Background: The TNF gene is regulated in a cell type-specific fashion.
Results: A TNF locus site that interacts with the nuclear matrix and proteins that affect DNA torsional stress is selectively accessible in monocytic cells.
Conclusion: Monocyte-specific accessibility of the site provides a mechanism of cell type-specific TNF gene regulation.
Significance: Cell type-specific chromatin organization elucidates control of TNF regulation and dysregulation.
Keywords: DNA Topoisomerase, Gene Transcription, Monocytes, Nuclear Matrix, Tumor Necrosis Factor (TNF)
Abstract
Regulation of TNF gene expression is cell type- and stimulus-specific. We have previously identified highly conserved noncoding regulatory elements within DNase I-hypersensitive sites (HSS) located 9 kb upstream (HSS−9) and 3 kb downstream (HSS+3) of the TNF gene, which play an important role in the transcriptional regulation of TNF in T cells. They act as enhancers and interact with the TNF promoter and with each other, generating a higher order chromatin structure. Here, we report a novel monocyte-specific AT-rich DNase I-hypersensitive element located 7 kb upstream of the TNF gene (HSS−7), which serves as a matrix attachment region in monocytes. We show that HSS−7 associates with topoisomerase IIα (Top2) in vivo and that induction of endogenous TNF mRNA expression is suppressed by etoposide, a Top2 inhibitor. Moreover, Top2 binds to and cleaves HSS−7 in in vitro analysis. Thus, HSS−7, which is selectively accessible in monocytes, can tether the TNF locus to the nuclear matrix via matrix attachment region formation, potentially promoting TNF gene expression by acting as a Top2 substrate.
Introduction
The TNF gene encodes a cytokine whose protein product has been implicated in a variety of immunopathological processes (1). TNF, an immediate-early response gene, is induced by a variety of stimuli in a cell type- and inducer-specific manner (2–7). The gene thus presents an important model system in which to dissect mechanisms of inducible eukaryotic gene transcription in general. Previously, we have shown that cell type- and inducer-specific enhanceosomes are recruited to the proximal TNF promoter and that this region of the upstream flanking sequence is sufficient for TNF transcription (2–7).
More recently, we identified two highly conserved, noncoding, distal regulatory elements in the TNF locus, which undergo activation-dependent intrachromosomal interactions in T cells and enhance TNF gene transcription (8). First identified by DNase I hypersensitivity assay (DHA),3 these elements, which are located 9 kb upstream (HSS−9) and 3 kb downstream (HSS+3) of the TNF transcription start site, bind NF-AT and NF-κB proteins and function as enhancers of TNF transcription in T cells. Furthermore, upon T cell activation, HSS−9 and HSS+3 interact with the TNF promoter and with each other, generating a higher order chromatin structure that can sequester the TNF gene from the neighboring and closely positioned LT-α and LT-β genes (8). These intrachromosomal interactions involve bringing NF-AT-containing enhancer complexes into close proximity to each other and, by circularizing the TNF gene, creating a configuration favoring reinitiation of TNF transcription (8, 9).
Here, we report the discovery of a novel cell type-specific DNase I-hypersensitive site 7 kb upstream of the TNF gene that is evident in murine and human monocytes but is not accessible to DNase I digestion in T cells. This element, HSS−7, does not function as a classical transcriptional enhancer but contains a highly conserved AT-rich noncoding region that binds acetylated histones, topoisomerase IIα (Top2), and HMGA1a (high mobility group protein A1a) in vitro and in vivo and that acts as a substrate for Top2 cleavage in vitro. Consistent with these observations, HSS−7 functions as a nuclear MAR in monocytes, and activation of TNF gene expression is suppressed by etoposide, a Top2 inhibitor. Thus, a region of the TNF locus that is accessible in monocytes tethers the locus to the nuclear matrix. These data demonstrate cell type-specific chromatin reorganization of the TNF locus and are consistent with sequestration of the TNF locus in a gene expression domain in monocytic cells. Furthermore, these data suggest the hypothesis that the action of Top2 at HSS−7 may promote TNF gene transcription by relieving torsional stress induced by transcription in the vicinity of the TNF locus, which would provide an additional, novel level of TNF gene regulation.
EXPERIMENTAL PROCEDURES
DHA, Luciferase, ChIP, and RNase Protection Assays
Previously described protocols were used to perform DHAs (8, 10–12), luciferase assays, and ChIP assays (2, 5, 6). The TNF-Luc-HSS−7 reporter, based on the TNF-Luc-HSS−9 and TNF-Luc-HSS+3 vectors used previously (8), was prepared by PCR amplification of a 308-bp region encompassing HSS−7 from Balb/c genomic DNA using primers 5′-GTGTCGACATACTCCTGATGGCTGTTA-3′ and 5′-GTGTCGACTTGGGCCTGAGAACA-3′ (with SalI sites underlined), subcloning the fragment into pCR®-XL-TOPO® (Invitrogen) for sequencing, and inserting the SalI digestion product from this vector into the SalI site of the pGL3-TNF-Luc reporter (8). Antibodies to unacetylated histone H3, unacetylated histone H4, acetylated histone H3 (Lys-9/Lys-14), acetylated histone H4 (Lys-8), topoisomerase IIα, HMG-I/HMG-Y, and isotype controls (Santa Cruz Biotechnology) were used for ChIP assays. Cytokine mRNA levels were analyzed by multiprobe RPA. RNA was hybridized overnight against 32P-labeled antisense RNA probes for TNF, LT-α, LT-β, and L32, which had been synthesized in vitro from separate templates according to standard techniques (2, 13, 14). J774 cells were treated with etoposide (Sigma) at increasing concentrations of 0.1, 0.2, and 0.4 mg/ml.
MAR Assay
The MAR assay was adapted from Refs. 15 and 16. Briefly, nuclear matrices were prepared by lysing J774 cells with buffer containing 0.5% Triton X-100, 10 mm PIPES (pH 6.8), 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 1 mm EGTA, 1 mm PMSF, and protease inhibitor mixture. The matrices were digested overnight with EcoRI, BamHI, HindIII, PstI, and NspI. The digested preparations were spun down (to pellet the MAR-containing matrices) and digested with proteinase K. The DNA was phenol/chloroform-extracted and ethanol-precipitated prior to PCR with primers specific for HSS−7 (underlined in Fig. 6E) and HSS−9 region 1 (8). PCR products were resolved on a 1% Tris acetate/EDTA-agarose gel and stained with EtBr.
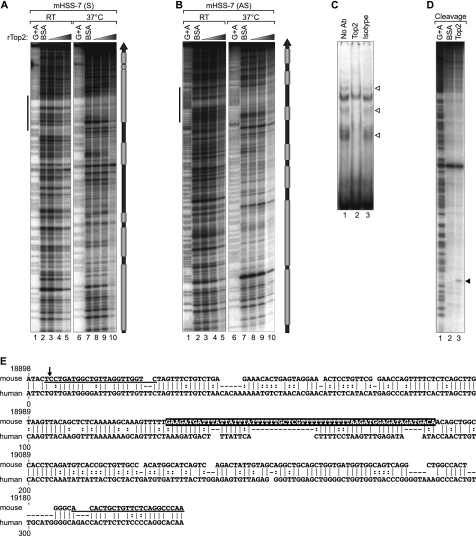
FIGURE 6.
HSS−7 is a substrate for Top2 in vitro. A and B, quantitative DNase I footprinting analysis of Top2 binding to HSS−7. 308-bp double-stranded DNA probes containing murine HSS−7 radiolabeled on the sense (mHSS−7 (S); A) and antisense (mHSS−7 (AS); B) strands were incubated with increasing concentrations of recombinant human Top2 protein (rTop2) at room temperature (RT) or at 37 °C. DNase I-digested probes were resolved by 8% denaturing PAGE and subjected to autoradiography. Gray boxes to the right of A and B denote Top2-binding positions. C, EMSA performed by incubating a 64-bp HSS−7 probe (sequence highlighted in E, with its position indicated by vertical bars to the left of A and B) with nuclear extracts from LPS-stimulated J774 cells. The EMSA was performed with an isotype control or anti-Top2 antibody; disrupted Top2 complexes are indicated by arrowheads. D, concentrated and purified recombinant Drosophila Top2 (lane 3) was utilized in an in vitro cleavage reaction with 308-bp radiolabeled DNA containing the murine HSS−7 region and resolved by 8% denaturing PAGE. E, PipMaker alignment of mouse (308 bp) and human (288 bp) HSS−7 sequences. Dashes represent gaps in the alignment, vertical lines represent aligned identical nucleotides, and dots indicate a purine or pyrimidine change in the sequence. The numbers at the top of the alignment correspond to positions of the mouse sequence in the TNF locus as in Fig. 2. The sequences of primers used in MAR and ChIP assays are underlined.
Top2 Cleavage Assay
Top2 cleavage of HSS−7 was adapted from Refs. 17 and 18 and performed utilizing purified Drosophila topoisomerase II (a kind gift from Dr. Tao-Shih Hsieh, Department of Biochemistry, Duke University). Briefly, a 308-bp probe spanning the murine HSS−7 region was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase. The probe was incubated at 37 °C with buffer containing 50 mm Tris-HCl, 150 mm NaCl, 10 mm MgCl2, 5 mm DTT, 30 μg/ml BSA, 2 mm ATP, and either BSA or purified topoisomerase protein. After a 10-min incubation, SDS was added to a concentration of 1%, and the samples were incubated for another 20 min. The reactions were stopped by the addition of Sarkosyl to 0.8%, bromphenol blue to 0.02%, and glycerol to 4%. The ethanol-precipitated samples were resolved on an 8% denaturing polyacrylamide gel.
DNase I Footprinting and EMSA
DNase I footprinting of HSS−7 was performed with recombinant human topoisomerase IIα (a kind gift from Dr. Caroline Austin, NuVenture/Institute for Cell and Molecular Biosciences, University of Newcastle). Briefly, a 308-bp fragment of the mouse HSS−7 region was labeled with [γ-32P]ATP at one end using T4 polynucleotide kinase and incubated with increasing amounts of recombinant protein at room temperature or at 37 °C as shown in Fig. 6. Note that probes incubated at 37 °C were mildly denatured (70 °C for 2 min) and slowly cooled down to 37 °C before mixing them with the protein. After a 30-min incubation, the samples were digested with 0.3 units of DNase I for exactly 1 min, purified, resolved on an 8% denaturing polyacrylamide gel, and exposed overnight to film as described previously (19). EMSAs were performed as described previously (19) using [γ-32P]ATP end-labeled oligonucleotide (5′-GAAGATGATTTATTATTTATTTTTTGCTCGTTTTTTTTTTTTTTTAAGATGGAGATAGATGACA-3′), and J774 nuclear extracts were stimulated with LPS for 3 h. For EMSAs, the anti-topoisomerase IIα antibody (Abcam) was preincubated with nuclear extracts for 30 min before adding the labeled probe.
RESULTS
DNase I Cleavage Pattern at the TNF Genomic Locus in Monocytes Is Distinct from That in T Cells
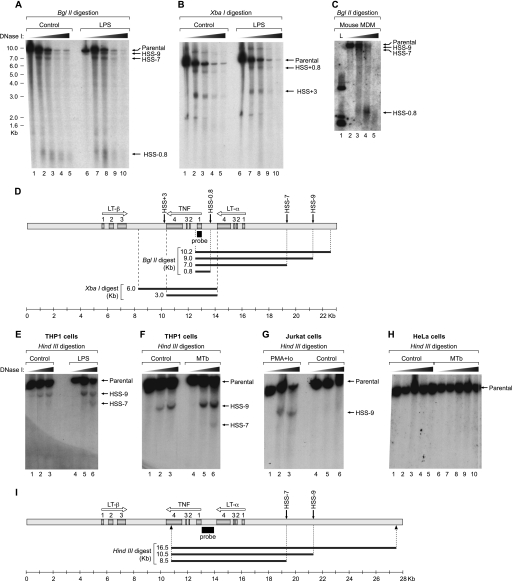
To characterize the DNase I hypersensitivity profile of the TNF locus (23.5 kb) in monocytes, we first prepared DNA from the mouse monocytic cell line J774 in the presence and absence of LPS stimulation and digested the gene-dense TNF locus with BglII or XbaI. We observed all three hypersensitive sites previously identified in T cells, HSS−0.8 (corresponding to the TNF promoter), HSS−9, and HSS+3, in the monocytic cells (Fig. 1, A, B, and D) (8). Strikingly, however, we also discovered a new DNase I-hypersensitive site in monocytes located 7 kb upstream of the TNF gene (HSS−7) that was not accessible to DNase I digestion in murine T cells (8). We confirmed that HSS−7 is present in primary murine bone marrow-derived macrophages (Fig. 1C). Consistent with these findings in murine cells, HSS−7 was clearly evident in a human monocytic cell line (THP-1) (Fig. 1, D and E) but not in a human T cell line (Jurkat) (Fig. 1F). As expected, it also was not evident in human HeLa cells (Fig. 1H), which do not produce TNF (2).
FIGURE 1.
DNase I hypersensitivity profile of the TNF locus in monocytes. Nuclei from unstimulated and LPS- or Mycobacterium tuberculosis (MTb)-stimulated cells were digested with increasing concentrations of DNase I. Purified DNAs from murine J774 cells (A and B), from mouse bone marrow-derived macrophages (MDM; C), and from THP-1 (E and F), Jurkat (G), and HeLa (H) human cell lines were digested with BglII (A and C), XbaI (B) or HindIII (E–H); resolved on a 0.9% agarose gel; and subjected to Southern blotting using a 32P-labeled probe (black boxes) spanning the first exon of TNF in murine cells (D) or the full TNF promoter in human cells (I). The TNF, LT-α, and LT-β genes (arrows) and exons (gray boxes) in the TNF locus and the projected restriction fragments from the DHAs are represented in D and I. PMA/Io, phorbol 12-myristate 13-acetate/ionomycin. L, 1 kb ladder, Invitrogen.
Computational Analysis of HSS−7 Reveals MAR Potential
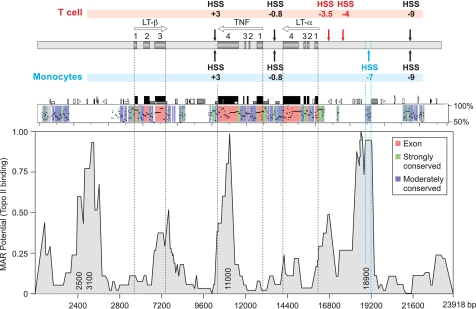
Computational analysis of HSS−7 using PipMaker (20) showed that HSS−7 coincided with a highly conserved noncoding region (Fig. 2). Notably, the previously identified HSS−9, HSS+3, and HSS−0.8 (the TNF promoter) (8, 21, 22) sites are also all located within highly conserved noncoding regions of the TNF locus (Fig. 2). However, unlike any of these regions (and the TNF locus as a whole), which have a balanced AT/GC content, HSS−7 has a particularly high AT content (55 and 59% in mouse and human, respectively) (see Fig. 6E). A total of four SNPs are present in the region corresponding to HSS−7 in the human TNF locus, only one of which, a single AT base pair deletion, occurs within an AT-rich sequence (23). Similarly, in the regions corresponding to murine HSS+3 and HSS−9, there are one and three SNPs, respectively, none of which disrupt transcription factor-binding sites (23).
FIGURE 2.
HSS−7 sequence conservation and MAR potential. Upper, diagram of the TNF, LT-α, and LT-β genes in the TNF locus relative to hypersensitive sites present in T cells (above) and monocytes (below), with genes and exons labeled as described in the legend to Fig. 1. Middle, PipMaker (20) alignment of the murine and human TNF loci showing percent identity to the human sequence (y axis), exons (red), and strongly (green) and moderately (blue) conserved noncoding regions. Lower, MAR potential of the aligned 23.5-kb fragment shown in PipMaker (with nucleotide positions on the x axis) as analyzed by MAR-Wiz. MAR potential as a function of predicted binding to Top2 (threshold value = 0.6) is shown on the y axis. Numbers inside the peaks indicate the position at which the highest MAR potential in the area is located. (1 is the highest possible value for the MAR potential.) MAR-Wiz peaks located in non-conserved regions (2500 and 3100 nucleotides) or in the TNF 3′-UTR (11,000 nucleotides) or below the threshold level (300, 7200, and 16,800 nucleotides) were not considered putative MAR sites. Topo II, topoisomerase IIα.
Unlike HSS+3 and HSS−9, which bind to NF-AT and NF-κB proteins (8), HSS−7 contains no obvious consensus transcription factor-binding sites. Consistent with this observation, we found that HSS−7 did not enhance but rather repressed the levels of basal and LPS-induced transcription mediated by the TNF promoter in luciferase assays in J774 cells (supplemental Fig. 1). Thus, in contrast to our findings in T cells, where HSS−9 and HSS+3 are able to act as NF-AT-dependent enhancers (8), HSS−7 does not function as an enhancer in monocytes.
Notably, however, MARs are known to have high AT content (24) and to mediate the binding of DNA to the nuclear matrix, which is the protein scaffold that remains after the nuclear membrane and soluble proteins have been extracted from nuclei (25). Furthermore, MARs define gene expression domains and serve as essential locus control regions (26, 27). We thus next probed the TNF locus utilizing MAR-Wiz (FutureSoft) to determine whether HSS−7 has MAR potential. Strikingly, this analysis revealed peaks that coincide with HSS−7, consistent with its having high MAR potential, whereas HSS−9 and HSS+3 did not display MAR potential (Fig. 2). Taken together, the presence of evolutionary sequence conservation, the prediction data generated by MAR-Wiz, and the cell type-specific accessibility to DNase cleavage suggest that HSS−7 functions as a MAR of the TNF locus in monocytic cells.
HSS−7 Is a MAR
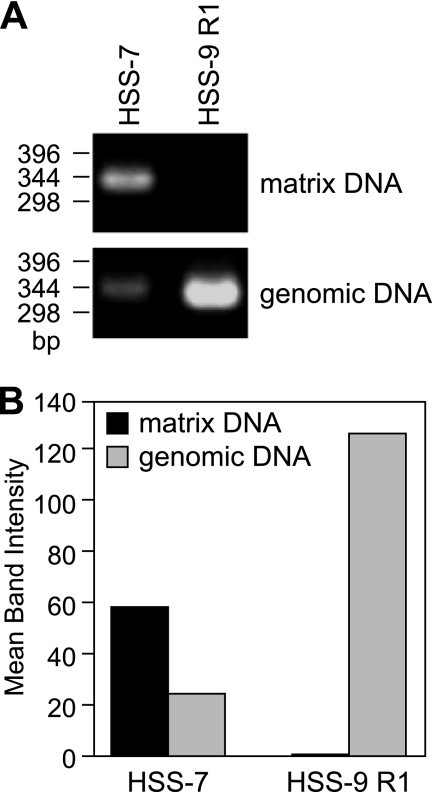
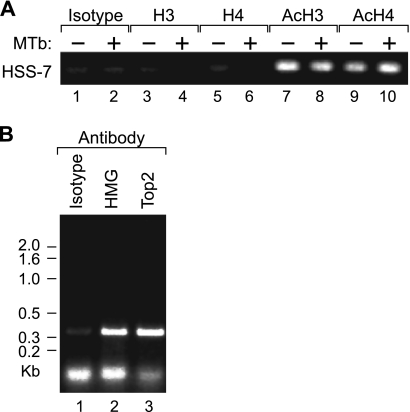
We thus next prepared nuclear matrices (15, 16) from purified J774 nuclei and examined whether HSS−7 is enriched in the nuclear matrix fraction of monocytic cells. We digested the matrices with a combination of enzymes (EcoRI, BamHI, HindIII, PstI, and NspI) that cleave DNA at positions immediately upstream and downstream of HSS−7, isolated DNA by proteinase K treatment, and then PCR-amplified HSS−7 with specific primers. Given that HSS−9 and HSS−7 are both present in monocytic cells (Fig. 1), HSS−9 was used as an experimental control. We also cleaved DNA at positions upstream and downstream of HSS−9 and used specific primers for HSS−9. As shown in Fig. 3, consistent with the MAR-Wiz analysis, HSS−7 was highly enriched in the nuclear matrix fraction of J774 cells, whereas HSS−9 could not be amplified from the same matrix fraction (Fig. 3).
FIGURE 3.
HSS−7 associates with the nuclear matrix. A, nuclear matrices were isolated from J774 cells and digested with EcoRI, BamHI, HindIII, PstI, and NspI. The matrix fraction was then separated from the non-matrix-associated fraction by centrifugation. Purified matrix DNA and genomic DNA were amplified by PCR utilizing primers specific for HSS−7 and HSS−9 region 1. B, densitometric analysis of the bands is shown.
Acetylated Histones, Topoisomerase IIα, and HMGA1a Are Recruited to HSS−7 in Vivo
Given that HSS−7 is accessible to DNase I cleavage, we speculated that it may have an impact upon chromatin organization of the TNF locus through favoring a constitutively open DNA conformation and the recruitment of acetylated histones. To test whether HSS−7 interacts with these proteins in vivo, we next performed a ChIP assay using formaldehyde-cross-linked chromatin from J774 monocytes with antibodies specific for unacetylated histone H3, unacetylated histone H4, acetylated histone H3, and acetylated histone H4 and found that HSS−7 was immunoprecipitated by both the anti-acetylated histone 3 and anti-acetylated histone 4 antibodies in unstimulated and stimulated J774 cells (Fig. 4A). We note that these results should be taken as a test for the presence of acetylated histones H3 and H4 in the vicinity of HSS−7, similar to previous studies of gene regulatory regions (for example, see Refs. 28 and 29), rather than a quantitative assessment of histone acetylation. AT-rich DNA like HSS−7 can also regulate transcription by recruiting the structural transcription factor family of HMG proteins and the DNA-unwinding protein Top2. Both of these proteins can in turn also associate with the nuclear matrix (30). As shown in Fig. 4B, HMGA1a, also known as HMG-I/HMG-Y, and Top2 are also recruited to HSS−7 in vivo in J774 murine monocytic cells.
FIGURE 4.
HSS−7 associates with acetylated histones, HMGA1a, and Top2 in vivo. ChIP assays were performed using antibodies prepared against peptides comprising the N-terminal tails of histone H3 acetylated at Lys-9 and Lys-14 (anti-acetylated histone H3), histone H4 acetylated at Lys-8 (anti-acetylated H4), or histones H3 and H4 in an unmodified state (anti-unacetylated histones H3 and H4, respectively). A, ChIP assay with unstimulated and Mycobacterium tuberculosis (MTb)-stimulated J774 cells using the anti-acetylated histone H3 (AcH3) or H4 (AcH4) antibody. B, ChIP assay with unstimulated J774 cells and the anti-HMGA1a or anti-Top2 antibody. Immunoprecipitation with the anti-unacetylated histone H3 (H3) or H4 (H4) antibody (A) and an isotype control antibody (A and B) served as negative controls. DNA was amplified with primers flanking HSS−7 (underlined in Fig. 6E).
Inhibition of Top2 Suppresses Induction of Endogenous TNF Gene Expression
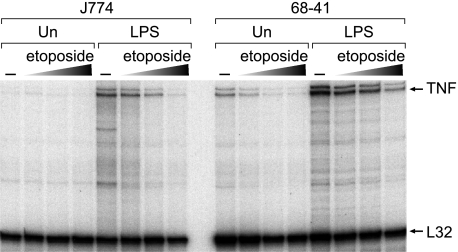
To characterize further the impact of Top2 upon TNF expression in vivo, we performed RPAs in J774 cells and, for comparison, in the murine 68-41 T cell line (8) with a Top2 inhibitor and chemotherapy agent, etoposide. Importantly, etoposide is not a general inhibitor of inducible gene expression, as it has been shown to activate or repress transcription in different contexts (31–36). In RPAs, we observed that following LPS treatment of J774 cells and phorbol 12-myristate 13-acetate/ionomycin treatment of 68-41 cells, expression of TNF mRNA was strongly inhibited by etoposide (Fig. 5). In contrast, etoposide did not affect mRNA expression of the ribosomal protein L32 housekeeping gene (Fig. 5) in either cell line. These data are consistent with Top2 playing a role in TNF transcription in monocytes, with a potential functional interaction between Top2 and HSS−7. These data also indicate that, in addition to the mechanism of activation-dependent higher order chromatin interactions between the TNF promoter and distal enhancers (8), a role exists for Top2 in TNF transcription in T cells.
FIGURE 5.
Top2 inhibitor etoposide inhibits LPS-induced TNF gene expression in monocytic cells and T cells. Left panel, RPA of endogenous expression of the TNF gene and, as a control, the ribosomal protein L32 gene in unstimulated (Un) and LPS (1 μg/ml final concentration)-stimulated J774 cells. J774 cells were pretreated with increasing concentrations of etoposide (0.1, 0.2, and 0.4 mg/ml) as indicated. Right panel, RPA of endogenous TNF and L32 gene expression in unstimulated and phorbol 12-myristate 13-acetate/ionomycin (20 ng/ml and 1 μm final concentrations, respectively)-stimulated 68-41 T cells pretreated with etoposide as described for the left panel.
Topoisomerase IIα Binds to Multiple Sites in HSS−7
To localize Top2 binding to specific regions in HSS−7, we performed a quantitative DNase I in vitro footprinting assay with recombinant human Top2 protein and a 308-bp probe spanning the entire sequence of HSS−7. We incubated HSS−7 with increasing amounts of recombinant Top2 at room temperature and analyzed both DNA strands to increase the resolution of the footprints. As shown in Fig. 6 (A and B, lanes 1–5), weak DNA protection from DNase I digestion by Top2 was observed at multiple places along the probes.
Because Top2 is known to have binding preferences for conflicting DNA structures, including Holliday junctions, cruciforms, and hairpins (37), we reasoned that Top2 binding to the HSS−7 probe would be enhanced by exposing the probe to mild denaturing conditions prior to protein binding, which would transiently separate the strands and allow “breathing” of the helix. Strikingly, when the labeled probe was first denatured by heating it to 70 °C, followed by incubation with Top2 at 37 °C, the binding of Top2 to HSS−7 was significantly enhanced and was detected at seven distinct regions of HSS−7 (Fig. 6, A and B, compare lanes 6–10 with lanes 1–5).
Notably, when we aligned the mouse HSS−7 sequence (308 bp) with the entire human genome using nucleotide BLAST (BLASTN), we found that the central region of the sequence aligned with multiple intergenic regions (supplemental Fig. 2). This region of HSS−7 contains a 64-bp sequence (highlighted in Fig. 6E) that bound to Top2 in the footprinting analysis (Fig. 6, A and B). When we used a probe matching this 64-bp sequence in an EMSA with J774 nuclear extracts, we detected three distinct complexes that specifically reacted with the anti-Top2 antibody (Fig. 6C, lanes 1 and 2), indicating that native Top2 binds to this region.
HSS−7 Is a Substrate for Cleavage by Topoisomerase IIα in Vitro
To determine whether HSS−7 is capable of being cleaved by Top2, we used a recombinant Drosophila Top2 enzyme previously shown to be able to cleave DNA in vitro (38). We incubated Top2 with a 308-bp radiolabeled probe encompassing HSS−7 in buffer containing ATP and Mg2+. Strikingly, Top2 cleaved HSS−7 at a location that falls within the Top2-binding region detected in the footprinting analysis (Fig. 6D, lane 3). In contrast, a control reaction containing BSA instead of Top2 showed no cleavage of HSS−7 (Fig. 6D, lane 2). These data thus indicate that HSS−7 is cleaved by Top2.
DISCUSSION
In this study, we have identified HSS−7, a highly conserved cell type-specific DNase I-hypersensitive site in monocytes (but not in T cells) that is located 7 kb upstream of the TNF gene. HSS−7 is an AT-rich noncoding region that binds to proteins involved in altering chromatin organization and DNA architecture and topology such as acetylated histones, HMGA1a, and Top2. HSS−7 does not exhibit the properties of a transcriptional enhancer, but it functions as a MAR that can be cleaved by Top2. This interaction between Top2 and HSS−7 presents a potential molecular mechanism to facilitate TNF induction by LPS in monocytes, given that the Top2 inhibitor etoposide inhibits TNF gene expression.
Type II topoisomerases are DNA topology-modifying enzymes that are critical in gene transcription and that relieve DNA supercoiling in a two-stage process. First, they cleave both strands of the DNA double helix and pass another uncut double-stranded DNA molecule through the cleaved segment. This is followed by religation of the cleaved segment in an ATP- and Mg2+-dependent manner (37). Top2 recognizes DNA targets with high potential for the formation of secondary stem-loop structures, including hairpins, Holliday junctions, cruciforms, and crossovers (37). AT-rich areas that have the potential of forming secondary stem-loop structures are found across the murine and human genomes. Given the high AT-rich content of HSS−7 and its accessibility to DNase I cleavage and, in particular, our finding that preincubation heat denaturation promotes Top2 binding to an HSS−7 oligonucleotide probe, the probability that HSS−7 can adopt such a secondary structure is high. Indeed, a computational analysis using Mfold (39) predicting structures that can potentially be formed by HSS−7 revealed a variety of stem-loop geometries that Top2 would recognize (supplemental Fig. 3).
As transcription proceeds, the separation of strands of the DNA duplex results in the creation of a bubble that alters the local topology of DNA by creating positive supercoils ahead of the bubble and negative supercoils behind it (40). We note that the TNF gene is located ∼15 kb downstream of the NFKBIL1 gene (which encodes a divergent member of the IκB family of proteins, whose function remains to be elucidated), which itself occupies >12 kb of the chromosome's length. Activation of murine bone marrow-derived macrophages by the Toll-like receptor ligand LPS, via TLR4, leads to overexpression of the NFKBIL1 gene (41) as well as TNF (1). It is interesting to speculate that the transcription bubble created by the polymerase II complex moving along 12 kb of the chromosomal double helix occupied by NFKBIL1 would create a number of positive writhes, or supercoils, toward the LT-α and TNF genes in LPS-stimulated monocytes. As a substrate of Top2, HSS−7 could resolve the conflicting DNA topology generated by transcription of the upstream gene and relieve the stress of excessive positive supercoiling and thus promote TNF gene transcription in activated monocytes. This is consistent with our observation that LPS-induced TNF gene expression in monocytes is suppressed by the Top2 inhibitor etoposide. Notably, our BLAST analysis of HSS−7 indicated that many similar sequences are present in intergenic regions (supplemental Fig. 2), indicating that these conserved sequences may play a specific role in gene transcription and have functions in other gene loci similar to the role HSS−7 plays in the TNF locus in monocytes.
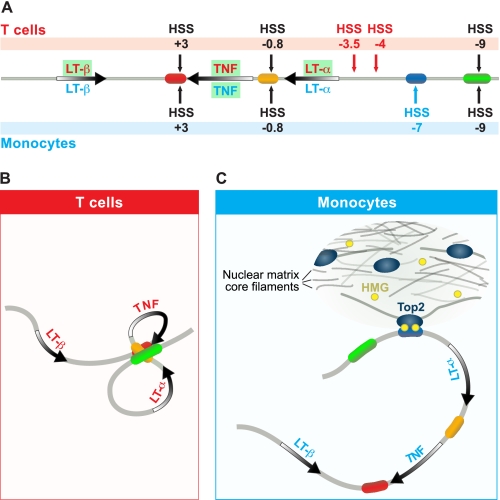
We note that although the TNF locus-flanking genes, NFKBIL1 (upstream) and LST1 (downstream), are ubiquitously expressed in both T lymphocytes and monocytes, the three closely positioned genes in the TNF locus, LT-α, TNF, and LT-β, have different patterns of expression depending on cell type (Fig. 7A). In stimulated T cells, LT-α, TNF, and LT-β are all active, whereas in stimulated monocytes, only the TNF gene is expressed, and both the LT-α and LT-β genes are silent (search of microarray-based expression profiling using ArrayExpress, available on the European Informatics Institute Database). A number of differences in chromatin organization at the TNF locus are evident in monocytes and T cells, and these can elucidate the mechanisms underlying the cell type-specific expression of genes in the TNF locus. There is differential DNA accessibility at the TNF locus in T cells and monocytes; for example, in T cells, hypersensitive sites are present upstream of the LT-α gene (HSS−3.5 and HSS−4), whereas HSS−7 does not appear (Fig. 7A). Strikingly, in T cells, the TNF promoter and the distal enhancers at HSS−9 and HSS+3, all of which activate transcription in an NF-AT-dependent fashion, undergo activation-dependent intrachromosomal interactions (Fig. 7B). The resulting double-loop TNF locus conformation would enhance efficient and relatively independent transcription of each gene by isolating TNF and LT-α in two separate loops (8). In contrast, in monocytes, the selective DNase I sensitivity of HSS−7 exposes a MAR, suggesting that, in monocytes, the TNF transcriptional unit is defined in a cell type-specific manner by the tethering of the locus to the nuclear matrix by HSS−7, which could selectively sequester TNF in a region of active transcription (Fig. 7C). This could be further enhanced by relief of transcription-induced topological stress via the Top2/HSS−7 interaction. Given that etoposide inhibits TNF expression in monocytes and T cells, we are characterizing additional AT-rich regions in the TNF locus that are accessible in T cells and may thus functionally interact with Top2. However, although the expression of NFKBIL1 and LST1 also provides a source of topological stress in T cells, the expression of LT-α and LT-β, as well as TNF, in T cells suggests that, in monocytes, the Top2/HSS−7 interaction plays a more selective role in TNF expression.
FIGURE 7.
Model of organization of the TNF locus in activated J774 cells. A, diagram of the TNF locus, with active genes in the specific cell types shown in green. B, predicted TNF locus structure in activated T cells based on studies from Ref. 8, where TNF, LT-α, and LT-β genes are all active. C, predicted TNF locus structure in monocytes, where LT-α and LT-β genes are silent. In monocytes, HSS−7 interacts with the nuclear matrix, associating with Top2 and HMGA1a proteins and sequestering TNF in a region where its transcription is selectively promoted by the relief of torsional stress through Top2.
Supplementary Material
Acknowledgments
We thank Robert Barthel for the DHAs that are presented in Fig. 1 (E–H). We are indebted to Drs. Caroline Austin and Tao-Shih Hsieh for the generous gifts of recombinant human topoisomerase IIα protein and Drosophila topoisomerase II, respectively. We are grateful to Renate Hellmiss for graphic artwork and James Falvo for helpful discussions, critical reading, and invaluable assistance in the preparation of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM076685 (to A. E. G.). This work was also supported by GlaxoSmithKline (to A. E. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- DHA
- DNase I hypersensitivity assay
- HSS
- hypersensitive site
- LT
- lymphotoxin
- MAR
- matrix attachment region
- RPA
- RNase protection assay
- HMG
- high mobility group.
REFERENCES
- 1. Falvo J. V., Tsytsykova A. V., Goldfeld A. E. (2010) Curr. Dir. Autoimmun. 11, 27–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barthel R., Tsytsykova A. V., Barczak A. K., Tsai E. Y., Dascher C. C., Brenner M. B., Goldfeld A. E. (2003) Mol. Cell. Biol. 23, 526–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsytsykova A. V., Goldfeld A. E. (2002) Mol. Cell. Biol. 22, 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsytsykova A. V., Goldfeld A. E. (2000) J. Exp. Med. 192, 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Falvo J. V., Uglialoro A. M., Brinkman B. M., Merika M., Parekh B. S., Tsai E. Y., King H. C., Morielli A. D., Peralta E. G., Maniatis T., Thanos D., Goldfeld A. E. (2000) Mol. Cell. Biol. 20, 2239–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai E. Y., Falvo J. V., Tsytsykova A. V., Barczak A. K., Reimold A. M., Glimcher L. H., Fenton M. J., Gordon D. C., Dunn I. F., Goldfeld A. E. (2000) Mol. Cell. Biol. 20, 6084–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falvo J. V., Brinkman B. M., Tsytsykova A. V., Tsai E. Y., Yao T. P., Kung A. L., Goldfeld A. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3925–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tsytsykova A. V., Rajsbaum R., Falvo J. V., Ligeiro F., Neely S. R., Goldfeld A. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16850–16855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cockerill P. N. (2008) Sci. Signal. 1, pe15. [DOI] [PubMed] [Google Scholar]
- 10. Barthel R., Goldfeld A. E. (2003) J. Immunol. 171, 3612–3619 [DOI] [PubMed] [Google Scholar]
- 11. Hostert A., Tolaini M., Festenstein R., McNeill L., Malissen B., Williams O., Zamoyska R., Kioussis D. (1997) J. Immunol. 158, 4270–4281 [PubMed] [Google Scholar]
- 12. Cockerill P. N. (2000) Methods Mol. Biol. 130, 29–46 [DOI] [PubMed] [Google Scholar]
- 13. Goldfeld A. E., McCaffrey P. G., Strominger J. L., Rao A. (1993) J. Exp. Med. 178, 1365–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsytsykova A. V., Falvo J. V., Schmidt-Supprian M., Courtois G., Thanos D., Goldfeld A. E. (2007) J. Biol. Chem. 282, 11629–11638 [DOI] [PubMed] [Google Scholar]
- 15. Kumar P. P., Bischof O., Purbey P. K., Notani D., Urlaub H., Dejean A., Galande S. (2007) Nat. Cell Biol. 9, 45–56 [DOI] [PubMed] [Google Scholar]
- 16. Lei J. X., Liu Q. Y., Sodja C., LeBlanc J., Ribecco-Lutkiewicz M., Smith B., Charlebois C., Walker P. R., Sikorska M. (2005) Cell Death Differ. 12, 1368–1377 [DOI] [PubMed] [Google Scholar]
- 17. Lee M. P., Sander M., Hsieh T. (1989) J. Biol. Chem. 264, 21779–21787 [PubMed] [Google Scholar]
- 18. Lee M. P., Sander M., Hsieh T. S. (1989) J. Biol. Chem. 264, 13510–13518 [PubMed] [Google Scholar]
- 19. Tsai E. Y., Yie J., Thanos D., Goldfeld A. E. (1996) Mol. Cell. Biol. 16, 5232–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwartz S., Zhang Z., Frazer K. A., Smit A., Riemer C., Bouck J., Gibbs R., Hardison R., Miller W. (2000) Genome Res. 10, 577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leung J. Y., McKenzie F. E., Uglialoro A. M., Flores-Villanueva P. O., Sorkin B. C., Yunis E. J., Hartl D. L., Goldfeld A. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6614–6618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baena A., Mootnick A. R., Falvo J. V., Tsytskova A. V., Ligeiro F., Diop O. M., Brieva C., Gagneux P., O'Brien S. J., Ryder O. A., Goldfeld A. E. (2007) PLoS ONE 2, e621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. The International HapMap Consortium (2003) Nature 426, 789–796 [DOI] [PubMed] [Google Scholar]
- 24. Boulikas T. (1995) Int. Rev. Cytol. 162A, 279–388 [DOI] [PubMed] [Google Scholar]
- 25. Bode J., Stengert-Iber M., Kay V., Schlake T., Dietz-Pfeilstetter A. (1996) Crit. Rev. Eukaryot. Gene Expr. 6, 115–138 [DOI] [PubMed] [Google Scholar]
- 26. Moltó E., Fernández A., Montoliu L. (2009) Brief. Funct. Genomic Proteomic. 8, 283–296 [DOI] [PubMed] [Google Scholar]
- 27. Bode J., Goetze S., Heng H., Krawetz S. A., Benham C. (2003) Chromosome Res. 11, 435–445 [DOI] [PubMed] [Google Scholar]
- 28. Balakrishnan L., Milavetz B. (2007) J. Mol. Biol. 371, 1022–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balakrishnan L., Milavetz B. (2007) J. Mol. Biol. 365, 18–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chattopadhyay S., Pavithra L. (2007) Subcell. Biochem. 41, 213–230 [PubMed] [Google Scholar]
- 31. Mondal N., Parvin J. D. (2001) Nature 413, 435–438 [DOI] [PubMed] [Google Scholar]
- 32. Jiao W., Lin H. M., Timmons J., Nagaich A. K., Ng S. W., Misteli T., Rane S. G. (2005) Cancer Res. 65, 4067–4077 [DOI] [PubMed] [Google Scholar]
- 33. Mitchell R., Chiang C. Y., Berry C., Bushman F. (2003) Mol. Ther. 8, 674–687 [DOI] [PubMed] [Google Scholar]
- 34. Nagashima M., Shiseki M., Miura K., Hagiwara K., Linke S. P., Pedeux R., Wang X. W., Yokota J., Riabowol K., Harris C. C. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9671–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lehar S. M., Nacht M., Jacks T., Vater C. A., Chittenden T., Guild B. C. (1996) Oncogene 12, 1181–1187 [PubMed] [Google Scholar]
- 36. Horiguchi-Yamada J., Fukumi S., Saito S., Nakayama R., Iwase S., Yamada H. (2002) Anticancer Res. 22, 3827–3832 [PubMed] [Google Scholar]
- 37. Wang J. C. (2009) Untangling the Double Helix: DNA Entanglement and the Action of the DNA Topoisomerases, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 38. Sander M., Hsieh T. S. (1985) Nucleic Acids Res. 13, 1057–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zuker M. (2003) Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu L. F., Wang J. C. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramsey S. A., Klemm S. L., Zak D. E., Kennedy K. A., Thorsson V., Li B., Gilchrist M., Gold E. S., Johnson C. D., Litvak V., Navarro G., Roach J. C., Rosenberger C. M., Rust A. G., Yudkovsky N., Aderem A., Shmulevich I. (2008) PLoS Comput. Biol. 4, e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.