Background: LAPTM5 is a membrane protein on the intracellular vesicles and the molecular mechanism for its post-translational regulation has been unclear.
Results: ITCH E3 ligase promoted ubiquitination-mediated degradation and prevented LAPTM5-mediated cell death.
Conclusion: LAPTM5 is a substrate of the ITCH-mediated degradation and its protein level is negatively regulated by ITCH.
Significance: This system might act as a negative regulator in tumorigenesis by preventing LAPTM5-mediated cell death.
Keywords: Cell Death, E3 Ubiquitin Ligase, Protein Degradation, Tumor, Vesicles
Abstract
LAPTM5 (lysosomal-associated protein multispanning transmembrane 5) is a membrane protein on the intracellular vesicles. We have previously demonstrated that the accumulation of LAPTM5-positive vesicles was closely associated with the programmed cell death occurring during the spontaneous regression of neuroblastomas. Although the accumulation of LAPTM5 protein might occur at the post-translational level, the molecular mechanism has been unclear. Here, we found that the level of LAPTM5 protein is regulated negatively by the degradation through ubiquitination by ITCH, an E3 ubiquitin ligase. ITCH directly binds to the PPxY motif of LAPTM5 via its WW domains and promotes ubiquitination through a HECT-type ligase domain. Overexpression of ITCH led to the degradation of LAPTM5 protein, and conversely, knockdown of ITCH by siRNA resulted in the stabilization of LAPTM5 protein. In addition, the inhibition of ITCH enhanced the cell death occurred by accumulation of LAPTM5 in neuroblastoma cells. These findings suggest that LAPTM5 is a novel substrate in terms of degradation by the ubiquitin ligase ITCH, and this system might act as a negative regulator in the spontaneous regression of neuroblastomas by preventing LAPTM5-mediated cell death.
Introduction
Ubiquitination, the process by which ubiquitin is conjugated to Lys side chains of proteins, has emerged as the mechanism by which most intracellular proteins are targeted for lysosomal and proteasomal degradation, the two major proteolytic systems of the cell (1). The specificity of the ubiquitination reaction is achieved by the E3 ubiquitin ligases (E3), which mediate the transfer of ubiquitin from E2 ubiquitin-conjugating enzymes (E2) to substrates.
ITCH or AIP4 (atrophin-1-interacting protein 4, hereafter referred to as ITCH) belongs to the Nedd4-like family, which is characterized by a distinct modular domain architecture, with each member consisting of a calcium membrane targeting domain, two to four WW domains conferring substrate specificity, and a HECT-type ligase domain coordinating with E2 and providing the catalytic E3 activity (2, 3). ITCH contributes to many cellular processes by targeting the regulators of multiple signaling pathways (3).
We have identified previously that ITCH was highly expressed by a mechanism of gene amplification in anaplastic thyroid carcinoma (4). Additionally, it has been reported that ITCH was highly expressed in human cancers, including lymphomas, bladder cancer, and breast cancer, and may contribute to tumorigenesis by targeting tumor suppressors including p63, p73, and LATS (3–6). LAPTM5 (lysosomal-associated protein multispanning transmembrane 5) is a membrane protein on the intracellular vesicles and regulates vesicle trafficking such as endocytosis (7–9). We have recently suggested that the accumulation of LAPTM5-positive vesicles was closely associated with the programmed cell death occurring during the spontaneous regression of neuroblastomas (NBs)3 and that the treatment with proteasomal and/or lysosomal inhibitors results in the accumulation of LAPTM5-positive vesicles and enhances LAPTM5-mediated cell death in NB cells (10). Thus, LAPTM5 protein is degraded by a proteasomal and/or lysosomal pathway; however, its molecular mechanism remains unknown.
ITCH is involved in proteasomal and/or lysosomal degradation by interacting with the PPxY motif of substrate through a WW domain and by promoting ubiquitination through a HECT ligase domain (3). LAPTM5 also has a PPxY motif within its C terminus. Hence, we examined whether LAPTM5 is a substrate for the protein degradation through ubiquitination by ITCH. We showed the physical interaction of these molecules through the WW domain of ITCH and PPxY motif of LAPTM5. This interaction resulted in the ubiquitination and the protein degradation of LAPTM5 in the proteasomal and/or lysosomal pathway. Moreover, inhibition of ITCH led to the stabilization of LAPTM5, resulting in the enhancement LAPTM5-mediated cell death in NB cells. Our findings suggest that ITCH negatively regulates the level of LAPTM5 protein through ubiquitin-mediated degradation.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293, 8305C, and GOTO cells were obtained from the ATCC. The KYSE410 cell line established from surgically resected tumors (11) was kindly provided by Dr. Yutaka Shimada (University of Toyama). The HEK293 and 8305C cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. The GOTO and KYSE410 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cells were transfected with each plasmid DNA or siRNA by Lipofectamine 2000 or Lipofectamine RNAiMAX (Invitrogen), respectively, according to the manufacturer's instructions.
Antibodies and Reagents
Mouse monoclonal and rabbit polyclonal anti-Myc tag (Cell Signaling Technology), mouse monoclonal anti-β-actin (Sigma), mouse monoclonal anti-FLAG tag (Sigma), goat and rabbit polyclonal anti-ITCH (Santa Cruz Biotechnology), mouse monoclonal anti-cathepsin D (Chemicon), and rabbit polyclonal anti-LAPTM5 (10) antibodies were used for Western blot and/or immunofluorescence analyses. For the treatment with drugs in cell culture, cycloheximide (Sigma), bafilomycin A1 (Sigma), ALLN (Calbiochem), NH4Cl (Wako), and MG132 (Calbiochem) were used.
Plasmids, Site-directed Mutagenesis, and Synthetic siRNA
Human ITCH and LAPTM5 cDNAs were obtained by PCR and inserted into pCMV-3Tag1A, pCMV-Tag3B (Stratagene), pGEX4T-1 (GE Healthcare), or pEXP5-NT (Invitrogen). The expression vector for catalytic inactivate mutant of ITCH or PPPY-deleted mutant of LAPTM5 were constructed by PCR using primers with mutation or deletion. The expression vector for a pCMV-3Tag1A-ITCH C830A truncation mutant and the constructs deleted each WW domain, including ΔWW1 (288–318), ΔWW2 (320–349), ΔWW3 (400–429), or ΔWW4 (438–469), were generated using the GeneTailor site-directed mutagenesis system (Invitrogen). The pcDNA/His-Ub plasmid was kindly provided by Dr. Tamotsu Nishida (Mie University). Primer sequences used in the construction of vectors are provided in supplemental Table 1. The siRNAs for ITCH and luciferase as a control were synthesized by Sigma (4).
Recombinant Protein
The GST-fused protein with the intact or inactivated form of ITCH (GST-ITCH or GST-ITCH C830A) was expressed in Escherichia coli. GST-fused proteins were purified with glutathione-Sepharose 4B (GE Healthcare) (12). Recombinant proteins were dialyzed against 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, and 0.6% CHAPS. The concentrations of recombinant proteins were determined by Coomassie Brilliant Blue R-250 staining. The LAPTM5 protein fused with His6 was purified by the MembraneMax system following the manufacturer's recommendations (Invitrogen).
Immunoprecipitation
Cells were lysed in buffer (10 mm Tris-HCl, pH 7.8, 150 mm NaCl, 3 mm MgCl2, and 1% Nonidet P-40) with a protease inhibitor mixture tablet (Roche Applied Science) on ice, and the lysate was sonicated. After centrifugation at 15,000 rpm for 15 min at 4 °C, the supernatant was incubated with protein A-Sepharose (GE Healthcare) and the antibody for 6 h at 4 °C. Beads were washed five times with wash buffer (10 mm Tris-HCl, pH 7.8, 150 mm NaCl, 3 mm MgCl2, 0.1% Nonidet P-40, and 200 mm phenylmethylsulfonyl fluoride), resuspended in sample buffer, and analyzed by SDS-PAGE.
Western Blot Analysis
Cell lysates and immunoprecipitates were subjected to SDS-PAGE, and proteins were transferred to PVDF membranes (GE Healthcare). The membrane was blocked in TBS containing 5% nonfat milk and 0.1% Tween 20 and incubated with the primary antibody at room temperature overnight, followed by the HRP-conjugated secondary antibody for 1 h. Afterward, the proteins of interest were visualized using the ECL chemiluminescence system (Thermo) on LAS-3000 (Fuji Film). In some experiments, the intensity of detected bands was measured using MultiGauge software (Fuji Film). The dilutions for used primary antibodies were followed; rabbit anti-LAPTM5 (1/1000), goat anti-ITCH (1/1000), rabbit anti-ITCH (1/500), anti-FLAG tag (1/1000), anti-Myc tag (1/1000), and anti-β-actin (1/5000).
Ubiquitination Assay
For ubiquitination assay, pcDNA/His-Ub, Myc-LAPTM5, and 3×FLAG-ITCH or 3×FLAG-ITCH C830A were transiently transfected in HEK293 cells. After 24 h, the cells were treated with ALLN (10 μm) and bafilomycin A1 (50 nm) for 6 h. Then, they were lysed in buffer A (6 m guanidinium chloride, 0.1 m Na2HPO4/NaH2PO4 pH 8.0, and 10 mm imidazole). Lysates were affinity-purified with nickel-nitrilotriacetic acid (Ni-NTA) superflow (Qiagen) and were analyzed by immunoblotting (13). For the in vitro ubiquitination assay, His6-LAPTM5 trapped on Ni-NTA superflow (Qiagen) was mixed with 1 μg of ubiquitin-activating enzyme (BostonBiochem), 1.8 μg of UbcH7 (BostonBiochem), 15 μg of biotinylated ubiquitin (BostonBiochem), and 500 ng of GST-ITCH or GST-ITCH C830A. The mixture was incubated at 25 °C for 30 min in the presence of 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2, 2 mm dithiothreitol, and 2 mm ATP in a final volume of 50 μl. The resin was washed three times with 10 mm Tris-HCl, pH 7.4, 3 mm MgCl2, 200 mm NaCl, 0.2 mm phenylmethylsulfonyl fluoride, and 0.1% Nonidet P-40. The ubiquitinated proteins on the resin were subjected to SDS-PAGE and detected by Western blot using peroxidase-conjugated avidin (ExtrAvidin; Sigma) or anti-LAPTM5 antibody.
Recombinant Adenovirus
The replication-defective recombinant adenovirus was constructed with the adenovirus expression vector kit (Takara) following the manufacturer's recommendations (10). Viral titers were measured in pfu/ml by a limiting dilution method using the HEK293 cells. Cells were infected with each multiplicity of infection (pfu/cell).
Half-life Determination
HEK293 cells were transiently co-transfected with Myc-LAPTM5 and 3×FLAG empty vector, 3×FLAG-ITCH WT, or 3×FLAG-ITCH C830A. After 24 h, cycloheximide was added at a final concentration of 100 μg/ml to inhibit protein synthesis. Cells were harvested at the indicated time points for Western blot analysis.
Immunofluorescence Analysis
Cells were fixed in 10% TCA, permeabilized with 0.2% Triton X-100 for 10 min, and treated with blocking solution (1% BSA/0.01% Triton X-100 in PBS) for 1 h, and then incubated with the primary antibody at room temperature overnight. The bound antibody was visualized using a FITC-conjugated antibody (Jackson ImmunoResearch Laboratories) or Alexa Fluor 594-conjugated antibody (Invitrogen). For immunofluorescence analysis in KYSE410, cells were fixed in methanol at −20 °C for 20 min, permeabilized with 0.5% Triton X-100 for 10 min, and treated with blocking solution (3% BSA/0.05% Triton X-100 in PBS) for 1 h and then incubated with anti-LAPTM5 antibody at room temperature overnight. The bound antibody was visualized using an Alexa Fluor 488-conjugated antibody (Invitrogen). After nuclear staining by DAPI (Sigma), the cells were observed under a LSM510 confocal microscope (Carl Zeiss). The dilutions for used antibodies were as follows: rabbit anti-LAPTM5 (1/1000), mouse anti-cathepsin D (1/2000), goat anti-ITCH (1/1000), and secondary antibodies (1/2000).
Cell Death Assay
The number of dead cells was measured by the trypan blue exclusion method. The results from six independent experiments were presented as the mean and S.D. Differences were tested with a two-sided test (Student's t test).
Acridine Orange (AO) Uptake Analysis
Cells were stained with 5 μg/ml of AO (Sigma) for 30 min at 37 °C. Then, the detached cells were removed, and the attached cells were collected by trypsinization and washed twice with PBS. The cells untaken AO were detected by flow cytometry using an Accuri Cytometer (Becton Dickinson) and were analyzed by FlowJo software. Differences were tested with a two-sided test (Student's t test).
RESULTS
Interaction with ITCH and LAPTM5 through WW Domain of ITCH and PPxY Motif of LAPTM5
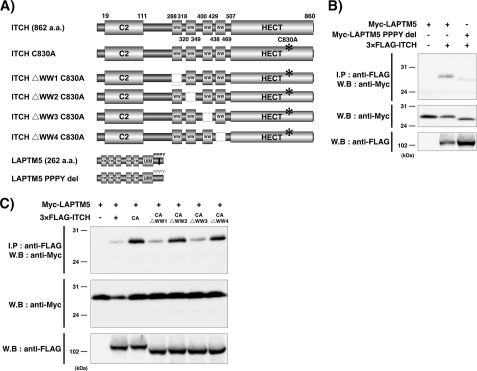
ITCH contains four WW domains that mediate the binding to PPxY motif of substrates (Fig. 1A) (14). LAPTM5 contains five transmembrane domains, a ubiquitin-interacting motif, and PPxY motifs (256–259 amino acids, PPPY) (Fig. 1A). To determine whether ITCH and LAPTM5 interact through these domains, we first performed an immunoprecipitation Western blot analysis. As shown in Fig. 1B, when the Myc-tagged LAPTM5 WT and 3×FLAG-tagged ITCH expression vector were co-transfected into HEK293 cells, these proteins were interacted. In contrast, the interaction between the Myc-tagged LAPTM5 with deletion for the PPPY motif and 3×FLAG-tagged ITCH was nearly abolished (Fig. 1B). Furthermore, when the Myc-tagged LAPTM5 WT was co-transfected with 3×FLAG-tagged ITCH C830A (a point mutant with an inactivated E3 ligase), amount of Myc-tagged LAPTM5 protein was remarkably increased in compared with that in co-transfection with 3×FLAG-tagged ITCH WT, suggesting that ITCH negatively regulates the level of LAPTM5 protein (Fig. 1C). Next, to determine the essential domains among the four WW domains of ITCH for ITCH-LAPTM5 interaction, we generated the deleted mutants for each WW domain in the 3×FLAG-tagged ITCH C830A expression vector (Fig. 1A). On co-transfection with the deleted mutant for the first or third WW domain and Myc-tagged LAPTM5 WT expression vector, the ITCH-LAPTM5 interaction was decreased remarkably (Fig. 1C). These results suggest that ITCH interacts with PPPY within the C terminus of LAPTM5 through the first and third WW domains and contributes to the down-regulation of LAPTM5 protein level.
FIGURE 1.
ITCH and LAPTM5 interact through WW domains and the PPxY motif. A, schematic representation of the motif for ITCH, LAPTM5, and respective deletion mutants. The mutant with C830A results in the production of an inactivated ITCH protein. Four deletion mutants for each WW domain were generated from the ITCH C830A expression vector. The expression vector with deletion within the PPxY motif of LAPTM5 was generated. B, significance of PPxY motif for LAPTM5-ITCH interaction. HEK293 cells were transiently co-transfected with Myc-tagged LAPTM5 alone, or 3×FLAG-ITCH and Myc-tagged LAPTM5 WT, or PPPY deletion mutant (PPPY del). After 24 h, cell lysates were prepared in lysis buffer and immunoprecipitated with anti-FLAG antibody. The whole cell lysates or immunoprecipitates were subjected to SDS-PAGE and immunoblotted (W.B) using the indicated antibodies. C, significance of the HECT and WW domains of ITCH for LAPTM5-ITCH interaction. HEK293 cells were transiently co-transfected with Myc-tagged LAPTM5 alone, or Myc-tagged LAPTM5 and 3×FLAG-ITCH, 3×FLAG-ITCH C830A, 3×FLAG-ITCH C830A with a deletion in each WW domain. After 24 h, cell lysates were prepared in lysis buffer and immunoprecipitated (I.P) with anti-FLAG antibody. The whole cell lysates or immunoprecipitates were subjected to SDS-PAGE and immunoblotted using the indicated antibodies. a.a., amino acids. CA, C830A.
ITCH Negatively Affects Stability of LAPTM5
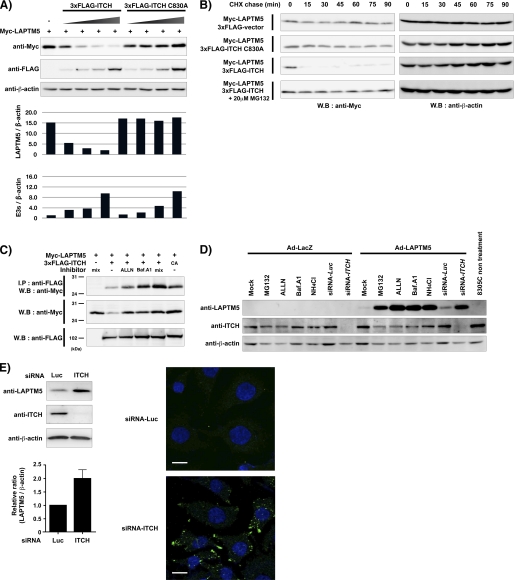
Next, we examined whether ITCH contributes to the protein degradation of LAPTM5 via its E3 ligase activity. When the different amounts of 3×FLAG-tagged ITCH WT expression vector was co-transfected transiently with Myc-tagged LAPTM5 expression vector in HEK293 cells, the level of LAPTM5 protein gradually decreased as correlated with the amount of ITCH protein, whereas it did not decrease in the 3×FLAG-tagged ITCH C830A mutant expression vector (Fig. 2A). Furthermore, we examined the effects of ITCH on LAPTM5 turnover. HEK293 cells were co-transfected with Myc-tagged LAPTM5 and 3×FLAG-tagged empty vector, 3×FLAG-tagged ITCH WT, or 3×FLAG-tagged ITCH C830A mutant expression vector, and after 24 h, the cells were treated with cycloheximide to block protein synthesis. As shown in Fig. 2B, co-expression with ITCH WT and LAPTM5 resulted in a striking decrease in the level of LAPTM5 protein even at 15 min after the treatment with cycloheximide, indicating that ITCH promoted the protein degradation of LAPTM5. In contrast, ITCH C830A mutant could not promote the protein degradation of LAPTM5. Furthermore, the treatment with MG132 completely rescued the ITCH-mediated down-regulation of LAPTM5.
FIGURE 2.
ITCH negatively regulates LAPTM5 stability. A, down-regulation of LAPTM5 protein by ITCH. HEK293 cells were transiently co-transfected with Myc-tagged LAPTM5 alone, or Myc-tagged LAPTM5 and 3×FLAG-ITCH or 3×FLAG-ITCH C830A with different amounts of plasmid DNA (0.25∼2.0 μg). The whole cell lysates were subjected to SDS-PAGE and immunoblotted (W.B) with mouse anti-Myc tag, anti-FLAG tag, or anti-β-actin antibody. The graphs indicate the ratio for the intensity of each band (upper, Myc-tagged LAPTM5/β-actin; lower, 3×FLAG-ITCH or 3×FLAG-ITCH C830A/β-actin). E3s, 3×FLAG-ITCH or 3×FLAG-ITCH C830A. B, the effects of ITCH for LAPTM5 protein turnover. HEK293 cells were transiently co-transfected with Myc-tagged LAPTM5 and 3×FLAG-ITCH or 3×FLAG-ITCH C830A. After 24 h, cells were incubated with 100 μg/ml cycloheximide (CHX) with or without 20 μm MG132 and then lysed at the indicated times (15–90 min). The whole cell lysates were subjected to SDS-PAGE and immunoblotted with rabbit anti-Myc tag antibody or anti-β-actin antibody. CA, C830A. C, effect of the treatment with proteasomal and/or lysosomal inhibitors for the LAPTM5-ITCH interaction. HEK293 cells were transiently co-transfected with Myc-tagged LAPTM5 alone or Myc-tagged LAPTM5 and 3×FLAG-ITCH or 3×FLAG-ITCH C830A. On the next day, cells were treated with ALLN (10 μm) and/or bafilomycin A1 (50 nm, Baf.A1) for 6 h before being harvested. Then cell lysates were prepared in lysis buffer and immunoprecipitated with anti-FLAG antibody. The whole cell lysates or immunoprecipitates were subjected to SDS-PAGE and immunoblotted using the indicated antibodies. D, effect of siRNA-mediated ITCH knockdown or the treatment with proteasomal/lysosomal inhibitors for LAPTM5 stability. 8305C cells were infected with Ad-LacZ or Ad-LAPTM5 and after 24 h treated with MG132 (5 μm), ALLN (10 μm), bafilomycin A1 (50 nm), or NH4Cl (25 mm) for 24 h. 8305C cells were transfected with luciferase (Luc) siRNA or ITCH siRNA and after 4 h were infected with Ad-LacZ or Ad-LAPTM5 for 2 days. The whole cell lysates were subjected to SDS-PAGE and immunoblotted with anti-LAPTM5, goat anti-ITCH, or anti-β-actin antibody. E, effect of siRNA-mediated ITCH knockdown in KYSE410 cells. Left panel, cells were transfected with luciferase siRNA or ITCH siRNA, and after 2 days, the whole cell lysates were subjected to SDS-PAGE and immunoblotted with anti-LAPTM5, rabbit anti-ITCH, or anti-β-actin antibody. The graph indicates the relative increase of LAPTM5 protein level in siRNA-ITCH-transfected cells to the level in luciferase siRNA-transfected cells. Vertical lines indicate S.D. for four independent experiments. Right panel, cells were transfected with luciferase siRNA or ITCH siRNA. After 2 days, the cells were fixed, immunoreacted with rabbit anti-LAPTM5 antibody, and visualized using Alexa Fluor 488-conjugated anti-rabbit IgG (green). Bar, 10 μm.
In our previous study, we demonstrated that LAPTM5 protein was accumulated by the treatment with proteasomal or lysosomal inhibitors, suggesting that this protein is degraded through these systems (10). In consist with this notion, when 3×FLAG-tagged ITCH WT and Myc-tagged LAPTM5 expression vectors were transiently co-transfected with the treatment of ALLN (a proteasomal inhibitor) or bafilomycin A1 (a lysosomal inhibitor) in HEK293 cells, amount of expressed LAPTM5 protein was increased in compared with that in no treatment of those inhibitors, as well as that in the co-transfection with 3×FLAG-tagged ITCH C830A instead of ITCH WT (Fig. 2C). Additionally, we examined the effect of ITCH inhibition by specific siRNAs in 8305C cells (anaplastic thyroid cancer cell line), which was highly expressed by gene amplification (4). When adenoviral LAPTM5 (Ad-LAPTM5) was infected in siRNA-ITCH-transfected 8305C cells, the level of exogenously expressed LAPTM5 was remarkably increased, compared with that in luciferase siRNA-transfected 8305C cells, as well as by treatment with proteasomal inhibitors (MG132 and ALLN) or lysosomal inhibitors (bafilomycin A1 and NH4Cl) in Ad-LAPTM5-infected 8305C cells (Fig. 2D). Furthermore, we showed that the siRNA-mediated inhibition of ITCH leads to the stabilization of the LAPTM5 protein in KYSE410 cells, an esophageal squamous cell carcinoma cell line, which is highly expressed endogenous LAPTM5 (Fig. 2E). Taken together, these findings suggest that ITCH negatively regulates the stability of LAPTM5.
Co-localization between ITCH and LAPTM5 on Intracellular Vesicles
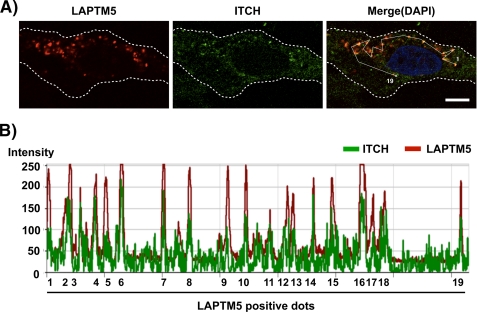
We then examined the intracellular distribution of ITCH and the exogenously expressed LAPTM5 in 8305C cells by immunofluorescence analysis. It has been reported that LAPTM5 was located on the intracellular transport vesicles such as endosomes (7–10, 15). When Ad-LAPTM5 was infected in 8305C cells, the intracellular distribution of exogenously expressed LAPTM5 exhibited as the speckled pattern (Fig. 3A). In this setting, endogenous ITCH was also frequently co-localized with its LAPTM5-positive dots (Fig. 3, A and B). These results strongly suggest that ITCH negatively regulates the level of LAPTM5 protein by directly interacting with LAPTM5 on the intracellular vesicles.
FIGURE 3.
Intracellular distribution of LAPTM5 and ITCH in 8305C cells. A, 8305C cells were plated on coverslips in 24-well plates (1 × 105/well) and were infected the next day with Ad-LacZ or Ad-LAPTM5. After 2 days, the cells were fixed, immunoreacted with anti-LAPTM5 antibody and goat anti-ITCH antibody, and visualized using Alexa Fluor 594-conjugated anti-rabbit (red) or FITC-conjugated anti-goat IgG antibody (green). The images were obtained by confocal microscopy. Bar, 10 μm. B, the fluorescence intensity distribution for LAPTM5 and ITCH on 19 spots that a white line passed is shown in the lower panel.
ITCH Promotes Ubiquitination of LAPTM5
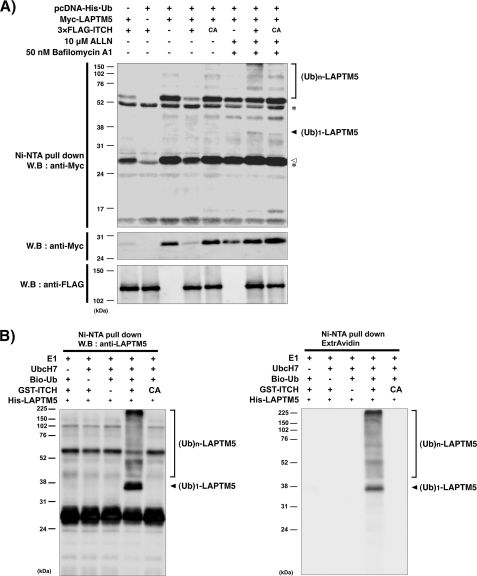
To examine whether ITCH ubiquitinates LAPTM5, we performed ubiquitination assays using transfected cells and in vitro. Myc-tagged LAPTM5, pcDNA-His-ubiquitin, and 3×FLAG-tagged ITCH or 3×FLAG-tagged ITCH C830A mutant expression vectors were co-expressed in HEK293 cells, and after 24 h, the cells were treated with ALLN and bafilomycin A1. Then, His-ubiquitin was purified by pulldown using Ni-NTA beads under the denaturing conditions, and its eluate was immunoblotted. As shown in Fig. 4A, the mono- and polyubiquitin signals were augmented by ITCH WT with these inhibitors, but not without inhibitors or by the inactive ITCH C830A mutant, suggesting that ITCH can ubiquitinate LAPTM5 protein. Next, we confirmed using in vitro ubiquitination assays whether LAPTM5 protein was ubiquitinated directly by ITCH. Consistent with the result of ubiquitination assay using HEK293 cells, we showed that both the mono- and polyubiquitination of LAPTM5 were directly catalyzed through its HECT domain of ITCH (Fig. 4B). These results indicate that ITCH acts as an ubiquitin ligase for LAPTM5.
FIGURE 4.
ITCH promotes ubiquitination of LAPTM5. A, ubiquitination assay for LAPTM5 in HEK293 cells. The cells were transfected with the indicated plasmids. After 24 h, cells were treated with ALLN (10 μm) and bafilomycin A1 (50 nm) for 6 h before being harvested. Ni-NTA pulldown was performed, and its eluate or whole cell lysates were subjected to SDS-PAGE and immunoblotted (W.B) using the indicated antibodies. A white arrowhead indicates Myc-LAPTM5 generated by the nonspecific binding with Ni-NTA. An asterisk indicates the nonspecific band. B, ubiquitination assay for LAPTM5 in vitro. The ubiquitination reaction was performed as described under “Experimental Procedures”. The ubiquitinated proteins in the reaction were subjected to SDS-PAGE and immunoblotted using ExtrAvidin (right panel) or anti-LAPTM5 (left panel) antibody. Ub, ubiquitin; CA, C830A.
Inhibition of ITCH Enhances LAPTM5-mediated Lysosomal Cell Death in Neuroblastoma Cells
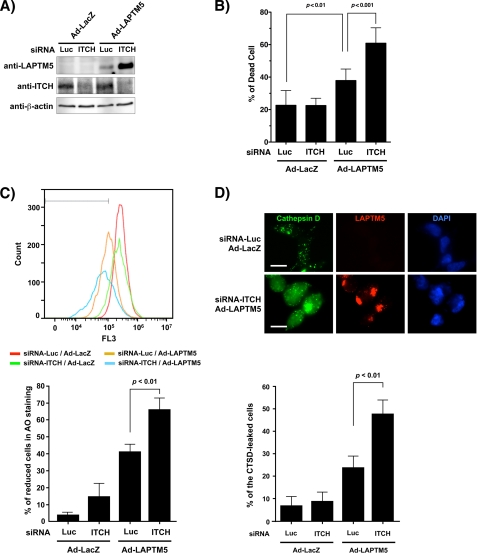
We demonstrated previously that the accumulation of LAPTM5 protein could induce the non-apoptotic cell death in the caspase-independent manner in GOTO cells, a neuroblastoma cell line (10). Furthermore, we have suggested that this LAPTM5-mediated cell death was the lysosomal cell death attributed by lysosomal destabilization with the leakage of cathepsin D from lysosomes into cytosols through lysosomal membrane permeabilization (10). Therefore, we examined the effect of ITCH inhibition for LAPTM5-mediated lysosomal cell death. When Ad-LAPTM5 was infected in luciferase siRNA-transfected GOTO cells, cell death was induced in compared with the infection by Ad-LacZ (p = 0.0065) (Fig. 5, A and B). In siRNA-ITCH-transfected GOTO cells, the expressed LAPTM5 protein was more remarkably accumulated than in luciferase siRNA-transfected GOTO cells, and the frequency of cell death was enhanced significantly (p = 0.0008) (Fig. 5, A and B). Because lysosomal cell death is triggered by the leakage of enzymes from the lysosome into the cytosol through lysosomal membrane permeabilization, we examined the diminution in AO uptake by FACS analysis as an indicator for lysosomal membrane permeabilization and the subcellular distribution of cathepsin D, a lysosomal enzyme (16, 17). Although overexpression of LAPTM5 induced the reduction of AO uptake in luciferase siRNA-transfected GOTO cells, its LAPTM5-induced AO reduction was significantly enhanced in siRNA-ITCH transfected GOTO cells (p = 0.0055 in Ad-LAPTM5-infected and siRNA-ITCH transfected GOTO cells compared with Ad-LAPTM5-infected and luciferase siRNA-transfected GOTO cells; Fig. 5C). As correlation with the reduction of AO uptake, the frequency of cells that cathepsin D was localized in cytoplasm by leaking from lysosomes was significantly enhanced in Ad-LAPTM5-infected and siRNA-ITCH transfected GOTO cells, compared with Ad-LAPTM5-infected and luciferase siRNA-transfected GOTO cells (p = 0.0083; Fig. 5D). These results suggest that ITCH can prevent LAPTM5-mediated lysosomal cell death in GOTO cells, and thus, the expression of ITCH may contribute to the tumorigenesis of neuroblastomas.
FIGURE 5.
Inhibition of ITCH enhances LAPTM5-mediated cell death in GOTO cells. A, GOTO cells were plated in six-well plates (2 × 105/well) and were transfected the next day with luciferase (Luc) siRNA or ITCH siRNA, and infected with Ad-LacZ or Ad-LAPTM5 after 4 h for 4 days. The whole cell lysates were subjected to SDS-PAGE and immunoblotted (W.B) with anti-LAPTM5, rabbit anti-ITCH, or anti-β-actin antibody. B, enhancement by inhibition of ITCH for the frequency of dead cells. GOTO cells were treated as indicated in A. After 4 days, dead cells were counted using the trypan blue exclusion method, and the values were indicated as the percentages. Vertical lines; S.D. for six experiments. C, AO uptake analysis. GOTO cells (2 × 105/well) were plated in six-well plates and were treated the next day as indicated in A. After 4 days, the cells were stained with AO (5 mg/ml, 30 min). The detached cells were removed, and the attached cells were collected by trypsinization and washed twice with PBS. Upper panel, the intensity of staining was measured by FACS using a channel of FL3; the range containing >99.9% of the cells without AO fluorescence was gated. Lower panel, the percentage of cells with a reduction in AO fluorescence. Vertical lines, S.D. for three independent experiments. D, representative images for cathepsin D (CTSD) staining. GOTO cells (5 × 104/well) were plated on coverslips in 24-well plates and the next day were treated as indicated in A. After 4 days, the cells were fixed in 10% TCA, reacted with an anti-cathepsin D and anti-LAPTM5 antibody, and visualized with an Alexa Fluor 488-conjugated mouse IgG and Alexa Fluor 594-conjugated rabbit IgG antibody, respectively. Upper panel, the representative images of cathepsin D staining. Bar, 10 μm. Lower panel, the percentage of cells that cathepsin D is stained in cytoplasm by leaking from lysosomes was measured. Vertical lines, S.D. for three independent experiments.
DISCUSSION
LAPTM5 is a membrane protein having five transmembrane domains and located on endosomes, lysosomes, or the intracellular vesicles transported from the Golgi to lysosomes (8, 9, 15). A key finding in this study is that the ITCH E3 ligase catalyzes the ubiquitination of LAPTM5 by directly interacting with PPPY of LAPTM5 via its WW domain and negatively regulates the level of LAPTM5 protein. We have demonstrated previously that LAPTM5 protein is degraded in the proteasomal and lysosomal pathways (10). It has been demonstrated that LAPTM5 has an essential role for the lysosomal degradation of T cell and B cell receptors through the transport of endosomes to lysosomes (8, 9). In addition, NEDD4-1, a HECT-type E3 ligase, which belongs to the Nedd4 family along with ITCH, could also interact with the PPPY motif of LAPTM5 via its WW domain, and the interaction of these proteins is critical for the transport of intracellular vesicles from the Golgi to lysosome (7). Thus, LAPTM5 protein is degraded by not only the proteasomal system but also the lysosomal system through transport into lysosome of LAPTM5-positive vesicles. However, it has been known that ITCH promotes not only the proteasomal degradation but also the lysosomal degradation of membrane proteins on the endosome, such as Melan-A, ErbB4, and CXCR4, through ubiquitination of those substrates (18–20). In general, the receptors for growth factors and chemokines are internalized by endocytosis in the response to stimuli, and the generated endosomes are transported into the lysosomes for removal or into the cellular membrane for recycling (21). Taken together, ITCH may associate with not only the proteasomal degradation of LAPTM5 protein but also the lysosomal removal of LAPTM5-positive vesicles by ubiquitinated LAPTM5 protein. Although we found that ITCH catalyzes the poly- and monoubiquitination of LAPTM5 in this study, it is unclear for its physiological significance. This differential status for ubiquitination (mono or poly) may contribute to the mark for proteasomal or lysosomal degradation. Indeed, it has been suggested that monoubiquitination by ITCH promotes the lysosomal degradation of the substrates such as ErbB4 or CXCR4 (19, 20).
NB is a malignant tumor occurring within the abdomen (the adrenal medulla affected in ∼50% of NB patients) in children (22–24). The clinical behavior of NB is heterogeneous: the tumors found in patients under 1 year of age are favorable due to spontaneous regression via programmed cell death; the tumors found in older children often grow rapidly to become fatal, despite intensive chemotherapy (22–24). Thus, to develop a breakthrough in therapy against advanced tumors, which cannot regress spontaneously, it is important to understand the molecular mechanisms for the spontaneous regression of NBs. Recently, we found that the accumulation of LAPTM5-positive vesicles often occurred in dying NB cells within regressing area on the tissue section from favorable NB tumors and that the exogenous expression of LAPTM5 led to the gradual accumulation of LAPTM5-positive vesicles and induced cell death in NB cells (10). Importantly, we have observed that those LAPTM5-positive vesicles accumulated remarkably by the treatment with a proteasomal or lysosomal inhibitors, strongly suggesting that the dysregulation of these degradation systems might be associated with the induction of cell death occurred by the accumulation of LAPTM5-positive vesicles in NB cells. In this study, we demonstrated that the inhibition of ITCH led to the accumulation of LAPTM5 protein and enhanced LAPTM5-mediated cell death in NB cells. Thus, inactivation of ITCH may be a trigger for spontaneous regression of NB tumor by inducing LAPTM5-mediated cell death in NB cells. Further examination will be required to clarify the physiological association with ITCH and LAPTM5 for tumorigenesis of NB tumors.
We showed the stabilization of LAPTM5 protein by ITCH inhibition in KYSE410 cells, an esophageal squamous cell carcinoma cell line, which is expressed endogenous LAPTM5 at the transcriptional level, but we could not detect the cell death in this case. This result suggests that ITCH contributes to the degradation of LAPTM5 protein at the physiological condition, and it may be needed the chronic inhibition of ITCH to induce the cell death occurred by accumulation of endogenous LAPTM5. Furthermore, we could not detect the increase in the level of endogenous LAPTM5 protein by ITCH inhibition in 8305C cells showing amplification and overexpression of ITCH (4) due to the low level of LAPTM5 mRNA in this cells (data not shown). Thus, the function of ITCH as a negative regulator of LAPTM5 protein might be exerted under specific conditions where the transcription of LAPTM5 is activated. Indeed, our preliminary data have showed that LAPTM5 was transcriptionally up-regulated by stimuli, including cell differentiation and cellular stress. Thus, to further characterize LAPTM5-mediated cell death, it is important to determine the cellular condition for the accumulation of LAPTM5 protein.
Supplementary Material
Acknowledgments
We thank Ayako Takahashi and Rumi Mori for technical assistance.
This work is supported by a grant-in-aid for Scientific Research on Priority Areas and Innovative Areas, Young Scientists (B), Scientific Research (A) and (C), and a Global Center of Excellence Program from the Japanese Ministry of Education Core Research for Evolutional Science and Technology of Japan Science and Technology Corporation, and New Energy and Industrial Technology Development Organization.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- NB
- neuroblastoma
- Ni-NTA
- nickel-nitrilotriacetic acid
- AO
- acridine orange
- ITCH
- itchy E3 ubiquitin protein ligase homolog (mouse)
- HECT
- Homologous to the E6-AP carboxyl terminus
- LATS
- large tumor suppressor.
REFERENCES
- 1. Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2. Perry W. L., Hustad C. M., Swing D. A., O'Sullivan T. N., Jenkins N. A., Copeland N. G. (1998) Nat. Genet. 18, 143–146 [DOI] [PubMed] [Google Scholar]
- 3. Melino G., Gallagher E., Aqeilan R. I., Knight R., Peschiaroli A., Rossi M., Scialpi F., Malatesta M., Zocchi L., Browne G., Ciechanover A., Bernassola F. (2008) Cell Death Differ. 15, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 4. Ishihara T., Tsuda H., Hotta A., Kozaki K., Yoshida A., Noh J. Y., Ito K., Imoto I., Inazawa J. (2008) Cancer Sci. 99, 1940–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang P., Wang C., Gao K., Wang D., Mao J., An J., Xu C., Wu D., Yu H., Liu J. O., Yu L. (2010) J. Biol. Chem. 285, 8869–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Salah Z., Melino G., Aqeilan R. I. (2011) Cancer Res. 71, 2010–2020 [DOI] [PubMed] [Google Scholar]
- 7. Pak Y., Glowacka W. K., Bruce M. C., Pham N., Rotin D. (2006) J. Cell Biol. 175, 631–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ouchida R., Kurosaki T., Wang J. Y. (2010) J. Immunol. 185, 294–301 [DOI] [PubMed] [Google Scholar]
- 9. Ouchida R., Yamasaki S., Hikida M., Masuda K., Kawamura K., Wada A., Mochizuki S., Tagawa M., Sakamoto A., Hatano M., Tokuhisa T., Koseki H., Saito T., Kurosaki T., Wang J. Y. (2008) Immunity 29, 33–43 [DOI] [PubMed] [Google Scholar]
- 10. Inoue J., Misawa A., Tanaka Y., Ichinose S., Sugino Y., Hosoi H., Sugimoto T., Imoto I., Inazawa J. (2009) PLoS One 4, e7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimada Y., Imamura M., Wagata T., Yamaguchi N., Tobe T. (1992) Cancer 69, 277–284 [DOI] [PubMed] [Google Scholar]
- 12. Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S., Tsukita S. (1998) J. Cell Biol. 140, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishida T., Terashima M., Fukami K., Yamada Y. (2007) Biochem. J. 405, 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qiu L., Joazeiro C., Fang N., Wang H. Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y. C. (2000) J. Biol. Chem. 275, 35734–35737 [DOI] [PubMed] [Google Scholar]
- 15. Adra C. N., Zhu S., Ko J. L., Guillemot J. C., Cuervo A. M., Kobayashi H., Horiuchi T., Lelias J. M., Rowley J. D., Lim B. (1996) Genomics 35, 328–337 [DOI] [PubMed] [Google Scholar]
- 16. Boya P., Andreau K., Poncet D., Zamzami N., Perfettini J. L., Metivier D., Ojcius D. M., Jäättelä M., Kroemer G. (2003) J. Exp. Med. 197, 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boya P., Kroemer G. (2008) Oncogene 27, 6434–6451 [DOI] [PubMed] [Google Scholar]
- 18. Lévy F., Muehlethaler K., Salvi S., Peitrequin A. L., Lindholm C. K., Cerottini J. C., Rimoldi D. (2005) Mol. Biol. Cell 16, 1777–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sundvall M., Korhonen A., Paatero I., Gaudio E., Melino G., Croce C. M., Aqeilan R. I., Elenius K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. (2003) Dev. Cell 5, 709–722 [DOI] [PubMed] [Google Scholar]
- 21. David R. (2011) Nat. Rev. Mol. Cell Biol. 12, 135. [DOI] [PubMed] [Google Scholar]
- 22. Brodeur G. M. (2003) Nat. Rev. Cancer 3, 203–216 [DOI] [PubMed] [Google Scholar]
- 23. Schwab M., Westermann F., Hero B., Berthold F. (2003) Lancet Oncol. 4, 472–480 [DOI] [PubMed] [Google Scholar]
- 24. Maris J. M., Hogarty M. D., Bagatell R., Cohn S. L. (2007) Lancet 369, 2106–2120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.