Background: Nhx1/Vps44 is proposed to be a Class E gene involved in formation of the multivesicular body (MVB). However, this hypothesis has not been tested.
Results: Nhx1 is not required for cargo sorting or MVB formation and shows synthetic phenotypes with select ESCRT mutants.
Conclusion: Nhx1 functions independently of the ESCRT pathway.
Significance: Nhx1 may have a post-ESCRT role in endosomal membrane fusion.
Keywords: Endosomal pH Function, Membrane Trafficking, Transporters, Vacuolar Acidification, Yeast, ESCRT Complex, Na+/H+ Exchanger, Multivesicular Body, Vacuole Protein Sorting (VPS)
Abstract
The multivesicular body (MVB) is an endosomal intermediate containing intralumenal vesicles destined for membrane protein degradation in the lysosome. In Saccharomyces cerevisiae, the MVB pathway is composed of 17 evolutionarily conserved ESCRT (endosomal sorting complex required for transport) genes grouped by their vacuole protein sorting Class E mutant phenotypes. Only one integral membrane protein, the endosomal Na+ (K+)/H+ exchanger Nhx1/Vps44, has been assigned to this class, but its role in the MVB pathway has not been directly tested. Herein, we first evaluated the link between Nhx1 and the ESCRT proteins and then used an unbiased phenomics approach to probe the cellular role of Nhx1. Select ESCRT mutants (vps36Δ, vps20Δ, snf7Δ, and bro1Δ) with defects in cargo packaging and intralumenal vesicle formation shared multiple growth phenotypes with nhx1Δ. However, analysis of cellular trafficking and ultrastructural examination by electron microscopy revealed that nhx1Δ cells retain the ability to sort cargo into intralumenal vesicles. In addition, we excluded a role for Nhx1 in Snf7/Bro1-mediated cargo deubiquitylation and Rim101 response to pH stress. Genetic epistasis experiments provided evidence that NHX1 and ESCRT genes function in parallel. A genome-wide screen for single gene deletion mutants that phenocopy nhx1Δ yielded a limited gene set enriched for endosome fusion function, including Rab signaling and actin cytoskeleton reorganization. In light of these findings and the absence of the so-called Class E compartment in nhx1Δ, we eliminated a requirement for Nhx1 in MVB formation and suggest an alternative post-ESCRT role in endosomal membrane fusion.
Introduction
The multivesicular body (MVB)3 pathway solves the topological problem of degrading integral membrane proteins (and accompanying lipid) by sorting monoubiquitylated cargo into intralumenal vesicles, which are then delivered via the MVB to the vacuole/lysosome for enzymatic degradation (1). In mammals, this pathway is responsible for receptor down-regulation and may be exploited by retroviruses, such as HIV, for particle biosynthesis (2). In yeast, 17 soluble and evolutionarily conserved vacuole protein sorting (VPS) Class E proteins have been organized into ESCRTs (endosomal sorting complexes required for transport) of ubiquitin-tagged cargo to the vacuole (reviewed in Ref. 1). Deletion of each of these ESCRT genes results in accumulation of endosomal cargo in a large aberrant structure adjacent to the vacuole called the Class E compartment. The integral membrane protein Vps44/Nhx1 functions as a cation/proton exchanger and has also been included in this class, although its role in MVB formation is unclear. The initial characterization of this pathway relied predominantly on methods of physical network identification, such as the Gal4-based yeast two-hybrid system, protein complex isolation, identification, and crystallization efforts, which, for the most part, do not accommodate hydrophobic proteins (1, 3, 4). This could potentially explain the omission of Nhx1/Vps44 from the current model of this pathway, as it is the only membrane embedded protein in the VPS Class E gene set. It is also possible that Nhx1 does not contribute to MVB formation and functions at a separate step in the delivery of cargo to the vacuole.
Nhx1 is a Na+ (K+)/H+ exchanger (5) that confers tolerance to osmotic, salt, and low pH stress (6–8). It is orthologous to human NHE6 and NHE9, which are linked to mental retardation, autism, attention deficit hyperactivity disorder, and epilepsy (9–11), although the cellular basis underlying disease phenotypes is not clear. Yeast Nhx1 co-localizes with the syntaxin Pep12 in an endosomal compartment directly abutting the vacuolar membrane in live cells, suggestive of a role for pH and ion gradients in vacuolar biogenesis (12). Consistent with this hypothesis, NHX1 was found to be allelic to VPS44 and shown to be required for proper processing and localization of carboxypeptidase Y (CPY) to the vacuole. In nhx1Δ, as in other VPS Class E mutants, endosomal cargo is retained in a prevacuolar compartment (13).
Regulation of endosomal pH by Nhx1 was directly linked to cargo trafficking by experiments showing that manipulation of compartmental pH by weak acids or bases could simulate or ameliorate trafficking defects, respectively. Thus, reversing hyperacidic compartmental pH in nhx1Δ mutants by the addition of a weak base alleviates a subset of nhx1Δ phenotypes, including hygromycin sensitivity, defective FM 4-64 efflux, and abnormal retention of Ste3-GFP in the prevacuolar compartment (8). Although the accumulated evidence points to an important function in late endosomal/MVB trafficking, the role of Nhx1, if any, in MVB formation and its relation to the ESCRT pathway remain unclear. To address this problem, we first identified late-acting ESCRT mutants that closely resembled nhx1Δ in cation-related growth defects and then performed a detailed analysis of cation homeostasis, cargo trafficking, and intralumenal vesicle (ILV) formation in single and double mutants lacking Nhx1 and late ESCRT components. In an independent approach, we undertook a genome-wide unbiased screen for single gene deletions that phenocopied nhx1Δ to identify a limited subset of proteins implicated in late endosome/MVB-vacuole fusion. Together, these approaches clarify the role of Nhx1 in endosome trafficking and vacuole biogenesis.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
All Saccharomyces cerevisiae strains used were derivatives of BY4741 or BY4742 (ResGen). Double knock-out strains were made by first transforming nhx1Δ BY4741 mutants with the linearized plasmid pAG25 (14) to replace the KanR cassette with the NatR cassette, conferring resistance to nourseothricin in place of kanamycin. Select ESCRT mutants (BY4742, MATα, KanR) and nhx1Δ (BY4741, MATa, NatR) were mated, and the resulting diploids underwent sporulation when incubated in agitated liquid sporulation medium (1% sodium acetate and 0.005% zinc acetate) with required supplements (5 mg/liter each uracil, histidine, leucine, lysine, and methionine) for 4 days at 25 °C. Tetrads were dissected, and spores resistant to Geneticin (200 μg/ml) and nourseothricin (100 μg/liter) were matched to the BY4742 background. For electron microscopy, the strains used were SEY6210 (NHX1) (15) and GOY91 (SEY6210 leu2-3,112::pBHY11, nhx1Δ:HIS3) (this work). For the ubiquitin blot experiments, the strains used were TVY1 (pep4Δ) (16), MWY6 (pep4Δ doa4C571S) (17), and MWY31 (SEY6210 leu2-3,112::pBHY11, nhx1Δ:HIS3, pep4Δ:KAN) (this work).
Yeast Growth Assays
Strains were grown at 30 °C in arginine/phosphate/glucose (APG) medium, a synthetic minimal medium containing 10 mm arginine, 8 mm phosphoric acid, 2% (w/v) glucose, 2 mm MgSO4, 1 mm KCl, 0.2 mm CaCl2, and trace minerals and vitamins (6). The pH was adjusted by the addition of phosphoric acid to 4.0 or 2.7 as specified. Where indicated, NaCl, KCl, or hygromycin B was added to the specified concentration. Seed cultures were grown in synthetic complete medium to saturation, washed three times with water, and used to seed 200 μl of APG medium in 96-well plates at ∼0.05 A600 nm units/ml. Growth was monitored between 17 and 24 h by taking A600 nm measurements at 30 °C.
A library of 4828 S. cerevisiae strains with deletion of each nonessential gene in a haploid background (BY4742, MATα) was purchased from ResGen. las17Δ (RLY157) and the isogenic WT strain (RLY138) (kind gifts from Dr. Rong Li, Harvard Medical School, Boston, MA) (18) were added to the collection of mutants. Fifty-two 96-well plates representing the collection of mutant strains were thawed, and 5 μl of each culture was used to inoculate 200-μl seed cultures grown in synthetic complete medium at 30 °C. 4 μl of each seed culture was then used to inoculate 96-well plates, each containing 200 μl of APG medium. Growth (A600 nm) was read at 19 and 48 h on a FLUOstar Optima multimode plate reader with FLUOstar Optima Version 1.20 software (BMG Labtech, Durham, NC). Immediately prior to recording, cultures were rapidly resuspended using an electromagnetic microtiter plate shaker (Union Scientific, Randallstown, MD), and all recordings were made at 30 °C.
A600 nm values were background-subtracted and normalized to average WT growth (mean calculated from 52 separate cultures spanning all microtiter plates per screen). Strains showing <20% of WT growth (<1% of the collection) under control conditions (APG medium, pH 4.0) were omitted from further analysis. Of the strains that were included, low A600 nm values of the seed cultures did not correlate with growth defects observed under experimental conditions. For each screen, conditional growth values were normalized to A600 nm values obtained under control conditions so that percentage values <100 indicate sensitivity and those >100 indicate tolerance. For data sets showing normal distributions, means ± S.D. were calculated, and strains showing growth greater than or less than 2 S.D. from the mean were considered to have a significant change in growth. Similar statistical analyses were performed on data sets showing bimodal distributions (800 mm NaCl or KCl growth screens at the 19-h measurement), whereby the data sets were divided into poorly growing and robust populations. Mutant strains in the robust population with growth values 2 S.D. greater than the mean were considered significantly tolerant to the condition, and all mutant strains in the poorly growing population were considered significantly sensitive with the exception of those with growth values 2 S.D. greater than the mean. Significant growth sensitivity or tolerance was confirmed (n > 3) for all mutants originally identified by screening experiments. Data were organized, plotted, and analyzed using Microsoft Excel X, Insightful S-PLUS 6.2, and SPSS 12.0 software. The Munich Information Center for Protein Sequences (MIPS) Database was used to obtain annotated FunCat (functional categorization) and cellular location of genes identified in the phenocopy and chemical synthetic lethality screens.
CPY Secretion
CPY secretion from yeast cultures was detected as follows. Freshly grown seed cultures were washed with water, resuspended to a starting A600 nm of 0.05 units/ml in synthetic complete medium, and grown at 30 °C for 21 h. Cultures from 2.0 A600 nm units of cells were centrifuged, and supernatants were applied to Millipore Immobilon membranes using a dot-blot apparatus. The membrane was dried overnight, and CPY was detected by immunoblotting using anti-CPY monoclonal antibody (1:1000 dilution; Molecular Probes) and horseradish peroxidase-coupled sheep anti-mouse antibody (1:10,000 dilution; Amersham Biosciences).
Rim101 Processing
Yeast strains carrying plasmid pKJB11 (gift of Dr. Aaron Mitchell, Carnegie Mellon University) (19) were grown overnight in selective medium at 30 °C and used to inoculate fresh cultures at A600 nm = 0.25. After two doublings, cells were pelleted, resuspended at A600 nm = 50 in 3× Laemmli buffer, vortexed with glass beads, and boiled for 5 min. After centrifugation, 30 μl of the supernatant was fractionated on a 10% SDS-polyacrylamide gel and transferred to nitrocellulose. For V5 epitope detection, the membrane was probed with horseradish peroxidase-coupled anti-V5 antibody (1:5000 dilution in PBS/Tween; Invitrogen). Peroxidase activity was visualized using ECL detection reagents (Thermo Scientific, Rockford, IL).
Fluorescence Microscopy
GFP and FM 4-64 fluorescence was performed at a magnification of ×100 using a Carl Zeiss Axioplan 2 microscope equipped with an oil immersion objective (numerical aperture of 1.40). Fluorescence microscopic images were acquired with a Photometrics CoolSNAP FX digital camera and processed using Scanalytics IPLab software and Adobe Photoshop CS2 software. GFP-Bro1 or GFP-carboxypeptidase S (CPS) was introduced by transforming cells with either pGO249 or pMB118, respectively (20, 21). Nhx1-GFP has been described (12). Strains were grown to logarithmic phase at 30 °C in synthetic medium before observation at room temperature in synthetic medium. Vacuolar membrane staining was performed using 40 μm FM 4-64 for 30 min in yeast extract/peptone/dextrose medium as described (22).
Ubiquitin (Ub)-CPS Immunoprecipitation
Denatured immunoprecipitations were performed to detect Ub-CPS as described previously (23, 24). 0.5 A600 nm units were resolved by SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting using rabbit anti-CPS polyclonal antiserum (25) or anti-Ub monoclonal antibodies (Zymed Laboratories Inc.). The vacuolar protease gene PEP4 was deleted in all strains to reduce the nonspecific cleavage of Ub from CPS after cellular lysis.
Electron Microscopy
Yeast cells were high pressure frozen and freeze-substituted as described previously (17, 26) and embedded at −60 °C in Lowicryl HM20 (Polysciences). Plastic blocks were trimmed and cut into 80-nm thin sections with a Leica microtome, placed on rhodium-plated copper slot grids, and imaged on a Philips CM10 transmission electron microscope.
RESULTS
Loss of Nhx1 Is Phenocopied by Select ESCRT Pathway (VPS Class E) Mutants
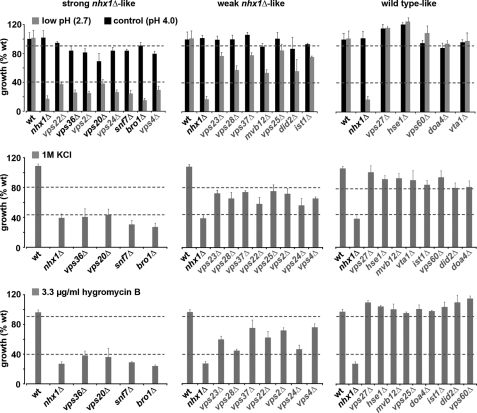
To map Nhx1 function in the MVB pathway, we initially identified mutants harboring single deletions in each of the 17 other VPS class E genes or three additional genes reported to regulate the MVB pathway (reviewed in Ref. 27) that shared growth phenotypes with nhx1Δ. Logarithmic phase growth of nhx1Δ is sensitive to three conditions: (i) acid pH (8), (ii) high salt (8), and (iii) hygromycin B (28). Eight mutants showed growth sensitivity to acid stress similar to nhx1Δ (Fig. 1, upper panel). However, only half of these mutants also shared sensitivity to high salt (identical results were observed with either KCl or NaCl) (Fig. 1, middle panel) and hygromycin B (lower panel). These four strains harbor deletions in VPS36, VPS20, SNF7, and BRO1, which function together at a late stage of the MVB pathway. It is noteworthy that deleting ESCRT genes that function farther upstream or downstream of these components generally resulted in progressively weaker nhx1Δ-like phenotypes, although differences within ESCRT-II components suggest more complicated and previously unappreciated relationships that warrant further study (supplemental Table S1). Vps36 is a component of ESCRT-II that binds and recruits ESCRT-III for cargo transfer (1). Vps20 and Snf7 compose a subcomplex of ESCRT-III that has a reported role in the membrane scission event required for ILV formation (29). Snf7 also recruits Bro1 to ESCRT-III, where it promotes deubiquitylation of cargo by Doa4 to coordinate cargo packaging into budding ILVs (21, 24). These findings raised the possibility that Nhx1 may function together with the ESCRT-III complex when cargo packaging and ILV budding occur. Alternatively, Nhx1 and the late-acting ESCRT component may have independent functions that affect the same cellular processes that underlie their shared phenotypes. To distinguish between these possibilities, we directly evaluated ESCRT function, including cargo sorting and MVB formation, in the nhx1Δ mutant.
FIGURE 1.
Select ESCRT mutants phenocopy nhx1Δ growth. Growth of BY4742 (WT), nhx1Δ, and isogenic single gene deletions in the ESCRT pathway was monitored in defined (APG) medium adjusted to the indicated pH (4.0 or 2.7) or supplemented with KCl (1 m) or hygromycin B (3.3 μg/ml). Growth (A600 nm) was normalized to control pH (4.0; upper panel) or to medium without KCl (middle panel) or without hygromycin B (lower panel) for each strain. Results are average of triplicates and representative of at least three independent experiments. Similar results were obtained with NaCl in place of KCl (not shown). Strains are grouped on basis of similarity to nhx1Δ as seen by placement of horizontal gray dashed lines.
Nhx1 Is Not Essential for Cargo Deubiquitylation, Packaging, or Formation of ILVs
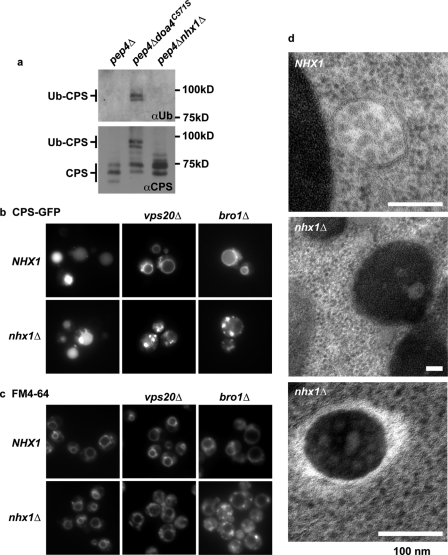
Cargo destined for vacuole degradation enters the MVB pathway when monoubiquitylated. Once sent to the endosome, ESCRT-III concentrates and packages the Ub-tagged cargo into ILVs. At the final step, the Ub signal is cleaved by the deubiquitylase Doa4 when ESCRT is disassembled (by Vps4, a AAA-ATPase) immediately prior to ILV scission (27). This maintains a cellular pool of ubiquitin required for proper protein degradation. We showed that nhx1Δ phenocopies ESCRT deletion strains that function after Ub-cargo recruitment and sequestration but upstream of Doa4 recruitment and function (vps36Δ, vps20Δ, snf7Δ, or bro1Δ). These mutations prevent cargo deubiquitylation, causing Ub-cargo to accumulate on limiting membranes (17). Thus, if Nhx1 has a role in a late stage of MVB formation, we would expect impairment of cargo deubiquitylation in the null strain. As a control for loss of deubiquitylation, we introduced a C571S mutation in Doa4, which blocks Ub transfer and prevents removal of Ub from the cargo CPS (Fig. 2a) (30). We observed that CPS deubiquitylation persisted in nhx1Δ cells as seen by the absence of Ub staining on the Western blot (Fig. 2a). These results do not exclude the formal possibility that the cargo was deubiquitylated without being incorporated into ILVs.
FIGURE 2.
nhx1Δ cells form MVBs. a, CPS is efficiently deubiquitylated in nhx1Δ. Immunoprecipitates of CPS were evaluated in an isogenic pep4Δ background by Western blotting using anti-Ub (upper panel) or anti-CPS (lower panel) antibody. Ub-CPS could not be deubiquitylated in the strain carrying the C571S mutation in Doa4. Multiple CPS bands (lower panel) represent glycosylation. b, localization of GFP-CPS to the vacuolar lumen occurs in the wild-type (NHX1) and nhx1Δ mutant strains but not in ESCRT mutant strains vps20Δ and bro1Δ. Similar results were obtained with snf7Δ and vps36Δ (not shown). Note the prominent punctate accumulation of GFP-CPS in strains carrying nhx1Δ. c, FM 4-64 staining of the vacuolar limiting membrane in wild-type (NHX1) and mutant strains as in b. Punctate accumulation of FM 4-64 in the mutant strains was exacerbated in double mutants of nhx1Δ and ESCRT mutants vps20Δ and bro1Δ. Similar results were obtained with snf7Δ and vps36Δ (not shown). d, electron micrographs of wild-type cells and representative cells from nhx1Δ showing MVBs. Scale bars = 100 nm. Note the dense background in nhx1Δ MVBs.
To determine whether CPS is properly processed through the MVB pathway, we examined if deletion of NHX1 affects the cellular location of GFP-CPS (Fig. 2b). CPS requires proper sorting as well as ILV formation to reach the vacuole lumen, where it is processed and performs its cellular function. As reported previously for vps36Δ and snf7Δ (24), we observed that sorting of CPS into ILVs was blocked in vps20Δ and bro1Δ, resulting in accumulation at the limiting membrane of the vacuole (Fig. 2b). In contrast, nhx1Δ cells showed CPS localizing to the vacuole lumen, albeit with prominent retention in enlarged endosomal compartments (also detected by FM 4-64 staining) (Fig. 2c). Thus, Nhx1 is not required for CPS sorting or lumenal targeting, which implies that ILV formation can occur. To confirm the existence of ILVs in nhx1Δ cells, we examined the ultrastructure of MVBs by electron microscopy (24). Unlike snf7Δ (31) or vps36Δ and vps20Δ,4 nhx1Δ cells appeared to have intact MVBs containing clearly defined ILVs (Fig. 2d). One distinction is, however, that MVBs in nhx1Δ cells were electron-dense. Given that alkaline vacuoles (vma mutants) appear abnormally clear in electron micrographs, electron-dense MVBs in nhx1Δ are consistent with lumenal hyperacidification reported previously (8, 32). In summary, these data demonstrate that Nhx1 is not essential for cargo sorting or packaging and ILV formation in the MVB pathway.
Nhx1 Is Not Required for ESCRT-mediated Rim101 Signaling
Another function of the ESCRT pathway is in the Rim101-mediated cell signaling response to alkaline stress required for filamentation in the fungal pathogen Candida albicans (33). When activated by upstream signal factors, Rim20 binds to Snf7, in complex with Vps20 and associated with Vps36, as a necessary step in the recruitment of the protease required for Rim101 cleavage and activation (34). Thus, raising the external pH induces Rim101 cleavage in wild-type cells, whereas disruption of VPS4, the AAA-ATPase required for ESCRT disassembly, causes constitutive activation of Rim101 signaling due to accumulation of the Vps20-Snf7 complex on the endosomal membrane (35), as shown in Fig. 3a. Given the disruption in cellular pH homeostasis in nhx1Δ (8, 32), we asked whether Rim101 signaling is altered in nhx1Δ. However, Nhx1 was not required for Rim101 processing in response to alkaline pH, nor did it alter constitutive activation of Rim101 processing in vps4Δ (Fig. 3a), thus ruling out a role for Nhx1 in ESCRT-mediated Rim signaling. Vps4 is also required for release of Bro1 from endosomal membranes, as seen by altered distribution of Bro1-GFP in vps4Δ. Our results demonstrate that loss of Nhx1 did not alter the distribution of Bro1-GFP, nor did it interfere with Vps4-mediated release of Bro1-GFP from endosomal compartments (Fig. 3b). Taken together, the results show that Nhx1 is not required for late ESCRT function.
FIGURE 3.
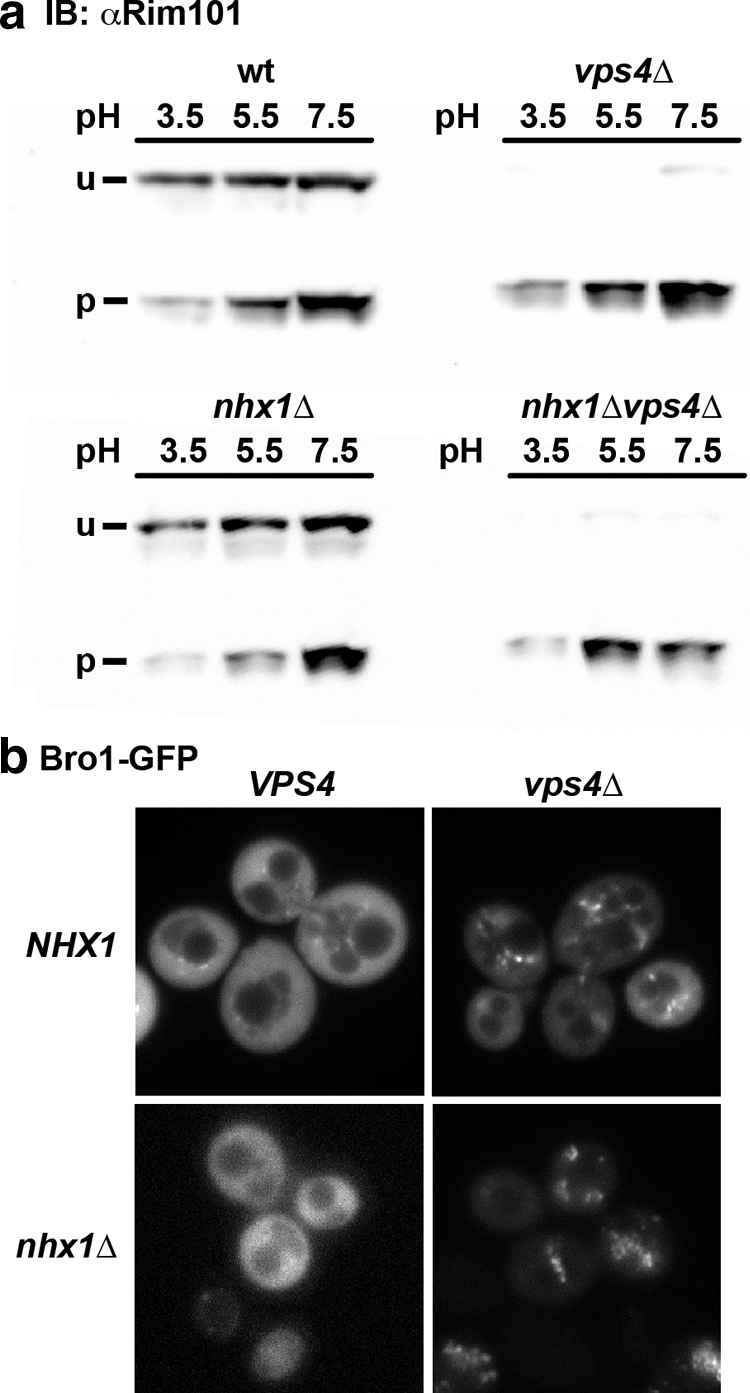
Bro1 localization and Rim101 cleavage are unaffected by NHX1 deletion. a, WT or the nhx1Δ, vps4Δ, or nhx1Δvps4Δ mutant were grown in medium adjusted to the indicated pH and analyzed for V5 epitope-tagged Rim101 by Western blotting. Note the increase in the proteolytically processed (p) band relative to the unprocessed (u) band at more alkaline pH. Alkaline pH-induced or constitutive (in vps4Δ) Rim101 processing was not affected by nhx1Δ. IB, immunoblot. b, Bro1-GFP localization in nhx1Δ is similar to that in WT. Endosomal accumulation of Bro1-GFP occurred in both vps4Δ and nhx1Δvps4Δ mutants.
Nhx1 Functions Independently of ESCRT
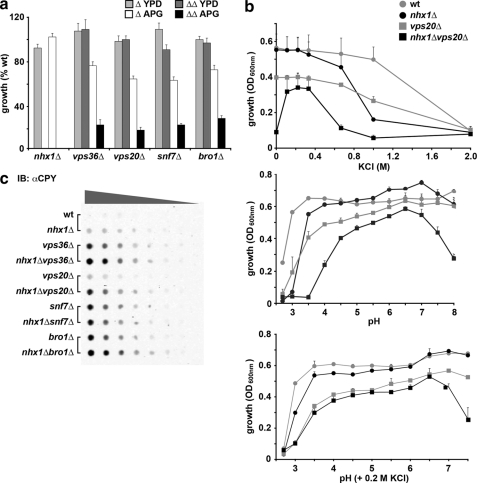
Having established that ESCRT activity is not directly impacted in nhx1Δ, we wanted to determine whether ESCRT affects Nhx1 function. We performed genetic epistasis experiments by making double mutant strains harboring nhx1Δ and each of the following ESCRT gene deletions: vps36Δ, vps20Δ, snf7Δ, or bro1Δ. We then examined growth and trafficking phenotypes with the expectation that loss of a gene found most downstream in a common pathway would not alter phenotypes conferred by deleting upstream genes. Thus, if NHX1 is epistatic to late ESCRT genes, the phenotypes of the double mutants would be similar to those of nhx1Δ. We first observed that the double mutants grew very poorly in the minimal medium APG (Fig. 4a). Further analysis revealed that this growth defect was due in part to KCl restriction in this medium (e.g. nhx1Δvps20Δ) (Fig. 4b). Importantly, growth sensitivity to high KCl and low pH phenotypes conferred by nhx1Δ and individual ESCRT mutants (see Fig. 1) were additive in the double mutants (Fig. 4b). Double mutants also gained sensitivity to alkaline pH, which, unlike acid pH sensitivity, could not be rescued by the addition of KCl.
FIGURE 4.
Additive phenotypes of nhx1Δ and ESCRT mutations. a, growth of the indicated single gene deletion strains (Δ) and double mutants with nhx1Δ (ΔΔ) was measured in yeast extract/peptone/dextrose (YPD) medium or in defined APG medium relative to isogenic BY4742 (WT). Note the severe growth impairment of the ΔΔ strains in APG medium. b, impaired growth of the nhx1Δvps20Δ double mutant shown in a was corrected to single mutant (vps20Δ) levels by the addition of KCl to APG medium (upper panel). Further addition of KCl resulted in acute growth inhibition. Growth of the double mutant was more sensitive to low and high pH (middle panel), with the addition of KCl (0.2 m) correcting the growth sensitivity at low pH to single mutant (vps20Δ) levels (lower panel). The average of triplicate experiments is shown. Similar results were observed for other ESCRT mutants shown in a. c, missorting of CPY to the extracellular medium was increased in double mutants. Serial dilutions of culture supernatant were applied to filters and probed with anti-CPY antibody as described under “Experimental Procedures.” IB, immunoblot.
Evaluation of trafficking phenotypes confirmed these findings. Proper targeting of the acid hydrolase CPY to its resident compartment, the vacuole, requires a functional endocytic pathway. Missorting and abnormal secretion of CPY are the defining phenotypes of all VPS mutants. As reported previously, defects in nhx1Δ and ESCRT pathway mutants resulted in the extracellular appearance of CPY (Fig. 4c). We found that double mutants secreted more CPY than the parental single gene mutant strains (Fig. 4c). For example, although CPY secretion from both nhx1Δ and vps20Δ mutants was observed to be modest, there was a substantial increase in the double mutant. Interestingly, this additive effect was difficult to detect in only one example, snf7Δ. We showed previously that the vacuolar pH in snf7Δ is more alkaline, in contrast to other ESCRT pathway mutants that show an acidic shift, such as nhx1Δ (36). Thus, the acidification resulting from loss of Nhx1 in snf7Δ may have been attenuated relative to other double mutants. It is known that CPY secretion in nhx1Δ mutants is proportional to lumenal pH and can be corrected, in part, by the addition of a weak base (37). We also found that the vacuolar lumen-targeting defect of GFP-CPS was strikingly exacerbated in double mutants: a small portion of GFP-CPS accumulated on limiting membranes but was largely trapped within aberrant compartments, suggesting that vacuole targeting or fusion was impaired, although we noted that normal vacuolar morphology was retained (Fig. 2, b and c). These structures were also apparent in nhx1Δvps4Δ cells where Bro1-GFP accumulates (Fig. 3b). Together, these results confirm a role for Nhx1 in endosome (or MVB)-to-vacuole trafficking but eliminate a requirement for MVB formation.
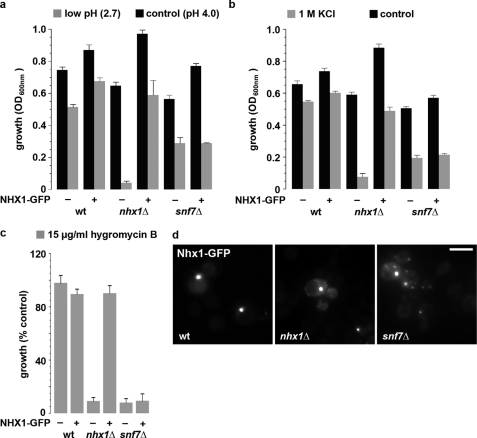
Next, we evaluated whether multicopy expression of NHX1 could rescue ESCRT mutant phenotypes. Whereas Nhx1-GFP could fully restore growth sensitivity to low pH, high KCl, or hygromycin B in nhx1Δ, it failed to suppress these phenotypes in ESCRT mutants (Fig. 5, a–c). Nhx1-GFP localized to an endosomal compartment closely abutting the vacuolar membrane when expressed in the wild type or nhx1Δ as reported previously (12). The endosomal distribution of Nhx1-GFP appeared to be retained in ESCRT mutants, albeit with a shift toward multiple puncta rather than a predominant single compartment as seen in most wild-type cells (Fig. 5d) (36). The lack of phenotypic suppression and additive growth phenotypes led to the conclusion that Nhx1 and ESCRT function in independent processes.
FIGURE 5.
Multicopy expression of NHX1 does not suppress snf7Δ phenotypes. NHX1-GFP (2μ plasmid) suppressed growth impairment of the nhx1Δ but not snf7Δ mutant at low pH (a), in 1 m KCl (b), or in 15 μg/ml hygromycin B (c). Growth data are the average of triplicates and representative of several independent experiments. d, Nhx1-GFP localized to endosome-like puncta in the WT and nhx1Δ and snf7Δ mutants. Similar results were observed for vps20Δ, bro1Δ, and vps36Δ (not shown).
Limited Number of Mutants Phenocopy nhx1Δ in an Unbiased Genome-wide Screen
NHX1 has been considered to be a VPS Class E gene based on cargo retention in a prevacuolar compartment. Conceptually, this could occur through one of two mechanisms: impaired invagination for ILV formation resulting in the aberrant “Class E” compartment as seen in ESCRT pathway mutants or diminished consumption of the limiting membrane through compromised endosome-vacuole fusion. Our observations thus far appeared to eliminate the first possibility. Therefore, we considered whether Nhx1 could have a role in MVB consumption instead of formation. The previous observation of a physical and functional interaction of Nhx1 with Gyp6, a GTPase-activating protein for the endosomal Rab ortholog Ypt6, supported a role in late endosome fusion (32). To seek independent evidence for similar functions, we extended the nhx1Δ phenocopy screen to the viable single gene deletion ResGen library of 4828 strains using four time-sensitive conditions shown previously to compromise the growth of nhx1Δ. Of these, growth sensitivities to hygromycin B and low pH were fairly selective, with significant defects in growth observed in only 170 and 186 mutants at late exponential phase (19 h) in the presence of 15 μg/ml hygromycin B or at pH 2.7, respectively, representing 3.5 and 3.8% of all strains screened (supplemental Table S2). Many more mutants showed growth sensitivity to salt stress at the 19-h time point: 737 and 1174 in the presence of 800 mm NaCl or KCl, respectively. However, the vast majority of these, including nhx1Δ, recovered growth by 48 h (supplemental Table S2).
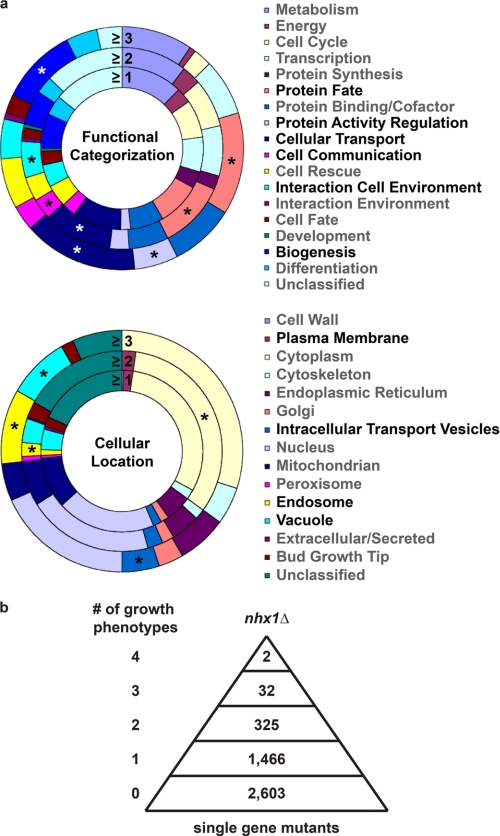
Each condition revealed a unique gene profile associated with distinct cellular mechanisms for survival, confirming the diverse physiology underlying these phenotypes. However, there was one cellular location, the vacuole, and one functional category, cellular transport, that were common to all phenotypes. These categories represent the effector compartment and the annotated function of Nhx1, respectively. Genes with multiple shared null phenotypes were enriched for vacuole membrane trafficking function, further supporting the significance of this observation (Fig. 6a). Of a total 1868 null strains identified with at least one growth phenotype similar to nhx1Δ, 326 displayed growth sensitivity to at least two conditions, 217 of which were sensitive to both NaCl and KCl, characteristic of involvement in the cellular response to osmotic shock. Fewer strains (Fig. 6b and Table 1) showed growth sensitivity to at least three conditions, including some late-acting ESCRT mutants, consistent with the data shown in Fig. 1. Approximately 60% of this gene set had established roles in cellular transport compared with ∼17% of the yeast genome (p = 4.9 × 10−7). Notable in this highly selective set (Table 1) were genes with known functions in endosome trafficking and fusion, including the Rab GTPase Ypt7, its upstream activator Mon1 (guanine nucleotide exchange factor) (38), and its downstream effector Vps41 (39). Thus, an unbiased genome-wide approach suggests a functional role for Nhx1 in endosome fusion.
FIGURE 6.
nhx1Δ phenocopy screen of single gene deletions. a, distribution of functional categories (upper) or cellular location (lower) as annotated by the MIPS Database of genes identified by mutant growth sensitivity to at least one (≥1) or more (≥2, ≥3) of four conditions as described under “Results.” *, p < 0.01. Note that functions and locations that are significantly enriched closely overlap with those of Nhx1, including cellular transport (function) and the endosome/vacuole (location). b, increasing selectivity of mutants with multiple shared nhx1Δ phenotypes. Of the ResGen yeast collection of 4828 viable single gene deletions, 1825 strains shared at least one of four phenotypes with nhx1Δ. Most had only one phenotype, with the number of mutants sharing multiple phenotypes falling steeply. Only two mutants (las17Δ and rcy1Δ) shared all four nhx1Δ phenotypes under the screen conditions.
TABLE 1.
Genes deletions that phenocopy nhx1Δ
The viable single gene deletion yeast collection was screened for growth sensitivity to four nhx1Δ phenotypes: 15 μg/ml hygromycin B (HygB), pH 2.7, 800 mm KCl, and 800 mm NaCl. Gene deletions showing sensitivity to at least three of these four conditions after 19 h growth are listed according to their annotated function. GEF, guanine nucleotide exchange factor; WASP, Wiskott-Aldrich syndrome protein; PtdIns(3,5)P2, phosphatidylinositol 3,5-bisphosphate.
| Gene | Function | HygB | pH 2.7 | KCl | NaCl |
|---|---|---|---|---|---|
| MVB formation (three) | |||||
| BRO1 | Recruits deubiquitylase Doa4 to endosome, regulates membrane scission | + | + | + | |
| VPS4 | AAA-ATPase, ESCRT-III disassembly | + | + | + | |
| VPS36 | ESCRT-II subunit, Ub-dependent protein sorting | + | + | + | |
| Vacuole fusion (four) | |||||
| MON1 | GEF for vacuole Rab GTPase Ypt7 | + | + | + | |
| VAM7 | SNAP25 ortholog, Q-SNARE | + | + | + | |
| VPS41 | Ypt7 effector, component of HOPS | + | + | + | |
| YPT7 | Vacuole Rab GTPase | + | + | + | |
| Endosome trafficking and fusion (six) | |||||
| PEP7/VAC1 | Ortholog of mammalian EEA1 | + | + | + | |
| RCY1 | Effector for Rab GTPases Ypt31 and Ypt32 | + | + | + | + |
| SYS1 | Arl3 receptor on Golgi, multicopy suppressor of ypt6Δ | + | + | + | |
| VPS1 | Dynamin-like, possible role in membrane fission | + | + | + | |
| VPS9 | GEF for endosomal Rab GTPase Vps21 | + | + | + | |
| VPS35 | Component of retromer | + | + | + | |
| Actin cytoskeleton (three) | |||||
| LAS17/BEE1 | WASP ortholog, Arp2/3-mediated actin nucleation | + | + | + | + |
| SAC6 | Fimbrin ortholog, actin filament stability | + | + | + | |
| VRP1 | Verprolin (WIP ortholog) binds WASP to remodel actin | + | + | + | |
| Membrane lipid distribution (three) | |||||
| OPI1 | Transcription factor, pH-sensitive regulation of phospholipid biosynthesis genes | + | + | + | |
| SAC3 | PtdIns(3,5)P2 phosphatase required for endocytic trafficking | + | + | + | |
| ERG24 | Ergosterol biosynthesis | + | + | + | |
| Cell signaling (four) | |||||
| ARG82 | Inositol polyphosphate multikinase | + | + | + | |
| CNB1 | Calcineurin B | + | + | + | |
| REF2 | Regulator of Glc7 protein phosphatase I, involved in cation homeostasis | + | + | + | |
| STE11 | MEK kinase (MAPKKK) | + | + | + | |
| Other (11) | |||||
| DHH1 | Cytoplasmic DEx(D/H) box helicase | + | + | + | |
| HSP31 | Possible chaperone and cysteine protease | + | + | + | |
| NST1 | Unknown function, mediates salt stress | + | + | + | |
| OCT1 | Mitochondrial intermediate (metallo)peptidase | + | + | + | |
| PDB1 | Pyruvate dehydrogenase β-subunit | + | + | + | |
| RAI1 | Activator of exoribonuclease Rat1 | + | + | + | |
| SPT20 | Subunit of SAGA transcriptional regulation complex | + | + | + | |
| UIP5 | Unknown function, putative U box ubiquitin ligase | + | + | + | |
| VPH2 | Required for V-ATPase activity | + | + | + | |
| YDR455C | Dubious ORF, overlaps NHX1* | + | + | + | |
| YEL007W | Unknown function, C. albicans ortholog mediates white opaque switching | + | + | + | |
DISCUSSION
Nhx1 Is Not a Typical VPS Class E Gene and Is Not Required for MVB Formation
More than 50 genes are known to be required for CPY trafficking and processing, resulting in the VPS mutant phenotype. These have been grouped into Classes A–F based on vacuolar morphology, with the underlying assumption that genes contributing to the same step in the CPY trafficking pathway would share a common morphological phenotype (40). The VPS classification of NHX1 has been problematic, in keeping with its obscure role in endosome trafficking. Originally, NHX1 was assigned to Class A because of the absence of an obvious defect in vacuolar morphology (40). Subsequently, Bowers et al. (13) showed that nhx1Δ mutants retained cargo in an enlarged prevacuolar compartment, as did other Class E mutants. However, they noted that the Class E phenotype of nhx1Δ was weak because some cargo was returned to the Golgi (Vps10) or delivered to the vacuole (Vma2). Overall, evidence from light microscopic analysis of nhx1Δ supported inclusion in the Class E set. Electron microscopic analysis of a typical Class E mutant, vps28Δ, revealed an aberrant multilamellar structure consisting of stacks of curved membrane cisternae (41). This Class E compartment resulted from failure of the limiting membrane to invaginate and form cargo-containing ILVs, characteristic of the MVB. It was then shown that VPS Class E proteins associated to form distinct complexes, ESCRT-0, -I, -II, and -III, essential for MVB formation (reviewed in Ref. 1). Given its classification with other ESCRT genes, it seemed reasonable to assume that Nhx1 was somehow involved in the MVB pathway, yet despite intensive analysis of the ESCRT pathway, Nhx1 remained “the most mysterious in terms of its role in budding” (42).
We have shown that Nhx1 is not required for cargo sorting, deubiquitylation, or formation of ILVs. Consistent with our findings, Nhx1 is not required for in vitro formation of MVBs from reconstituted ESCRT complexes and giant unilamellar vesicles (29). The absence of an aberrant Class E compartment as determined by ultrastructural analysis in nhx1Δ suggests, at least semantically, that NHX1 is not a Class E gene.
Nhx1 May Function in Organelle Transport and Heterotypic Endosome-Vacuole Fusion
Gene deletions that share multiple growth phenotypes with nhx1Δ offer novel insight into potential shared functions. One such example is Rcy1, a downstream effector of Rab GTPases Ypt31 and Ypt32 responsible for Snc1 (R-SNARE)-mediated fusion of the endosome to the trans-Golgi network membrane (43). Another Rab effector, Vps41, was included in this selective set, along with its cognate Rab partner Ypt7 and guanine nucleotide exchange factor protein Mon1, which function together to regulate endosome-vacuole fusion. Previously, we showed that the Rab GTPase-activating protein Gyp6 is a negative regulator of Nhx1 function (32). Gyp6 is one of a family of Rab GTPase-activating proteins that interact promiscuously with the Ypt/Rab family of GTPases, including Ypt6 and Ypt7, to terminate GTP signaling (44). Ypt6 is localized to the Golgi, where it is likely to regulate incoming and outgoing vesicle trafficking (45). Thus, Ypt6, on the Golgi, and Ypt31/32, possibly on the endosome, may coordinate function with Nhx1 to mediate retrograde membrane fusion events. Ypt7, the other target of Gyp6, is the Rab GTPase responsible for membrane fusion at the vacuole (46), although YPT7 overexpression affects retrograde trafficking as well (47). Also included in this pathway and selective phenocopy set is Vam7, a Q-SNARE responsible for membrane fusion at the vacuole and endosomes (48). Although predominantly found on endosomes (12, 13), Nhx1 promotes initiation of the first rounds of homotypic vacuolar fusion, a requirement that could be overcome by excess amounts of Vam7 (49). As Ypt7 and Vps41 are proposed to regulate endosome-vacuole fusion as well as homotypic vacuole fusion (50), we speculate that Nhx1 function drives Rab-mediated endosome fusion events either with the vacuole (anterograde) or Golgi (retrograde).
It has been proposed that reorganization of the actin cytoskeleton during early and late stages of the endocytic pathway allows vesicle formation as well as vesicle movement away from the originating membrane along actin comets (e.g. Ref. 51). In this context, we found several mutants defective in organization of the actin cytoskeleton to phenocopy nhx1Δ (vrp1Δ, sac3Δ, sac6Δ, and las17Δ; p = 1.45 × 10−3). Under the control of the small GTPases Rho1 and Cdc42, Vrp1 is proposed to drive actin reorganization prior to trans-SNARE pairing required for homotypic vacuole fusion, presumably to strip actin from the intermembrane space, permitting trans-bilayer contact (52). Similarly, Las17/Bee1, an ortholog of mammalian Wiskott-Aldrich syndrome protein, recruits Vrp1 to rearrange the organellar actin cytoskeletal coat, accommodating organellar fusion and endocytosis (53–55). There is ample evidence linking mammalian plasma membrane Na+/H+ exchangers to the actin cytoskeleton. Thus, H+ transport by NHE1 is stimulated by Rho signaling (56) and drives Cdc42 activation (57) and actin remodeling required for plasma membrane deformation, thereby permitting mammalian cell migration, differentiation, and division (58). Given that H+ transport by Nhx1 drives endocytic trafficking (8) and membrane fusion (49) in yeast, it remains to be determined whether spatial and temporal control of actin reorganization is an evolutionarily conserved function of Na+/H+ exchangers required for remodeling membranes at numerous sites within the cell.
In summary, detailed analysis of the MVB pathway failed to reveal a requirement for Nhx1. On the other hand, an unbiased phenomics approach identified a limited number of gene products that could function in concert with Nhx1 to mediate membrane trafficking or fusion at the endosome and vacuole. While this study was in review, Kanazawa and co-workers (59) used a different but complementary approach of yeast lysates and an indirect assay of dye uptake to demonstrate a modest (30%) decrease in ILV formation in the absence of Nhx1. Similarly, they found that cargo sorting of GFP-CPS did occur in nhx1Δ with scant impairment. All things considered, we hypothesize that rather than being required for ILV formation, ion translocation by Nhx1 coordinates actin reorganization and Rab signaling to permit endosomal membrane fusion events. Phenotype analysis suggests that ESCRT-III might be involved not only in ILV formation but also in Rab signaling/endolysosome fusion. Further studies will be needed to clarify the role of pH and ion gradients in Nhx1-mediated endosome fusion.
Supplementary Material
Acknowledgments
We thank Yongqiang Zhang for making the double knock-out strains used in this study, Matthew West for electron microscopy, and Brooke Weckselblatt for experimental assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK054214 (to R. R.) and GM065505 (to G. O.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
G. Odorizzi and M. A. Wemmer, unpublished data.
- MVB
- multivesicular body
- VPS
- vacuole protein sorting
- CPY
- carboxypeptidase Y
- CPS
- carboxypeptidase S
- ILV
- intralumenal vesicle
- APG
- arginine/phosphate/glucose
- Ub
- ubiquitin.
REFERENCES
- 1. Babst M. (2005) Traffic 6, 2–9 [DOI] [PubMed] [Google Scholar]
- 2. Gould S. J., Booth A. M., Hildreth J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10592–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. von Schwedler U. K., Stuchell M., Müller B., Ward D. M., Chung H. Y., Morita E., Wang H. E., Davis T., He G. P., Cimbora D. M., Scott A., Kräusslich H. G., Kaplan J., Morham S. G., Sundquist W. I. (2003) Cell 114, 701–713 [DOI] [PubMed] [Google Scholar]
- 4. Bowers K., Lottridge J., Helliwell S. B., Goldthwaite L. M., Luzio J. P., Stevens T. H. (2004) Traffic 5, 194–210 [DOI] [PubMed] [Google Scholar]
- 5. Brett C. L., Donowitz M., Rao R. (2005) Am. J. Physiol. Cell Physiol. 288, C223–C239 [DOI] [PubMed] [Google Scholar]
- 6. Nass R., Cunningham K. W., Rao R. (1997) J. Biol. Chem. 272, 26145–26152 [DOI] [PubMed] [Google Scholar]
- 7. Nass R., Rao R. (1999) Microbiology 145, 3221–3228 [DOI] [PubMed] [Google Scholar]
- 8. Brett C. L., Tukaye D. N., Mukherjee S., Rao R. (2005) Mol. Biol. Cell 16, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gilfillan G. D., Selmer K. K., Roxrud I., Smith R., Kyllerman M., Eiklid K., Kroken M., Mattingsdal M., Egeland T., Stenmark H., Sjøholm H., Server A., Samuelsson L., Christianson A., Tarpey P., Whibley A., Stratton M. R., Futreal P. A., Teague J., Edkins S., Gecz J., Turner G., Raymond F. L., Schwartz C., Stevenson R. E., Undlien D. E., Strømme P. (2008) Am. J. Hum. Genet. 82, 1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morrow E. M., Yoo S. Y., Flavell S. W., Kim T. K., Lin Y., Hill R. S., Mukaddes N. M., Balkhy S., Gascon G., Hashmi A., Al-Saad S., Ware J., Joseph R. M., Greenblatt R., Gleason D., Ertelt J. A., Apse K. A., Bodell A., Partlow J. N., Barry B., Yao H., Markianos K., Ferland R. J., Greenberg M. E., Walsh C. A. (2008) Science 321, 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franke B., Neale B. M., Faraone S. V. (2009) Hum. Genet. 126, 13–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nass R., Rao R. (1998) J. Biol. Chem. 273, 21054–21060 [DOI] [PubMed] [Google Scholar]
- 13. Bowers K., Levi B. P., Patel F. I., Stevens T. H. (2000) Mol. Biol. Cell 11, 4277–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goldstein A. L., McCusker J. H. (1999) Yeast 15, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 15. Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. (1988) Mol. Cell. Biol. 8, 4936–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gerhardt B., Kordas T. J., Thompson C. M., Patel P., Vida T. (1998) J. Biol. Chem. 273, 15818–15829 [DOI] [PubMed] [Google Scholar]
- 17. Richter C., West M., Odorizzi G. (2007) EMBO J. 26, 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li R. J. (1997) Cell Biol. 136, 649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barwell K. J., Boysen J. H., Xu W., Mitchell A. P. (2005) Eukaryot. Cell 4, 890–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Davies B. A., Azmi I. F., Payne J., Shestakova A., Horazdovsky B. F., Babst M., Katzmann D. J. (2010) Mol. Biol. Cell 21, 3396–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Odorizzi G., Katzmann D. J., Babst M., Audhya A., Emr S. D. (2003) J. Cell Sci. 116, 1893–1903 [DOI] [PubMed] [Google Scholar]
- 22. Vida T. A., Emr S. D. (1995) J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katzmann D. J., Babst M., Emr S. D. (2001) Cell 106, 145–155 [DOI] [PubMed] [Google Scholar]
- 24. Luhtala N., Odorizzi G. (2004) J. Cell Biol. 166, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cowles C. R., Snyder W. B., Burd C. G., Emr S. D. (1997) EMBO J. 16, 2769–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nickerson D. P., West M., Odorizzi G. (2006) J. Cell Biol. 175, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nickerson D. P., Russell M. R., Odorizzi G. (2007) EMBO Rep. 8, 644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gaxiola R. A., Rao R., Sherman A., Grisafi P., Alper S. L., Fink G. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 1480–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wollert T., Hurley J. H. (2010) Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Papa F. R., Hochstrasser M. (1993) Nature 366, 313–319 [DOI] [PubMed] [Google Scholar]
- 31. Wemmer M., Azmi I., West M., Davies B., Katzmann D., Odorizzi G. (2011) J. Cell Biol. 192, 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ali R., Brett C. L., Mukherjee S., Rao R. (2004) J. Biol. Chem. 279, 4498–4506 [DOI] [PubMed] [Google Scholar]
- 33. Xu W., Smith F. J., Jr., Subaran R., Mitchell A. P. (2004) Mol. Biol. Cell 15, 5528–5537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis D. A. (2009) Curr. Opin. Microbiol. 12, 365–370 [DOI] [PubMed] [Google Scholar]
- 35. Hayashi M., Fukuzawa T., Sorimachi H., Maeda T. (2005) Mol. Cell. Biol. 25, 9478–9490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brett C. L., Kallay L., Hua Z., Green R., Chyou A., Zhang Y., Graham T. R., Donowitz M., Rao R. (2011) PLoS ONE 6, e17619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mukherjee S., Kallay L., Brett C. L., Rao R. (2006) Biochem. J. 398, 97–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nordmann M., Cabrera M., Perz A., Bröcker C., Ostrowicz C., Engelbrecht-Vandré S., Ungermann C. (2010) Curr. Biol. 20, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 39. Brett C. L., Plemel R. L., Lobinger B. T., Vignali M., Fields S., Merz A. J. (2008) J. Cell Biol. 182, 1141–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Raymond C. K., Howald-Stevenson I., Vater C. A., Stevens T. H. (1992) Mol. Biol. Cell 3, 1389–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rieder S. E., Banta L. M., Köhrer K., McCaffery J. M., Emr S. D. (1996) Mol. Biol. Cell 7, 985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hurley J. H., Emr S. D. (2006) Annu. Rev. Biophys. Biomol. Struct. 35, 277–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen S. H., Chen S., Tokarev A. A., Liu F., Jedd G., Segev N. (2005) Mol. Biol. Cell 16, 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Strom M., Vollmer P., Tan T. J., Gallwitz D. (1993) Nature 361, 736–739 [DOI] [PubMed] [Google Scholar]
- 45. Luo Z., Gallwitz D. (2003) J. Biol. Chem. 278, 791–799 [DOI] [PubMed] [Google Scholar]
- 46. Haas A., Scheglmann D., Lazar T., Gallwitz D., Wickner W. (1995) EMBO J. 14, 5258–5270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Balderhaar H. J., Arlt H., Ostrowicz C., Bröcker C., Sündermann F., Brandt R., Babst M., Ungermann C. (2010) J. Cell Sci. 123, 4085–4094 [DOI] [PubMed] [Google Scholar]
- 48. Schwartz M. L., Merz A. J. (2009) J. Cell Biol. 185, 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qiu Q. S., Fratti R. A. (2010) J. Cell Sci. 123, 3266–3275 [DOI] [PubMed] [Google Scholar]
- 50. Peplowska K., Markgraf D. F., Ostrowicz C. W., Bange G., Ungermann C. (2007) Dev. Cell 12, 739–750 [DOI] [PubMed] [Google Scholar]
- 51. Chang F. S., Stefan C. J., Blumer K. J. (2003) Curr. Biol. 13, 455–463 [DOI] [PubMed] [Google Scholar]
- 52. Eitzen G., Thorngren N., Wickner W. (2001) EMBO J. 20, 5650–5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dewar H., Warren D. T., Gardiner F. C., Gourlay C. G., Satish N., Richardson M. R., Andrews P. D., Ayscough K. R. (2002) Mol. Biol. Cell 13, 3646–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eitzen G., Wang L., Thorngren N., Wickner W. (2002) J. Cell Biol. 158, 669–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fratti R. A., Jun Y., Merz A. J., Margolis N., Wickner W. (2004) J. Cell Biol. 167, 1087–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tominaga T., Barber D. L. (1998) Mol. Biol. Cell 9, 2287–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Frantz C., Karydis A., Nalbant P., Hahn K. M., Barber D. L. (2007) J. Cell Biol. 179, 403–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Denker S. P., Barber D. L. (2002) J. Cell Biol. 159, 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mitsui K., Koshimura Y., Yoshikawa Y., Matsushita M., Kanazawa H. (2011) J. Biol. Chem. 286, 37625–37638 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.