Background: C1 is the main physiological cleavage fragment of PrP, but its role in disease is unknown.
Results: C1 is not toxic when expressed in mice and delays the onset of disease and PrPSc formation when co-expressed with WT PrP.
Conclusion: C1 is a dominant-negative inhibitor of PrPSc formation.
Significance: Modulation of C1 cleavage may represent a therapeutic strategy for combating PrPSc infection.
Keywords: Cell Biology, Infectious Diseases, Neurodegenerative Diseases, Prions, Transgenic Mice, Cleavage, Dominant-negative, Proteolytic
Abstract
The cellular prion protein (PrPC) undergoes constitutive proteolytic cleavage between residues 111/112 to yield a soluble N-terminal fragment (N1) and a membrane-anchored C-terminal fragment (C1). The C1 fragment represents the major proteolytic fragment of PrPC in brain and several cell types. To explore the role of C1 in prion disease, we generated Tg(C1) transgenic mice expressing this fragment (PrP(Δ23–111)) in the presence and absence of endogenous PrP. In contrast to several other N-terminally deleted forms of PrP, the C1 fragment does not cause a spontaneous neurological disease in the absence of endogenous PrP. Tg(C1) mice inoculated with scrapie prions remain healthy and do not accumulate protease-resistant PrP, demonstrating that C1 is not a substrate for conversion to PrPSc (the disease-associated isoform). Interestingly, Tg(C1) mice co-expressing C1 along with wild-type PrP (either endogenous or encoded by a second transgene) become ill after scrapie inoculation, but with a dramatically delayed time course compared with mice lacking C1. In addition, accumulation of PrPSc was markedly slowed in these animals. Similar effects were produced by a shorter C-terminal fragment of PrP(Δ23–134). These results demonstrate that C1 acts as dominant-negative inhibitor of PrPSc formation and accumulation of neurotoxic forms of PrP. Thus, C1, a naturally occurring fragment of PrPC, might play a modulatory role during the course of prion diseases. In addition, enhancing production of C1, or exogenously administering this fragment, represents a potential therapeutic strategy for the treatment of prion diseases.
Introduction
Transmissible spongiform encephalopathies such as Creutzfeldt-Jakob disease and bovine spongiform encephalopathy are fatal neurodegenerative disorders whose pathology is associated with propagation of prions, novel infectious agents whose transmission is based on changes in protein conformation rather than inheritance of nucleic acid sequence (1). Prion propagation depends on conversion of an endogenous cellular glycoprotein (PrPC)2 into an aggregated, protease-resistant isoform (PrPSc) that is rich in β-sheet structure (1–4). PrPC is synthesized on endoplasmic reticulum-attached ribosomes and transits the secretory pathway to the cell surface, where most molecules are attached to the outer leaflet of the lipid bilayer via a C-terminal glycosylphosphatidylinositol (GPI) anchor (5). Most of the protein resides in lipid rafts on the plasma membrane, although some molecules are constitutively endocytosed via clathrin-coated pits and are then recycled back to the cell surface (6–9).
After its synthesis, PrPC is known to undergo proteolytic processing in at least three sites. One cleavage (sometimes referred to as the α-cleavage) occurs between residues 111/112 to yield a soluble N-terminal fragment called N1 and a GPI-anchored, C-terminal fragment called C1 (10–13). The N1/C1 cleavage occurs constitutively in 10–50% of the molecules, but it can be stimulated by activators of protein kinase C (10, 13–15). There is disagreement about the cellular site and proteases responsible for the α-cleavage, with endosomal/lysosomal compartments, late compartments of the secretory pathway, and the cell surface (mediated by a disintegrin and metalloproteases (ADAMs)) having all been suggested (6, 15–18). A second cleavage occurs between residues 89/90, generating a soluble N2 fragment and a GPI-anchored C2 fragment (10). This so-called β-cleavage occurs at low levels under normal conditions, possibly catalyzed by reactive oxygen species acting on cell-surface PrP, but it is enhanced during generation of PrPSc (10, 19–21). A third cleavage, catalyzed by members of the ADAM protease family, occurs near the site of GPI anchor attachment (residue 230), shedding most of the polypeptide chain into the extracellular medium (11, 22, 23). Additional proteolytic cleavages may also occur at low levels (24). The proteolytic fragments generated by these different cleavage reactions may have a role in the physiological functions of PrPC, such as protection against oxidative stress (20), although this remains unclear in part because of uncertainty about the normal biological role of PrPC.
C1 is quantitatively the major proteolytic fragment of PrP present in brain and many cells types (10–12, 20, 25). In this study, we sought to investigate the role of C1 by the creation of transgenic mice that express this fragment in the presence and absence of endogenous PrP. We demonstrate that although C1 itself is not inherently toxic nor convertible to PrPSc after inoculation of Tg(C1) animals, it acts as a potent dominant-negative inhibitor, significantly delaying scrapie illness and decreasing PrPSc production from wild-type PrP. These results indicate that the C1 cleavage product acts as a physiologically generated inhibitor of prion propagation, and therefore increasing production of this fragment represents a potential therapeutic strategy for treatment of prion diseases.
EXPERIMENTAL PROCEDURES
Construction of Transgenic Mice
A cDNA encoding murine C1(Δ23–111) was generated by PCR amplification. The following primers were used: 5′ (5′-TCCGAAAGCTTCTCGAGGCCGCCACCATGGCGAACCTTGGCTACTGGCTGCTGGCCCTCTTTGTGACTATGTGGACTGATGTCGGCCTCTGCAGGCCCATGATCCATTTTGGC-3′) and 3′ (5′-CGGACTCTAGACTCGAGTCATCATCCCACGATCAGGAAGAT-3′).Cloning of a cDNA encoding PrP(Δ23–134) has been described elsewhere (30). The 5′ primers contain HindIII and XhoI restriction sites along with a Kozak consensus sequence. The 3′ primer incorporated XhoI and XbaI sites for the initial cloning into the pcDNA 3.1(+) Hygro plasmid and subsequent insertion into the transgenic vector. The resulting PCR product was digested with HindIII and XbaI and cloned into pcDNA 3.1(+) Hygro (Invitrogen).
To create the transgenic mouse vector, a fragment encoding the C1 sequence was released from the pcDNA 3.1(+) Hygro/C1 plasmid by digestion with XhoI and ligated into the XhoI site of MoPrP.Xho (26). Colony PCR was performed to select clones containing the insert in the correct orientation, using the following primers: P1 (5′-AACCGAGCTGAAGCATTCTGCC-3′) and P4 (5′-CACGAGAAATGCGAAGGAACAAGC-3′) (27). The transgene was released from the recombinant plasmid by NotI restriction digestion, purified on GFX PCR DNA columns (GE Healthcare), and injected into the pro-nuclei of fertilized eggs from mice on the C57BL6×CBA background. Transgenic founders were bred initially to Tga20+/+ mice on a C57BL6/CBA/129 background (obtained from the European Mouse Mutant Archive (EMMA)) and were then back-crossed to Prn-p0/0 mice on a pure C57BL6 background (from EMMA).
Genotyping of transgenic mice was performed by PCR analysis of tail DNA prepared using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN). Genotyping was performed using primers P1 and P4 (27). These primers amplify both the C1 and Tga20 transgenes, which can be distinguished from each other by size. P2 (5′-CTTCAGCCTAAATACTGGGCAC-3′) and P4 (5′-CACGAGAAATGCGAAGGAACAAGC-3′) primers (27) were used to amplify the Prn-p allele. All Tg(C1) mice used in this study were heterozygous for the C1 transgene (i.e. Tg(C1+/−)).
Histology
Animals were perfused transcardially with 4% paraformaldehyde, after which brains were removed and post-fixed in the same solution. Paraffin sections of brain were stained with hematoxylin and eosin or GFAP as described previously (28).
Biochemical Procedures
10% w/v brain homogenates were generated by mechanical dissociation of single hemispheres using plastic pestles (South Jersey Precision Tool and Mold Inc., Vineland, NJ) in phosphate-buffered saline (PBS) containing a protease inhibitor mixture (Roche Applied Science). For deglycosylation, 20 μg of protein was treated with PNGase F for 3 h at 37 °C. Western blots were performed using anti-PrP antibody 6H4 or D18 (kind gift of Dennis Burton (Scripps Institute, La Jolla, CA)).
For proteinase K treatment, 10% w/v brain homogenates prepared as above were centrifuged for 5 min at 2,300 × g. The supernatant was diluted 1:10 in lysis buffer (10 mm Tris-HCl, pH 7.4, 0.5% Triton X-100, 0.5% sodium deoxycholate) and incubated with 20 μg/ml of proteinase K (PK) at 37 °C for 1 h prior to Western blotting with anti-PrP antibody 6H4.
RESULTS
Creation of Transgenic Mice Expressing the C1 Fragment of PrP
To investigate the properties of the C-terminal cleavage product, C1, in the absence of full-length PrP or the N1 fragment, we generated transgenic mice (designated Tg(C1)) that express a PrP molecule deleted for residues 23–111 (Fig. 1A) under control of a modified Prn-p promoter (26). After cleavage of the N-terminal signal peptide (residues 1–22) and attachment of the C-terminal GPI anchor (at residue 230) during biosynthesis, the transgenically encoded protein was predicted to correspond to the C1 fragment that is endogenously generated from full-length PrP by proteolytic cleavage.
FIGURE 1.
Schematic illustration of PrP constructs, and analysis of PrP expression in transgenic mice. A, schematic of wild-type (WT), C1 (Δ23–111), and PrP(Δ23–134). Structural domains of PrP are indicated by the colored blocks: SS, signal sequence (blue); PBD, polybasic domain (yellow); OR, octapeptide repeats (orange); CC, charged cluster (red); HD, hydrophobic domain (green); GPI, GPI attachment signal (purple). The dotted lines indicate deleted regions. B, Western blot analysis of protein expression. Brain samples from mice of the indicated genotypes were normalized for total protein, treated with or without PNGase F to remove N-linked oligosaccharides (+ and − lanes, respectively), and subjected to Western blotting with anti-PrP antibody 6H4. Filled and open arrowheads to the left of lane 2 indicate the positions of cleavage products C1 and C2, respectively. The upper band in lane 8 (asterisk) represents residual, mono-glycosylated C1 that was not completely shifted by treatment with PNGase. Molecular size markers are given in kDa.
Founders were bred onto the Prn-p0/0 genetic background to obtain Tg(C1)/Prn-p0/0 mice that expressed the C1 fragment in the absence of any WT PrP. Of five transgene-positive founders obtained, three transmitted the transgene to their progeny. Western blot analysis of brain homogenates revealed detectable PrP in only one of the lines (Fig. 1B). Quantification with the Odyssey infrared imaging system revealed that this line expressed the C1 protein at a level ∼7 times that of WT PrP in Prn-p+/+ mice and intermediate between the levels of WT PrP in Tga20+/+ and Tga+/0 mice (data not shown, see Fig. 1B). The transgenically encoded protein exhibited three different glycoforms, analogous to WT PrP (Fig. 1B, lane 7). When samples were treated with PNGase to remove N-linked oligosaccharides, the protein migrated at the same position as the C1 fragment generated endogenously in the brains of Prn-p+/+ and Tga20 mice (Fig. 1B, filled arrowhead).
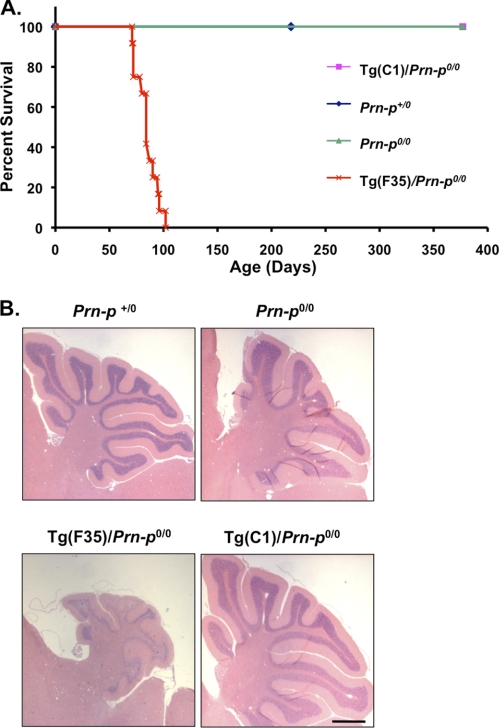
We observed Tg(C1)/Prn-p0/0 mice for signs of spontaneous illness. We noted no clinical symptoms or increased mortality for up to 1 year (Fig. 2A, pink line/squares). In contrast, Tg(F35)/Prn-p0/0 mice expressing PrP deleted for residues 32–134 at even lower levels (∼2 times) (29) showed neurological symptoms, including ataxia, kyphosis, hyper-activity, hind limb paralysis, and tail clasp by 30.21 ± 2.6 days, and succumbed by 100 days (Fig. 2A, red line/crosses). The brains of Tg(C1)/Prn-p0/0 animals showed no histological abnormalities at 1 year, in contrast to dramatic cerebellar degeneration in Tg(F35) mice (Fig. 2B). Tg(C1) mice on the Prn-p+/0 and Prn-p+/+ genetic backgrounds were also clinically and neurohistologically normal (data not shown).
FIGURE 2.
Tg(C1) mice on the Prn-p0/0 background do not develop spontaneous neurological illness. A, survival was monitored in mice of the following genotypes, with the number of animals indicated in parentheses: Tg(C1)/Prn-p0/0 (10), Prn-p+/0 (8), Prn-p0/0 (7), and Tg(F35)/Prn-p0/0 (12). B, cerebellar sections from 365 day-old mice of the indicated genotypes were stained with hematoxylin and eosin. Scale bar (applicable to all panels) is 1 mm.
Tg(C1) Mice Are Resistant to Infection with Scrapie and Cannot Propagate PrPSc
Tg(C1)/Prn-p0/0 mice were inoculated intracerebrally with the RML strain of scrapie and observed for clinical symptoms. Animals remained healthy for at least 1 year after inoculation and showed no increase in mortality, similar to Prn-p0/0 lacking the C1 transgene (Fig. 3A, green line/squares and blue line/diamonds). In contrast, scrapie-inoculated Prn-p+/+ mice became terminally ill at 171.8 ± 16.2 days (Fig. 3A, red line/crosses). The brains of inoculated Tg(C1)/Prn-p0/0 mice did not display any histological abnormalities based on hematoxylin/eosin or anti-GFAP staining, similar to inoculated Prn-p0/0 mice (Fig. 3B). We did not detect protease-resistant PrP in brain homogenates prepared from inoculated Tg(C1)/Prn-p0/0 mice (Fig. 3C, lane 4), although homogenates from infected Prn-p+/+ animals displayed the characteristic PrP(27–30) fragment (Fig. 3C, lane 2). As expected, no protease-resistant PrP was detected in the brains of inoculated Prn-p0/0 mice (data not shown).
FIGURE 3.
Scrapie-inoculated Tg(C1)/Prn-p0/0 mice do not develop clinical illness or histopathology, and do not accumulate protease-resistant PrP. A, survival was monitored in mice of the following genotypes after RML inoculation, with the number of animals indicated in parentheses: Prn-p0/0 (9), Tg(C1)/Prn-p0/0 (8), Prn-p+/+ (8). B, sections from the cerebellum (panels 1 and 2) or hippocampus (panels 3 and 4) from 365 day-old mice of the indicated genotypes were stained with hematoxylin and eosin or anti-GFAP antibody, respectively. Insets show hippocampal sections stained with DAPI to reveal cell nuclei. Scale bar in panel 4 (applicable to all panels) is 1 mm. C, Western blotting for protease-resistant PrP. Brain homogenates containing equivalent amounts of protein from mice of the indicated genotypes were treated with or without 20 μg/ml proteinase K for 1 h at 37 °C (+ and − lanes, respectively), and were subjected to Western blotting with anti-PrP antibody 6H4.
C1 Inhibits Disease Progression and PrPSc Accumulation in Mice Co-expressing WT PrP
The observation that Tg(C1)/Prn-p0/0 mice are resistant to scrapie infection and cannot produce PrPSc led us to wonder whether C1 might act as a dominant-negative inhibitor of prion propagation in mice co-expressing WT PrP encoded either by the endogenous Prn-p gene or the Tga20 transgene. To test this hypothesis, we intracerebrally inoculated Tg(C1)/Prn-p+/+ and Tg(C1)/Tga20+/0 mice with RML scrapie. We observed that survival time was significantly longer in these mice than in the corresponding Prn-p+/+ and Tga20+/0 mice that lacked the C1 transgene (Fig. 4A). The mean survival time in Tg(C1)/Prn-p+/+ mice was 229.3 ± 18.5 days compared with 171.8 ± 16.2 days in Prn-p+/+ mice and 112.1 ± 10.9 days in Tg(C1)/Tga20+/0 mice compared with 76.9 ± 6.2 in Tga20+/0 mice. Importantly, we found that co-expression of mutant PrP with wild type did not impact expression levels of either protein and did not change WT PrP cleavage (Fig. 4B).
FIGURE 4.
C1 prolongs survival time in mice expressing WT PrP. A, survival was monitored in mice of the following genotypes after scrapie inoculation, with the number of animals indicated in parentheses: Prn-p+/+ (8), Tg(C1)/Prn-p+/+ (8), Tga20+/0 (16), and Tg(C1)/Tga20+/0 (8). Significant differences were found between the following groups as determined by an unpaired student's t test: Tg(C1)/Prn-P+/+ versus Prn-P+/+ (p < 0.0001); Tg(C1)/Tga20+/0 versus Tga20+/0 (p < 0.0001). B, brain homogenates containing equal amounts of total protein from mice of the indicated genotypes were treated with or without PNGase F to remove N-linked oligosaccharides (+ and − lanes, respectively), and subjected to Western blotting with anti-PrP antibody D18. Filled and open arrowheads indicate the positions of cleavage products C1 and C2, respectively. Molecular size markers are given in kDa.
We also analyzed the effect of the C1 transgene on scrapie-induced pathology, in particular the extent of spongiform change and astrogliosis. Although survival times were longer in Tg(C1)/Prn-p+/+ and Tg(C1)/Tga20+/0 mice, at the terminal stage of disease the degree of spongiform change in the cerebellum, hippocampus, and brainstem in these animals was comparable with that seen in Prn-p+/+ and Tga20+/0 mice, respectively (Fig. 5, A–H, and data not shown). GFAP staining was also similar in terminally ill Tg(C1)/Prn-p+/+ and Prn-p+/+ mice (Fig. 5, I and J). Interestingly, however, GFAP staining was much more intense in Tg(C1)/Tga20+/0 mice than in Tga20+/0 mice at the terminal stage (Fig. 5, K and L). Uninfected animals of all genotypes lacked detectable GFAP staining (data not shown).
FIGURE 5.
Effect of C1 on scrapie-induced pathology in terminally ill mice expressing WT PrP. Mice of the indicated genotypes were taken for histological analyses at the terminal stage of illness, with the age at sacrifice given in parentheses: Tg(C1)/Prn-p+/+ (233d), Prn-p+/+ (158d), Tg(C1)/Tga20+/0 (120d), Tga20+/0 (65d). Sections from the cerebellum were stained with hematoxylin and eosin (A–H) and sections from the hippocampus with anti-GFAP antibody (I–L). Areas within the cerebellar white matter outlined by the boxes in A–D are shown at higher magnification in E–H. Scale bars, 1 mm (A–D and I–L) and 50 μm (E–H).
To determine the effect of C1 expression on accumulation of PrPSc, brain homogenates were treated with PK, and the amount of protease-resistant PrP was analyzed by Western blotting. Brains were collected from Tg(C1)/Prn-p+/+ mice at two different time points as follows: during the pre-symptomatic stage (180 days post-inoculation (p.i.)) and at the terminal stage (250 days p.i.). At the earlier time point, there was substantially less PK-resistant PrP in Tg(C1)/Prn-p+/+ mice than in terminally ill Prn-p+/+ mice at 152 days p.i. (Fig. 6A). However, by the time Tg(C1)/Prn-p+/+ mice reached the terminal phase, PrPSc had accumulated to a level comparable with that seen in terminally ill Prn-p+/+ mice (Fig. 6B). This result indicates that expression of C1 slows, but does not prevent, accumulation of PrPSc, which eventually reaches levels seen in mice lacking the C1 transgene.
FIGURE 6.
C1 inhibits accumulation of protease-resistant PrP in animals expressing WT PrP. Uninoculated or scrapie-inoculated mice (− and + lanes, respectively) of the indicated genotypes were sacrificed at the times shown below each lane. Brain homogenates containing equivalent amounts of protein were treated with (lower blots) or without (upper blots) 20 μg/ml proteinase K for 1 h at 37 °C, and were subjected to Western blotting with anti-PrP antibody 6H4. A shows samples from presymptomatic animals, and B and C show samples from terminally ill mice. Samples from two different mice at 180d, 80d, and 120d are shown. Actin is shown as a loading control in all panels.
Interestingly, the presence of C1 had an even more profound effect on accumulation of PrPSc in Tga20+/0 mice. Even at the terminal stage, Tg(C1)/Tga20+/0 mice (120 days p.i.) contained significantly less PrPSc in their brains than terminally ill Tga20+/0 mice (80 days p.i.) (Fig. 6C). Despite this dramatic reduction in the amount of PrPSc, the brains of terminally ill Tg(C1)/Tga20+/0 mice still contained infectious scrapie prions, as demonstrated by the ability of brain samples to transmit disease to Tga20+/+ indicator mice. Incubation times after inoculation of Tga20+/+ mice with 1% brain homogenates derived from Tg(C1)/Tga20+/0 mice were indistinguishable from those for homogenates derived from Tga20+/0 mice lacking the C1 transgene (62. 3 ± 5.3 versus 61.6 ± 6.7 days, respectively, p > 0.7, by Student's t test). Moreover, the recipient mice accumulated similar amounts of protease-resistant PrP in their brains after inoculation with both sets of samples (data not shown). This result implies that, although Tg(C1)/Tga20+/0 mice accumulate greatly reduced levels of PrPSc in their brains, these animals accumulate infectious prions with the ability to propagate in Tga20+/+ host mice expressing WT PrP.
A Shorter C-terminal Fragment of PrP Also Acts as Dominant-negative Inhibitor
We created Tg(Δ23–134) mice that synthesize a form of PrP that, after biosynthetic processing, corresponds to residues 135–230. This fragment lacks a hydrophobic domain (residues 112–134) that is present at the N terminus of the C1 fragment (Δ23–111) (Fig. 1A). Tg(Δ23–134) mice remain healthy and, unlike Tg(F35) mice expressing PrP(Δ32–134), do not develop spontaneous neurological illness (30).
To see if the shorter C-terminal fragment produced in Tg(Δ23–134) mice was capable of sustaining prion propagation, we inoculated two lines, L and H, that express the truncated protein at 0.2 and 1× levels, respectively, with RML scrapie prions. Like Tg(C1) mice, Tg(Δ23–134)/Prn-p0/0 mice did not exhibit any clinical illness for >1 year after inoculation (Fig. 7A, pink line/squares and green line/triangles). Moreover, the brains of inoculated animals did not display any abnormalities by hematoxylin/eosin staining (Fig. 7B) or GFAP histochemistry (data not shown) and did not contain any PK-resistant PrP by Western blotting (data not shown).
FIGURE 7.
Scrapie-inoculated Tg(Δ23–134)/Prn-p0/0 mice do not develop clinical Illness or histopathology. A, survival was monitored in mice of the following genotypes after scrapie inoculation, with the number of animals indicated in parentheses: Prn-p0/0 (9), Tg(PrPΔ23–134L)/Prn-p0/0 (7), Tg(PrPΔ23–134H)/Prn-p0/0 (9), Prn-p+/+ (8). Data from RML-inoculated Prn-p0/0 and Prn-p+/+ mice shown in Fig. 4 are reproduced here to allow for direct comparison. B, mice were taken for histological analysis at 1 year of age, at which point they remained healthy. Cerebellar sections were stained with hematoxylin and eosin. Scale bar (applicable to all panels) is 1 mm.
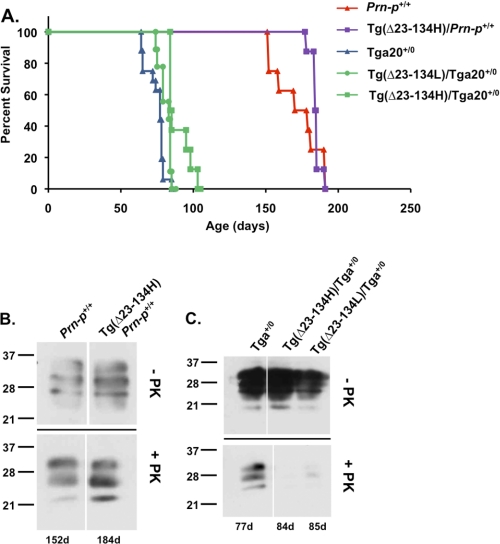
To determine whether PrP(Δ23–134) has an inhibitory effect on scrapie propagation similar to that of C1, we inoculated Tg(Δ23–134)/Prn-p+/+ and Tg(Δ23–134)/Tga20+/0 mice with RML prions. In both kinds of mice, the time to terminal disease was increased by the presence of PrP(Δ23–134), compared with the corresponding mice lacking the truncated protein (Fig. 8A). The survival time in Tg(Δ23–134H)/Prn-p+/+ mice was 184.5 ± 3.5 days compared with 171.8 ± 16.2 days in Prn-p+/+ mice and 93.9 ± 8.1 days in Tg(Δ23–134H)/Tga20+/0 mice or 83 ± 3.8 days in Tg(Δ23–134L)/Tga20+/0 mice compared with 76.9 ± 6.2 in Tga20+/0 mice. Expression of Δ23–134 in the H (1×) line led to a statistically significant increase in survival time in both Prn-p+/+ and Tga20+/0 mice, although this prolongation was not as dramatic as for C1, possibly because the latter was expressed at higher levels (7×). The effect of PrP(Δ23–134) was dose-dependent, with a more marked prolongation of survival for the H line than for the L line in Tga20+/0 mice.
FIGURE 8.
Expression of PrP(Δ23–134) prolongs survival and reduces protease-resistant PrP. A, survival was monitored in mice of the following genotypes after scrapie inoculation, with the number of animals indicated in parentheses: Prn-p+/+ (8), Tg(Δ23–134H)/Prn-p+/+ (8), Tga20+/0 (16), Tg(Δ23–134L)/Tga20+/0 (9), and Tg(Δ23–134H)/Tga20+/0 (8). Significant differences were found between the following groups: Tg(Δ23–134H)/Prn-p+/+ versus Prn-p+/+ (p < 0.0001); and Tg(Δ23–134H)/Tga20+/0 versus Tga20+/0 (p < 0.0001) when analyzed either by Kruskal-Wallis one way analysis of variance with Dunn's Multiple Comparison Test, or by Students t test. C, animals of the indicated genotypes were sacrificed at the terminal stage of illness after scrapie inoculation. Brain homogenates containing equivalent amounts of protein were treated with (lower blots) or without (upper blots) 20 μg/ml proteinase K for 1 h at 37 °C, and were subjected to Western blotting with anti-PrP antibody 6H4.
We examined the effect of PrP(Δ23–134) on the accumulation of PrPSc by Western blotting of PK-treated brain homogenates. Parallel to what we observed in mice expressing C1, the amount of protease-resistant PrP was comparable in terminally ill Tg(Δ23–134)/Prn-p+/+ mice and Prn-p+/+ mice (Fig. 8B). In contrast, Tg(Δ23–134H)/Tga20+/0 mice accumulated much less protease-resistant PrP than Tga20+/0 mice, even at the terminal stage of disease (Fig. 8C).
DISCUSSION
In this study, we have used transgenic mice expressing the C1 cleavage fragment of PrPC to investigate the role of this naturally occurring proteolytic product in the normal biology of PrPC as well as its role in prion illness. In transgenic mice expressing PrP(Δ23–111) (C1) on a Prn-p0/0 background, we observed no spontaneous clinical or histological abnormalities, indicating that C1 alone is not inherently neurotoxic. We also demonstrate that although C1 is not itself convertible to a PK-resistant form, scrapie-inoculated mice co-expressing C1 and WT PrP show a significant delay in scrapie illness as well as decreased PrPSc formation. These results have important implications for the role of the physiologically generated C1 fragment and its use as a target for therapeutic intervention in prion diseases.
C1 Is Not Neurotoxic
Transgenic mice expressing PrP with specific deletions in the N-terminal half of the protein display spontaneous neurodegenerative phenotypes that are suppressed by co-expression of WT PrP. This phenomenon is observed for PrP molecules harboring deletions of amino acids 32–121, 32–134, 94–134, and 105–125 (28, 29, 31). On the Prn-p0/0 background, animals expressing these truncated forms of PrP exhibit ataxia, accompanied by extensive degeneration of the cerebellar granule neurons, as well as degenerative changes in white matter areas of the brain and spinal cord. Ectopic expression in the brain of the PrP paralog Doppel (Dpl), which is structurally homologous to N-terminally truncated PrP, also results in a neurodegenerative illness that is overcome by the presence of WT PrP (32–35). Taken together, these results indicate that deletion of the highly conserved central region of PrP (particularly residues 105–125) endows the protein with potent neurotoxic activity. This activity may be related in some way to the normal function of PrPC, because it is reversible by co-expression of WT PrP. We have recently shown that the neurotoxicity of deleted forms of PrP may be related to their ability to induce ion channels or pores in the cell membrane (36, 37).
Because the C1 fragment is missing a portion of the central domain implicated in these neurotoxic phenomena, we wondered whether expression of the C1 fragment alone, in the absence of WT PrP, might produce a neurodegenerative illness. We found that Tg(C1)/Prn-p0/0 mice displayed no clinical or histological signs of neurological disease, despite the fact that the truncated protein was expressed at supraphysiological expression levels (7 times). Thus, the C1 fragment (Δ23–111) does not display intrinsic neurotoxic activity. This result is consistent with another study showing that mice expressing PrP(Δ111–134) display spontaneous neurodegenerative illness, although mice expressing PrP(Δ94–110) do not (31, 38). Taken together, the available data indicate that deletion of hydrophobic residues between 111 and 125 is required to endow PrP with neurotoxic activity. We have recently shown that a nine-amino acid, polybasic segment at the extreme N terminus (residues 23–31), is crucial for the neurotoxic and channel-inducing activities of PrP molecules carrying central region deletions (30, 37). The Δ23–111 fragment expressed in Tg(C1) mice also lacks this element. The C1 fragment of PrP has been reported to have a p53-dependent pro-apoptotic function in cell culture assays (39), but our results indicate that this effect is not likely to come into play in an in vivo context.
Tg(C1)/Prn-p0/0 Mice Are Resistant to Scrapie
Tg(C1)/Prn-p0/0 mice inoculated with RML scrapie prions do not develop clinical or neuropathological signs of illness and do not accumulate protease-resistant PrP in their brains. This result is consistent with previous in vitro and in vivo studies demonstrating that formation of PrPSc is dependent on the presence of the central domain of PrP, which is lacking in C1. For example, although the majority of the N terminus (up to residue 90) is not essential to the conversion process (40, 41), more C-terminal deletions of residues 114–121, 95–107, or 108–121 render PrP incapable of conversion to PrPSc in cultured cells and transgenic mice (42, 43). It is therefore likely that the central domain of PrP is a part of the molecule that undergoes conformational changes during the formation of PrPSc (44), making C1 incapable of conversion into the misfolded conformer.
C1 Is a Dominant-negative Inhibitor of Scrapie-induced Disease and PrPSc Production
Although C1 itself was incapable of sustaining PrPSc production and development of neurological illness, it had a prominent inhibitory effect on disease progression and PrPSc accumulation in mice co-expressing C1 and WT PrP. Scrapie-inoculated Tg(C1)/Prn-p+/+ or Tg(C1)/Tga20+/0 mice had a significantly prolonged survival time compared with corresponding Prn-p+/+ or Tga20+/0 mice lacking the C1 transgene. The increase in survival amounted to 33 and 45%, respectively, for the Prn-p+/+ or Tga20+/0 genotypes. The presence of C1 also markedly slowed the accumulation of protease-resistant PrP in the brain. There was substantially less PrPSc in Tg(C1)/Prn-p+/+ mice at pre-clinical time points compared with Prn-p+/+ mice, although at the terminal stage the amount of PrPSc was similar with or without C1. Tg(C1)/Tga20+/0 mice, however, accumulated less PrPSc than Tga20+/0 mice, even at the terminal stage of illness. It is noteworthy that C1 PrP was expressed at supra-physiological levels in these mice, likely accentuating the dominant-negative effects that would be produced by endogenous levels of C1.
These results imply that C1 has a dominant-negative effect on the production of PrPSc, which is correlated with a delay in development of clinical symptoms. Based on these results and previous literature (42, 45–48), we hypothesize that C1 inhibits an early stage of the PrPC-PrPSc conversion process, potentially because C1 competes with WT PrP for binding to PrPSc seeds. This model is supported by the fact that antibodies recognizing C-terminal residues are capable of inhibiting PrPSc formation in cells (49, 50) and interacting specifically with PrPSc (51, 52).
Previous studies have documented the dominant-negative effects on PrPSc formation of PrP molecules carrying deletions or point mutations. For example, substitutions at residues 167, 171, 214, or 218 of PrPC inhibited production of PrPSc in scrapie-infected neuroblastoma cells that express endogenous WT PrP (48). Experiments in transgenic mice have confirmed and extended these results. Perrier et al. (53) generated transgenic mice expressing PrP harboring either the Q167R or Q218K mutations. After scrapie inoculation, transgenic mice on the Prn-p0/0 background showed no clinical or histopathological abnormalities and did not produce PrPSc. Moreover, the Q167R and Q218K mutants significantly delayed disease development on the Prn-p+/+ background and decreased accumulation of PrPSc. In addition to these point mutations, short deletions within the central region of the protein (Δ112–119, Δ114–121, and Δ105–125) led to a dominant-negative effect in infected cells and mice, respectively (42, 54). In vitro conversion reactions showed that the dominant-negative effect of point mutations at residues 218 or 171 does not require the presence of auxiliary molecules (55, 56), implying a direct interaction between the C terminus of PrP and the PrPSc seed. Collectively, these results demonstrate that modifications in the C-terminal half of the protein (after 111) can significantly impact the generation of PK-resistant PrP. However, this study documents for the first time that deletions within the N terminus of PrP(Δ23–111 or Δ23–134) can also lead to a dominant-negative effect on the conversion of WT PrP into PrPSc.
PrPSc Formation Is Not Correlated with Clinical Illness
Even when terminally ill, scrapie-infected Tg(C1)/Tga20+/0 mice display greatly reduced levels of protease-resistant PrP compared with mice without the C1 transgene. Nevertheless, the brains of these animals contain substantial infectivity as determined by transmission to indicator mice. These results demonstrate a lack of correlation between development of clinical disease, PrPSc formation, and accumulation of infectivity in Tg(C1)/Tga20+/0 mice.
There are many examples of related phenomena in the literature (27, 57–63), including a recent study (64) concluding that prion propagation and neurodegeneration occur in distinct chronological and mechanistic phases, with the latter phase taking much longer and being more closely tied to levels of PrPC. Taken together, our results and those of others suggest that prion toxicity is attributable to accumulation of a critical level of a specific toxic PrP species (PrPtoxic) that is distinct from infectious PrPSc. The C1 fragment may have dominant-negatively inhibited formation of both kinds of PrP in Tg(C1)/Tga20+/0 mice, although it appears to have produced a greater delay in formation of PrPSc, which was present at low levels in terminally ill animals. Based on our studies in Tg(PG14) mice, we have hypothesized that PrPtoxic consists of small oligomers of PrP (65), similar to the nonfibrillar oligomers of Aβ that have been hypothesized to be synaptotoxic in Alzheimer disease (66, 67).
C1 Is a Physiologically Generated Inhibitor of PrPSc
Tg(C1) mice express a PrP fragment that occurs naturally in the brain as a result of proteolytic cleavage of PrP (10–12). Our results therefore indicate that a dominant-negative inhibitor of prion replication is generated normally in the brain. This observation has two major implications. First, it is possible that alterations in the levels of C1 might take place during the course of prion disease and influence disease progression. For example, there is evidence that β-cleavage predominates over α-cleavage during prion infection (10). If this shift in cleavage preference causes a reduction in the levels of the inhibitory C1 fragment, it might contribute to a feedback acceleration of PrPSc formation.
A second implication of our study is that manipulation of C1 levels might represent a therapeutic strategy for treating prion diseases. Although there is some disagreement about the proteases responsible for the α-cleavage (14, 15, 68), it is possible that pharmacologically activating these proteases, or enhancing delivery of PrPC to the cellular compartments where they reside, might create more C1, which would dominantly inhibit formation of PrPSc and therefore ameliorate disease. In addition, exogenous administration of C1 or peptide fragments derived from it might also have therapeutic benefit. Supporting the feasibility of this approach, several synthetic PrP peptides, including 119–136, 166–179, and 200–223, inhibited conversion in both cell-free and cell culture systems (45, 69). Additionally, transgenic or lentivirally driven expression of dominant-negative PrP mutants has been shown to inhibit or delay disease progression in mice (53, 70). The dominant-negative lentiviral vector was capable of prolonging survival even when administered after the onset spongiosis in scrapie-infected mice (70). The reversibility of scrapie-associated pathogenesis in this and other studies (71–73) suggests that increased expression of the dominant-negative C1 protein may represent a promising treatment for established prion diseases.
Acknowledgments
We thank Cheryl Adles and Su Deng for mouse colony maintenance and genotyping. We acknowledge Aimin Li for help with primer design and cloning, and advice on the course of the project. We appreciate the kind gift of D18 antibody from Dennis Burton (Scripps Institute, La Jolla, CA). We are grateful to Heather True and Emiliano Biasini for critically reading and commenting on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants NS40975, NS052526, NS056376, and NS065244 (to D. A. H.).
- PrPC
- cellular prion protein
- GPI
- glycosylphosphatidylinositol
- PrP
- prion protein
- GFAP
- glial fibrillary acidic protein
- PNGase
- peptide:N-glycosidase
- p.i.
- post-inoculation.
REFERENCES
- 1. Prusiner S. B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguzzi A., Baumann F., Bremer J. (2008) Annu. Rev. Neurosci. 31, 439–477 [DOI] [PubMed] [Google Scholar]
- 3. Pan K. M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E., Prusiner S. B. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10962–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Safar J., Roller P. P., Gajdusek D. C., Gibbs C. J., Jr. (1993) J. Biol. Chem. 268, 20276–20284 [PubMed] [Google Scholar]
- 5. Stahl N., Borchelt D. R., Hsiao K., Prusiner S. B. (1987) Cell 51, 229–240 [DOI] [PubMed] [Google Scholar]
- 6. Shyng S. L., Huber M. T., Harris D. A. (1993) J. Biol. Chem. 268, 15922–15928 [PubMed] [Google Scholar]
- 7. Sunyach C., Jen A., Deng J., Fitzgerald K. T., Frobert Y., Grassi J., McCaffrey M. W., Morris R. (2003) EMBO J. 22, 3591–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Magalhães A. C., Silva J. A., Lee K. S., Martins V. R., Prado V. F., Ferguson S. S., Gomez M. V., Brentani R. R., Prado M. A. (2002) J. Biol. Chem. 277, 33311–33318 [DOI] [PubMed] [Google Scholar]
- 9. Shyng S. L., Heuser J. E., Harris D. A. (1994) J. Cell Biol. 125, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., Autilio-Gambetti L. (1995) J. Biol. Chem. 270, 19173–19180 [DOI] [PubMed] [Google Scholar]
- 11. Harris D. A., Huber M. T., van Dijken P., Shyng S. L., Chait B. T., Wang R. (1993) Biochemistry 32, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 12. Mangé A., Béranger F., Peoc'h K., Onodera T., Frobert Y., Lehmann S. (2004) Biol. Cell 96, 125–132 [DOI] [PubMed] [Google Scholar]
- 13. Vincent B., Paitel E., Frobert Y., Lehmann S., Grassi J., Checler F. (2000) J. Biol. Chem. 275, 35612–35616 [DOI] [PubMed] [Google Scholar]
- 14. Laffont-Proust I., Faucheux B. A., Hässig R., Sazdovitch V., Simon S., Grassi J., Hauw J. J., Moya K. L., Haïk S. (2005) FEBS Lett. 579, 6333–6337 [DOI] [PubMed] [Google Scholar]
- 15. Vincent B., Paitel E., Saftig P., Frobert Y., Hartmann D., De Strooper B., Grassi J., Lopez-Perez E., Checler F. (2001) J. Biol. Chem. 276, 37743–37746 [DOI] [PubMed] [Google Scholar]
- 16. Walmsley A. R., Watt N. T., Taylor D. R., Perera W. S., Hooper N. M. (2009) Mol. Cell. Neurosci. 40, 242–248 [DOI] [PubMed] [Google Scholar]
- 17. Taraboulos A., Raeber A. J., Borchelt D. R., Serban D., Prusiner S. B. (1992) Mol. Biol. Cell 3, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B., Avraham D., Laszlo L., Prusiner S. B. (1995) J. Cell Biol. 129, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McMahon H. E., Mangé A., Nishida N., Créminon C., Casanova D., Lehmann S. (2001) J. Biol. Chem. 276, 2286–2291 [DOI] [PubMed] [Google Scholar]
- 20. Watt N. T., Taylor D. R., Gillott A., Thomas D. A., Perera W. S., Hooper N. M. (2005) J. Biol. Chem. 280, 35914–35921 [DOI] [PubMed] [Google Scholar]
- 21. Yadavalli R., Guttmann R. P., Seward T., Centers A. P., Williamson R. A., Telling G. C. (2004) J. Biol. Chem. 279, 21948–21956 [DOI] [PubMed] [Google Scholar]
- 22. Parkin E. T., Watt N. T., Turner A. J., Hooper N. M. (2004) J. Biol. Chem. 279, 11170–11178 [DOI] [PubMed] [Google Scholar]
- 23. Borchelt D. R., Rogers M., Stahl N., Telling G., Prusiner S. B. (1993) Glycobiology 3, 319–329 [DOI] [PubMed] [Google Scholar]
- 24. Taguchi Y., Shi Z. D., Ruddy B., Dorward D. W., Greene L., Baron G. S. (2009) Mol. Biol. Cell 20, 233–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiménez-Huete A., Lievens P. M., Vidal R., Piccardo P., Ghetti B., Tagliavini F., Frangione B., Prelli F. (1998) Am. J. Pathol. 153, 1561–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Borchelt D. R., Davis J., Fischer M., Lee M. K., Slunt H. H., Ratovitsky T., Regard J., Copeland N. G., Jenkins N. A., Sisodia S. S., Price D. L. (1996) Genet. Anal. Biomol. Eng. 13, 159–163 [DOI] [PubMed] [Google Scholar]
- 27. Chiesa R., Piccardo P., Ghetti B., Harris D. A. (1998) Neuron 21, 1339–1351 [DOI] [PubMed] [Google Scholar]
- 28. Li A., Christensen H. M., Stewart L. R., Roth K. A., Chiesa R., Harris D. A. (2007) EMBO J. 26, 548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shmerling D., Hegyi I., Fischer M., Blättler T., Brandner S., Götz J., Rülicke T., Flechsig E., Cozzio A., von Mering C., Hangartner C., Aguzzi A., Weissmann C. (1998) Cell 93, 203–214 [DOI] [PubMed] [Google Scholar]
- 30. Westergard L., Turnbaugh J. A., Harris D. A. (2011) J. Neurosci. 31, 14005–14017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baumann F., Tolnay M., Brabeck C., Pahnke J., Kloz U., Niemann H. H., Heikenwalder M., Rülicke T., Bürkle A., Aguzzi A. (2007) EMBO J. 26, 538–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rossi D., Cozzio A., Flechsig E., Klein M. A., Rülicke T., Aguzzi A., Weissmann C. (2001) EMBO J. 20, 694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakaguchi S., Katamine S., Nishida N., Moriuchi R., Shigematsu K., Sugimoto T., Nakatani A., Kataoka Y., Houtani T., Shirabe S., Okada H., Hasegawa S., Miyamoto T., Noda T. (1996) Nature 380, 528–531 [DOI] [PubMed] [Google Scholar]
- 34. Moore R. C., Mastrangelo P., Bouzamondo E., Heinrich C., Legname G., Prusiner S. B., Hood L., Westaway D., DeArmond S. J., Tremblay P. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 15288–15293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamaguchi N., Sakaguchi S., Shigematsu K., Okimura N., Katamine S. (2004) Biochem. Biophys. Res. Commun. 319, 1247–1252 [DOI] [PubMed] [Google Scholar]
- 36. Solomon I. H., Huettner J. E., Harris D. A. (2010) J. Biol. Chem. 285, 26719–26726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Solomon I. H., Khatri N., Biasini E., Massignan T., Huettner J. E., Harris D. A. (2011) J. Biol. Chem. 286, 14724–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A. D., Toyka K. V., Nave K. A., Weis J., Aguzzi A. (2010) Nat. Neurosci. 13, 310–318 [DOI] [PubMed] [Google Scholar]
- 39. Sunyach C., Cisse M. A., da Costa C. A., Vincent B., Checler F. (2007) J. Biol. Chem. 282, 1956–1963 [DOI] [PubMed] [Google Scholar]
- 40. Fischer M., Rülicke T., Raeber A., Sailer A., Moser M., Oesch B., Brandner S., Aguzzi A., Weissmann C. (1996) EMBO J. 15, 1255–1264 [PMC free article] [PubMed] [Google Scholar]
- 41. Flechsig E., Shmerling D., Hegyi I., Raeber A. J., Fischer M., Cozzio A., von Mering C., Aguzzi A., Weissmann C. (2000) Neuron 27, 399–408 [DOI] [PubMed] [Google Scholar]
- 42. Hölscher C., Delius H., Bürkle A. (1998) J. Virol. 72, 1153–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Muramoto T., DeArmond S. J., Scott M., Telling G. C., Cohen F. E., Prusiner S. B. (1997) Nat. Med. 3, 750–755 [DOI] [PubMed] [Google Scholar]
- 44. Peretz D., Williamson R. A., Matsunaga Y., Serban H., Pinilla C., Bastidas R. B., Rozenshteyn R., James T. L., Houghten R. A., Cohen F. E., Prusiner S. B., Burton D. R. (1997) J. Mol. Biol. 273, 614–622 [DOI] [PubMed] [Google Scholar]
- 45. Horiuchi M., Baron G. S., Xiong L. W., Caughey B. (2001) J. Biol. Chem. 276, 15489–15497 [DOI] [PubMed] [Google Scholar]
- 46. Rigter A., Langeveld J. P., Timmers-Parohi D., Jacobs J. G., Moonen P. L., Bossers A. (2007) BMC Biochem. 8, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Horiuchi M., Caughey B. (1999) EMBO J. 18, 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaneko K., Zulianello L., Scott M., Cooper C. M., Wallace A. C., James T. L., Cohen F. E., Prusiner S. B. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Enari M., Flechsig E., Weissmann C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9295–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Peretz D., Williamson R. A., Kaneko K., Vergara J., Leclerc E., Schmitt-Ulms G., Mehlhorn I. R., Legname G., Wormald M. R., Rudd P. M., Dwek R. A., Burton D. R., Prusiner S. B. (2001) Nature 412, 739–743 [DOI] [PubMed] [Google Scholar]
- 51. Moroncini G., Kanu N., Solforosi L., Abalos G., Telling G. C., Head M., Ironside J., Brockes J. P., Burton D. R., Williamson R. A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10404–10409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Solforosi L., Bellon A., Schaller M., Cruite J. T., Abalos G. C., Williamson R. A. (2007) J. Biol. Chem. 282, 7465–7471 [DOI] [PubMed] [Google Scholar]
- 53. Perrier V., Kaneko K., Safar J., Vergara J., Tremblay P., DeArmond S. J., Cohen F. E., Prusiner S. B., Wallace A. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13079–13084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Norstrom E. M., Mastrianni J. A. (2005) J. Biol. Chem. 280, 27236–27243 [DOI] [PubMed] [Google Scholar]
- 55. Lee C. I., Yang Q., Perrier V., Baskakov I. V. (2007) Protein Sci. 16, 2166–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Geoghegan J. C., Miller M. B., Kwak A. H., Harris B. T., Supattapone S. (2009) PLoS Pathog. 5, e1000535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lasmézas C. I., Deslys J. P., Robain O., Jaegly A., Beringue V., Peyrin J. M., Fournier J. G., Hauw J. J., Rossier J., Dormont D. (1997) Science 275, 402–405 [DOI] [PubMed] [Google Scholar]
- 58. Hsiao K. K., Groth D., Scott M., Yang S. L., Serban H., Rapp D., Foster D., Torchia M., Dearmond S. J., Prusiner S. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9126–9130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Manson J. C., Jamieson E., Baybutt H., Tuzi N. L., Barron R., McConnell I., Somerville R., Ironside J., Will R., Sy M. S., Melton D. W., Hope J., Bostock C. (1999) EMBO J. 18, 6855–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Barron R. M., Thomson V., Jamieson E., Melton D. W., Ironside J., Will R., Manson J. C. (2001) EMBO J. 20, 5070–5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hsiao K. K., Scott M., Foster D., Groth D. F., DeArmond S. J., Prusiner S. B. (1990) Science 250, 1587–1590 [DOI] [PubMed] [Google Scholar]
- 62. Collinge J., Owen F., Poulter M., Leach M., Crow T. J., Rossor M. N., Hardy J., Mullan M. J., Janota I., Lantos P. L. (1990) Lancet 336, 7–9 [DOI] [PubMed] [Google Scholar]
- 63. Collinge J., Palmer M. S., Sidle K. C., Gowland I., Medori R., Ironside J., Lantos P. (1995) Lancet 346, 569–570 [DOI] [PubMed] [Google Scholar]
- 64. Sandberg M. K., Al-Doujaily H., Sharps B., Clarke A. R., Collinge J. (2011) Nature 470, 540–542 [DOI] [PubMed] [Google Scholar]
- 65. Chiesa R., Harris D. A. (2001) Neurobiol. Dis. 8, 743–763 [DOI] [PubMed] [Google Scholar]
- 66. Walsh D. M., Klyubin I., Fadeeva J. V., Cullen W. K., Anwyl R., Wolfe M. S., Rowan M. J., Selkoe D. J. (2002) Nature 416, 535–539 [DOI] [PubMed] [Google Scholar]
- 67. Selkoe D. J. (2008) Behav. Brain Res. 192, 106–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cissé M. A., Sunyach C., Lefranc-Jullien S., Postina R., Vincent B., Checler F. (2005) J. Biol. Chem. 280, 40624–40631 [DOI] [PubMed] [Google Scholar]
- 69. Chabry J., Priola S. A., Wehrly K., Nishio J., Hope J., Chesebro B. (1999) J. Virol. 73, 6245–6250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Toupet K., Compan V., Crozet C., Mourton-Gilles C., Mestre-Francés N., Ibos F., Corbeau P., Verdier J. M., Perrier V. (2008) PLoS ONE 3, e2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mallucci G., Dickinson A., Linehan J., Klöhn P. C., Brandner S., Collinge J. (2003) Science 302, 871–874 [DOI] [PubMed] [Google Scholar]
- 72. Mallucci G. R., White M. D., Farmer M., Dickinson A., Khatun H., Powell A. D., Brandner S., Jefferys J. G., Collinge J. (2007) Neuron 53, 325–335 [DOI] [PubMed] [Google Scholar]
- 73. White M. D., Farmer M., Mirabile I., Brandner S., Collinge J., Mallucci G. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10238–10243 [DOI] [PMC free article] [PubMed] [Google Scholar]