of the major S. cerevisiae transporters responsible for polyamine import that include SAM3, AGP2, GAP1, and DUR3 genes. BLAST homology analysis did not identify SAM3 in C. albicans, although general amino acid permeases Gap4 (orf19.4456) and Gap12 (orf19.1799) showed 42% amino acid sequence identities to S. cerevisiae Sam3 protein. Other C. albicans genes with high homology to SAM3 included orf19.6993 GAP2 (41%), orf19.4304 GAP1 (36%), and orf19.6659 GAP6 (37%). C. albicans AGP2 (orf19.4679) was found to have 40% amino acid identity with S. cerevisiae AGP2 (as well as having 24% identity with S. cerevisiae Sam3p). Interestingly, the majority of S. cerevisiae GAP family members are amino acid permeases regulated in response to antifungal drugs such as ketoconazole, caspofungin, or flucytosine. Based on their homology to the S. cerevisiae family of Gap permeases, C. albicans GAP family members are likely to be related amino acid permeases that include orthologs to S. cerevisiae Sam3, Gap1, and Agp2.
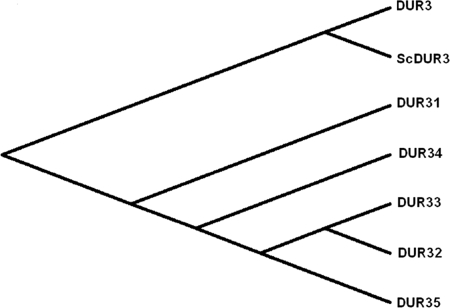
In contrast, a Candida genome data base search of S. cerevisiae DUR3 identified six related members of C. albicans Dur proteins. To find the nearest ortholog of ScDur3p, clustal analysis was performed by optimal alignment of the amino acid sequences using CLUSTAL X software. All C. albicans Dur proteins showed homology to S. cerevisiae Dur3p ranging from 26 to 54%. C. albicans Dur1,2 protein showed high homology to the S. cerevisiae Dur1,2 protein, but was very distinct from other Dur proteins of C. albicans. Amino acid sequence alignment showed orf19.781 to be the ortholog of ScDur3p in C. albicans instead of orf19.6656, which is also annotated as the CaDur3 protein. We found that the current nomenclature used for C. albicans Dur transporters in the Candida genome data base is quite complex and often not a true representation of relationships among the Dur protein family members. Bensen et al. (43) first referred to C. albicans orf19.6656 as DUR3, and it became the standard name. Later Braun et al. (44) and Navarathna et al. (45) referred to C. albicans orf19.781 (as well as orf19.6656) as DUR3. Our homology analyses illustrate the pitfalls in the existing nomenclature. We found C. albicans orf19.781 to be most closely related to S. cerevisiae Dur3p and not to the annotated C. albicans Dur3p (orf19.6656). The alignment of amino acid sequences of orf19.6656 and orf19.781 showed only 16% amino acid sequence identities, which confirms that orf19.6656 and orf19.781 each code for two significantly different proteins. Thus, we propose designating C. albicans orf19.781 as Dur3 because it is the true ortholog of the S. cerevisiae Dur3 protein and is a major inducible urea transporter (45), and designating C. albicans orf19.6656 as Dur31. Our analysis clearly shows Dur1,2 is not part of the Dur membrane transporter family but instead are intracellular enzymes involved in the Degradation of urea (hence the DUR nomenclature). Hence, to prevent future confusion with the nomenclature of C. albicans Dur transporter family members, we propose a novel nomenclature based upon their topologies, structures, conserved domain motifs, and homology with other Dur proteins. Under our new proposed nomenclature, C. albicans Dur transporter family members now consist of only 6 members and are named Dur3, Dur31, Dur32, Dur33, Dur34, and Dur35 (Fig. 1).
FIGURE 1.
Phylogenetic analysis of C. albicans DUR transporter family members and relationships with the S. cerevisiae Dur3 protein. Members of the C. albicans Dur transporter family are shown with Dur3p homologous protein sequences from the yeast S. cerevisiae. C. albicans Dur3 protein (orf19.781) has the closest phylogenetic relationship to S. cerevisiae polyamine transporter Dur3. C. albicans Dur proteins Dur32 (orf19.5017), Dur33 (orf19.7205), and Dur35 (orf19.5915) are closely related to each other but are most distantly related to Dur3; whereas Dur31 (orf19.6656) and Dur34 (orf19.5677) are intermediately related to these groups.
The evolutionary relationship among C. albicans Dur family proteins was analyzed at two levels: amino acid sequence and functional similarity. Dur1,2 protein is more divergent from other members of Dur transporter family showing only 9–15% similarity and were not included for phylogenetic analysis. Alignment of Dur proteins showed 9–84% amino acid sequence homology. Amino acid sequence alignment and phylogenetic analysis of Dur transporter proteins showed high similarity between C. albicans Dur3 and S. cerevisiae Dur3 protein (Fig. 1). Among C. albicans Dur proteins, Dur32, Dur33, and Dur35 showed close relationships with 80–84% genetic similarity, and are grouped together in one cluster; thus indicating a functional similarity among Dur32, Dur33, and Dur35 proteins.
The deduced amino acid sequences were used to generate hydropathy plots for Dur proteins shown in Fig. 1, and all were found to contain 13 to 15 putative transmembrane domains in an α-helical conformation (data not shown). Because 13–15 transmembrane domains have been reported as a special feature of the SSS family of proteins, our results showed that C. albicans Dur transporter proteins fit with the classical SSS family of integral membrane proteins.
DUR3 and -31 Deletion Reduces Sensitivity to Hst 5
To examine the possible role of individual Dur proteins in polyamine and Hst 5 uptake in C. albicans, we constructed a series of isogenic C. albicans null mutants for Dur protein family members from the parental strain CAF4-2. Based upon the phylogenetic tree of Dur proteins on amino acid sequences, one member from each cluster was selected to construct gene deletion null mutants. Independent homozygous mutants for Dur3, Dur31, Dur33, and Dur34 proteins, double gene knock-out mutants for Dur3/Dur31, a Dur3 restoration strain (dur3/DUR3), and an Dur3 overexpression strain (DUR3 O/E) were constructed (Table 1).
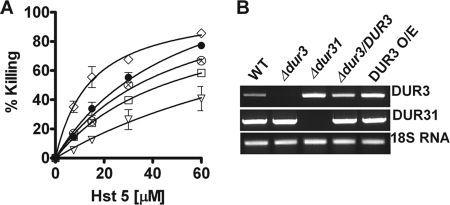
Deletion of DUR3 resulted in significant loss of Hst 5 sensitivity (Fig. 2A), which was reversed upon gene restoration (data not shown); whereas Δdur31 cells had reduced sensitivity only at the highest Hst 5 concentration tested. Interestingly, cells with deletion of both DUR3 and DUR31 had much greater resistance to Hst 5 than either single deletion mutants, suggesting additive effects resulting from loss of both genes in Δdur3/Δdur31 cells. As expected, DUR3 O/E was significantly more sensitive to Hst 5. No differences in Hst 5 toxicity were found in Δdur33 and Δdur34 strains compared with CAF4-2 (data not shown); thus these two Dur family transporters are unlikely to be involved in Hst 5 killing. Therefore, we did not examine these strains further.
FIGURE 2.
A, polyamine transporter mutants DUR3 O/E (♢), Δdur3 (□), Δdur31 (⊗), and Δdur3/Δdur31 (▿) were altered in sensitivity to Hst 5 compared with wild-type CAF4-2 (●) cells. Colony forming units were determined after a 1-h exposure to different concentrations of Hst 5, and percent killing was calculated from the cfu in Hst 5-treated cells compared with control cells. C. albicans Δdur3 and Δdur3/Δdur31 were found to be resistant to Hst 5 mediated killing, whereas the DUR3 overexpression strain was hypersensitive to Hst 5. The Δdur31 strain was resistant only at a 60 μm Hst 5 dose. B, reverse transcription-PCR analysis was performed to determine the expression levels of DUR3 and DUR31 genes in WT and DUR mutants. The expression level of DUR3 was elevated in DUR3 O/E as well as in Δdur31 cells compared with WT. Thus, increased expression of DUR3 in Δdur31 cells masks changes in sensitivity to Hst 5. There were no differences in expression levels of DUR31 in any strain examined.
To identify a reason for the disparity between the lack of Hst 5 sensitivity of Δdur31 cells compared with Δdur3/Δdur31 cells, we examined expression levels of DUR3 and DUR31 genes in these strains (Fig. 2B). We found that levels of DUR3 transcripts were elevated in Δdur31 cells almost to the level of the DUR3 overexpression strain. However, deletion of DUR31 did not result in an increase in DUR3 transcripts. Thus the small reduction in Hst 5 killing in Δdur31 cells is likely a result of the compensatory increase in DUR3. However, whereas the phenotype of Δdur31 cells is masked by higher transcription of DUR3, DUR31 also contributes to Hst 5 sensitivity, because killing of Δdur31 cells was not as high as the DUR3 O/E strain (both strains having equivalent levels of DUR3 transcripts). Overall, these data point to a role of both Dur3 and its closely related family member Dur31 in Hst 5 sensitivity.
C. albicans Cells Cultured with Added Polyamines Are Resistant to Hst 5 Killing
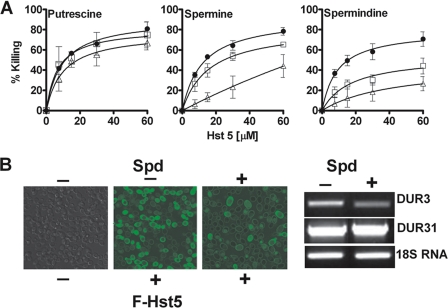
Because polyamine uptake and transporter expression levels are repressed in the presence of high extracellular polyamines (34), an initial screening was performed to ascertain if cells grown with polyamines were altered in Hst 5 intracellular translocation and cytotoxicity. For these experiments, C. albicans (CAF4-2) cells were grown in medium containing putrescine, spermidine, and spermine (1 or 2 mm), followed by a thorough washing to remove all extracellular polyamine before addition of Hst 5. Cells grown in the presence of spermidine or spermine, but not putrescine, were resistant to Hst 5 mediated killing (Fig. 3A). Hst 5 induced killing was proportionally reduced with increasing concentrations of polyamines in growth medium. In contrast, no significant inhibition of killing was observed when cells were grown with added putrescine (Fig. 3A). Cells cultured in medium supplemented with 2 mm spermidine showed the highest inhibition of killing with Hst 5; resulting in only 26% killing as compared with 70% killing in cells grown in the absence of spermidine (Hst 5 at 60 μm). Although cells cultured with spermine showed resistance to Hst 5 killing, the loss in killing activity by Hst 5 was less pronounced. Addition of spermine (2 mm) to cultures decreased the subsequent Hst 5 killing by a maximum of 50%, whereas only 1 mm spermidine was required to reduce killing to this level.
FIGURE 3.
Uptake and killing activity of Hst 5 is reduced in cells cultured with spermidine and spermine. A, wild-type CAF4-2 cells were grown either in growth medium only (●) or in growth medium supplemented with putrescine, spermine, or spermidine (1 mm (□) and 2 mm (Δ)). Addition of putrescine to growth medium did not affect the sensitivity of C. albicans cells to Hst 5 mediated killing (p > 0.05). The presence of either spermine or spermidine in the growth medium significantly increased the resistance of C. albicans to Hst 5 toxicity, with the highest inhibition of killing in cells grown in spermidine. B, intracellular translocation of FITC-Hst 5 (30 μm) was reduced in cells grown in medium with added spermidine (Spd) (2 mm) (right panel) as compared with control cells grown in medium without the addition of spermidine (middle panel). Transcript levels of DUR3 were reduced in CAF4-2 cells grown in the presence of 2 mm spermidine, whereas there was no difference in the expression level of DUR31.
To determine whether the observed decrease in Hst 5 toxicity in cells cultured in the presence of spermidine was due to reduced Hst 5 uptake, CAF4-2 cells grown in the presence and absence of 2 mm spermidine were treated with FITC-Hst 5 (which retains biological activity) and observed by time-lapse confocal microscopy. Substantial uptake of FITC-Hst 5 was observed in cells grown without spermidine compared with cells grown in the presence of spermidine (Fig. 3B). Reduction in intracellular uptake of Hst 5 in cells grown in spermidine medium (2 mm) was not due to altered cell wall binding, because these cells had abundant cell wall binding of Hst 5 despite reduced cytosolic levels of Hst 5 (Fig. 3B). To confirm that cells cultured in the presence of spermine or spermidine had repression of DUR3 or DUR31, RT-PCR was performed on cells cultured as for the Hst 5 assays. Interestingly, C. albicans DUR3 transcript levels were substantially reduced when cells were grown with spermidine, however, there was no change in expression of DUR31 (Fig. 3B). Because Hst 5 killing in cells grown in spermidine was more pronounced than cells with DUR3 deletion and was similar to Δdur3/Δdur31 cells (Fig. 2A), this suggested that culturing C. albicans in the presence of polyamines not only repressed expression of Dur3 transporters, but also reduced the substrate affinities of Dur3 and perhaps Dur31 transporters.
Inhibition of Polyamine Biosynthesis by the Spermidine Synthase Inhibitor CHA
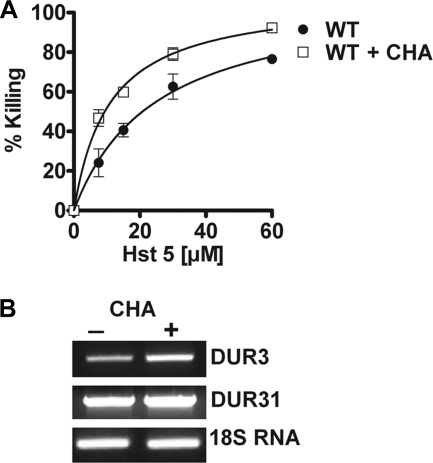
To rule out the possibility that added exogenous spermidine might affect Hst 5 binding with the cell surface, we examined cells grown in the presence of spermine synthase inhibitor for their sensitivity to Hst 5. Addition of cyclohexylamine blocks intracellular synthesis of polyamines, and forces cells to rely on their uptake from the medium. Therefore, we expected that C. albicans cells cultured with 5 mm cyclohexylamine would have increased Hst 5-mediated killing as a result of up-regulation of Dur transporters. Indeed, we observed that in the presence of CHA, cells showed significantly higher killing by Hst 5 at all concentrations tested (Fig. 4A). Increased sensitivity of cells to Hst 5 was similar to that found with the DUR3 O/E strain; and indeed transcripts of DUR3 but not DUR31 were found to be elevated upon treatment of C. albicans cells with CHA (Fig. 4B). Collectively these data show that the expression levels of DUR3 are directly proportional to Hst 5 killing, thus implicating Dur3p as a major intracellular transporter for Hst 5. Also, DUR3 transcriptional regulation appears to be highly responsive to cellular requirements for polyamines, whereas DUR31 transcripts are stable.
FIGURE 4.
Inhibition of polyamine biosynthesis by the spermidine synthase inhibitor CHA increased cell sensitivity to Hst 5. A, WT cells were cultured overnight in medium with or without 5 mm CHA, and then tested for Hst 5 sensitivity. WT cells cultured with CHA showed significantly (p < 0.01) increased Hst 5 mediated killing as compared with cells grown in medium only, which was accompanied by (B) an increase in the expression level of DUR3 but not DUR31 in 5 mm CHA-treated cells; thus indicating the role of Dur3p for Hst 5 toxicity.
Spermidine Uptake in C. albicans Is Concentration and Energy Dependent
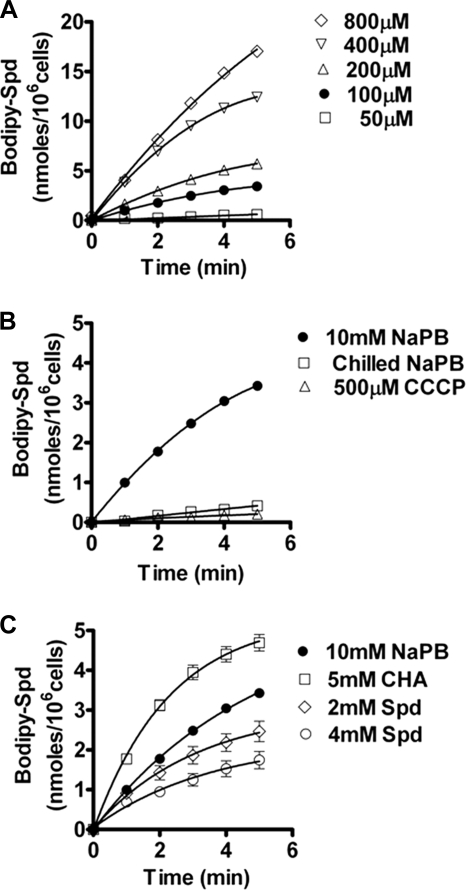
To better define spermidine transport in the context of its role in Hst 5 uptake by Dur transporters, we examined whether spermidine uptake was affected by conditions that are known to inhibit Hst 5 uptake. Because Hst 5 intracellular transport and killing of C. albicans is abolished by incubating cells at 4 °C (19) or with membrane protonophores including CCCP (14, 17), we tested whether spermidine uptake was also prevented under these conditions. Spermidine uptake was measured using spermidine conjugated with the fluorophore dye BODIPY 630/650 (BODIPY-Spd). Initial studies of C. albicans cells following incubation with BODIPY-Spd for 10 min and visualization by confocal microscopy showed uniform cytosolic distribution of red fluorescence, providing unambiguous evidence of internalization of the BODIPY-Spd conjugate (data not shown). To perform uptake and competition assays, a microtiter plate assay was developed and optimized for C. albicans cell numbers, growth phase, and concentration of BODIPY-Spd. Values for BODIPY-Spd uptake by C. albicans cells were calculated based on fluorescence units as described under “Experimental Procedures.” A concentration-dependent uptake of BODIPY-Spd by wild-type cells was observed that was linear within the first 5 min (Fig. 5A). Based on this dose curve, 100 μm BODIPY-Spd was selected for use in subsequent experiments as it allowed easily measurable uptake and did not alter Hst 5 cell wall binding. Next, we quantified BODIPY-Spd uptake under conditions that inhibit Hst 5 uptake (low temperature or protonophores), and found that chilling cells at 4 °C or pretreatment with 500 μm CCCP completely inhibited uptake of BODIPY-Spd (Fig. 5B). Furthermore, pretreatment of cells with CHA resulting in increased Hst 5 killing also increased spermidine uptake; although culturing cells in spermidine that reduced Hst 5 killing also decreased subsequent spermidine uptake (Fig. 5C). Thus both Hst 5 and spermidine uptake by C. albicans are dependent upon the same conditions, suggesting that a similar mechanism is involved in intracellular translocation.
FIGURE 5.
Spermidine uptake in C. albicans is concentration and energy dependent. A, C. albicans CAF4-2 cells were grown to mid-log phase, and 1 × 106 cells in 10 mm sodium phosphate buffer (NaPB) were added to each microtiter plate well for spermidine uptake assays using 50, 100, 200, 400, and 800 μm BODIPY-labeled spermidine (Bodipy-Spd). Uptake was calculated on the basis of fluorescence units of BODIPY-Spd and normalized by subtraction from the fluorescent count values from control wells containing only BODIPY-Spd as well as from the readings at time 0. Cells demonstrated a concentration-dependent linear uptake of BODIPY-Spd within 5 min. B, wild-type CAF4-2 cells were preincubated with 500 μm CCCP in NaPB or chilled at 4 °C before addition of BODIPY-X-Spd (100 μm). Uptake of BODIPY-X-Spd was significantly inhibited by more than 90% in chilled cells or cells pretreated with CCCP. C, culture of wild-type CAF4-2 cells with spermidine (2–4 mm) significantly reduced BODIPY-X-Spd uptake by 25–50%, whereas growth of cells with 5 mm CHA significantly increased BODIPY-X-Spd uptake by more than 50%. These changed in BODIPY-X-Spd uptake that positively correlate with the altered transcript levels of DUR3 (see Figs. 3 and 4). Data shown are the mean ± S.E. of triplicate independent experiments.
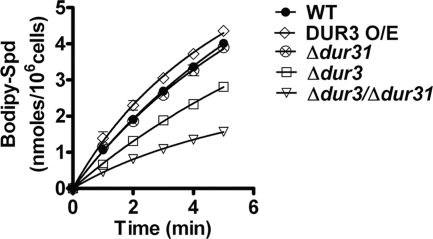
To determine the role of each Dur transporter in spermidine uptake, quantitative transport of BODIPY-Spd was measured for each DUR deletion mutant and compared with parental CAF4-2 cells (Fig. 6). The uptake of BODIPY-Spd was decreased by one-third in the Δdur3 strain and further reduced by two-thirds in the double deletion mutant Δdur3/Δdur31 (both significant using the Dunn multiple comparison test at p < 0.01). As expected the Δdur31 strain did not have significantly reduced spermidine uptake due to overexpression of DUR3 (Fig. 2B), whereas the DUR3 O/E strain had significantly higher spermidine uptake when compared with wild-type CAF4-2 cells. Replacement of DUR3 to the Δdur3 deletion strain (Δdur3::DUR3) restored spermidine transport to wild-type levels (data not shown). Altogether, these data demonstrate that Dur3 and Dur31 proteins are major transporters used by C. albicans cells for spermidine uptake, and show that spermidine uptake and Hst 5 killing require the same Dur transporters.
FIGURE 6.
Spermidine uptake is most reduced by DUR3 and DUR31 gene deletions in C. albicans cells. C. albicans cells were grown to mid-log phase, and 1 × 106 cells in 10 mm sodium phosphate buffer (NaPB) were added to each microtiter plate well for spermidine uptake assays using 100 μm BODIPY-labeled spermidine (Bodipy-Spd). Uptake for each strain was calculated for 5 min after addition of BODIPY-X-Spd, and normalized by subtraction from the fluorescent count values from control wells containing only BODIPY-X-Spd as well as from the readings at time 0. Spermidine uptake was increased in the DUR3 overexpression strain; but was most significantly (p < 0.01) reduced in the double deletion strain Δdur3/Δdur31 and significantly (p < 0.05) reduced in Δdur3 cells when compared with WT CAF4-2 cells. Cells with deletion of DUR31 alone did not differ from WT. Data shown are the mean ± S.E. of triplicate independent determinations.
Dur3p and Dur31p Are Preferential Transporters for Spermidine and Hst 5
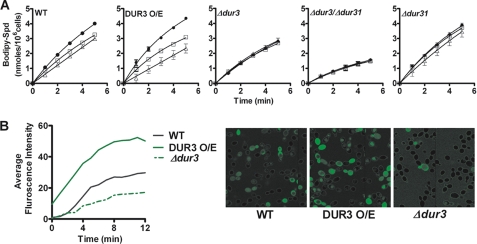
To directly test the ability of Hst 5 to utilize Dur transporters for its uptake, competition experiments were carried out. We chose to test both 15.5 and 31 μm Hst 5 in competition assays, as these are relevant physiological concentrations characterized in killing experiments (Fig. 2A). Addition of Hst 5 along with BODIPY-Spd (100 μm) to WT CAI4-2 cells resulted in reduced spermidine uptake that was more pronounced with higher (31 μm) Hst 5 concentrations (Fig. 7A), thus proving that Hst 5 utilizes the same transporters as spermidine for its entry into the cells. C. albicans cells (DUR3 O/E) overexpressing Dur3 had even more reduction in spermidine uptake in the presence of Hst 5 compared with WT cells. We observed that both the single deletion mutant Δdur3 and the double deletion mutant Δdur3/Δdur31 had a complete loss of competition at both 15.5 and 31 μm Hst 5 (Fig. 7A), showing that Hst 5 competes for spermidine uptake with both Dur3 and Dur31 transporters. Competition was restored in the Dur3 complementation strain (data not shown). Deletion of the Dur31 protein resulted in partial inhibition at 31 μm Hst 5 and no inhibition at 15.5 μm Hst 5, as expected due to overexpression of the remaining DUR3 gene.
FIGURE 7.
Hst 5 competes for spermidine uptake in C. albicans cells and is dependent upon Dur3 and Dur31. A, uptake of BODIPY-X-Spd (100 μm) was measured in the presence of 15.5 and 31 μm Hst 5 in wild-type and mutant strains of the DUR gene family. Uptake of BODIPY-X-Spd (●) was calculated in the presence of 15.5 μm Hst 5 (□) or 31 μm Hst 5 (Δ) based upon fluorescence units. Deletion of Dur3 protein (Δdur3) and Δdur3/Δdur31 double deletion resulted in complete loss of competitive inhibition of BODIPY-X-Spd by Hst 5 at both doses (31 and 15.5 μm), whereas the Δdur31 deletion mutant showed partial loss of Hst 5 inhibition. The DUR3 O/E strain showed higher competition for Hst 5. Data shown are the mean ± S.E. of triplicate independent determinations. B, cellular uptake of FITC-Hst 5 (30 μm) in WT, Δdur3, and DUR3 O/E was compared using time-lapse confocal microscopy. Intracellular translocation of FITC-Hst 5 was substantially reduced in Δdur3 cells as compared with wild-type cells, whereas in DUR3 O/E cells there was increased intracellular translocation of FITC-Hst 5.
To further confirm the involvement of Dur3 in the uptake of Hst 5, we measured the intracellular accumulation of FITC-Hst 5 using time-lapse confocal microscopy in Δdur3 and DUR3 O/E C. albicans cells (Fig. 7B). Cells overexpressing Dur3 had more rapid uptake and higher intracellular accumulation of Hst 5, whereas Δdur3 cells had substantially reduced uptake of Hst 5. Double deletion mutants Δdur3/Δdur31 had virtually no uptake of Hst 5 during the time course of the experiment (data not shown). Collectively, these data demonstrate that Dur3p and Dur31p are plasma membrane transporters involved in intracellular translocation of Hst 5 in C. albicans.
To determine the kinetic parameters for spermidine uptake and Hst 5 competition, Km and Vmax values for spermidine uptake for C. albicans strains bearing DUR3 and DUR31 mutations were calculated and compared with parental WT values (Table 2). The measured Km values were calculated using 106 cells, and predominantly reflects changes in transporter-ligand affinity for the whole cell. The Km value for spermidine uptake in WT cells was 3.6 ± 0.7 μm, which increased significantly (p < 0.01) in single Dur3 or Dur31 transporter deletion mutant strains tested. Both Δdur3 and Δdur31 strains showed more than a 2-fold decrease in affinity for spermidine, whereas the Δdur3/Δdur31 mutant showed more than a 10-fold decrease in affinity. Hence, our results illustrate that Dur3 and Dur31 are high affinity transporters for spermidine in C. albicans. Correspondingly, the Vmax value was reduced by half for the Δdur3 deletion mutant, and restored upon gene replacement. Although Δdur31 mutants did not show significantly reduced uptake of spermidine thus demonstrating the compensatory role of DUR3 in the Δdur31 mutant, the Vmax value for the Δdur3/Δdur31 mutant was reduced by 6-fold to 2.1 nmol/min/106 cells, illustrating that both transporters are major transporters governing the rate of total cellular uptake of spermidine. The remaining intracellular transport of spermidine observed in Δdur3/Δdur31 cells (∼17% of WT levels) is likely due to other Dur family members or possibly other related transporters such as Agp2.
TABLE 2.
Kinetics of spermidine uptake and Hst 5 inhibition in C. albicans strains
| Strains | Vmax | Substrate, spermidine, Km | Inhibitor, histatin 5, Ki |
|---|---|---|---|
| nmol/min/106 cells | μm | μm | |
| CAF4-2 | 12.5 ± 1.5 | 3.6 ± 0.7 | 1.2 ± 0.2 |
| Δdur3 | 6.4 ± 1.3a | 7.5 ± 0.5a | 9.9 ± 0.6a |
| Δdur31 | 9.6 ± 0.7 | 7.8 ± 0.1a | 5.3 ± 0.2a |
| Δdur3/Δdur31 | 2.1 ± 0.2a | 31.9 ± 0.6a | 84.3 ± 0.1a |
| Δdur3/DUR3 | 10.1 ± 1.0 | 4.1 ± 0.3 | NDb |
| DUR3 O/E | 14.1 ± 2.8 | 2.8 ± 0.1 | 0.4 ± 0.1a |
a p value <0.01 compared with CAF4-2.
b ND, not determined.
Next we evaluated the ability of Hst 5 to act as a competitive inhibitor of spermidine uptake by measuring Ki values in each strain (Table 2). The Ki value for Hst 5 in CAF4-2 (WT) cells was 1.2 ± 0.2 μm. Assays conducted with multiple concentrations of Hst 5 showed increased Km values and unchanged Vmax values indicating that Hst 5 is a competitive inhibitor for spermidine in WT cells.
In contrast, the Ki value for Hst 5 showed more than an 8-fold increase upon deletion of Dur3p (Δdur3); and increased by 80-fold in the double deletion mutant (Δdur3/Δdur31). As expected, Hst 5 competition in the DUR3 overexpression strain was significantly reduced to 0.4 ± 0.1 μm. Our results confirm the ability of Hst 5 to compete with spermidine uptake (Fig. 7A) and reveals Dur31p and Dur3p as the preferential spermidine transporters used by Hst 5 for its intracellular translocation in C. albicans cells.
DISCUSSION
Many cationic peptides exert their antimicrobial effects by insertion or intercalation within the plasma membranes of target cells to induce pore formation or membrane perturbation. These toxin peptides are considered to be membrane-lytic. However, human salivary Hsts represent a unique class of peptides whose antifungal mechanism originates from the cytosol following intracellular import. Therefore, identification of their route of entry is crucial for understanding their mechanism of action and ultimately their potential as adjunctive therapeutic agents. This is the first report demonstrating that a small cationic human antifungal protein utilizes native fungal transporters to gain entry into their target cells to exert toxic effects. In this study, we show that a major route of entry of Hst 5 into C. albicans cells is via the Dur family of polyamine transporters.
The discovery of this intracellular transport mechanism explains many of the energy-dependent requirements of Hst 5 activity. It is known that Hst 5 toxicity is reduced upon energy depletion of fungal cells by sodium azide or the protonophores CCCP and dinitrophenol. Incubation of S. cerevisiae cells with azide strongly reduced putrescine uptake by Dur3p (34) because polyamine transport systems are dependent on energy derived from membrane-associated ATPases. Another important determinant of polyamine transport in S. cerevisiae is the Ser/Thr protein kinase named polyamine transport protein kinase 2 (Ptk2p), which is an activator of plasma membrane permeability to various cations including polyamines (46). S. cerevisiae Dur3p is activated by phosphorylation of Thr250, Ser251, and Thr684 by Ptk2p (34). In turn, Ptk2p activity is induced by the glucose-dependent activation of the Pma1p H+-ATPase, thereby increasing the proton motive force that energizes the transport of cationic compounds and polyamines (47). Treatment of yeast cells with energy inhibitors of Pma1p such as the protonophores CCCP and dinitrophenol, as well as azide, inhibits Pma1p function and permease activity, thus accounting for the loss of spermidine uptake we observed in C. albicans cells. Our finding that Hst 5 utilizes polyamine transporters for its intracellular uptake now explains the loss of Hst 5 uptake and killing following incubation of C. albicans with these agents. The requirement of ATPases for polyamine permease activity may also explain the finding that addition of the organic solvent dimethyl sulfoxide (10–40%) to C. albicans cells reduced Hst 5 toxicity (20), because this solvent also inhibits ATPase activity (48). Thus, the observation that dimethyl sulfoxide reduces Hst 5 toxicity is likely not to be a result of increasing membrane rigidity as previously suggested (20), but rather due to loss of polyamine permease activity needed for Hst 5 uptake.
A similar intracellular transport mechanism has been characterized for the antineoplastic drug bleomycin in human and S. cerevisaie cells. Bleomycin is a hydrophilic glycopeptide antibiotic produced by Streptomyces verticillus and used as a genotoxic drug because of its ability to damage tumor DNA. A specialized form of bleomycin called bleomycin-A5 is transported into S. cerevisaie cells by the polyamine permease Agp2, because this bleomycin variant contains a spermidine residue as its terminal amine substituent (49). The therapeutic utility of bleomycin-A5 in cancer patients is based upon the functional similarity of the human high affinity l-carnitine transporter hCT2 with S. cerevisaie Agp2, both of which transport bleomycin-A5 intracellularly to damage DNA (50). This raises the interesting possibility that adding a spermidine or spermine moiety to Hst 5 or other antifungal drugs may improve intracellular delivery through polyamine transporters.
S. cerevisiae uses only four plasma membrane permeases, (Dur3, Sam3, Agp2, and Gap1) to catalyze the uptake of extracellular polyamines, although Dur3 and Sam3 primarily contribute to uptake activity. In contrast, C. albicans has six Dur family members, as well as an Agp2 permease and multiple Sam3/Gap1 orthologs, suggesting considerable functional redundancy among these transporters. The relative abundance of these permeases suggest that regulation of the intracellular polyamine uptake is critical in C. albicans and may play a crucial role in its ability to colonize and survive in various ecological niches. Therefore, it is expected that these permeases have a role in virulence. Interestingly, the Dur3 deletion mutant was not attenuated in systemic virulence (45). However, our results found that although deletion of DUR3 reduced spermidine uptake by half, deletion of both DUR3 and DUR31 was required for the most extensive reduction in spermidine transport. Therefore, we expect that C. albicans Δdur3/Δdur31 mutants would have reduced virulence in vivo where environmental polyamine uptake is crucial for fungal survival. We are currently carrying out experiments to evaluate this possibility.
Our finding that Hst proteins are recognized as polyamines in respect to intracellular transport in fungal cells also points toward other potential interactions between Hsts and cells based upon known interactions with polyamines. We have identified cell wall Hsp70 chaperone proteins in C. albicans that bind Hst 5 and facilitate its uptake, although they do not function themselves as transporters. We speculate that these cell wall proteins serve as initial binding partners before passing Hsts to the actual transporter. Evidence to support this hypothesis is provided by yeast two-hybrid screening of the S. cerevisiae Hsp90-proteome. Strong interactions with a large group of membrane transporters including AGP2 were found (51), suggesting that Hsp chaperones could function to pass substrates to transporter proteins. Thus Candidal Hsp 70 proteins may chaperone Hst 5 from the outer cell wall to the cell membrane where Hst 5 is transferred to membrane localized Dur permeases.
Structural similarities between Hst 5 and polyamines may account for the means by which Hst 5 exerts its toxic effect in target fungal cells. There is substantial evidence that intracellular polyamines function as gating molecules for potassium (K+) channels in both eukaryotic and bacterial cells (52, 53). Polyamines function as modulators of voltage-dependent inward rectification by blocking or gating the K+ channel pore from the intracellular aspect. In respect to Hst 5, we found that one of the initial effects of this peptide upon entry into fungal cells is efflux of K+ ions and ATP as well as the accompanying loss of the cells ability to regulate ion homeostasis. Hst 5 binding to the C. albicans K+ channel Trk1p produces a leakage pathway, through either Trk1 protein itself or a larger complex involving Trk1p. Our model is that Trk1p is distorted by Hst 5 binding so that it permits the efflux of larger anions such as ATP, then causing cellular osmotic stress response, cell volume loss, and eventual cell cycle arrest. Thus it is possible that Hst 5 is a polyamine analog in its interactions with fungal K+ channels as well as with Dur permeases.
Because our evidence shows that Hst 5 functions as a spermidine analog for uptake in fungal cells, other interesting possibilities are raised. In S. cerevisiae, addition of spermidine can alter gene expression involved in various metabolic pathways and activate other transport systems in the absence of the preferred transporter (54, 55); thus it is possible that the presence of Hst 5 modulates C. albicans metabolism or carrier affinities. Also, intracellular Hst 5 may be recognized as contributing to high levels of cellular polyamines, and then be excreted from the cell via polyamine TPO permeases. S. cerevisiae expresses polyamine excretion permeases on the plasma membrane (FLU1, TPO1, TPO4, and TPO5) that have important roles in detoxification and drug excretion. We have identified at least four C. albicans plasma membrane homologues of S. cerevisiae TPO and FLU1 polyamine exporters, thus it is possible that the Hst 5 export from fungal cells may be a major route of acquired resistance to the peptide that has been observed upon prolonged exposure to this peptide. This is an area of active investigation in our laboratory.
The concept that the functionality of Hst 5 in C. albicans is based upon its recognition as a polyamine homologue leads to a unified understanding of its cytotoxic activities within the fungal cell. Hst 5 energy-dependent uptake through Dur transporters, intracellular potassium channel targets, and regulation of intracellular concentration are consistent with the known functions of polyamines. These findings also provide guided strategies to improve cationic peptide delivery to target cells by linkage with small polyamine carriers.
This work was supported, in whole or in part, by National Institutes of Health Grant DE10641 from the NIDCR (to M. E.).
- Hst 5
- histatin 5
- Spd
- spermidine
- YPD
- yeast extract/peptone/dextrose
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone
- CHA
- cyclohexylamine
- NaPB
- sodium phosphate buffer
- STR
- second target sequence
- DUR3 O/E
- Dur3 overexpression strain.
REFERENCES
- 1. Oppenheim F. G., Xu T., McMillian F. M., Levitz S. M., Diamond R. D., Offner G. D., Troxler R. F. (1988) J. Biol. Chem. 263, 7472–7477 [PubMed] [Google Scholar]
- 2. Xu T., Levitz S. M., Diamond R. D., Oppenheim F. G. (1991) Infect. Immun. 59, 2549–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rothstein D. M., Spacciapoli P., Tran L. T., Xu T., Roberts F. D., Dalla Serra M., Buxton D. K., Oppenheim F. G., Friden P. (2001) Antimicrob. Agents Chemother. 45, 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jang W. S., Li X. S., Sun J. N., Edgerton M. (2008) Antimicrob. Agents Chemother. 52, 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hancock R. E., Lehrer R. (1998) Trends Biotechnol. 16, 82–88 [DOI] [PubMed] [Google Scholar]
- 6. Lear J. D., Wasserman Z. R., DeGrado W. F. (1988) Science 240, 1177–1181 [DOI] [PubMed] [Google Scholar]
- 7. Shai Y. (2002) Biopolymers 66, 236–248 [DOI] [PubMed] [Google Scholar]
- 8. Brewer D., Hunter H., Lajoie G. (1998) Biochem. Cell Biol. 76, 247–256 [DOI] [PubMed] [Google Scholar]
- 9. Raj P. A., Marcus E., Sukumaran D. K. (1998) Biopolymers 45, 51–67 [DOI] [PubMed] [Google Scholar]
- 10. Situ H., Balasubramanian S. V., Bobek L. A. (2000) Biochim. Biophys. Acta 1475, 377–382 [DOI] [PubMed] [Google Scholar]
- 11. Ruissen A. L., Groenink J., Helmerhorst E. J., Walgreen-Weterings E., Van't Hof W., Veerman E. C., Nieuw Amerongen A. V. (2001) Biochem. J. 356, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Den Hertog A. L., Wong Fong Sang H. W., Kraayenhof R., Bolscher J. G., Van't Hof W., Veerman E. C., Nieuw Amerongen A. V. (2004) Biochem. J. 379, 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li X. S., Sun J. N., Okamoto-Shibayama K., Edgerton M. (2006) J. Biol. Chem. 281, 22453–22463 [DOI] [PubMed] [Google Scholar]
- 14. Koshlukova S. E., Lloyd T. L., Araujo M. W., Edgerton M. (1999) J. Biol. Chem. 274, 18872–18879 [DOI] [PubMed] [Google Scholar]
- 15. Baev D., Rivetta A., Vylkova S., Sun J. N., Zeng G. F., Slayman C. L., Edgerton M. (2004) J. Biol. Chem. 279, 55060–55072 [DOI] [PubMed] [Google Scholar]
- 16. Vylkova S., Sun J. N., Edgerton M. (2007) Purinergic Signal. 3, 91–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jang W. S., Bajwa J. S., Sun J. N., Edgerton M. (2010) Mol. Microbiol. 77, 354–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baev D., Li X., Edgerton M. (2001) Microbiology 147, 3323–3334 [DOI] [PubMed] [Google Scholar]
- 19. Xu Y., Ambudkar I., Yamagishi H., Swaim W., Walsh T. J., O'Connell B. C. (1999) Antimicrob. Agents Chemother. 43, 2256–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Veerman E. C., Valentijn-Benz M., Nazmi K., Ruissen A. L., Walgreen-Weterings E., van Marle J., Doust A. B., van't Hof W., Bolscher J. G., Amerongen A. V. (2007) J. Biol. Chem. 282, 18831–18841 [DOI] [PubMed] [Google Scholar]
- 21. Mochon A. B., Liu H. (2008) PLoS Pathog. 4, e1000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pao S. S., Paulsen I. T., Saier M. H., Jr. (1998) Microbiol. Mol. Biol. Rev. 62, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bauer B. E., Wolfger H., Kuchler K. (1999) Biochim. Biophys. Acta 1461, 217–236 [DOI] [PubMed] [Google Scholar]
- 24. Pasrija R., Banerjee D., Prasad R. (2007) Eukaryot. Cell 6, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Decottignies A., Goffeau A. (1997) Nat. Genet. 15, 137–145 [DOI] [PubMed] [Google Scholar]
- 26. Sanglard D., Ischer F., Calabrese D., Majcherczyk P. A., Bille J. (1999) Antimicrob. Agents Chemother. 43, 2753–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jungwirth H., Kuchler K. (2006) FEBS Lett. 580, 1131–1138 [DOI] [PubMed] [Google Scholar]
- 28. Igarashi K., Kashiwagi K. (1999) Biochem. J. 344, 633–642 [PMC free article] [PubMed] [Google Scholar]
- 29. Woolridge D. P., Vazquez-Laslop N., Markham P. N., Chevalier M. S., Gerner E. W., Neyfakh A. A. (1997) J. Biol. Chem. 272, 8864–8866 [DOI] [PubMed] [Google Scholar]
- 30. Shah P., Briles D. E., King J., Hale Y., Swiatio E. (2009) Exp. Biol. Med. 234, 403–409 [DOI] [PubMed] [Google Scholar]
- 31. Tomitori H., Kashiwagi K., Asakawa T., Kakinuma Y., Michael A. J., Igarashi K. (2001) Biochem. J. 353, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aouida M., Leduc A., Poulin R., Ramotar D. (2005) J. Biol. Chem. 280, 24267–24276 [DOI] [PubMed] [Google Scholar]
- 33. Uemura T., Kashiwagi K., Igarashi K. (2005) Biochem. Biophys. Res. Commun. 328, 1028–1033 [DOI] [PubMed] [Google Scholar]
- 34. Uemura T., Kashiwagi K., Igarashi K. (2007) J. Biol. Chem. 282, 7733–7741 [DOI] [PubMed] [Google Scholar]
- 35. Fonzi W. A., Irwin M. Y. (1993) Genetics 134, 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morschhäuser J., Michel S., Staib P. (1999) Mol. Microbiol. 32, 547–556 [DOI] [PubMed] [Google Scholar]
- 37. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 38. Saitou N., Nei M. (1987) Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 39. Choi J. H., Jung H. Y., Kim H. S., Cho H. G. (2000) Bioinformatics 16, 1056–1058 [DOI] [PubMed] [Google Scholar]
- 40. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 41. Spyropoulos I. C., Liakopoulos T. D., Bagos P. G., Hamodrakas S. J. (2004) Bioinformatics 20, 3258–3260 [DOI] [PubMed] [Google Scholar]
- 42. Finn R. D., Mistry J., Schuster-Böckler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Eddy S. R., Sonnhammer E. L., Bateman A. (2006) Nucleic Acids Res. 34, D247-D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bensen E. S., Martin S. J., Li M., Berman J., Davis D. A. (2004) Mol. Microbiol. 54, 1335–1351 [DOI] [PubMed] [Google Scholar]
- 44. Braun B. R., van Het Hoog M., d'Enfert C., Martchenko M., Dungan J., Kuo A., Inglis D. O., Uhl M. A., Hogues H., Berriman M., Lorenz M., Levitin A., Oberholzer U., Bachewich C., Harcus D., Marcil A., Dignard D., Iouk T., Zito R., Frangeul L., Tekaia F., Rutherford K., Wang E., Munro C. A., Bates S., Gow N. A., Hoyer L. L., Köhler G., Morschhäuser J., Newport G., Znaidi S., Raymond M., Turcotte B., Sherlock G., Costanzo M., Ihmels J., Berman J., Sanglard D., Agabian N., Mitchell A. P., Johnson A. D., Whiteway M., Nantel A. (2005) PLoS Genet. 1, 36–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Navarathna D. H., Das A., Morschhaeuser J., Nickerson K. W., Roberts D. D. (2011) Microbiology 157, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nozaki T., Nishimura K., Michael A. J., Maruyama T., Kakinuma Y., Igarashi K. (1996) Biochem. Biophys. Res. Commun. 228, 452–458 [DOI] [PubMed] [Google Scholar]
- 47. Goossens A., de La Fuente N., Forment J., Serrano R., Portillo F. (2000) Mol. Cell. Biol. 20, 7654–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ferreira-Pereira A., Alves-Ferreira M., de Carvalho-Alves P. C. (1994) J. Biol. Chem. 269, 12074–12079 [PubMed] [Google Scholar]
- 49. Aouida M., Leduc A., Wang H., Ramotar D. (2004) Biochem. J. 384, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aouida M., Poulin R., Ramotar D. (2010) J. Biol. Chem. 285, 6275–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Millson S. H., Truman A. W., King V., Prodromou C., Pearl L. H., Piper P. W. (2005) Eukaryot. Cell 4, 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oliver D., Baukrowitz T., Fakler B. (2000) Eur. J. Biochem. 267, 5824–5829 [DOI] [PubMed] [Google Scholar]
- 53. Cheng W. W., Enkvetchakul D., Nichols C. G. (2009) J. Gen. Physiol. 133, 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chattopadhyay M. K., Chen W., Poy G., Cam M., Stiles D., Tabor H. (2009) Yeast 26, 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kaouass M., Gamache I., Ramotar D., Audette M., Poulin R. (1998) J. Biol. Chem. 273, 2109–2117 [DOI] [PubMed] [Google Scholar]