Background: Proinsulin, an intermediate in insulin biosynthesis, is refractory to crystallization.
Results: Synchrotron-based hydroxyl radical footprints of proinsulin were obtained in relation to classical structures of insulin.
Conclusion: Molecular models based on footprinting provide evidence for native self-assembly and an ensemble of C-domain orientations.
Significance: Footprinting promises to enable analysis of toxic aggregation of clinical proinsulin variants in neonatal diabetes mellitus.
Keywords: Diabetes, Mass Spectrometry (MS), Molecular Modeling, Protein Structure, Radicals, Footprinting, Proinsulin
Abstract
Mutations in the insulin gene can impair proinsulin folding and cause diabetes mellitus. Although crystal structures of insulin dimers and hexamers are well established, proinsulin is refractory to crystallization. Although an NMR structure of an engineered proinsulin monomer has been reported, structures of the wild-type monomer and hexamer remain undetermined. We have utilized hydroxyl radical footprinting and molecular modeling to characterize these structures. Differences between the footprints of insulin and proinsulin, defining a “shadow” of the connecting (C) domain, were employed to refine the model. Our results demonstrate that in its monomeric form, (i) proinsulin contains a native-like insulin moiety and (ii) the C-domain footprint resides within an adjoining segment (residues B23–B29) that is accessible to modification in insulin but not proinsulin. Corresponding oxidation rates were observed within core insulin moieties of insulin and proinsulin hexamers, suggesting that the proinsulin hexamer retains an A/B structure similar to that of insulin. Further similarities in rates of oxidation between the respective C-domains of proinsulin monomers and hexamers suggest that this loop in each case flexibly projects from an outer surface. Although dimerization or hexamer assembly would not be impaired, an ensemble of predicted C-domain positions would block hexamer-hexamer stacking as visualized in classical crystal lattices. We anticipate that protein footprinting in combination with modeling, as illustrated here, will enable comparative studies of diabetes-associated mutant proinsulins and their aberrant modes of aggregation.
Introduction
Insulin, central to the hormonal control of metabolism, has long provided a model for the development of biophysical techniques (1). Renewed interest in the folding of insulin and its biosynthetic precursor (proinsulin) has been stimulated by the discovery of clinical mutations associated with permanent neonatal onset diabetes mellitus (DM)2 (2–4). Although such patients are heterozygous, the remaining wild-type insulin allele fails to enable metabolic homeostasis. A variety of evidence suggests that such DM-associated mutations impair the nascent folding of the variant proinsulin and block in trans the biosynthesis of the wild-type hormone. Disulfide-coupled protein misfolding disrupts trafficking from the endoplasmic reticulum, leading to β-cell dysfunction and eventual cell death (5). This syndrome (which accounts for almost half of the cases of DM presenting within the first year of life) has motivated structural studies of proinsulin. Although an extensive crystallographic database of insulin structures has been obtained over the past four decades (1), proinsulin has proven refractory to crystallization.3 We therefore sought to establish an alternative approach to the biophysical characterization of proinsulin. The present study thus employed mass spectrometry (6) in combination with synchrotron-based hydroxyl radical modification (7, 8).
Insulin is a globular protein containing two chains, designated A (21 residues) and B (30 residues). The hormone is the product of single-chain precursor proinsulin (86 residues), in which a connecting domain (C; 35 residues) links the C terminus of the B-domain to the N terminus of the A-domain. Excision of the C-domain by specific proteases (prohormone convertases) occurs in post-Golgi vesicles; the mature hormone is stored within secretory granules of pancreatic β-cells as zinc-stabilized hexamers (9). A recent NMR structure of an engineered proinsulin monomer (designated DKP-proinsulin (10))4 provided evidence that the precursor consists of a folded insulin-like moiety with a disordered C-domain projecting from one surface. Although the range of positions accessible to the C-domain was not well defined in the NMR structure (10),5 the results are in accordance with an analogy between the C-domain and hairs on the head of Medusa (comprised by the A/B-domains (11)). We hypothesized that the ensemble of C-domain conformations is not truly random but instead populates an envelope of accessible conformations along a well organized insulin surface.
To test this hypothesis, we sought to define the “shadow” of the connecting domain as projected on the surface of the insulin moiety by MS-based hydroxyl radical footprinting (7, 8, 12). In this approach, hydroxyl radicals, generated by radiolysis of water, oxidize solvent-accessible and reactive amino acid side chains on the surface of proteins (8, 12). Potential changes in accessibility of multiple side chains (as probed by increases or decreases in respective rates of oxidation rates as a function of ligand binding, protein assembly, or conformational changes) provide powerful tools for analysis of protein structures. We thus anticipated that sites of rapid modification on the surface of insulin, if by contrast protected in proinsulin, would define a C-domain footprint. An advantage of such an approach would be its broad applicability irrespective of oligomeric state. Unlike x-ray crystallography or conventional NMR spectroscopy, MS-based footprinting would in principle extend to non-native modes of protein aggregation characteristic of diverse diseases of protein misfolding.
In this study, we present the MS hydroxyl radical footprinting of human insulin (HI) and proinsulin (HPI) under two conditions: monomer and hexamer. Our analysis utilized classical crystal structures as a foundation for analysis of the additional C-domain; our results enabled a model of the HPI hexamer to be refined. In combination with the recent NMR study of DKP-proinsulin ((10), a coherent dynamic model of the wild-type HPI has been obtained as a monomer and hexamer. We anticipate that these studies will provide a foundation for comparative studies of disease-associated mutant proinsulins (13, 14).
EXPERIMENTAL PROCEDURES
Protein Preparation
Solutions of HI and HPI were prepared as T-like zinc-free monomers as described (15). Solutions of HI and HPI hexamer were prepared as a T6 zinc-stabilized hexamers according to a published protocol (1). Footprinting was undertaken at protein concentrations of 5 μm in 10 mm sodium cacodylate (pH 7.4).
Radiolysis
Each sample (volume 5 μl) was exposed to the X-28C x-ray white beam at the National Synchrotron Light Source, Brookhaven National Laboratory for 0, 8, 15, and 20 ms at ambient temperature. Exposure times were controlled by using an electronic shutter (Vincent Associates, Rochester, NY). Experiments were performed at ring energy 2.8 GeV with beam currents ranging between 195 and 225 mA. X-ray beam parameters were optimized using Alexa Fluor 488 fluorophore assay as described (16). To quench Met oxidation unrelated to primary hydroxyl radical attack, a buffer consisting of 10 mm Met-NH2·HCl (pH 7.0) was added immediately after irradiation.
Proteolysis and MS Analysis
After exposure, proteins were reduced, alkylated, and subjected to proteolysis by modified trypsin (Promega) at an enzyme-to-protein ratio of 1:20 w/w at 37 °C overnight. The digestion reaction was terminated by freezing. The digests (∼1 pmol) were loaded onto a PepMap reverse-phase trapping column (300 μm × 5 mm C18) to preconcentrate and wash away excess salts using a nano HPLC UltiMate-3000 (Dionex) column switching technique; reverse-phase separation was then performed on a C18, PepMap column (75 μm × 15 cm). Buffer A (100% water and 0.1% formic acid) and buffer B (20% water, 80% acetonitrile, and 0.1% formic acid) were employed in a linear gradient. Proteolytic peptides eluting from the column with an acetonitrile gradient (2% per min) were directed to an LTQ-FT mass spectrometer (Thermo Fisher Scientific) equipped with a nanospray ion source and with the needle voltage of 2.4 kV. Mass spectra were acquired in data-dependent experiments such as MS, and tandem MS spectra were acquired in the positive-ion mode, with the following acquisition cycle: a full scan recorded in the Fourier transform analyzer at resolution (R) 100,000 followed by MS/MS of the eight most intense peptide ions in the linear trap quadrupole (LTQ) analyzer. Dose-response curves were obtained by plotting the fraction unmodified for each peptide as a function of exposure time. Mass spectra were acquired in the positive ion mode; detected ion currents were utilized to determine the extent of oxidation by separate quantitation of the unmodified proteolytic peptides and their radiolytic products. Sites of oxidation were determined from tandem MS/MS data. MS/MS spectra of the peptide mixtures were searched against the human database for modifications (oxidation) of the tryptic peptides from HI using the Mascot search engine (Matrix Science Co.). Interpretation of MS/MS mass spectra of modified peptides was manually verified and correlated with hypothetical MS/MS spectra predicted for proteolysis products.
Calculation of Modification Rates
Integrated areas for each unmodified and modified peptide ion were calculated from selected ion current chromatograms. All peak areas obtained for multiple modified species with single or multiple modifications in the same peptide were added into the sum total modified. The extent of modification was then calculated from the ratio of the integrated peak area under the ion signals for the unmodified peptides to the sum of those for the unmodified peptide and their radiolytic products (sum total modified). Background modification seen for some peptides in the unexposed sample was subtracted from the totals. The fraction unmodified peptide was fit to the equation Y = Y0 e−kt using Origin 6.0 (MicroCal Software, Inc., Northampton, MA), where Y and Y0 are the fraction of unmodified peptide at a time t and 0 s, respectively, and k is a first-order rate constant. Dose-response curves were generated by plotting the fraction unmodified for each peptide as a function of x-ray exposure time.
Modeling of HPI
Cartesian coordinates of both insulin and DKP-proinsulin were retrieved from the Protein Data Bank (17). The proposed hexameric configuration of HPI was constructed based on the crystal structure of the T6 HI hexamer (1.5 Å resolution; Protein Data Bank (PDB) entry 4INS) (1). The solution structure of DKP-proinsulin has been solved by NMR spectroscopy (PDB entry 2KQP) (10). Mathematical averaging of coordinates of 20 model of HPI derived by NMR was performed; the average geometry was relaxed by energy minimization. Sites of mutation in DKP-proinsulin (residues B10, B28, and B29)6 were reverted to wild type using MODELLER (18). In this procedure, the positions were sequentially substituted with concomitant local conformational optimization of the mutant side chain. Optimization was achieved using a combination of energy minimization employing a conjugate gradient algorithm and simulated annealing with molecular dynamics. Rotation and translation matrices were computed to align this average structure of HPI to each chain of the HI hexamer. LSQMAN (19) software was used to perform brute force alignment of the HPI protomer with each of the six chains of the HI hexamer; for these calculations, the minimum length of fragment to match was 30 with a step size of 15.
Side-chain Solvent Accessibility
To verify the correlation of the side-chain reactivity and solvent accessibility (SA) for modified residues for HI monomer and hexamer, SA surface areas of side chains were calculated in Å2 using the VADAR computer program (PENCE, University of Alberta, Edmonton, Canada). The crystal structure of porcine insulin hexamer in the T6 form (PDB entry 4INS) and the NMR structure of DKP-proinsulin (PD entry 2KQP) were utilized for this analysis. Models of HI monomers and dimers were extracted from protomers in the crystallographic hexamer. Similarly, the SA of the modeled HPI hexamer was also calculated.
RESULTS AND DISCUSSION
Radiolytic Footprinting Strategy
Samples of HI and HPI as T-like monomers (15, 20) and T6 Zn2+-stabilized hexamers (1) were exposed to a focused x-ray beam and rapidly quenched (see above). On the time scale of irradiation, the primary products were oxidative modifications of side chains of reactive amino acids without protein cross-linking or cleavage (21). Following digestion with trypsin, the four peptide products of HI and six peptide products of HPI (covering 99% of their respective sequences) were analyzed by LC-MS. Tandem MS analysis was employed to confirm the sequence identities and define specific sites of oxidative modification.
The extent of side-chain modifications was evaluated as a function of exposure time to yield first-order rate constants (7, 12). Such rate constants typically correlate with measures of SA of amino acid side chains within a protein structure (8, 12). The pattern of modification (i.e. increased accessibility of some protein regions and protection of others) is thus relevant to potential structures and conformational rearrangements.
Consistency of Footprinting and Crystallography
We first analyzed HI by hydroxyl radical footprinting to demonstrate its consistency with available crystallographic data; a model hexamer was provided by Protein Data Bank code entry 4INS. Four tryptic peptides and their oxidative products, as derived from the HI monomer and hexamer, were analyzed quantitatively by MS-coupled nano-HPLC; 16 independent MS experiments (two sets for each sample) were performed. The four HI-derived peptides from the hexamer each exhibited significant decreases in oxidation rates relative to the monomer (Table 1). The largest changes from monomer to hexamer were observed within peptides B11–B22: in particular, residues TyrB16, LeuB17, and ArgB22 exhibited a 4.8-fold decrease in oxidation rate. These observations suggest that the latter reactive side chains are protected in the HI hexamer relative to HI monomer (8, 12, 22, 23) in accordance with the burial of this surface in the dimer interface (5–14-fold). Residue PheB1 (within peptides B1–B10) experienced the next largest decrease in oxidation rate on hexamer assembly (3.5-fold), whereas the rate constants for the side chains of HisB5, CysB7, and HisB10 within the same peptide dropped by 2.0-fold. The crystal structure of the HI hexamer revealed a large decrease in SA for PheB1 residue (27-fold), whereas HisB10 showed a 2.4-fold decrease. The side chains of PheB24, PheB25, and TyrB26 within segment B23–B29 each showed decreases in oxidation by 1.8-fold in the HI hexamer; ProB28 side chain within the same peptide exhibited a drop in modification rate by 2.4-fold. This segment participates in dimerization; the SAs of TyrB26 and PheB24 were reduced by 9-fold and to 0 Å2, respectively, and the ProB28 SA dropped by 5-fold in the HI hexamer relative to the HI monomer. Footprinting analysis of the monomeric and hexameric forms of HI further showed a modest decrease in modification rates (1.6-fold) for residues CysA7, ThrA8, IleA10, LeuA13, TyrA14, and TyrA19 in the A1–A21 segment. Although SAs for the rest of the oxidized side chains within peptides A1–A21 exhibited no changes upon hexamer formation, the SA for the LeuA13 side chain (as calculated from the crystal structure) decreased from 44 Å2 (monomer) to 0.7 Å2 (hexamer). In striking accord, residue LeuA13 of the A-chain was susceptible to oxidation only in the monomeric form; its modification was not detected in the hexamer. In the crystal structure of the insulin hexamer, only LeuA13 became buried within the A1–A21 region.
TABLE 1.
Peptide-specific rate constants in hydroxyl radical footprints
Rate constants for the oxidation of peptides from HI and HPI are shown as monomers and hexamers. Column 1 indicates the position and sequence of observed peptides; column 2 indicates observed oxidized residues. Columns 3 and 6 indicate SA (Å2) of the oxidized residues, as determined by crystallographic data for the HI monomer and hexamer, respectively. Columns 7 and 10 indicate SA (Å2) of the oxidized residues, as determined from NMR data for DKP-proinsulin and from computational modeling of HPI hexamer, respectively. Columns 4, 5, 8, and 9 represent respective rate constants for the oxidation of HI monomer, HI hexamer, wild-type HPI monomer, and HPI hexamer. Amino acid residues are represented with single-letter codes.
Control footprints of HI as a zinc-free monomer and T6 zinc hexamer are thus consistent with crystallographic data. Such footprinting reliably probes the changes in side-chain accessibility on the formation of interfaces. To the extent that discrepancies were observed in the quantitative extent of change on hexamer assembly (i.e. wherein changes in rate are attenuated relative to calculated decreases in surface area), such effects may reflect conformational fluctuations in solution that are dampened in a crystal lattice.
Footprint of C-domain
Oxidized residues in monomeric HPI are depicted in Fig. 1. Rates of oxidation were calculated for all HPI peptides as shown in Table 1. SA values calculated from the average of 20 NMR models of DKP-proinsulin and from a computational model of the HPI hexamer are also shown in Table 1. A computational model for an HPI monomer was also constructed using the average of 20 NMR structures of DKP-proinsulin (see “Experimental Procedures”). Side chains of PheB1, HisB5, CysB7, and HisB10 within HPI peptides B1–B10 exhibited insignificant decreases in oxidation rates (22%) relative to the corresponding insulin-derived peptide. Peptides B11–B22 of HPI similarly exhibited no changes in oxidation (within the experimental error) at sites TyrB16, LeuB17, and ArgB22. A similar correspondence of oxidation rates between HI and HPI monomers was observed at residues CysA7, ThrA8, IleA10, LeuA13, TyrA14, and TyrA19. These footprinting data were consistent with the SA values derived from the T6 crystal structure of HI, the solution structure of DKP-proinsulin, and a modeled structure of wild-type HPI.
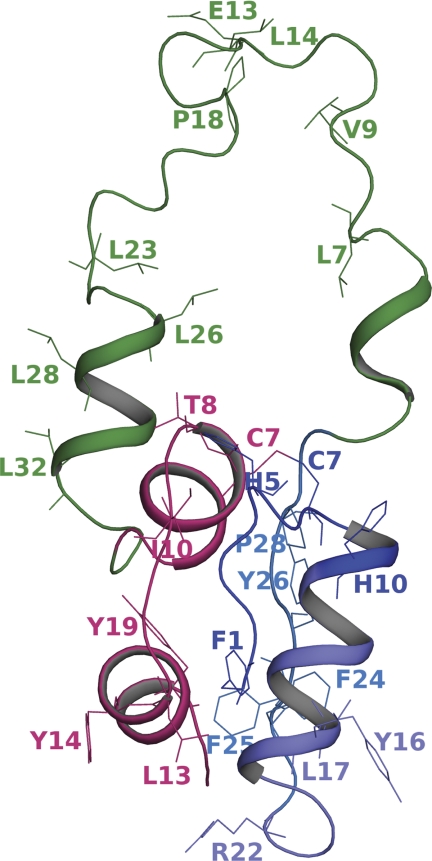
FIGURE 1.
Domain organization of proinsulin and sites of modification. The modeled structure of wild-type proinsulin depicting the amino acid residues undergoing modification in protein footprinting experiments is shown. The A-chain is in magenta, and the C-chain is in green. The three peptide fragments in the B-chain are rendered in different shades of blue.
Although the core insulin moieties of HI and HPI exhibit similar footprints as zinc-free monomers, significant changes in the oxidation rates were observed within peptides B23–B29 (i.e. near the C terminus of the B-chain in HI and near the BC junction of HPI). In particular, rates of oxidation of the side chains of PheB24, PheB25 and TyrB26 were attenuated by ∼1.5-fold relative to HI; oxidation for ProB28 was decreased by 2-fold. Because of the correlation between SA and reaction rates (8, 12, 22, 23), these findings suggest that reactive side chains in peptides B23–B29 are more buried (experienced greatest decrease in SA) in HPI than in HI. These overall results are consistent with the NMR structure of DKP-proinsulin as an engineered monomer (10). In the latter, as in our model wild-type structure, the presence of the C-domain is associated with a decrease in the SA of the side chain of TyrB26 (by 2.3- and 2.7-fold, respectively).
Local differences at sites of substitution in DKP-proinsulin might occur relative to the structure of wild-type HPI in the monomeric state. Indeed, whereas ProB29 in the NMR structure augmented SA relative to a crystallographic protomer of HI by 2.5-fold, the observed oxidation rate for ProB28 in the wild-type HPI was decreased by 2-fold relative to its oxidation rate in the HI monomer. These footprinting data thus suggest that the orientation of ProB29 in DKP-proinsulin differs from that of ProB28 in the wild-type prohormone. The decreased B28 SA value calculated based on our model structure was consistent with the protein footprinting observations. These observations suggest the utility of MS-based footprinting in assessing engineered models and in particular highlighting the appropriateness of modeling at sites of mutational differences. Because Pro can alter main-chain trajectories, it positioning relative to the BC junction (i.e. at positions B28 or B29) might be associated with changes in C-domain trajectories.
Evidence for the flexibility of the C-domain in the HPI monomer is provided by the large oxidation rate of peptides C2–C34 and C3–C34 (exceeded only by peptides A1–A21). Oxidation of 26% of the amino acid side chains in the C-domain (LeuC7, ValC9, GluC13, LeuC14, ProC18, LeuC23, LeuC26, LeuC28, and LeuC32) was detected; these probes are evenly distributed in the C-domain sequence. Our footprinting results thus suggest that C-domain residues exhibit a uniformly high solvent exposure. The slight difference in the oxidation rates between peptides C2–C34 and C3–C34 presumably resulted from minor differences in the efficiency of ionization.
Footprinting studies of HPI and HI as zinc-free monomers thus support a structural model in which the insulin moiety of HPI is similar to that of HI. The surface of peptides B23–B29 is accessible to hydroxyl radical modification in HI but protected in HPI; this difference defines the footprint of the flexible C-domain on the B-domain of HPI. No such shadow was detected on the A-domain surface.
Assembly of HPI Hexamer
A model for a T6-like zinc-stabilized HPI hexamer was constructed based on the classical T6 insulin hexamer using NMR constraints obtained in studies of DKP-insulin (see “Experimental Procedures”) as depicted in Fig. 2. MS-based footprinting of the HPI hexamer was undertaken in relation to both the HPI monomer and the HI hexamer; rates of oxidation are shown in Table 1. We observed that respective oxidation rates decreased for the side chains of PheB1 by 1.9-fold and for the side chains of HisB5, CysB7, and HisB10 by 1.5-fold within the B1–B10 segment. These data indicate that the PheB1 exhibited the largest change in SA within this peptide. These findings are consistent with the SA values of these side chains (as calculated from the crystal structure for the HI hexamer and our modeled structure of the HPI hexamer), which also showing the largest drop in the SA for PheB1 in this segment (Table 1). In the HI hexamer, however, respective modification rates were decreased for the side chains of B1 by 3.5-fold and for the side chains of B5, B7, and B10 by 2-fold. We thus observed greater relative N-terminal protection upon hexamer assembly of HI than of HPI.
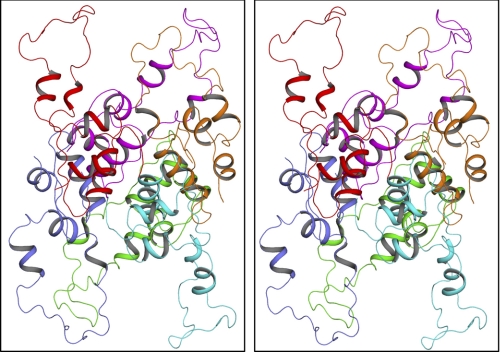
FIGURE 2.
Molecular modeling of the proinsulin hexamer. The stereo view (wall-eyed representation) of the HPI hexamer with different chains shown as a graphic in different colors is displayed. The model was reconstructed based on the crystal coordinates of insulin hexamer. The model shows that the C-domain in each protomer is absolutely exposed in accord with experimentally observed radiolytic modifications of various amino acid residues in this region.
Peptides B11–B22 of the HPI hexamer exhibited decreased oxidation by 4.3-fold relative to the HPI monomer. The side chains of TyrB16, LeuB17, and ArgB22 (corresponding to the central α-helix and succeeding β-turn of the B-domain) were identified as sites of oxidation. The SA of TyrB16 and LeuB17 showed a similar decrease upon hexamer assembly of both HI and HPI in accordance with our footprinting data. In either HPI or HI, residues PheB24, PheB25, and TyrB26 in C-terminal peptides B23–B29 showed similar decreases in oxidation rate (1.6- and 1.8-fold, respectively). The SA of PheB24 and TyrB26 in such hexamers is negligible. Also, on hexamer assembly, ProB28 (which stabilizes the dimer interfaces within the HI hexamer via hydrophobic interactions with GlyB20, GluB21 and GlyB23 (24, 25)) exhibited a decrease in oxidation rate of 2.4- and 2.0-fold for HI and HPI hexamers, respectively. An intriguing discrepancy was inferred at position B28 as its SA, calculated from our HPI models, exhibited a less marked decrease in the HPI hexamer relative to that predicted by the HI T6 crystal structure. This disagreement may suggest either that the HI crystal structure underestimates C-terminal conformational excursions in the B-chain and/or that ProB28 (adjacent to the BC-domain junction) is indeed more exposed in the HPI hexamer then in the HI hexamer. The latter possibility would be of biological interest in relation to the mechanism of prohormone processing by prohormone convertases.
Residues A1–A21 within the A-chain exhibited no substantial changes in relative rates of oxidation on assembly of the HPI hexamer. Such small assembly-dependent changes are in accordance with the predominant role of the B-chain at component dimer and trimer interfaces. Interestingly, in comparison with peptides derived from HI and HPI monomers, the largest oxidation rate occurred in the A-chain. These observations suggest that at least some of the oxidated side chains (CysA7, ThrA8, IleA10, LeuA13, TyrA14, and TyrA19) are highly accessible to the solvent even in the folded monomers. Such exposure is consistent with the NMR features of DKP-proinsulin and with modeling of the wild-type HPI, in particular the flexibility of the N-terminal A-domain segment implied by rapid amide-proton exchange and attenuated helix-related 13Cα and 13Cβ chemical shifts (10). Peptides C2–C34 and C3–C34 of the HPI C-domain also showed no assembly-dependent changes in the oxidation rates. Specifically, the side chains of LeuC7, ValC9, GluC13, LeuC14, ProC18, LeuC23, LeuC26, LeuC28, and LeuC32 (which cover 26% of the peptide sequence and are evenly distributed in the C-domain) were detected to be oxidized in both forms of HPI. These residues exhibited the next largest increase in the oxidation rate after the A-chain peptide. In our model, the entire C-domain is highly accessible to solvent in both the monomer and the hexamer. In light of the correspondence between the core footprints of HI and HPI hexamers (see above) and given the comparable oxidation rates and SAs of the C-domain of both HPI forms, our findings together suggest that the C-domain must be located on the outer surface of the monomer in such a way that it does not interfere with the formation of the hexamer core. This conclusion is in accordance with the location of the AC and BC junctions on the exterior of the HI monomer (6); protrusion of the C-domain from this surface would also rationalize the failure of HPI hexamers to form crystal lattices similar to those of HI hexamers.
Overall, the present radiolytic footprinting studies are consistent with the computational model of the HPI hexamer. We therefore suggest that the HPI hexamer is similar to HI in its insulin-like core and mode of hexamer assembly, whereas the six C-domains project from the outer surface. The present study suggests the broad applicability of synchrotron-based MS footprinting techniques and molecular modeling in studies of proteins and protein assemblies that (like proinsulin) are refractory to crystallization.
Concluding Remarks
Dominant mutations in the insulin gene causing permanent neonatal DM occur in each region of preproinsulin (signal peptide and B, C, and A) (2, 4, 26). These mutations fall into two classes. First, imbalance in Cys pairing can arise due to the introduction or loss of a Cys codon, leading to the intermolecular disulfide bridging and aggregation (5, 27). Second, non-Cys-related mutations (mapping predominantly in the B-domain) can impair native pairing, presumably leading to aggregation of partially folded species (28–34). We anticipate that footprinting studies of DM-associated HPI variants in combination with computational modeling, as exemplified here, can provide structural insights into such toxic aggregates. Of particular utility may be the ability to monitor the oxidation rate of cystine A7–B7 as this bridge is nearly fully exposed at the surface of the HPI in its native state. Similarly, HisB5 (a hot spot for clinical mutations of the second class) provides a key footprinting probe associated with a critical long range hydrogen bond recently shown to facilitate native disulfide pairing (3, 35). Aberrant burial of protein surfaces promises to enable modeling of misfolded proteins that are otherwise refractory to x-ray crystallography or NMR. The set of oxidation rates obtained in the present study provides a database for the design of such translational studies.
Hydroxyl radical footprinting provides a general probe of protein structure and its changes on ligand binding and assembly (12, 36–39). Because site-specific surface accessibilities can in principle be inferred from analysis of reactive aliphatic and aromatic side chains, potential models of hydrophobic interfaces may be constrained even for the irregular aggregates characteristic of pathological misfolding. The mutant proinsulin syndrome associated with DM promises to provide a molecular laboratory for the development of this approach.
Acknowledgments
We thank Dr. N. F. Phillips for assistance with sample preparation and Dr. S. Gupta for assistance with sample irradiation at the Brookhaven National Laboratory (BNL). The National Synchrotron Light Source is supported by the Department of Energy under Contract DE-AC02-98CH10886.
This work was supported, in whole or in part, by National Institutes of Health Grants P30-EB-09998 and R01-EB-09688 from the National Institute for Biomedical Imaging and Bioengineering (to M. R. C.) and by National Institutes of Health Grants DK069764 and DK40949 (to M. A. W.) through the NIDDK.
The presence of the C-domain in HPI and other vertebrate proinsulins prevents crystallization under conditions that promote the crystallization of insulin, presumably due to steric perturbation at hexamer-hexamer interfaces (1).
DKP-insulin and DKP-proinsulin are analogs containing amino acid substitutions HisB10 → Asp, ProB28 → Lys, and LysB29 → Pro. The engineered monomer contained two mutations in the dimer interface (ProB28 → Lys and LysB29 → Pro) and one substitution in the trimer interface and zinc coordination site (HisB10 → Asp); the structural rationale for its use as an NMR model has been described (10, 15).
Because the reported heteronuclear NMR analysis of DKP-proinsulin (10) was unaccompanied by analysis of residual dipolar couplings, no orientational information was obtained regarding peptide units in the C-domain.
Residues 1–30 in HPI correspond to B-chain residues B1–B30 in HI; similarly, residues 66–86 correspond to A-chain residues A1–A21. Residues 31–65 (C-domain) may also be designated C1–C35.
- DM
- diabetes mellitus
- HI
- human insulin
- HPI
- human proinsulin
- SA
- solvent accessibility.
REFERENCES
- 1. Baker E. N., Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. M., Hubbard R. E., Isaacs N. W., Reynolds C. D., et al. (1988) Philos. Trans. R. Soc. Lond. B. Biol. Sci. 319, 369–456 [DOI] [PubMed] [Google Scholar]
- 2. Støy J., Edghill E. L., Flanagan S. E., Ye H., Paz V. P., Pluzhnikov A., Below J. E., Hayes M. G., Cox N. J., Lipkind G. M., Lipton R. B., Greeley S. A., Patch A. M., Ellard S., Steiner D. F., Hattersley A. T., Philipson L. H., Bell G. I. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15040–15044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edghill E. L., Flanagan S. E., Patch A. M., Boustred C., Parrish A., Shields B., Shepherd M. H., Hussain K., Kapoor R. R., Malecki M., MacDonald M. J., Støy J., Steiner D. F., Philipson L. H., Bell G. I., Hattersley A. T., Ellard S. (2008) Diabetes 57, 1034–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colombo C., Porzio O., Liu M., Massa O., Vasta M., Salardi S., Beccaria L., Monciotti C., Toni S., Pedersen O., Hansen T., Federici L., Pesavento R., Cadario F., Federici G., Ghirri P., Arvan P., Iafusco D., Barbetti F. (2008) J. Clin. Invest. 118, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu M., Hodish I., Rhodes C. J., Arvan P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15841–15846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Adams M. J., Haas D. J., Jeffery B. A., McPherson A., Jr., Mermall H. L., Rossmann M. G., Schevitz R. W., Wonacott A. J. (1969) J. Mol. Biol. 41, 159–188 [DOI] [PubMed] [Google Scholar]
- 7. Maleknia S. D., Brenowitz M., Chance M. R. (1999) Anal. Chem. 71, 3965–3973 [DOI] [PubMed] [Google Scholar]
- 8. Takamoto K., Chance M. R. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 [DOI] [PubMed] [Google Scholar]
- 9. Aguilar C. F., Cronin N. B., Badasso M., Dreyer T., Newman M. P., Cooper J. B., Hoover D. J., Wood S. P., Johnson M. S., Blundell T. L. (1997) J. Mol. Biol. 267, 899–915 [DOI] [PubMed] [Google Scholar]
- 10. Yang Y., Hua Q. X., Liu J., Shimizu E. H., Choquette M. H., Mackin R. B., Weiss M. A. (2010) J. Biol. Chem. 285, 7847–7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steiner D. F. (2004) Exp. Diabesity Res. 5, 7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kiselar J. G., Maleknia S. D., Sullivan M., Downard K. M., Chance M. R. (2002) Int. J. Radiat. Biol. 78, 101–114 [DOI] [PubMed] [Google Scholar]
- 13. Hua Q. X., Hu S. Q., Jia W., Chu Y. C., Burke G. T., Wang S. H., Wang R. Y., Katsoyannis P. G., Weiss M. A. (1998) J. Mol. Biol. 277, 103–118 [DOI] [PubMed] [Google Scholar]
- 14. Weiss M. A. (2009) J. Biol. Chem. 284, 19159–19163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weiss M. A., Hua Q. X., Lynch C. S., Frank B. H., Shoelson S. E. (1991) Biochemistry 30, 7373–7389 [DOI] [PubMed] [Google Scholar]
- 16. Gupta S., Sullivan M., Toomey J., Kiselar J., Chance M. R. (2007) J. Synchrotron Radiat. 14, 233–243 [DOI] [PubMed] [Google Scholar]
- 17. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 19. Kleywegt G. J. (1996) Acta Crystallogr. D Biol. Crystallogr. 52, 842–857 [DOI] [PubMed] [Google Scholar]
- 20. Dong J., Wan Z., Popov M., Carey P. R., Weiss M. A. (2003) J. Mol. Biol. 330, 431–442 [DOI] [PubMed] [Google Scholar]
- 21. Davies MJ, Dean R. T. (eds) (1997) Radical-mediated Protein Oxidation: From Chemistry to Medicine, Oxford University Press USA, New York, NY [Google Scholar]
- 22. Guan J. Q., Vorobiev S., Almo S. C., Chance M. R. (2002) Biochemistry 41, 5765–5775 [DOI] [PubMed] [Google Scholar]
- 23. Xu G., Chance M. R. (2007) Chem. Rev. 107, 3514–3543 [DOI] [PubMed] [Google Scholar]
- 24. Ye J., Chang W., Liang D. (2001) Biochim. Biophys. Acta 1547, 18–25 [DOI] [PubMed] [Google Scholar]
- 25. Zoete V., Meuwly M., Karplus M. (2004) J. Mol. Biol. 342, 913–929 [DOI] [PubMed] [Google Scholar]
- 26. Hayes M. G., Pluzhnikov A., Miyake K., Sun Y., Ng M. C., Roe C. A., Below J. E., Nicolae R. I., Konkashbaev A., Bell G. I., Cox N. J., Hanis C. L. (2007) Diabetes 56, 3033–3044 [DOI] [PubMed] [Google Scholar]
- 27. Izumi T., Yokota-Hashimoto H., Zhao S., Wang J., Halban P. A., Takeuchi T. (2003) Diabetes 52, 409–416 [DOI] [PubMed] [Google Scholar]
- 28. Weiss M. A., Hua Q. X., Jia W., Chu Y. C., Wang R. Y., Katsoyannis P. G. (2000) Biochemistry 39, 15429–15440 [DOI] [PubMed] [Google Scholar]
- 29. Nakagawa S. H., Hua Q. X., Hu S. Q., Jia W., Wang S., Katsoyannis P. G., Weiss M. A. (2006) J. Biol. Chem. 281, 22386–22396 [DOI] [PubMed] [Google Scholar]
- 30. Hua Q. X., Nakagawa S. H., Jia W., Hu S. Q., Chu Y. C., Katsoyannis P. G., Weiss M. A. (2001) Biochemistry 40, 12299–12311 [DOI] [PubMed] [Google Scholar]
- 31. Chu Y. C., Wang R. Y., Burke G. T., Chanley J. D., Katsoyannis P. G. (1987) Biochemistry 26, 6975–6979 [DOI] [PubMed] [Google Scholar]
- 32. Kristensen C., Kjeldsen T., Wiberg F. C., Schäffer L., Hach M., Havelund S., Bass J., Steiner D. F., Andersen A. S. (1997) J. Biol. Chem. 272, 12978–12983 [DOI] [PubMed] [Google Scholar]
- 33. Molven A., Ringdal M., Nordbø A. M., Raeder H., Støy J., Lipkind G. M., Steiner D. F., Philipson L. H., Bergmann I., Aarskog D., Undlien D. E., Joner G., Søvik O., Bell G. I., Njølstad P. R. (2008) Diabetes 57, 1131–1135 [DOI] [PubMed] [Google Scholar]
- 34. Zoete V., Meuwly M. (2006) J. Comput. Chem. 27, 1843–1857 [DOI] [PubMed] [Google Scholar]
- 35. Hua Q. X., Liu M., Hu S. Q., Jia W., Arvan P., Weiss M. A. (2006) J. Biol. Chem. 281, 24889–24899 [DOI] [PubMed] [Google Scholar]
- 36. Takamoto K., Kamal J. K., Chance M. R. (2007) Structure 15, 39–51 [DOI] [PubMed] [Google Scholar]
- 37. Kiselar J. G., Janmey P. A., Almo S. C., Chance M. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3942–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kiselar J. G., Mahaffy R., Pollard T. D., Almo S. C., Chance M. R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1552–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gupta S., Cheng H., Mollah A. K., Jamison E., Morris S., Chance M. R., Khrapunov S., Brenowitz M. (2007) Biochemistry 46, 9886–9898 [DOI] [PubMed] [Google Scholar]