Background: WT1 is expressed in a variety of solid tumors and correlates with poor prognosis.
Results: WT1 directly activates VEGF transcription and is required for optimal response to hypoxia.
Conclusion: WT1 is a key mediator of hypoxia-mediated tumor angiogenesis.
Significance: These data suggest a new role for WT1 in tumor growth and progression.
Keywords: Angiogenesis, Hypoxia, Transcription Factors, Tumor Suppressor Gene, Vascular Endothelial Growth Factor (VEGF), Sarcoma
Abstract
WT1 is a zinc finger transcription factor expressed at high levels in many types of solid tumors, and high WT1 expression is an adverse prognostic factor. How WT1 contributes to tumor growth and influences prognosis remains unclear. We investigated the hypothesis that WT1 up-regulates VEGF in solid tumors, augmenting the response to hypoxia. We found a correlation between levels of WT1 expression and VEGF expression in Ewing sarcoma cell lines. Transfecting WT1-null SK-ES-1 cells with WT1 up-regulated VEGF mRNA expression and resulted in increased angiogenic activity in vitro. Conversely, diminishing WT1 expression in WT1-positive cell lines using WT1-specific shRNA down-regulated VEGF mRNA expression and decreased angiogenic activity in vitro. Transient transfection assays demonstrated that WT1 can regulate the activity of the VEGF promoter, and chromatin immunoprecipitation assays showed that WT1 can bind directly to the VEGF promoter in intact cells. WT1 expression in Ewing sarcoma cells is up-regulated by hypoxia. Importantly, using shRNA to inhibit this up-regulation blunted the hypoxia-mediated increase in VEGF expression. Taken together, these data demonstrate that VEGF is a direct, bona fide WT1 target gene in sarcoma and that WT1 plays a key role in optimizing the response of tumor cells to hypoxia.
Introduction
Although originally identified as a tumor suppressor gene in children with Wilms tumor, subsequent work has demonstrated that WT1 is overexpressed in a wide variety of tumor types, including leukemia (1, 2), breast cancer (3), and a number of other solid tumors, including sarcomas (4, 5). This has raised the possibility that WT1 could have tumorigenic activity in the proper context. In soft tissue sarcoma, osteosarcoma, and breast cancer patients, WT1 expression confers a poor prognosis (6). The role of WT1 in tumorigenesis and the mechanism by which expression affects prognosis are both unclear; however, because the WT1 protein has transcriptional regulatory activity, effects on tumorigenesis and prognosis are probably related to altered expression of key WT1 target genes.
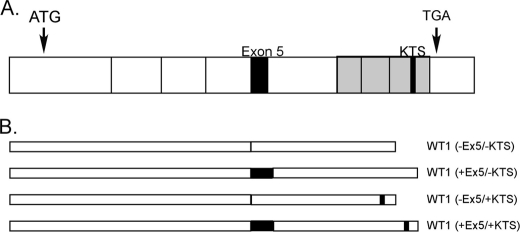
WT1 is a transcription factor containing four zinc fingers in a C-terminal DNA binding domain (7, 8). The WT1 mRNA is subject to alternative splicing at two independent sites (Fig. 1), resulting in the expression of four major protein isoforms (9). One of these alternate splicing events includes or excludes exon 5 from the mature mRNA. The other inserts a 9-bp sequence (termed the “KTS insert” after the amino acids encoded by the additional nucleotides (lysine, threonine, and serine)) between exons 9 and 10, which encode the third and fourth zinc fingers, changing the interactions between WT1 and DNA as well as affecting subcellular localization of the WT1 protein (10–12). Individual WT1 isoforms are named based on the presence or absence of exon 5 and the presence or absence of the KTS insert such that the isoform lacking both is designated WT1 (−Ex5/−KTS).3
FIGURE 1.
Schematic representation of WT1 mRNA. A, full-length mRNA. Each box represents an exon. The initiation (ATG) and termination (TGA) codons are noted. Alternatively spliced regions (exon 5 and the KTS insert) are indicated by the black boxes. The gray region indicates the DNA binding domain. B, the four possible protein isoforms are illustrated.
Our laboratory has investigated the hypothesis that different WT1 isoforms can have distinct effects on tumorigenesis. We found that induction of WT1 (−Ex5/−KTS) in mammary epithelial cells causes a cell cycle arrest and expression of genes such as albumin associated with a differentiated phenotype, whereas induction of WT1 (+Ex5/+KTS) results in an epithelial-to-mesenchymal transition and up-regulation of proliferation-associated genes such as B-raf (13). In the course of these investigations, we also found that WT1 (+Ex5/+KTS) up-regulates vascular endothelial growth factor (VEGF) in these cells. WT1 expression can be regulated by hypoxia, especially in coronary artery endothelial cells (14), and WT1 expression has been demonstrated in tumor endothelial cells as well (15). These observations led us to investigate the role of WT1 in the response of solid tumor cells to hypoxia. Our results demonstrate that WT1 is up-regulated by hypoxia in tumor cells, that WT1 directly regulates the expression of functional VEGF, and that suppression of the hypoxia-mediated up-regulation of WT1 blunts the hypoxia-mediated up-regulation of VEGF. These findings strongly implicate WT1 as a key mediator of the proangiogenic response of tumor cells to hypoxia, contributing to tumorigenicity and to the poor prognosis experienced by patients with WT1-expressing tumors.
EXPERIMENTAL PROCEDURES
Cell Culture
Ewing sarcoma cell lines MHH-ES, SK-ES-1, TC71, and RD-ES were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). NIH3T3 cells were propagated in DMEM (Invitrogen) supplemented with 10% fetal bovine serum. Cells stably transfected with both pNEB-R1 and pNEB-X1 Hygro plasmids (New England Biolabs, Ipswich, MA) were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Invitrogen), 0.5 mg/ml G418 (Mediatech Inc., Manassas, VA), and 0.8 mg/ml hygromycin (Mediatech Inc.). Cells were grown at 37 °C in atmospheric O2 (21%) or in a sealed incubator chamber (Billups-Rothenberg, Del Mar, CA) in 5% CO2, 1% O2, and 94% N2 (Airgas East, Linthicum Heights, MD).
Transfections
Cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For stable transfection of inducible shRNA, 2 × 106 MHH-ES cells were first transfected with 4.0 μg of the plasmid pNEB-R1 (New England Biolabs) containing the Rheo activator/receptor. Twenty-four hours after transfection, cells were seeded into 96-well plates in growth medium supplemented with 1.0 mg/ml G418. Stably transfected cells were screened by transiently transfecting pNEBX1-luc (New England Biolabs), a plasmid that expresses Gaussia luciferase under the control of the Rheo activator/receptor, followed by assays for luciferase activity after 48 h of growth in media with and without the RheoSwitch® Ligand (RSL; from New England Biolabs). Once stable transfection of the Rheo activator/receptor plasmid was confirmed, 2 × 106 of those cells were stably transfected with 4.0 μg of the pNEB-X1 Hygro plasmid (New England Biolabs) expressing either WT1 shRNA or a negative control shRNA under the control of the Rheo activator/receptor. Stable transfectants were selected with RPMI 1640 medium, 10% FBS, 0.5 mg/ml G418, and 1.0 mg/ml hygromycin. Stable transfection and inducible expression of the shRNA were confirmed using quantitative PCR following 48 h of growth in media with and without RSL.
Semiquantitative RT-PCR
Total RNA was harvested from cells with an RNeasy kit (Qiagen, Valencia, CA), and 1 μg of DNase I-treated RNA was reverse transcribed using iScript reverse transcriptase (Bio-Rad) according to the manufacturer's recommendations. PCR was performed after cDNA amplification using WT1-specific primers (see Table 1 for primer sequences) that span exon 5, and the products were resolved by electrophoresis on a 2% agarose gel. Amplification of the ribosomal RNA 36B4 served as a positive control for RNA integrity.
TABLE 1.
Primers used for RT-PCR and ChIP experiments
| Primer name | Sequence |
|---|---|
| WT1ex5fwd | 5′-GCGGCGCAGTTCCCCAACCA-3′ |
| WT1ex5rev | 5′-ATGGTTTCTCACCAGTGTGCTT-3′ |
| 36B4fwd | 5′-GATTGGCTACCCAACTGTTGCA-3′ |
| 36B4rev | 5′-CAGGGGCAGCAGCCACAAAGGC-3′ |
| VEGF-WT1–1fwd | 5′-CCTACAGACGTTCCTTAGTGCTG-3′ |
| VEGF-WT1–1rev | 5′-GTCAGTGACTGGGAGGGAAGAG-3′ |
| VEGF-WT1–2fwd | 5′-TAATTTATTTTTGCTTGCCATTCC-3′ |
| VEGF-WT1–2rev | 5′-CAAAAGCAGGTCACTCACTTTG-3′ |
| VEGF-WT1–3fwd | 5′-CTGACGGACAGACAGACAGACAC-3′ |
| VEGF-WT1–3rev | 5′-CCCAGAAGTTGGACGAAAAGTT-3′ |
| VEGF-WT1–4fwd | 5′-TGTTCTCGCTTCGGAGGAGC-3′ |
| VEGF-WT1–4rev | 5′-TGGAGCACTGTCTGCGCACA-3′ |
| VEGF-HRE-fwd | 5′-GCGGGTAGGTTTGAATCATC-3′ |
| VEGF-HRE-rev | 5′-CAGTGACTGGGAGGGAAGAG-3′ |
| VEGF-WT1–1Δfwd | 5′-CTGGCCTCCACCCGTTTCCACCAGCCCCCTG-3′ |
| VEGF-WT1–1Δrev | 5′-CAGGGGGCTGGTGGAAACGGGTGGAGGCCAG-3′ |
| VEGF-WT1–2Δfwd | 5′-GTCAGAGAGAGCGCGCTTTCGTGCGAGCAGCGAAA-3′ |
| VEGF-WT1–2Δrev | 5′-TTTCGCTGCTCGCACGAAAGCGCGCTCTCTCTGAC-3′ |
| VEGF-WT1–4Δfwd | 5′-CCGTGGTCCGCGCGTTTGAAGCCGAGCCGAG-3′ |
| VEGF-WT1–4Δrev | 5′-CTCGGCTCGGCTTCAAACGCGCGGACCACGG-3′ |
Quantitative RT-PCR
Primers specific to WT1, VEGF, and β-actin were obtained from SuperArray Bioscience (Fredrick, MD). Quantification of gene expression was performed using a Bio-Rad MyiQ single color real time PCR detection system with SYBR Green chemistry in 96-well plates for 40 cycles of 95 °C for 15 s and 60 °C for 1 min followed by melting curve analysis. The mean threshold cycle (Ct) of the triplicate samples was determined, and then the Ct for each sample was corrected against the Ct level of β-actin. Quantification of gene expression was performed by calculating ΔΔCt where ΔΔCt = (Ctsample − Ctactin)control − (Ctsample − Ctactin)treated. The -fold change in gene expression between two samples was then determined by calculating 2−ΔΔCt.
Chromatin Immunoprecipitation (ChIP)
Chromatin Immunoprecipitation was performed according to the manufacturer's recommendations (Active Motif, Carlsbad, CA). Briefly, cells were fixed for 10 min with 1% formaldehyde followed by enzymatic shearing of the fixed chromatin according to the manufacturer's protocol (Active Motif). ChIP was performed by incubating sheared chromatin with 2 μg of either WT1 (F-6X; Santa Cruz Biotechnology Inc., Santa Cruz, CA), RNA polymerase II, or a negative IgG control antibody (the latter two are included in the kit). Chromatin bound to the antibody was pulled down with magnetic Protein G-coated beads, washed, and eluted. Following cross-link removal, immunoprecipitated DNA was purified using a PCR purification kit (Qiagen) according to the manufacturer's instructions with the following modifications. After addition of a 5× volume of buffer PB and 10 μl of 3 m NaOAc, reactions were incubated for 30 min at room temperature on a rolling shaker. Additionally, two washes with buffer PE were performed before eluting the DNA from the PCR purification column. The purified products were then analyzed by real time PCR using primers (listed in Table 1) as described above.
Western Blotting
Total cellular protein was prepared with a QIAmp kit (Qiagen) according to the manufacturer's instructions. Samples were run on 4–12% bis-Tris gels in MES buffer followed by transfer onto polyvinylidene difluoride (PVDF) membranes and blocking overnight in TBS, 0.1% Tween 20, and 5% nonfat dry milk. WT1 primary antibody (Novus, Littleton, CO) was diluted 1:1000 into blocking solution, and the secondary antibody, mouse IgG (Invitrogen), was diluted 1:20,000. For loading controls, either HRP-conjugated β2-microglobulin antibody (Novus) diluted 1:5000 in blocking buffer or anti-GAPDH antibody (Santa Cruz Biotechnology Inc.) diluted 1:5000 followed by a 1:20,000 dilution of HRP-conjugated rabbit IgG (Invitrogen) was used. Immunoblotting was followed by visualization with ECL Plus (GE Healthcare) on x-ray film.
VEGF Promoter Mutants
Mutations in putative WT1 binding sites in the VEGF promoter were prepared using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) on the luciferase reporter plasmid pPVEGF-luc. Oligonucleotides that were used to generate the mutations are listed in Table 1. Mutations were confirmed by sequencing (3730xl DNA Analyzer, Applied Biosystems).
Luciferase Assays
VEGF promoter regulation by WT1 was assayed using a reporter vector expressing the cDNA (pGL2) for firefly luciferase driven by 3.3 kb of the VEGF promoter. NIH3T3 cells were transiently transfected with the reporter construct and either the empty expression vector as a control or an expression vector containing the cDNA for WT1 (+Ex5/+KTS) or WT1 (−Ex5/−KTS) under the control of the CMV immediate early promoter. A vector expressing Renilla luciferase was used as a control for transfection efficiency. Firefly luciferase expression was normalized to the Renilla luciferase activity, and -fold change in expression was calculated as the change in normalized firefly luciferase activity compared with the empty expression vector control. Luciferase assays were performed according to the manufacturer's (Promega, Madison WI) recommendations 24 h after transfection.
VEGF ELISAs
VEGF165 ELISAs on media from transiently transfected cells were performed according to the manufacturer's instructions (Invitrogen). Total protein for normalization of the ELISA was extracted from the transfected cells using the QIAmp kit (Qiagen) according to the manufacturer's protocol.
Tube Formation Assays
To assess angiogenic activity in vitro, human umbilical vein endothelial cell (HUVEC) tube formation assays were performed using an In Vitro Angiogenesis Assay kit (Millipore, Billerica, MA) according to the manufacturer's instruction. Briefly, 104 human umbilical vein endothelial cells/well were co-cultured with 3 × 103 cells from the Ewing sarcoma cell lines for 18 h, then fixed in 5% formaldehyde, and stained with 0.5% crystal violet. In some experiments, wells were treated with 0.5 μm VEGF-neutralizing antibody (R&D Systems, Minneapolis, MN). Tube formation was inspected under an inverted light microscope and quantified using a pattern index. The assay was done in duplicate or triplicate wells, and five to seven random high power fields were examined per well.
Statistical Analysis
Differences in gene expression and tube formation were tested for significance using a one-sided t test. Correlation between VEGF and WT1 mRNA expression was determined using standard linear regression. All statistical analyses were performed with Prism 4 software (GraphPad Software, Inc.).
RESULTS
VEGF mRNA Expression Correlates with WT1 Expression in Ewing Sarcoma Cell Lines
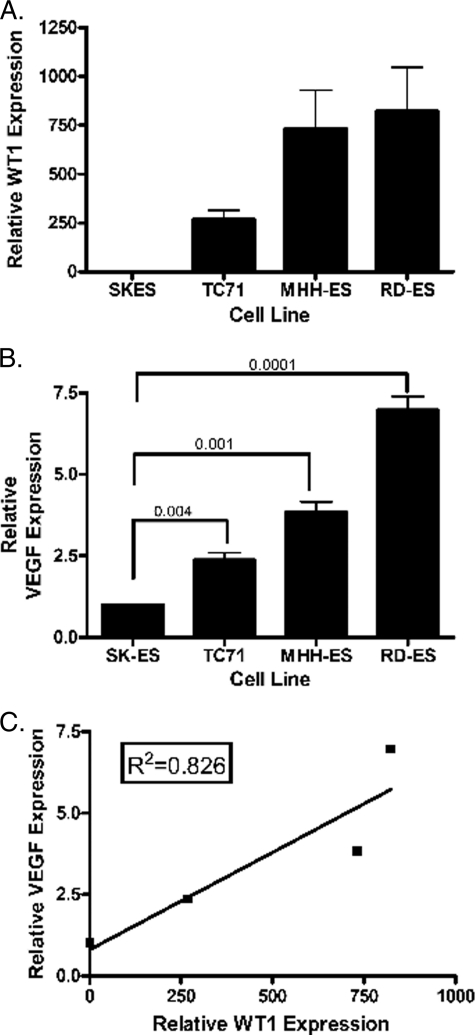
To investigate whether WT1 might affect VEGF expression in cells other than mammary epithelial cells, we measured WT1 and VEGF expression levels in Ewing sarcoma cell lines. TC71, RD-ES, and MHH-ES express significant amounts of both WT1 and VEGF mRNA. In contrast, the WT1-null Ewing sarcoma cell line SK-ES-1 expresses only very low levels of VEGF mRNA (Fig. 2, A and B). The difference between VEGF mRNA expression in SK-ES-1 and each of the other Ewing sarcoma cell lines is statistically significant with p values all <0.005. A plot of relative WT1 mRNA expression versus relative VEGF mRNA expression as determined by quantitative RT-PCR showed a correlation with R2 = 0.826 (Fig. 2C).
FIGURE 2.
WT1 and VEGF levels correlate. A, RNA was isolated from the indicated cell lines and analyzed by quantitative RT-PCR using WT1-specific primers designed to amplify all isoforms. The signal obtained from the WT1-null SK-ES-1 cell line was arbitrarily assigned a value of 1.0, and all other signals were compared with this using the ΔΔCt method. Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. B, quantitative RT-PCR was used to evaluate VEGF RNA expression in the indicated cell lines. The VEGF level of SK-ES-1 was arbitrarily set at 1.0. Numbers indicate p values of the differences between each WT1-expressing cell line and SK-ES-1. Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. C, relative VEGF mRNA expression (as determined by quantitative RT-PCR) was plotted against relative WT1 mRNA expression (as determined by quantitative RT-PCR). Standard linear regression analysis was performed to quantify the correlation between these values, and the coefficient of correlation was found to be R2 = 0.826.
WT1 Directly Regulates VEGF Expression in Ewing Sarcoma Cells
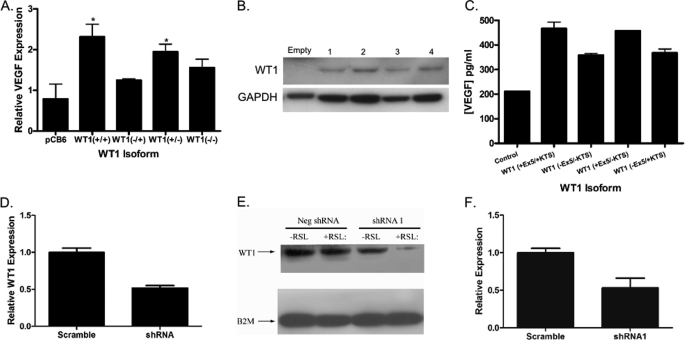
To determine whether the correlation between WT1 expression and VEGF expression reflects a direct effect of WT1 on VEGF mRNA levels, the WT1-null SK-ES-1 cell line was transiently transfected with plasmids containing the cDNA for each of the four major WT1 isoforms under the control of the constitutively active CMV immediate early promoter, and VEGF mRNA levels were measured by quantitative RT-PCR. WT1 (+Ex5/+KTS) caused a 2.9-fold increase in VEGF mRNA expression (p = 0.03) compared with cells transfected with the empty expression vector (Fig. 3A). WT1 (+Ex5/−KTS) also caused a statistically significant increase in VEGF expression (a 2.5-fold increase; p = 0.048). In contrast, although both WT1 (−Ex5/+KTS) and WT1 (−Ex5/−KTS) increased VEGF expression in this assay, neither reached the level of statistical significance. These differences reflect inherent differences between the isoforms as Western blotting confirmed equal expression of each WT1 isoform in transfected cells (Fig. 3B). Production of VEGF protein was confirmed by a VEGF165 ELISA performed on media from transfected cells. VEGF protein expression was significantly increased by each WT1 isoform (p < 0.01 for each isoform) with the largest increases also coming from WT1 (+Ex5/+KTS) and WT1 (+Ex5/−KTS) (Fig. 3C). In a complementary experiment, we used two different WT1-specific shRNA constructs. Transient transfection of the WT1-expressing MHH-ES cell line with either of these resulted in a 50% reduction of WT1 expression (p < 0.01 for each construct; Fig. 3D). Suppression of WT1 protein was confirmed by Western blotting (Fig. 3E). A corresponding 50% reduction in the level of VEGF mRNA compared with cells transiently transfected with a nonsense shRNA as measured by quantitative RT-PCR was also seen (p = 0.03; Fig. 3F). Taken together, these results demonstrate that modulation of WT1 expression directly results in a corresponding change in VEGF expression in Ewing sarcoma cell lines.
FIGURE 3.
Modulating WT1 expression in Ewing sarcoma cell lines affects VEGF expression. A, the WT1-null cell line SK-ES-1 was transiently transfected with either the empty pCB6 vector or the same vector containing cDNA for the indicated WT1 isoform. Relative VEGF mRNA expression was determined by quantitative RT-PCR. The * indicates p values <0.05. B, total cellular protein was isolated from SK-ES-1 cells transfected with either the empty pCB6 vector or the same vector containing cDNA for the indicated WT1 isoform (lane 1, WT1 (+Ex5/+KTS); lane 2, WT1 (−Ex5/−KTS); lane 3, WT1 (+Ex5/−KTS); lane 4, WT1 (−Ex5/+KTS)). Western blotting was performed with an antibody against WT1 (top panel) or GAPDH as a loading control (bottom panel). C, conditioned medium was collected from SK-ES-1 cells transfected with either the empty pCB6 vector or the same vector containing cDNA for the indicated WT1 isoform. VEGF165 protein was measured using an ELISA. The difference in VEGF protein concentration for each WT1 isoform is statistically significantly different from control with p < 0.01 for each. D, the WT1-expressing Ewing sarcoma cell line MHH-ES was transiently transfected with either a WT1-specific shRNA or a scrambled control. Relative expression of WT1 mRNA was determined by quantitative RT-PCR. The difference in WT1 expression is highly statistically significant (p < 0.002). E, total cellular protein was isolated from MHH-ES cells stably transfected with a plasmid containing an inducible promoter and either a control, scrambled RNA or an shRNA targeting WT1, and Western blotting was performed with an antibody against either WT1 (top panel) or β2-microglobulin (B2M) (lower panel) as a loading control. Addition of RSL, which activates the promoter, did not affect WT1 protein expression in the control cells but substantially diminished WT1 protein expression in cells with the WT1 shRNA. F, the WT1-expressing Ewing sarcoma cell line MHH-ES was transiently transfected with either a WT1-specific shRNA or a scrambled control. Relative expression of VEGF mRNA was significantly diminished by the WT1-specific shRNA (p = 0.03). Throughout this figure, error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. Neg, negative.
WT1 Directly Regulates VEGF Promoter
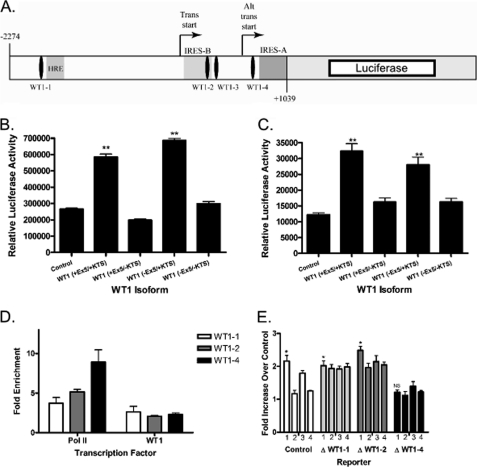
We next investigated whether the effect of WT1 on VEGF mRNA expression reflects a direct effect on the VEGF promoter. We created a promoter-reporter construct containing the firefly luciferase gene under the control of the VEGF promoter. The construct includes 3313 bp of the VEGF promoter from position −2274 to position +1039 relative to the main transcriptional start site (Fig. 4A). NIH3T3 cells were transiently transfected with this luciferase reporter and a plasmid containing the cDNA one of the WT1 isoforms (or a control plasmid with no cDNA insert) under the control of the constitutively active CMV immediate early promoter. In this assay, WT1 (+Ex5/+KTS) resulted in a 2.2-fold up-regulation of VEGF promoter activity compared with control (p < 0.0001; Fig. 4B), and WT1 (−Ex5/+KTS) caused a 2.8-fold increase in promoter activity (p < 0.0001). WT1 (+Ex5/−KTS) and WT1 (−Ex5/−KTS) had no significant effect on the activity of the luciferase construct. Similar results were seen when the WT1-null SK-ES-1 cell line was used instead of NIH3T3 cells (Fig. 4C). Although all four isoforms increased VEGF mRNA and protein expression (see Fig. 3), not all four isoforms activated the luciferase construct. This may simply be an artifact of the luciferase promoter/reporter assay system, which involves WT1 binding to episomal DNA; this is in contrast to the experiments in Fig. 3 that involve WT1 binding to intact, chromosomal DNA. Western blotting confirmed similar expression of each WT1 isoform in each cell line, thus ruling out differences in transfection efficiency as an explanation for these findings (data not shown, but see Fig. 3B).
FIGURE 4.
VEGF is a direct WT1 target gene. A, a schematic diagram of the VEGF promoter-luciferase reporter construct showing four putative WT1 binding sites, the HRE, and the dual internal ribosome entry site (IRES), and transcriptional start sites. Trans start, transcriptional start site; Alt trans start, alternate transcriptional start site. B, NIH3T3 cells were transfected with the VEGF promoter-luciferase reporter construct and either the empty pCB6 expression vector or pCB6 containing the cDNA for the indicated WT1 isoform. Relative luciferase activity is indicated on the y axis. The ** indicates values that are statistically significantly different from control with p < 0.0001. Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. C, SK-ES-1 cells were transfected with the VEGF promoter-luciferase reporter construct and either the empty pCB6 expression vector or pCB6 containing the indicated WT1 isoform. Relative luciferase activity is indicated on the y axis. The ** indicates values that are statistically significantly different from control with p < 0.005. Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. D, chromatin immunoprecipitation was performed using antibodies against RNA polymerase II (Pol II), WT1, or a control IgG. Precipitated DNA was analyzed by quantitative PCR using primers that flanked each WT1 binding site (WT1-1, WT1-2, WT1-3, and WT1-4). The graph shows the -fold enrichment in DNA immunoprecipitated by the indicated antibody compared with the control IgG. No data are presented for WT1-3 because we were unable to immunoprecipitate DNA that could be amplified with those primers. E, NIH3T3 cells were transfected with the indicated VEGF promoter-luciferase reporter construct and either the empty pCB6 expression vector or pCB6 containing one of the WT1 isoforms. On this graph, WT1 (+Ex5/+KTS) is column 1, WT1 (+Ex5/−KTS) is column 2, WT1 (−Ex5/+KTS) is column 3, and WT1 (−Ex5/−KTS) is column 4. As indicated by the *, the difference between WT1 (+Ex5/+KTS) and control reached statistical significance with p < 0.005 for control and the constructs with deletions of WT1−1 and WT1−2 but not for WT1−4 (NS represents not significant). Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times.
Promoter-reporter assays such as these do not conclusively prove that WT1 directly regulates the VEGF promoter. We therefore performed ChIP assays to determine whether endogenous WT1 directly binds to the endogenous VEGF promoter in Ewing sarcoma cells. WT1·histone complexes were immunoprecipitated from TC71 cells. We used four independent primer sets, one for each of the potential WT1 binding sites identified by inspection of the sequence of the VEGF promoter (Fig. 4A). Using a quantitative PCR approach, we were able to demonstrate WT1 binding to three of the four putative binding sites (WT1-1, WT1-2, and WT1-4; Fig. 4D). Our inability to demonstrate WT1 binding to WT1-3 could indicate that this is not a true WT1 binding site, or this may just be a DNA region that is unable to be amplified by PCR. The affinity of WT1 for each site was equivalent as judged by enrichment of WT1 binding to the site compared with a nonspecific IgG. As expected, the RNA polymerase II signal became stronger with increasing proximity to the transcriptional start site. Thus, WT1 binds to three of the four potential binding sites in the VEGF promoter in intact cells, consistent with our hypothesis that VEGF is a bona fide WT1 target gene.
To confirm the importance of WT1 binding to the VEGF promoter, we used site-directed mutagenesis to mutate each individual WT1 binding site in the promoter-reporter construct and repeated the luciferase assays in NIH3T3 cells. Mutation of sites WT1-1 and WT1-2 had no effect on the ability of WT1 to up-regulate promoter activity (Fig. 4E). In fact, deletion of binding sites WT1-1 and WT1-2 improved the ability of some isoforms to up-regulate promoter activity. In contrast, mutation of WT1-4 abolished the up-regulation of promoter activity by all WT1 isoforms (Fig. 4E). Taken together, these data demonstrate that WT1 binds the VEGF promoter in intact cells and up-regulates the activity of the VEGF promoter, and this up-regulation is dependent upon binding to site WT1-4.
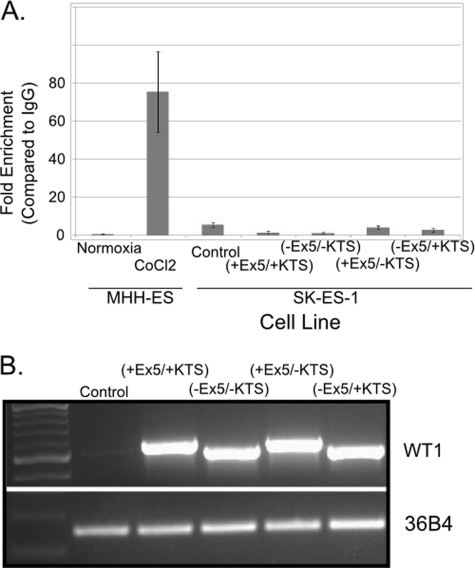
To rule out the possibility that WT1 stabilizes hypoxia-inducible factor (HIF)-1 and that HIF-1 binding to the VEGF hypoxia-response element (HRE) rather than WT1 binding to the WT1 binding sites is responsible for the transcriptional up-regulation of VEGF, we investigated whether an HIF-1·HRE complex could be immunoprecipitated from SK-ES-1 cells transiently transfected with WT1. SK-ES-1 cells were transfected with either an empty expression vector or with a cDNA encoding each of the WT1 isoforms. As a control, WT1-expressing MHH-ES cells under either normoxic or hypoxic (CoCl2 treatment) conditions were used. HIF-1·histone complexes were immunoprecipitated from these cells, and co-precipitated DNA was subjected to quantitative PCR with primers specific for the VEGF HRE (Table 1). As anticipated, normoxic MHH-ES cells showed no evidence of HIF-1 binding to the HRE, but a product corresponding to the HRE was amplified from immunoprecipitated material from CoCl2-treated cells (Fig. 5A). No such product was amplified from material from normoxic SK-ES-1 cells transfected with any WT1 isoform (Fig. 5A), ruling out the possibility that WT1 stabilizes HIF-1, resulting in up-regulation of VEGF transcription. Equal WT1 mRNA expression in transfected cells was confirmed by RT-PCR (Fig. 5B).
FIGURE 5.
WT1 does not stabilize HIF-1. A, chromatin was immunoprecipitated from MHH-ES cells grown under normoxia or hypoxia (1% O2) as well as from SK-ES-1 cells transfected with an empty expression vector (pCB6; labeled “Control”) or a pCB6 expression vector containing cDNA for the indicated WT1 isoform using antibody against HIF-1α or a control IgG. DNA was recovered from the immunoprecipitated chromatin and analyzed by quantitative PCR using primers surrounding the HRE in the VEGF promoter. The graph shows the relative enrichment of HRE in the HIF-1α immunoprecipitates compared with IgG control. Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. B, RNA was isolated from the same SK-ES-1 cells used for the experiment in A and reverse transcribed. RT-PCR was performed using primers that amplify WT1 (top) and the ribosomal RNA 36B4 (bottom). This experiment was repeated three times.
VEGF Up-regulated by WT1 Is Biologically Active
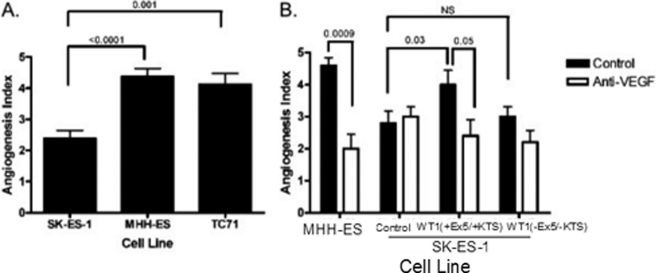
We next sought to determine whether the up-regulation of VEGF mRNA by WT1 leads to a corresponding increase in bioactive VEGF protein. To accomplish this, we investigated the angiogenic activity of Ewing sarcoma cell lines that express varying amounts of both WT1 and VEGF. We used tube formation by HUVECs as an in vitro measure of angiogenic activity. Briefly, after the cells were cultured, samples were coded by a laboratory member uninvolved with the experiments, and after 18 h, tube formation was quantified using the following angiogenesis index: 0 points for high powered field (hpf) with individual, well separated cells; 1 point for hpf with cells that appear to be aligning; 2 points for hpf with tubes but no evidence of sprouting; 3 points for hpf with evidence of sprouting; 4 points for hpf with closed polygons; and 5 points for hpf with a complete meshwork. A minimum of five high powered fields per well were evaluated. Because of the subjective nature of this assay, experiments were performed in a blinded fashion: the observer quantifying tube formation was blinded to the conditions in each culture. Co-culture of HUVECs with TC71 or MHH-ES cells, both of which express significant amounts of WT1 and VEGF, led to statistically significantly increased tube formation compared with HUVECs cultured with SK-ES-1 cells, which do not express WT1 and express only low levels of VEGF (p ≤ 0.001 for each cell line; Fig. 6A). Thus, in these cell lines, in vitro tube-forming activity parallels VEGF mRNA production.
FIGURE 6.
WT1 up-regulates angiogenic activity in vitro. A, HUVECs were co-cultured with the indicated Ewing sarcoma cell line, and the angiogenesis index was determined after 18 h. A minimum of five high powered fields were quantified, and the average ± S.E. are shown. Both WT1-expressing cell lines showed a statistically significant increase in angiogenic activity (p < 0.0001 for MHH-ES and p = 0.001 for TC71) compared with the WT1-null SK-ES-1 cell line. B, HUVECs were co-cultured with SK-ES-1 cells that had been transiently transfected with either the empty pCB6 expression vector or pCB6 containing the cDNA for either WT1 (+Ex5/+KTS) or WT1 (−Ex5/−KTS). Cells were cultured with or without 0.5 μm anti-VEGF antibody, and after 48 h, the angiogenesis index was determined. A minimum of five high powered fields were quantified, and the average ± S.E. are shown. MHH-ES cells were included as a positive control. The p values for key differences are shown. Throughout this figure, error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times.
Finally, we used this assay system to confirm that a WT1-mediated up-regulation of VEGF mRNA in the SK-ES-1 cell line translates into increased VEGF-mediated angiogenic activity. SK-ES-1 cells were transfected with either WT1 (+Ex5/+KTS), which significantly up-regulates VEGF expression; WT1 (−Ex5/−KTS), which only modestly affects VEGF expression; or a control plasmid without WT1. These cells were co-cultured with HUVECs, and tube formation was quantified as described. Only the cells transfected with WT1 (+Ex5/+KTS) had increased tube-forming activity (p = 0.03; Fig. 6B). To confirm that the increased tube-forming activity is due to increased VEGF expression, the experiment was repeated in the presence of a blocking antibody. As expected, inclusion of 0.5 μm VEGF-blocking antibody eliminated the WT1-mediated increase in angiogenic activity (p = 0.05; Fig. 6B). All angiogenic activity is not eliminated because there are proangiogenic molecules other than VEGF that can be produced by sarcoma cells that would be expected to cause background tube formation (16). Thus, our data demonstrate that WT1 (+Ex5/+KTS) up-regulates biologically active VEGF in Ewing sarcoma cell lines but do not preclude the possibility that other factors play a role in Ewing sarcoma angiogenesis.
WT1 Plays a Functional Role in Response to Hypoxia
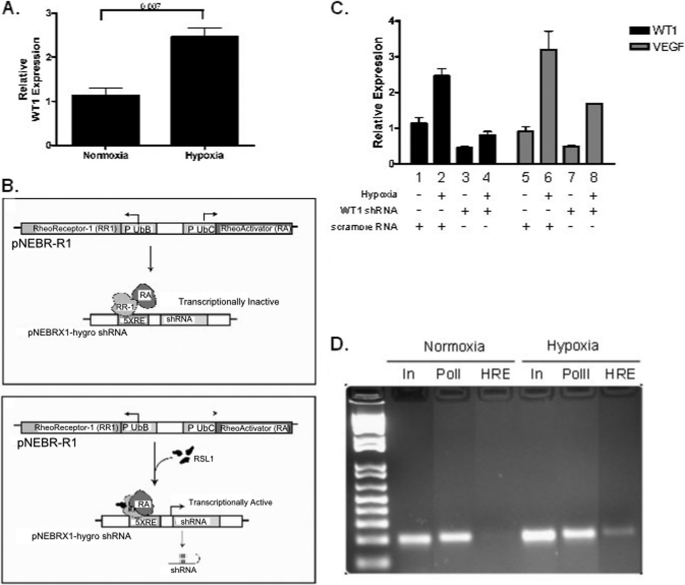
The results presented thus far demonstrate that WT1 directly regulates VEGF expression. WT1 can be up-regulated by hypoxia (17), and this prompted us to investigate whether up-regulation of WT1 is necessary for the robust increase in VEGF expression induced by hypoxia. As a first step, we investigated whether WT1 is up-regulated by hypoxia in the MHH-ES cell line. After the cells were incubated for 48 h under hypoxic conditions (1% O2), mRNA was harvested and analyzed for WT1 expression by quantitative PCR. As expected, hypoxia led to a statistically significant 2.2-fold increase in WT1 expression (p = 0.007; Fig. 7A).
FIGURE 7.
Inhibition of WT1 expression attenuates response to hypoxia. A, MHH-ES were cultured in 1% O2 for 48 h, and WT1 expression was determined by quantitative RT-PCR. Hypoxia caused a 2.2-fold increase in WT1 mRNA expression (p = 0.007). Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. B, schematic representation of the inducible promoter system illustrating how addition of RSL results in transcriptional activation of the WT1 shRNA. C, MHH-ES cells stably transfected with either a WT1-specific shRNA or a control scrambled RNA under the control of the RSL-inducible promoter were grown for 72 h in 1% O2 and then treated with RSL for 48 h to induce the shRNA or control. RNA was then harvested, and WT1 and VEGF expression was quantified using quantitative RT-PCR. Error bars represent S.E. of experiments done in triplicate. Experiments were repeated a minimum of three times. D, MHH-ES cells transfected with WT1-specific shRNA were grown under either normoxic or hypoxic (1% O2) conditions for 48 h. Chromatin immunoprecipitation was then performed using either an antibody against RNA polymerase II (PolII) or HIF-1 (HIF), and then those samples and unmanipulated input (In) DNA were subjected to PCR with primers surrounding the HRE in the VEGF promoter. RR-1, RheoReceptor-1; RA, RheoActivator.
Next, we needed a system to suppress the hypoxia-mediated up-regulation of WT1. For this, we utilized the RheoSwitch Mammalian Inducible Expression System to express inducible shRNA (Fig. 7B). This system utilizes a synthetic receptor consisting of two proteins, RheoReceptor-1 and RheoActivator that when expressed from constitutive promoters on plasmid pNEBR-R1 dimerize to form a holoreceptor. RheoReceptor-1 expresses a protein that includes an engineered ligand binding domain of an insect EcR nuclear receptor that is fused to a GAL4 DNA binding domain. The RheoActivator protein is an insect/mammalian retinoid X receptor hybrid ligand binding domain/viral activation domain VP16 fusion. The shRNAs were cloned into the pNEBR-X1 plasmid, which includes five tandem repeats of the GAL4 response element (5XRE). In the absence of ligand, the dimerized holoreceptor binds to the GAL4 elements and represses transcription. Transcription is induced when RSL alters RheoReceptor-1 protein conformation, resulting in stabilization of the receptor heterodimer on the 5XRE. The activated holoreceptor along with the VP16 activation domain recruit transcriptional coactivators and basal machinery to the promoter, producing a high level of transcriptional activity.
We used the inducible WT1 shRNA system to determine the effect of silencing WT1 on hypoxia-mediated up-regulation of VEGF. MHH-ES cells stably transfected with the WT1-specific inducible shRNA construct were incubated with RSL to induce shRNA expression. After 72 h to allow time for expression of the shRNA and down-regulation of WT1, cells were made hypoxic. After 48 h in a 1% O2 atmosphere, RNA was harvested, and WT1 and VEGF mRNA expression was quantified by quantitative PCR. As expected, hypoxia caused a 2.5-fold up-regulation of WT1 (p = 0.007) and a 3.5-fold up-regulation of VEGF in the control cells (p = 0.01; Fig. 7C). In contrast, induction of WT1 shRNA strongly attenuated both the hypoxia-mediated up-regulation of WT1 and the hypoxia-mediated up-regulation of VEGF (Fig. 7C). In normoxic cells, WT1 shRNA expression resulted in a 60% decrease in WT1 expression (p = 0.02) and a 47% decrease in VEGF mRNA expression (p = 0.04). When cells were made hypoxic in the presence of the WT1 shRNA, WT1 expression was unchanged compared with normoxic cells (Fig. 7C, compare columns 1 and 4). Similarly, in the presence of WT1 shRNA, hypoxic cells expressed 1.86-fold more VEGF than did control, normoxic cells (compare columns 5 and 8), but this was only 47% as much as the hypoxic control cells (compare columns 6 and 8; p = 0.05). Thus, although eliminating the hypoxia-mediated up-regulation of WT1 using WT1-specific shRNA did not reduce the relative response to hypoxia (there is still an increase between columns 7 and 8), there was an almost 50% decrease in the hypoxia-mediated up-regulation of VEGF mRNA under these conditions (columns 6 and 8), strongly implicating WT1 in the optimal tumor response to hypoxia.
To confirm that the cells were responding appropriately to hypoxia, we performed a ChIP assay using antibodies against HIF-1 and primers that recognize the HRE of the VEGF promoter (Table 1). Chromatin was isolated from MHH-ES cells transfected with WT1 shRNA and incubated in both normoxic and hypoxic conditions and immunoprecipitated with antibodies against HIF-1. As expected, in the normoxic cells, there was no appreciable HIF-1 bound to the VEGF HRE. In contrast, hypoxic cells had a substantial amount of HIF-1 bound to the VEGF HRE, confirming a normal response to hypoxia in these cells (Fig. 7D). Thus, although the WT1 shRNA attenuated induction of VEGF, it did not interfere with the ability of the cells to up-regulate HIF-1 DNA binding activity.
DISCUSSION
Our data demonstrate that WT1 up-regulates VEGF transcription, resulting in increased angiogenic activity. In Ewing sarcoma cell lines, we found a correlation between endogenous WT1 expression and VEGF expression. Up-regulation of WT1 in the low WT1-expressing cell lines led to a corresponding increase in VEGF expression (both mRNA and protein), whereas silencing of WT1 in the high WT1-expressing cell lines led to a corresponding decrease in VEGF expression. Finally, results from promoter-reporter, chromatin immunoprecipitation, and site-directed mutagenesis assays demonstrate that VEGF is a direct target of the transcriptional regulatory activity of WT1 and identify a specific sequence within the VEGF promoter (WT1-4 in Fig. 4) that is essential for the effect of WT1 on promoter activity.
One important aspect of our work is the confirmation that WT1 not only leads to increased VEGF transcription but also a corresponding increase in angiogenic activity. Our data demonstrate that transfection of WT1-null sarcoma cell lines leads directly to increased tube formation by co-cultured HUVECs, and that this effect is inhibited by a blocking anti-VEGF antibody. Thus, WT1 expression causes increased VEGF transcription, which in turn results in increased expression of bioactive VEGF protein and a proangiogenic phenotype.
Perhaps our most important finding is that endogenous WT1 expression is essential for an optimal response to hypoxia. After demonstrating that WT1 expression is induced by hypoxia in Ewing sarcoma cell lines, we found that hypoxia-mediated up-regulation of VEGF is attenuated by a WT1-specific shRNA that blocks induction of WT1 by hypoxia. Clearly, VEGF is up-regulated by hypoxia in WT1-null cells by a direct effect of HIF-1 on the activity of the VEGF promoter; however, our findings clearly place WT1 as a key mediator of the maximal induction of VEGF in response to hypoxia.
WT1 expression is an adverse prognostic factor in many types of sarcoma, but the biological basis for this finding is unclear. There are potential cell-autonomous explanations, such as regulation of apoptosis, as well as cell-nonautonomous explanations, such as an impact on angiogenesis. WT1 has a cell-autonomous antiapoptotic effect in some, but not all, cell types. Because we found that WT1 (+Ex5/+KTS), but not WT1 (−Ex5/−KTS), up-regulates VEGF expression in mammary epithelial cells, we hypothesized that WT1 expression affects the prognosis of sarcoma patients because it plays a key role in allowing tumor cells to adapt to hypoxia, adopt an angiogenic phenotype, and thus grow more aggressively. Work from other laboratories has implicated WT1 in the regulation of angiogenesis: WT1 is expressed in tumor endothelial cells (15) and is up-regulated in coronary artery endothelial cells in a mouse model of myocardial infarction (14). In prostate cancer cell lines, WT1 appears to regulate VEGF expression, although the magnitude and direction of this regulation are variable (18, 19). These data implicate WT1 in the response of tumor cells to hypoxia.
Taken together, our data confirm that WT1 is a hypoxia-responsive gene in Ewing sarcoma cell lines; that WT1 up-regulates VEGF transcription, leading to a corresponding increase in VEGF activity; and that attenuation of the hypoxia-mediated induction of WT1 blunts the up-regulation of VEGF by hypoxia. These findings provide at least one explanation for the correlation between WT1 expression and a poor outcome for patients with sarcoma: tumors with higher WT1 expression produce more VEGF, resulting in increased angiogenesis, which simultaneously allows for more rapid tumor growth and provides increased access for tumor cells to the vascular system, allowing for an increased propensity for metastasis, which is the usual cause of death in patients with sarcoma.
Acknowledgments
We thank Drs. Venu Raman, Alan Friedman, and Ido Paz-Priel for careful reading and thoughtful discussion of the manuscript.
This work was supported by grants from the Flight Attendant Medical Research Institute and The Children's Cancer Foundation (both to D. M. L.), Giant Foods, The Heather Brooke Foundation, and the Love for Luca Foundation.
- Ex5
- exon 5
- RSL
- RheoSwitch Ligand
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HUVEC
- human umbilical vein endothelial cell
- HRE
- hypoxia-response element
- hpf
- high powered field
- 5XRE
- five tandem repeats of the GAL4 response element
- HIF
- hypoxia-inducible factor.
REFERENCES
- 1. Miwa H., Beran M., Saunders G. F. (1992) Leukemia 6, 405–409 [PubMed] [Google Scholar]
- 2. Inoue K., Sugiyama H., Ogawa H., Nakagawa M., Yamagami T., Miwa H., Kita K., Hiraoka A., Masaoka T., Nasu K. (1994) Blood 84, 3071–3079 [PubMed] [Google Scholar]
- 3. Loeb D. M., Evron E., Patel C. B., Sharma P. M., Niranjan B., Buluwela L., Weitzman S. A., Korz D., Sukumar S. (2001) Cancer Res. 61, 921–925 [PubMed] [Google Scholar]
- 4. Carpentieri D. F., Nichols K., Chou P. M., Matthews M., Pawel B., Huff D. (2002) Mod. Pathol. 15, 1080–1086 [DOI] [PubMed] [Google Scholar]
- 5. Ueda T., Oji Y., Naka N., Nakano Y., Takahashi E., Koga S., Asada M., Ikeba A., Nakatsuka S., Abeno S., Hosen N., Tomita Y., Aozasa K., Tamai N., Myoui A., Yoshikawa H., Sugiyama H. (2003) Cancer Sci. 94, 271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Srivastava A., Fuchs B., Zhang K., Ruan M., Halder C., Mahlum E., Weber K., Bolander M. E., Sarkar G. (2006) Clin. Cancer Res. 12, 4237–4243 [DOI] [PubMed] [Google Scholar]
- 7. Call K. M., Glaser T., Ito C. Y., Buckler A. J., Pelletier J., Haber D. A., Rose E. A., Kral A., Yeger H., Lewis W. H. (1990) Cell 60, 509–520 [DOI] [PubMed] [Google Scholar]
- 8. Gessler M., Poustka A., Cavenee W., Neve R. L., Orkin S. H., Bruns G. A. (1990) Nature 343, 774–778 [DOI] [PubMed] [Google Scholar]
- 9. Haber D. A., Sohn R. L., Buckler A. J., Pelletier J., Call K. M., Housman D. E. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 9618–9622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drummond I. A., Rupprecht H. D., Rohwer-Nutter P., Lopez-Guisa J. M., Madden S. L., Rauscher F. J., 3rd, Sukhatme V. P. (1994) Mol. Cell. Biol. 14, 3800–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larsson S. H., Charlieu J. P., Miyagawa K., Engelkamp D., Rassoulzadegan M., Ross A., Cuzin F., van Heyningen V., Hastie N. D. (1995) Cell 81, 391–401 [DOI] [PubMed] [Google Scholar]
- 12. Stoll R., Lee B. M., Debler E. W., Laity J. H., Wilson I. A., Dyson H. J., Wright P. E. (2007) J. Mol. Biol. 372, 1227–1245 [DOI] [PubMed] [Google Scholar]
- 13. Simpson L. A., Burwell E. A., Thompson K. A., Shahnaz S., Chen A. R., Loeb D. M. (2006) Blood 107, 4695–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner K. D., Wagner N., Bondke A., Nafz B., Flemming B., Theres H., Scholz H. (2002) FASEB J. 16, 1117–1119 [DOI] [PubMed] [Google Scholar]
- 15. Wagner N., Michiels J. F., Schedl A., Wagner K. D. (2008) Oncogene 27, 3662–3672 [DOI] [PubMed] [Google Scholar]
- 16. Yoon S. S., Segal N. H., Park P. J., Detwiller K. Y., Fernando N. T., Ryeom S. W., Brennan M. F., Singer S. (2006) J. Surg. Res. 135, 282–290 [DOI] [PubMed] [Google Scholar]
- 17. Wagner K. D., Wagner N., Wellmann S., Schley G., Bondke A., Theres H., Scholz H. (2003) FASEB J. 17, 1364–1366 [DOI] [PubMed] [Google Scholar]
- 18. Graham K., Li W., Williams B. R., Fraizer G. (2006) Gene Expr. 13, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hanson J., Gorman J., Reese J., Fraizer G. (2007) Front. Biosci. 12, 2279–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]