Background: Cholesterol metabolism is critical in the chronic phase of Mycobacterium tuberculosis infection.
Results: A cholesterol metabolite structure and an enzyme activity responsible for its degradation were determined.
Conclusion: The igr operon encodes the enzymes that catalyze the final three steps in cholesterol side-chain degradation.
Significance: Insight into the function of enzymes encoded in the igr operon is important for understanding the role of cholesterol metabolism in pathogenesis.
Keywords: Cholesterol Metabolism, Enzymes, Isotopic Tracers, Lipid Structure, Microbiology, Mycobacterium Tuberculosis, Acyl-CoA Dehydrogenase, Steroid Degradation
Abstract
Mycobacterium tuberculosis, the bacterium that causes tuberculosis, imports and metabolizes host cholesterol during infection. This ability is important in the chronic phase of infection. Here we investigate the role of the intracellular growth operon (igr), which has previously been identified as having a cholesterol-sensitive phenotype in vitro and which is important for intracellular growth of the mycobacteria. We have employed isotopically labeled low density lipoproteins containing either [1,7,15,22,26-14C]cholesterol or [1,7,15,22,26-13C]cholesterol and high resolution LC/MS as tools to profile the cholesterol-derived metabolome of an igr operon-disrupted mutant (Δigr) of M. tuberculosis. A partially metabolized cholesterol species accumulated in the Δigr knock-out strain that was absent in the complemented and parental wild-type strains. Structural elucidation by multidimensional 1H and 13C NMR spectroscopy revealed the accumulated metabolite to be methyl 1β-(2′-propanoate)-3aα-H-4α-(3′-propanoic acid)-7aβ-methylhexahydro-5-indanone. Heterologously expressed and purified FadE28-FadE29, an acyl-CoA dehydrogenase encoded by the igr operon, catalyzes the dehydrogenation of 2′-propanoyl-CoA ester side chains in substrates with structures analogous to the characterized metabolite. Based on the structure of the isolated metabolite, enzyme activity, and bioinformatic annotations, we assign the primary function of the igr operon to be degradation of the 2′-propanoate side chain. Therefore, the igr operon is necessary to completely metabolize the side chain of cholesterol metabolites.
Introduction
Mycobacterium tuberculosis is the causative agent of tuberculosis, which accounts for ∼1.4 million deaths annually (1). M. tuberculosis establishes an infection in macrophages (Mφ)4 by preventing phagosome-lysosome fusion and by manipulating the host immune response. Additionally, infected Mφs orchestrate the formation of a granuloma, the hallmark pathologic lesion associated with a tuberculosis infection (2). Isolated within a granuloma, M. tuberculosis can persist for decades. The success of M. tuberculosis as a pathogen is attributable at least in part to the ability of the bacterium to utilize the available host-derived nutrients encountered during all stages of infection. Several lines of evidence indicate that M. tuberculosis shifts its metabolism to utilize preferentially host-derived lipid nutrients during an infection. Fatty acids, but not carbohydrates, are able to stimulate respiration in M. tuberculosis freshly harvested from mouse lungs (3). Additionally, mutants defective in the glyoxylate/methyl citrate pathways (icl) or gluconeogenesis (pckA) are attenuated in Mφ and/or murine infection models (4–6).
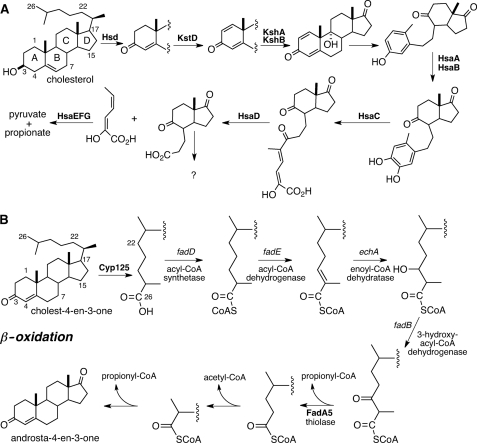
A large cholesterol degradation locus (∼83 genes) has been recently described in the M. tuberculosis genome, and this locus encodes many but not all of the enzymes necessary to degrade cholesterol (7–9). It is well established that M. tuberculosis can degrade cholesterol in vitro, and the bacillus requires several cholesterol degradation genes for full virulence in infection models (8, 10–12). The genes and encoded enzymes necessary for the metabolism of the A and B rings of cholesterol have been identified and characterized to varying extents (10, 13–19) (Fig. 1A). However, the fate of the C and D rings is unknown. Moreover, the side chain is predicted to be metabolized by β-oxidation (9, 20, 21) (Fig. 1B), but the genes encoding these enzymes have yet to be precisely mapped.
FIGURE 1.
M. tuberculosis cholesterol catabolic pathway. The enzymatic steps required for cholesterol metabolism are separated into steroid ring degradation (A) and side chain β-oxidation (B). Enzymes known to catalyze the depicted transformation are indicated in boldface. Catalytic steps for which the enzyme has not been identified are labeled with the predicted gene type.
The intracellular growth (igr) operon is located in the 83-gene cholesterol degradation locus and is required for in vitro growth on cholesterol as a sole carbon source but is not required for growth on fatty acids (8, 12). Growth of a Δigr knock-out strain of M. tuberculosis is attenuated in resting Mφs and early in the infection process in immunocompetent mice. However, this phenotype is minimized in INF-γ activated Mφ, which correlates with the onset of the adaptive immune response in mouse infections (22). Interestingly, the Δigr knock-out strain displays a cholesterol-sensitive phenotype in vitro when the bacteria enter a static growth phase in glycerol and glucose-containing media supplemented with low concentrations of cholesterol (0.1 mm) (12). This cholesterol-sensitive phenotype is characterized by the bacterium maintaining an intact respiratory electron transport chain in the absence of cell division (12). Moreover, the in vitro and in vivo cholesterol-sensitive phenotype of the Δigr knock-out can be suppressed by genetically disrupting yrbE4a/Rv3501c, which is the first gene in the operon that encodes the multisubunit cholesterol importer, Mce4 of M. tuberculosis (11, 12). These data suggest that disruption of the igr operon results in accumulation of a toxic cholesterol-derived metabolite and that this toxicity can be relieved by blocking cholesterol uptake.
Gene annotation and transcriptional regulation are consistent with the igr genes comprising an operon that encodes an incomplete β-oxidation sequence in sterol metabolism (22). The computationally annotated functions are a lipid transfer protein (ltp2/Rv3540c), two MaoC-like hydratases (Rv3541c and Rv3542c), two acyl-CoA dehydrogenases (fadE29/Rv3543c and fadE28/Rv3544c), and a cytochrome P450 (cyp125/Rv3545c). Only Cyp125 has been metabolically and enzymatically characterized. The annotated β-oxidation functions of the remainder of the igr operon have yet to be verified or specific substrates identified.
Heavy isotopes, either radioactive or stable, are essential tools for tracing the metabolic fate of carbon in living cells. Cholesterols labeled at either C4 or C26 with 14C or 13C are commercially available, and both have been used to analyze the final fate of these atoms in M. tuberculosis metabolism. The cholesterol C4 carbon is metabolized to CO2, and the C26 carbon is assimilated as lipid via propionyl-CoA. The use of these commercial reagents is limited to investigating A-ring and early side-chain metabolism by M. tuberculosis. The fates of the remaining carbons have not been elucidated, and late-stage intermediates of the pathway have not been identified or isolated.
Previous work indicated that cholest-4-ene-3-one is an in vitro substrate of Cyp125 (23). Recombinant Cyp125 catalyzes the oxygenation of cholesterol and cholest-4-ene-3-one to 3β-hydroxy-5-cholesten-26-oic acid or cholest-4-en-3-one-26-oic acid, respectively (18, 23). In the M. tuberculosis CDC1551 strain, the cyp125/Mt3649 gene is required for the bacterium to grow on cholesterol as a sole carbon source in vitro, whereas in strain M. tuberculosis H37Rv, the Δcyp125/Rv3545c mutant can grow on cholesterol as a sole carbon source due to the compensatory enzymatic activity of Cyp142/Rv3518c (24). The M. tuberculosis H37Rv Δigr mutant strain metabolizes C4 and C26 of cholesterol, indicating that the Cyp142/Rv3518c monooxygenase is functional in this mutant and that disruption of cholesterol degradation occurs after the C26 hydroxylation step (12).
To pursue a full analysis of cholesterol metabolism in the Δigr mutant strain of M. tuberculosis H37Rv, we required tools that trace the fate of the B-D rings and additional carbons of the sterol side chain. Here we describe the biosynthetic preparation of isotopically labeled [1,7,15,22,26-14C]cholesterol or [1,7,15,22,26-13C]cholesterol. The distribution of labels throughout the sterol ring system and side chain makes these reagents useful metabolic tracers to study cholesterol metabolism in M. tuberculosis.
In this study we employed lipoprotein (LDL) [1,7,15,22,26-14C]cholesterol and LDL [1,7,15,22,26-13C]cholesterol as tools to further investigate the cholesterol-derived metabolite profile of the M. tuberculosis H37Rv Δigr mutant strain and to aid in the isolation and structure elucidation of a key metabolite. Culture supernatants from the Δigr mutant accumulate a cholesterol-derived metabolite not observed in H37Rv wild-type or complemented strains. Multidimensional NMR and mass spectral analysis revealed the structure of this cholesterol-derived catabolite to be a late stage metabolic product: methyl 1β-(2′-propanoate)-3aα-H-4α(3′-propanoic acid)-7aβ-methylhexahydro-5-indanone, 1. Using synthetic substrates analogous to this metabolite, we verified the catalytic activity of the purified, recombinant FadE28-FadE29 protein complex, encoded in the igr operon, to be dehydrogenation of the 2′-propanoate-CoA side chain. We conclude the igr operon is required for degradation of the 2′-propanoate side chain fragment during metabolism of cholesterol by M. tuberculosis.
EXPERIMENTAL PROCEDURES
General Materials and Methods
M. tuberculosis (H37Rv) WT, Δigr, and complemented Δigr strains described in Chang et al. (22) were grown at 37 °C in Middlebrook 7H9 liquid media (BD Biosciences) supplemented with 10% oleate-albumin-dextrose-NaCl-catalase (OADC). High resolution mass spectrometry was performed on an LTQ-Orbitrap (Thermo Scientific) equipped with an electrospray source operating in positive ion mode with an ionization voltage of 1.8 kV, capillary voltage of 43 V, and tube lens of 150 V. Matrix-assisted laser desorption ionization (MALDI)-TOF spectra were acquired on a Bruker AutoFlex II spectrometer. NMR spectra were acquired on Bruker 800- or 900-MHz microcryoprobe or cryoprobe NMR spectrometers (New York Structural Biology Center). Coenzyme A, propionyl-CoA, and (RS)-[2-13C]-mevalonolactone were purchased from Sigma. (RS)-[2-14C]mevalonolactone was purchased from PerkinElmer Life Sciences. Vitamin D2 and 4,22-stigmastadien-3-one were purchased from Acros Organics and MP Biomedicals, respectively.
Isotopic Labeling of LDL Cholesterol
HepG2 human liver cells were grown to 80% confluence in HepG2 growth media (DMEM, 10% fetal calf serum, 20 mm l-glutamine, 100 IU/ml penicillin, 10 μg/ml streptomycin, and 10 mm HEPES). Media were then replaced with HepG2 growth media supplemented with 5 μm mevastatin and 0.8 μg/ml (RS)-mevalonolactone (natural abundance) or 0.8 μg/ml (RS)-[2-13C]mevalonolactone or 6.25 μCi of (RS)-[2-14C]-mevalonolactone. Cells were then grown for 4–5 days. Culture supernatants were then harvested and sterilized by filtration. Culture supernatant-derived LDL particles were concentrated, and unincorporated mevalonolactone was removed by ultrafiltration through a 100-kDa molecular weight cutoff filter. Removal of unincorporated mevalonolactone was performed by concentrating the supernatants 10-fold followed by two 100-ml washes with sterile PBS. The filter flow-through was monitored by scintillation counting, and the LDL was deemed free of [14C]mevalonolactone contamination when radioactive counts in the filter flow-through were no higher than background. LDL [13C]cholesterol was isolated in the same manner as the LDL [14C]cholesterol preparation, and the LDL [13C]cholesterol sample underwent the ultrafiltration procedure to remove unincorporated [13C]mevalonolactone.
Biochemical Analysis of LDL-derived 14C-labeled Lipids
Total lipids were extracted by the Bligh-Dyer method from the aqueous-soluble LDL particles (25). Briefly, the aqueous lipid solution was phase partitioned with 2:1 CHCl3:MeOH (v/v), and the organic solvent extractable material was dried by evaporation under a N2 stream. The dried lipids were resuspended in 25:24:4 CHCl3:MeOH:H2O (v/v/v), the phases were partitioned, the resulting organic solvent layer was saved to a new tube and dried by evaporation under a N2 stream. To analyze the apolar LDL lipids by TLC, the LDL lipid extracts were resuspended in CHCl3, and radioactivity was determined by scintillation counting. 2500 cpm of the apolar LDL-derived lipids was resolved by TLC on precoated F254 aluminum-backed silica plates (Sigma) in toluene:acetone (99:1, v/v). The TLC plates were visualized via molybdophosphoric acid staining and charring as described in Dobson et al. (26). Radiolabeled lipids were visualized by autoradiography of the TLC plates using phosphorimaging.
Characterization of LDL [1,7,15,22,26-13C]Cholesterol
Isolated LDL [13C]cholesterol was analyzed by MALDI-TOF MS in reflectron, positive ion mode with 2,5-dihydroxybenzoic acid matrix prepared in 0.1% TFA, 50% CH3CN. A [13C]DEPT135 NMR spectrum was acquired on a 800 MHz microcryoprobe in CDCl3 to confirm the positions of 13C labeling.
Metabolic Labeling of M. tuberculosis with LDL [14C]Cholesterol
The M. tuberculosis WT, Δigr, and complemented Δigr mutant cultures were grown in 7H9 OADC for 6 days, and the bacteria were resuspended in 7H9 OADC medium supplemented with LDL [14C]cholesterol (5000 cpm/ml). After 2 weeks of growth in the presence of LDL [14C]cholesterol, the bacterial cultures were centrifuged at 3000 rpm for 20 min to yield a cell pellet and culture supernatant. The supernatants were harvested by filtration through a 0.22 μm filter unit, and the resulting cell pellets and culture supernatants were used for lipid metabolite isolation.
Metabolic Labeling of M. tuberculosis with LDL [13C]Cholesterol
The M. tuberculosis WT, Δigr, and complemented Δigr mutant cultures were grown in 7H9 OADC for 6 days, and the cell pellet was resuspended in 7H9 OADC medium supplemented containing LDL [13C]cholesterol (50–60 μg/ml) of culture. Total cholesterol in the LDL [13C]cholesterol preparations was determined using the Amplex® Red cholesterol determination assay (Invitrogen). After 2 weeks of growth in the presence of LDL [13C]cholesterol, the bacterial cultures were centrifuged at 3000 rpm for 20 min to yield a cell pellet and culture supernatant. The supernatants were harvested by filtration through a 0.22-μm filter unit, and the resulting cell pellets and culture supernatants were used for lipid metabolite isolation.
Growth of M. tuberculosis in the Presence of Free Cholesterol
The M. tuberculosis WT, Δigr, and complemented Δigr mutant cultures were grown in 7H9 OADC for 6 days, and the cell pellet was resuspended in 7H9 OADC medium supplemented with 0.1 mm cholesterol added in a 1:1 tyloxapol:EtOH (v/v) solution as described (11). After 2 weeks of growth in the presence of free cholesterol, the bacterial cultures were centrifuged at 3000 rpm for 20 min to yield a cell pellet and culture supernatant. The supernatants were harvested by filtration through a 0.22-μm filter unit, and the resulting cell pellets and culture supernatants were used for lipid metabolite isolation.
Biochemical Extraction of Cholesterol-derived Bacterial Lipid Metabolites
The bacterial cell pellets were washed once in distilled H2O, and the pellet was extracted twice with 75 ml of EtOAc for 24 h. The organic layers were pooled and dried by evaporation under a N2 stream. The cell-free culture supernatants were extracted twice with 75 ml of EtOAc for 24 h. The organic layers were pooled and dried by evaporation under a N2 stream. In the case of LDL [14C]cholesterol labeling, the radioactivity in the bacterial metabolite samples was determined by scintillation counting.
LC/MS Analysis of Extracts
Extracts dissolved in MeOH were analyzed by microcapillary liquid chromatography-tandem mass spectrometry with a Dionex 3000 HPLC and a Thermo LTQ Orbitrap mass spectrometer equipped with a custom nanoLC electrospray ionization source. Analytes were separated on a column packed with 10 cm of 5 μm Magic C18 material (Agilent, Santa Clara, CA). A flow rate of 300 nl/min was used with a gradient from 0.1% formic acid, H2O (buffer A) to 0.1% formic acid, 98% CH3CN (buffer B). After analyte loading, the gradient was held constant at 100% buffer A for 5 min followed by a 30-min gradient to 40% buffer B. Then the gradient was switched from 40 to 80% buffer B over 5 min and held constant for 3 min. Finally, the gradient was changed from 80% buffer B to 100% buffer A over 1 min and then held constant at 100% buffer A for 15 more min. Application of a 1.8-kV distal voltage electrosprayed the eluted analytes directly into the ion trap mass spectrometer. Masses were recorded over a 50–1000 m/z range in positive mode.
Data were analyzed with the XCMS software package (27) and by manual inspection. An aligned peak list was generated by XCMS for each sample, including the integrated ion counts. Those ions with integrated intensity less than 1 × 105 were filtered from the sample. The integrated peak areas were then used to calculate -fold changes for pairwise comparisons of H37Rv:Δigr and complement:Δigr. Ions unique to H37Rv, complement, or Δigr were identified by generating a subset of those ions with -fold changes greater than 2.5. The subset was then reduced to only those ions derived from cholesterol by comparing the isotope distributions for the ion in natural abundance and LDL [13C]cholesterol samples (supplemental Table S1).
Purification and Characterization of Metabolite 1
Metabolite 1 was purified by reverse phase HPLC on a Phenomenex Luna C18 column (5 μm, 250 × 10 mm) from extracts of a 500-ml Δigr M. tuberculosis culture grown with unlabeled cholesterol. A flow rate of 3 ml/min was used with a gradient from H2O (buffer A) to MeOH (buffer B). The gradient was held at 100% A for 5 min, then changed to 50% A over 15 min. Next there was a linear change from 50% A to 20% A over 25 min, then to 0% A over 5 min. Finally, the gradient was held at 0% A for 10 min. Fractions containing the desired metabolite were combined and dried for NMR analysis in CD3OD on a 900-mHz cryoprobe spectrometer.
Recombinant Protein Purification and Analysis
The igr operon (Rv3545c-Rv3540c) was cloned from genomic DNA into pET28b (Novagen, Madison, WI) at NdeI and HindIII restriction sites to give construct igr-6. Constructs igr-5 and igr-3 were prepared by deletion of Rv3545c or deletion of Rv3545c/Rv3544c/Rv3543c, respectively, by PCR. Construct igr-1 was prepared by cloning Rv3540c from genomic DNA into pET28b at NdeI and XhoI restriction sites. Each construct introduced an N-terminal hexahistidine tag. Constructs were introduced into BL21(DE3) Escherichia coli, and single colonies were selected on LB plates supplemented with 30 μg/ml kanamycin and cultured in 2 × YT media at 37 °C. Expression was induced at A600 = 0.6–0.8 by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside, and cells were grown overnight at 25 °C. Harvested cells were suspended in 50 mm Tris-HCl, 300 mm NaCl, and 10 mm imidazole buffer at pH 8.0. Cells were lysed by French press, and cellular debris was removed by centrifugation at 125,000 × g for 1 h. Proteins were purified by immobilized metal affinity chromatography using Hisbind resin (Novagen) following the manufacturer's protocol. Eluted protein was analyzed by SDS-PAGE, and observed protein band identities were confirmed by in-gel tryptic digestion and MALDI mass fingerprinting. Briefly, excised bands were washed with CH3CN/H2O followed by reduction with 45 mm DTT in 100 mm ammonium bicarbonate at 56 °C for 45 min. Gel pieces were then covered in 55 mm iodoacetamide in 100 mm ammonium bicarbonate and incubated in the dark for 30 min. The samples were then dried and digested with trypsin in 50 mm ammonium bicarbonate overnight at 37 °C. Peptides were then extracted with 60% CH3CN, 0.1% TFA and dried. The MALDI sample was prepared using a C18-Zip-tip (Millipore). A saturated solution of matrix 3,5-dimethoxy-4-hydroxy-cinnamic acid was prepared in 29.9% EtOH, 70% H2O, 0.1% TFA. MALDI mass spectra were acquired on a Bruker Autoflex II TOF/TOF instrument in positive ion mode. Data were analyzed using Flex-Analysis software and MS-Bridge.
Substrate Synthesis
Carboxylic acids 1β-(2′-propanoic acid)-3aα-H-7aβ-methylhexahydro-4-indanone (28–30) and 3-oxo-4-pregnene-20-carboxylic acid (31) were prepared by ozonolysis of vitamin D2 and 4,22-stigmastadien-3-one, respectively. Briefly, the ozonolysis was performed in CH2Cl2 at −78 °C. The reaction was purged with ozone until the solution turned blue in color, then purged with O2 until the blue dissipated. 1% pyridine was added to the ozonolysis of 4,22-stigmastadiene-3-one to selectively ozonolyze the 22-ene, and the reaction was monitored by TLC (32). Dimethyl sulfide (10 eq) was added, and the reaction was allowed to warm to room temperature overnight. Solvent was removed under reduced pressure, and the product was redissolved in 10% H2O/CH3CN and chilled on ice. Sodium chlorite (10 eq) was added, and the reaction was stirred overnight at room temperature. The reaction was dried under reduced pressure, and the product was extracted from acidified brine with CH2Cl2. The products were purified by silica gel chromatography in 1:4 or 2:3 EtOAc:hexanes. 3-oxo-4-pregnene-20-carboxylic acid: 1H NMR (500 MHz, CDCl3) δ 9.5 (br, 1H, COOH), 5.8 (s, 1H, C4-H), 1.22 (d, 3H, J = 6.5 Hz, C21-H), 1.18(s, 3H, C19-H), 0.73 (s, 3H, C18-H); 13C NMR δ 203 (C3), 126 (C4), 175 (C5), 180 (C22); 1β-(2′-propanoic acid)-3aα-H-7aβ-methylhexahydro-4-indanone: 1H NMR (500 MHz, CDCl3) δ1.30 (d, 3H, J = 6.5 Hz, C3′-H), δ 0.69 (s, 3H, C8-H) 13C NMR δ 213 (C4), 184 (C1′). LC/electrospray ionization-MS indicated a purity of greater than 95%.
The carboxylic acids prepared above as well as butanoic acid, 3-methylbutanoic acid, and 2-methylpropanoic acid were converted to their corresponding acyl-CoA thioesters via the mixed anhydride method. Briefly, Et3N (2 eq) was added to the acid in CH2Cl2 and stirred for 10 min. Then ethyl chloroformate (2 eq) was added dropwise to the reaction on ice. The reaction was allowed to warm to room temperature and stirred for 2 h, then dried by a stream of N2. The mixed anhydride was dissolved in THF and filtered through glass wool into coenzyme A in H2O:THF (1:4), pH 8.0. Ellman's reagent was used to monitor the disappearance of free thiol, the reaction was then acidified to pH 4, and THF was removed. Remaining free acid was extracted into Et2O, and the product in the aqueous layer was purified by C18-reverse phase HPLC. The identities of CoA thioesters were confirmed by LC/MS.
Dehydrogenase Assay
FadE28-FadE29 (3.2 μg/ml) was assayed for dehydrogenase activity with ferrocenium hexafluorophosphate (250 μm) as the artificial electron acceptor and 100 μm substrate in 100 mm HEPES pH 7.4 (33, 34). The formation of product was followed spectrophotometrically at 300 nm and 25 °C. Specific activities were calculated from the initial velocity over the first 100 s of reaction and the extinction coefficient of ferrocenium hexafluorophosphate, 3.4 mm−1 cm−1. Assays were conducted in triplicate, and substrates tested include 1β-(2′-propanoyl-CoA)-3aα-H-7aβ-methylhexahydro-4-indanone 2, 3-oxo-4-pregnene-20-carboxyl-CoA 3, propanoyl-CoA, butyryl-CoA, isovaleryl-CoA, and isobutryrl-CoA. Product formation was confirmed by MALDI-TOF MS. Negative controls without substrate or without enzyme were conducted to check for background reduction of ferrocenium hexafluorophosphate.
RESULTS
Biosynthetic Preparation of Isotopically Labeled LDL Cholesterol
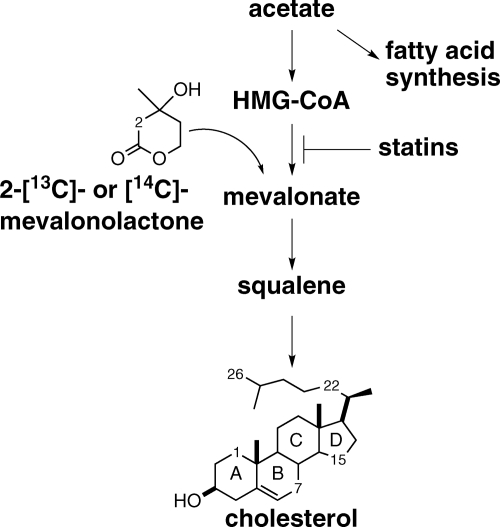
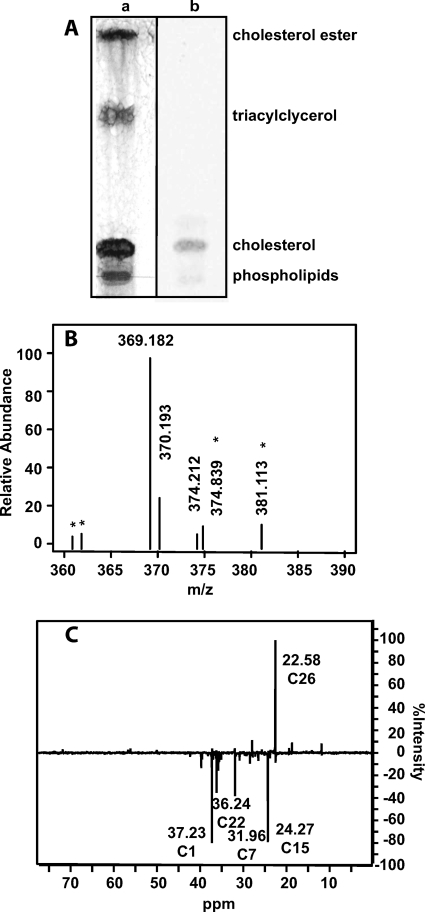
Mevalonate is the first dedicated precursor required for cholesterol biosynthesis (35–37). To metabolically label cholesterol in low density LDL particles, HepG2 (human liver) cells were cultured with either 2-[13C]- or [14C]mevalonolactone to produce LDL particles containing [1,7,15,22,26-13C]cholesterol or [1,7,15,22,26-14C]cholesterol (Fig. 2) (38). Mevalonolactone is the soluble and membrane-permeable form of mevalonate that is assimilated by cellular metabolism. Mevastatin, the HMG-CoA reductase inhibitor, was added to suppress the cellular conversion of unlabeled acetyl-CoA pools to mevalonate, which otherwise would be incorporated into cholesterol and reduce the heavy isotope incorporation. The 13C- or 14C-labeled cholesterol was isolated as soluble LDL particles from the HepG2 culture supernatant, and unincorporated mevalonolactone was removed by ultrafiltration. TLC analysis of the LDL lipids demonstrated that cholesterol and cholesterol esters contained 14C-labeled carbons (Fig. 3A). The LDL-derived phospholipid and triacylglycerol components contained no 14C label, suggesting that the label observed in cholesterol ester is likely not located in the esterified fatty acid. MALDI-TOF mass spectra of LDL-derived [13C]cholesterol contained the [M-H2O+H]+ dehydration product ion (m/z = 369) as well as an [M-H2O+H+5]+ ion (m/z = 374) that corresponds to the expected cholesterol labeled with five 13C atoms (Fig. 3B). The m/z = 369 ion had a natural isotope envelope for the +1 to +4 ions indicating that partially labeled cholesterol was not formed. The isotopic incorporation ranged from 10 to 30% depending on the preparation. DEPT135 NMR analysis was used to confirm the positions of the five 13C-labeled atoms (Fig. 3C). 13C resonances were compared with known cholesterol resonances, confirming labeling at the C1, C7, C15, C22, and C26 positions (39).
FIGURE 2.
13C- or 14C-metabolic labeling of LDL-derived cholesterol. Isotopically labeled 2-[13C] or [14C]mevalonolactone is converted into mevalonate and is incorporated into the cholesterol biosynthesis pathway, resulting in labeled [1,7,5,22,26-13C]cholesterol or [1,7,5,22,26-14C]cholesterol. HMG-CoA, 3-hydroxy-3-methylglutarate coenzyme A.
FIGURE 3.
Analysis of metabolically labeled LDL [13C]- or -[14C]cholesterol. A, shown is a TLC analysis of LDL [14C]cholesterol developed by charring (a) and autoradiography (b). B, shown is a MALDI-TOF MS spectrum of LDL [13C]cholesterol. Peaks labeled with asterisks are matrix ions. C, shown is a DEPT135 spectrum of isolated LDL [13C]cholesterol, confirming the position of isotopically labeled 13C carbons.
A 14C-labeled Cholesterol-derived Metabolite Accumulates in the Δigr Mutant When Supplied LDL [1,7,15,22,26-14C]Cholesterol
M. tuberculosis WT, Δigr, and complement strains were grown in 7H9 OADC media supplemented with LDL [14C]cholesterol or nonradioactive LDL cholesterol. After 2 weeks, extracts were isolated from the pellet and culture media of each culture, separately. LC/MS analysis of the bacterial lipid extracts from nonradioactive LDL cholesterol cultures confirmed that no free cholesterol remained in WT, Δigr, or complemented Δigr extracts. After 2 weeks of culture with LDL [14C]cholesterol, 14C counts were 2-fold higher for Δigr mutant extracts compared with WT or complemented Δigr strains, consistent with complete metabolism to CO2 or low molecular weight carbon in wild type. In the Δigr strain, the metabolism of cholesterol is incomplete. The majority of the remaining 14C counts (85%) in the lipid extracts from the Δigr strain was secreted into the culture supernatant. TLC analysis revealed that a new polar species was formed in the Δigr strain that was absent in extracts from the WT and complemented Δigr strains (supplemental Fig. S1).
Application of LDL [1,7,15,22,26-13C]Cholesterol as an Isotopic Tracer of Cholesterol Metabolism Intermediates by LC/MS
To characterize further the partially degraded cholesterol metabolites in Δigr M. tuberculosis, the bacteria were cultured with LDL [1,7,15,22,26-13C]cholesterol, and the extracts were analyzed by liquid chromatography and high resolution mass spectrometry using XCMS software for feature matching and detection. 13C-labeled, cholesterol-derived metabolites were identified by their unnatural 13C isotopomers. The presence of isotopomers was further confirmed by comparison to natural abundance LDL cholesterol metabolic profiles.
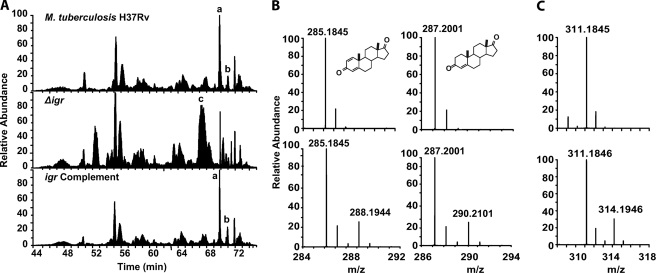
Wild-type M. tuberculosis Accumulates Androst-4-ene-3,17-dione (AD) and Androsta-1,4-diene-3,17-dione (ADD), Whereas the Δigr Mutant Does Not
Four species with 13C labels were observed in the WT and igr complemented strain that were not detected in Δigr extracts (Fig. 4A, supplemental Table S1A and Fig. S2). The two most prominent species, based on the peak area from the extracted ion chromatogram (A and B, Fig. 4A), were ADD and AD. AD and ADD are known intermediates of the cholesterol metabolism pathway and had been previously identified in culture supernatants of M. tuberculosis H37Rv cultured with cholesterol (8). The molecular formula, obtained from high resolution mass data and number of 13C labels match their structures (supplemental Table S1A). The remaining two metabolites observed in WT and igr complement extracts are keto and hydroxy oxidation products of AD and ADD (supplemental Table S1A). Interestingly, AD and ADD were not observed in the extracts isolated from the Δigr mutant, indicating that cholesterol metabolism is most likely blocked before AD and ADD formation in the Δigr mutant strain.
FIGURE 4.
LC/MS analysis of M. tuberculosis H37Rv, Δigr, and igr complement extracts cultured with LDL [13C]cholesterol and natural abundance LDL cholesterol. A, total ion chromatograms of M. tuberculosis H37Rv, Δigr, and igr complement are shown. Only the total ion chromatograms from samples cultured with LDL [13C]cholesterol are shown. Major metabolites are labeled, with a full list in supplemental Table S1. Metabolite peaks A and B were only observed in M. tuberculosis H37Rv and igr complement, and peak C was only observed in Δigr. B, MS of M. tuberculosis H37Rv and igr complement cholesterol derived metabolite peaks A and B, which are ADD and AD, respectively. C, MS of cholesterol-derived metabolite C that accumulates in the Δigr mutant and is absent in the H37Rv and igr complemented strains is shown. The top panels of B and C are from growth on natural abundance LDL cholesterol, and the bottom panels are from growth on isotopically labeled LDL [13C]cholesterol.
Identification of Cholesterol-derived Metabolites Present in Δigr Extracts and Absent in WT and igr-complemented Strains
Several cholesterol-derived metabolites unique to Δigr extracts were identified by the unnatural isotopomer distribution of their parent ions in high resolution mass spectra (supplemental Table S1B and Fig. S2). Peak C was the major component based on peak area of the extracted ion chromatogram (Fig. 3A, supplemental Table S1B), and the major ion had an m/z = 311.1846 (75%) and a minor ion with m/z = 309.1692 (25%). The remaining cholesterol-derived metabolites were present in 10–100-fold lower abundance.
Analysis of Δigr extracts cultured with unlabeled, free cholesterol dissolved in tyloxapol micelles indicated that the metabolic profile is unchanged and is not dependent on culturing with LDL. Free cholesterol was then used as the carbon source to grow sufficient quantities of Δigr cultures to isolate the main metabolite for spectroscopic and structural characterization.
Isolation and Structural Characterization of the Major Cholesterol Metabolite, 1, Formed by the Δigr Strain
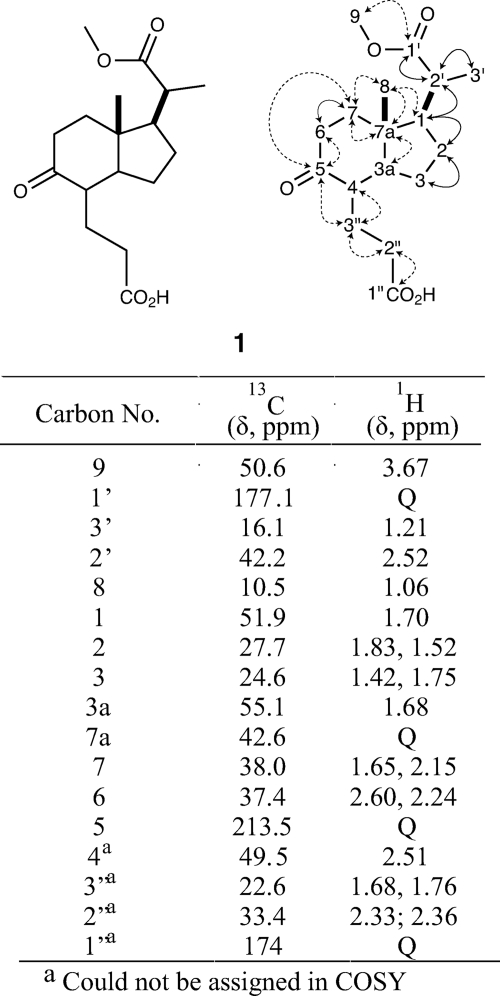
Ethyl acetate extraction of M. tuberculosis H37Rv Δigr cultures grown with free cholesterol followed by fractionation by C18 reverse phase HPLC yielded the major metabolite 1 in ∼85% purity. The metabolite structure was established by tandem mass spectrometry and 13C,1H COSY, 13C,1H HSQC, and HMBC NMR spectroscopy (Figs. 5 and 6).
FIGURE 5.
Structural characterization of Δigr metabolite 1. Structure of 1 was established from MS, tandem MS, and multidimensional NMR data. 1H and 13C NMR signals are listed. Dotted arrows represent HMBC correlations. Solid arrows represent COSY correlations. Only key correlations are displayed.
FIGURE 6.
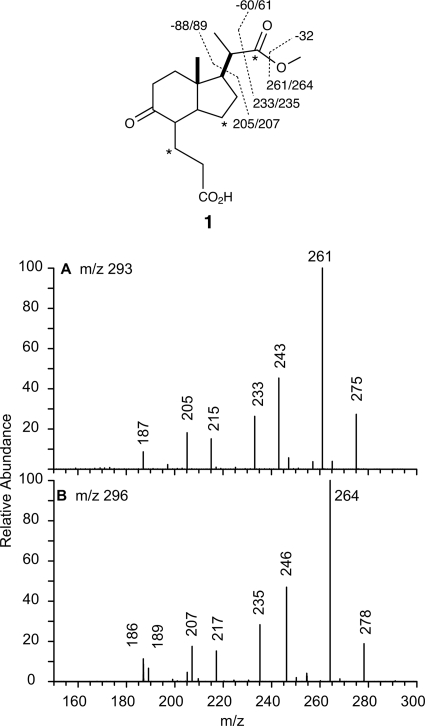
MS/MS/MS fragmentation analysis of Δigr metabolite 1. MS fragmentation data for the [MH-18]+ ion of metabolite 1 are shown. The loss of 18 Da is assumed to result from elimination of water from the enol tautomer. This fragmentation is not shown in the figure. A, [MH-18]+ = m/z 293 (natural abundance) is shown. B, [MH-18]+ = m/z 296 (three 13C labels) is shown. Sites of 13C labeling are indicated by asterisks.
The NMR spectra of 1 clearly mapped an indane carbon skeleton, presumably derived from the C and D rings of cholesterol. In the five-membered ring, the methine proton (δ1.70) correlated to C1 (δ51.8) in the 13C,1H HSQC. The C1 methine correlated to the C2 methylene (δ1.83; δ1.52) by COSY. These methylene protons correlated to C2 (δ27.7) in the 13C,1H HSQC spectrum. The C2 methylene coupled to the C3 (δ24.6) methylene protons (δ1.42; δ1.74). The quaternary carbon, C7a (δ42.6), in the 13C spectrum showed an HMBC correlation to C3a (δ55.1). C3a had an attached methine proton at δ1.68.
The six-membered ring was established from C7a (δ42.58) that correlated to the C8 (δ10.5) methyl protons (δ1.06) and C7 (δ37.9) methylene protons (δ1.65; 2.15) in the HMBC spectrum. The C7 methylene protons (δ1.65;2.15) coupled to the C6 (δ37.3) methylene protons (δ2.24;2.60). A carbonyl was observed in the 13C spectrum at δ213, consistent with a ketone at C5.
This C5 ketone correlated to protons at δ2.60, δ2.24, δ2.15, δ1.68, and δ1.76 in the HMBC. By 13C,1H HSQC, δ1.68 and δ1.76 were attached to C3′′ (δ22.6). Although no correlation to a proton at C4 was observed, this carbon was assigned to a signal of δ49.5 with an attached methine proton at δ2.51 by 13C,1H HSQC correlation. By HMBC, C4 correlated to C3′′ methylene protons (δ1.68; δ1.76). In the 13C,1H HSQC, C2′′ (δ33.4) showed a correlation to methylene protons (δ2.36; δ2.33). In the HMBC, C3′′ correlated to the C2′′ methylene protons. Reciprocally, correlations were observed for C2′′ and the C3′′ methylene protons. The C2′′ methylene protons correlated to C1′′ (δ174.7). Based on the molecular formula of 1, C1′′ is predicted to be a carboxylic acid. However, by HMBC, C1′′ also has long range correlations with a singlet at δ3.70, indicating that the carboxylic acid may be partially esterified. The C1′′-C3′′ propanoic acid group was assigned using long range correlations, making their assignment likely but not definite. COSY correlations were difficult to interpret due to co-purified metabolites of similar structure, primarily the dehydrated 3′-propenoate side chain (m/z = 309.1692), which we identified by mass spectral analysis. The NMR assignments made are consistent with published 1H and 13C data for 3aα-H-4α-(3′-propanoic acid)-7aβ-methylhexahydro-1,5-indadione (also known as DOHNAA) (20, 40).
The presence of a three-carbon 2′-propanoate side chain on the five-membered ring at C1 was readily established. The carboxylate side chain was isolated as the methyl ester. The 1H spectra showed two methyls at δ1.21 (s; 3H) and δ3.67 (d; 3H,), which correlated to C3′ (δ16.1) and C9 (δ50.64), respectively, in the 13C,1H HSQC. The C1′ carbonyl of the methyl ester was observed at δ177 in the 13C NMR spectrum and correlated to the methyl at δ3.67 and C2′ (δ42.2) methine at δ2.52 (dd, 1H) in the HMBC. The C2′ methine is coupled to the C3′ methyl (δ1.21) and C1 (δ51.8) methine (δ1.70) in the COSY spectrum. This assignment revealed that the side chain of cholesterol had been partially degraded, with a 2′-propanoate group remaining on what was formerly the D-ring.
In addition, the structure of the 2′-propanoate side chain was confirmed by tandem MS. Dehydrated parent ions (MH+ - 18) of 12C- and 13C-labeled metabolite 1 (m/z = 293 and 296, respectively) were further fragmented (Fig. 6). A loss of 32 and 50 Da was observed and attributed to the loss of MeOH and MeOH/CO, respectively. A loss of 51 Da was observed for 13C-labeled 1 upon loss of MeOH/CO. This 1-Da increase indicates a loss of 13C label and is consistent with the initial 13C-labeling of cholesterol at C22 of cholesterol. A loss of 88 and 89 Da from m/z 293 and m/z 296 indicates a loss of C4H8O2, which was assigned to the loss of the methyl 2′-propanoate side chain from C1 of 1. The minor metabolite (m/z = 309.1692) corresponds to the same structure with an additional unsaturation. This metabolite has an identical fragmentation and labeling pattern to metabolite 1. The NMR and mass spectral data are consistent with an additional unsaturation in the moiety substituted at C4, which is most likely a 3′-propenoic acid.
We observed ions corresponding to the diacid (m/z = 297.1691) and the dimethyl ester (m/z = 325.2004) forms of 1 in the mass spectral profiles (supplemental Table S1B). In addition, we observed the 3-oxo-4-pregnene-20-carboxylic acid precursor to 1, which has all four steroid rings intact (m/z = 345.2420). Last, we observed the β-oxidation precursor to 3-oxo-4-pregnene-20-carboxylic acid (m/z = 371.2574) that has a five-carbon side chain.
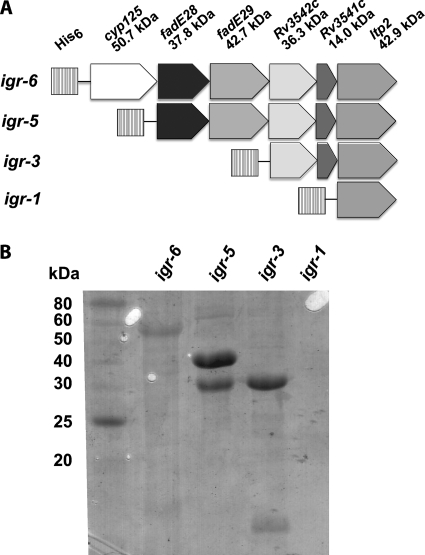
Proteins Encoded in the igr Operon Form Heteromeric Complexes
The native igr operon structure was used to heterologously express all six genes in E. coli using a single construct. The entire operon was cloned into expression vector pET28b, and the first open reading frame of the operon was expressed as an N-terminal His6-tagged protein for purification (Fig. 7A, igr-6). Expression of construct igr-6 resulted in isolation of soluble Cyp125 by immobilized metal ion affinity chromatography purification. Next, cyp125 was deleted to generate construct igr-5. Expression and purification by immobilized metal ion affinity chromatography resulted in co-isolation of FadE28 and FadE29 even though FadE29 did not contain a His6 tag. Therefore, these proteins are isolated as a complex. Then fadE28 and fadE29 were deleted to generate construct igr-3. Upon expression of igr-3 and purification by IMAC, N-His6-tagged Rv3542c and tagless Rv3541c were isolated, again indicating a protein complex was formed. Heterologous expression of His6-tagged ltp2 (igr-1) resulted in insoluble protein. Expression of fadE28, Rv3542c, or Rv3541c individually resulted in insoluble or unstable protein. Expression of fadE29 resulted in soluble, apoprotein (data not shown). Therefore, the protein expression data suggest that FadE28 forms a heteromeric complex with FadE29 and that likewise, Rv3542c forms a heteromeric complex with Rv3541c.
FIGURE 7.
Expression and purification of igr operon. A, constructs for expression of the igr operon in E. coli were prepared in vector pET28b. Each construct introduced an N-terminal His6 tag on the first gene product for purification. B, SDS-PAGE analysis of proteins purified by immobilized affinity chromatography from expression of constructs in A is shown. Protein identities were confirmed by trypsin digest and MALDI-TOF MS fingerprinting.
FadE28-FadE29 Catalyzes the Dehydrogenation of 2′-Propanoate-CoA Esters of Hexahydraoindanone and Pregenenone
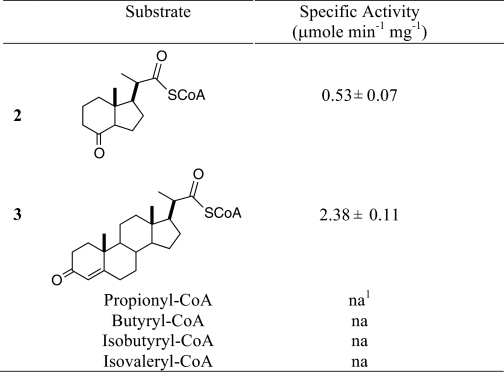
Purified FadE28-FadE29 complex was assayed for acyl-CoA dehydrogenase activity. Several CoA thioester substrates, including hexahydroindanone 2 and pregnenone 3, propionyl-CoA, butyryl-CoA, isobutyrl-CoA, and isovaleryl-CoA, were assayed. Each of these substrates was assayed at 100 μm with up to 80 μg/ml FadE28-FadE29. Oxidation was detected spectroscopically, and the formation of product was confirmed by MALDI-TOF mass spectrometry. FadE28-FadE29 catalyzed the dehydrogenation of 2 and 3 but not of propionyl-CoA, butyryl-CoA, isobutyryl-CoA, or isovaleryl-CoA. Negative controls without enzyme or without substrate were conducted, and no activity was detected. Thus, a short, straight, or branched fatty acid is insufficient as a substrate. The specific activities of FadE28-FadE29 using 2 and 3 were 0.53 ± 0.07 and 2.38 ± 0.11 μmol min−1 mg−1, respectively. FadE28-FadE29 shows a 5-fold preference for the pregnenone carbon skeleton over the hexahydroindanone skeleton under these assay conditions.
DISCUSSION
For intracellular pathogens to survive in the host they must be able to adapt to the environment and available nutrients. M. tuberculosis resides in host granulomas where cholesterol is abundant, and it has been demonstrated that M. tuberculosis is able to metabolize cholesterol. Its importance in pathogenesis is becoming increasingly clear (9). Several M. tuberculosis mutants show cholesterol sensitive phenotypes, including Δigr, which shows growth attenuation of the bacteria in MΦ and the mouse model. Herein, we investigated the functional role of the igr operon through 13C-metabolite profiling and biochemical assay of FadE28-FadE29, enzymes encoded in the operon.
13C-metabolite profiling is a powerful technique for studying carbon metabolism in a living system. The technique requires a 13C-labeled carbon source from which metabolites can be identified by MS or NMR isotopomer profiles. Here we report the biosynthetic preparation of LDL [1,7,15,22,26-13C]cholesterol and demonstrate its use as a tool to profile the degradation of cholesterol by Δigr H37Rv M. tuberculosis. The broad distribution of isotope labels allowed us to ascertain the metabolite profile of Δigr H37Rv M. tuberculosis compared with WT H37RV and igr complement strains. Methyl 1β-(2′-propanoate)-3aα-H-4α-(3′-propanoic acid)-7aβ-methylhexahydro-5-indanone, 1, accumulated in the Δigr mutant (Fig. 4). The hexahydroindanone skeleton has been isolated from several steroid-metabolizing bacterial species including species of Rhodococcus, Nocardia, Arthrobacter, and Streptomyces (20, 41–44). To our knowledge, this is the first report of the formation of hexahydroindanone species in M. tuberculosis.
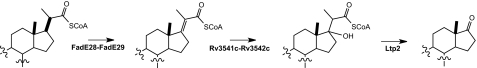
Interestingly, metabolite 1 contains a 2′-propanoate side chain at C1, indicating the igr operon is not essential for the first two cycles of β-oxidation required to produce this shortened side chain from cholesterol. The igr operon encodes an incomplete cadre of β-oxidation enzymes, as it lacks genes for two key enzymes: 3-hydroxyacyl-CoA-dehydrogenase and 3-ketoacyl-CoA-thiolase. Recombinant expression of the igr operon using the native structure of the operon in a single expression vector resulted in expression of two separate heterooligomeric complexes composed of FadE28-FadE29 and Rv3541c-Rv3542c. The latter has high similarity to MaoC like hydratases. We proposed that these protein complexes catalyze the acyl-CoA dehydrogenation and enoyl-CoA hydration, respectively, of the 2′-propanoate side chain to provide a quaternary alcohol. This alcohol would then readily undergo a retroaldol C1-C2′ cleavage reaction catalyzed by Ltp2 to form the ketone at C1 and propionyl-CoA (Fig. 8). This cleavage is favorable because the thermodynamically stable ketone is formed. In contrast, conventional β-oxidation of a fatty acid requires oxidation of the 3-hydroxyacyl-CoA and thiolase cleavage because direct formation of the less stable aldehyde through retroaldol cleavage is thermodynamically uphill.
FIGURE 8.
Proposed function of igr operon. The function of the igr operon was assigned to be degradation of the 2′-propanoate side chain. The proposed catalytic function of each enzyme or enzyme complex is shown.
The 3′-propanoate substituent at C4 is also hypothesized to be degraded by β-oxidation to yield acetyl-CoA and the formate substituent. The absence of the above-mentioned 3-hydroxyacyl-CoA-dehydrogenase and 3-ketoacyl-CoA-thiolase in the igr operon suggests that the igr enzymes do not catalyze the oxidation of this substituent. Consistent with this proposal, we observe minor metabolites corresponding to the C4-substituent β-oxidation intermediates in our NMR spectra. Moreover, van der Geize (45) recently reported that FadE30 of Rhodococcus equi is responsible for β-oxidation of the 3′-propanoate substituent at C4 of the 7aβ-methyl-hexahydro-5-indanone skeleton. R. equi FadE30 shares 68% amino acid identity with FadE30 (Rv3560c) from M. tuberculosis. Gene knock-out of R. equi fad30 blocks growth of the bacterium on 5-hydroxy-methylhexahydro-1-indanone propionate (5OH-HIP), whereas ΔfadE30 growth on AD accumulates 5OH-HIP. These results strongly suggest that fadE30 encodes the acyl-CoA dehydrogenase responsible for oxidation of the hexahydroindanone C4 substituent. R. equi FadE30 shares only 14 and 33% amino acid identity with FadE28 and FadE29, respectively. These low identities further support that the fadEs in the igr operon are not required for metabolism of C4 propanoate moiety.
To test our hypothesis that the igr operon enzymes catalyze the metabolism of the 2′-propanoate substituent derived from the side chain of cholesterol, two potential polycyclic hydrocarbon substrates bearing a 2′-propanoate-CoA side chain were synthesized in addition to four short straight or branched fatty acyl-CoA esters (Table 1). The CoA thioesters were prepared, and dehydrogenase activity was demonstrated for FadE28-FadE29 with both 2 and 3. No activity was observed for short chain substrates including propionyl-CoA, butyryl-CoA, isobutyrl-CoA, and isovaleryl-CoA, indicating that this enzyme is specific for steroid metabolites. Under the assay conditions tested thus far, the FadE28-FadE29 complex shows a 5-fold preference for the 4-ring steroid skeleton over the 2-ring indanone skeleton. This preference suggests that in vivo the side chain can be metabolized to AD before further ring degradation occurs. This order of metabolism is further supported by the isolation of AD and ADD from wild-type extracts (8) and the reported substrate specificity of the 3-keto-5α-steroid-Δ1-dehydrogenase, KstD (16). However, a recent report suggests that the KshA/KshB hydroxylase is more specific for partial side-chain degradation intermediates (46). Most likely, the ring system and side chains are degraded in tandem. In conclusion, we assign the primary function of the igr operon to be oxidation, hydration, and retro-aldo cleavage of the 2′-propanoate side chain to provide androst-4-ene-3,17-dione and its ring-degraded analogs during cholesterol metabolism. Future studies will address understanding the mechanistic details of these enzymes.
TABLE 1.
Specific activity data for FadE28-FadE29
Enzyme assays were conducted with ferrocenium hexafluorophosphate as the electron acceptor with 3.2 μg of enzyme and 100 μm substrate at 25 °C, pH 7.4.
1 na: no activity observed at FadE28-FadE29 concentrations up to 80 μg/mL.
Supplementary Material
Acknowledgments
We thank Antonius Köller (Stony Brook University Proteomics Center) for assistance with mass spectrometry, Michael Goger (New York Structural Biology Center) for assistance with nuclear magnetic resonance spectroscopy, and Eugenie Dubnau (Public Health Research Institute, University of Medicine and Dentistry of New Jersey) for helpful discussions in initiating this work.
This work was supported, in whole or in part, by National Institutes of Health Grant R21AI092455, R01HL53306, S10RR021008 (to N. S. S.), R01AI067027 (to D. G. R.), and S10RR025072 (to Orbitrap). This work was also supported by a DOE-GAANN fellowship (to S. T. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- Mφ
- macrophage
- igr
- intracellular growth
- OADC
- oleate-albumin-dextrose-NaCl catalase
- AD
- androst-4-ene-3,17-dione
- ADD
- androsta-1,4-diene-3,17-dione
- HSQC
- heteronuclear single quantum correlation
- HMBC
- heteronuclear multiple bond coherence.
REFERENCES
- 1. WHO (2011) Global Tuberculosis Control 2011, World Health Organization, WHO/HTM/TB/2011.16 [Google Scholar]
- 2. Russell D. G., Cardona P. J., Kim M. J., Allain S., Altare F. (2009) Nat. Immunol. 10, 943–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bloch H., Segal W. (1956) J. Bacteriol. 72, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marrero J., Rhee K. Y., Schnappinger D., Pethe K., Ehrt S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9819–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McKinney J. D., Höner zu Bentrup K., Muñoz-Elías E. J., Miczak A., Chen B., Chan W. T., Swenson D., Sacchettini J. C., Jacobs W. R., Jr., Russell D. G. (2000) Nature 406, 735–738 [DOI] [PubMed] [Google Scholar]
- 6. Muñoz-Elías E. J., Upton A. M., Cherian J., McKinney J. D. (2006) Mol. Microbiol. 60, 1109–1122 [DOI] [PubMed] [Google Scholar]
- 7. Van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., Eltis L. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nesbitt N. M., Yang X., Fontán P., Kolesnikova I., Smith I., Sampson N. S., Dubnau E. (2010) Infect. Immun. 78, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Griffin J. E., Gawronski J. D., Dejesus M. A., Ioerger T. R., Akerley B. J., Sassetti C. M. (2011) PLoS Pathog. 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yam K. C., D'Angelo I., Kalscheuer R., Zhu H., Wang J. X., Snieckus V., Ly L. H., Converse P. J., Jacobs W. R., Jr., Strynadka N., Eltis L. D. (2009) PLoS Pathog. 5, e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pandey A. K., Sassetti C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4376–4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang J. C., Miner M. D., Pandey A. K., Gill W. P., Harik N. S., Sassetti C. M., Sherman D. R. (2009) J. Bacteriol. 191, 5232–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lack N. A., Yam K. C., Lowe E. D., Horsman G. P., Owen R. L., Sim E., Eltis L. D. (2010) J. Biol. Chem. 285, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dresen C., Lin L. Y., D'Angelo I., Tocheva E. I., Strynadka N., Eltis L. D. (2010) J. Biol. Chem. 285, 22264–22275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sim E., Sandy J., Evangelopoulos D., Fullam E., Bhakta S., Westwood I., Krylova A., Lack N., Noble M. (2008) Curr. Drug Metab. 9, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Knol J., Bodewits K., Hessels G. I., Dijkhuizen L., van der Geize R. (2008) Biochem. J. 410, 339–346 [DOI] [PubMed] [Google Scholar]
- 17. Capyk J. K., D'Angelo I., Strynadka N. C., Eltis L. D. (2009) J. Biol. Chem. 284, 9937–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Capyk J. K., Kalscheuer R., Stewart G. R., Liu J., Kwon H., Zhao R., Okamoto S., Jacobs W. R., Jr., Eltis L. D., Mohn W. W. (2009) J. Biol. Chem. 284, 35534–35542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang X., Dubnau E., Smith I., Sampson N. S. (2007) Biochemistry 46, 9058–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miclo A., Germain P. (1992) Appl. Microbiol. Biotechnol. 36, 456–460 [DOI] [PubMed] [Google Scholar]
- 21. Sih C. J., Tai H. H., Tsong Y. Y., Lee S. S., Coombe R. G. (1968) Biochemistry 7, 808–818 [DOI] [PubMed] [Google Scholar]
- 22. Chang J. C., Harik N. S., Liao R. P., Sherman D. R. (2007) J. Infect Dis. 196, 788–795 [DOI] [PubMed] [Google Scholar]
- 23. Ouellet H., Guan S., Johnston J. B., Chow E. D., Kells P. M., Burlingame A. L., Cox J. S., Podust L. M., de Montellano P. R. (2010) Mol. Microbiol. 77, 730–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnston J. B., Ouellet H., Ortiz de Montellano P. R. (2010) J. Biol. Chem. 285, 36352–36360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 26. Dobson G., Minnikin D. E., Minniken S. M., Parlett J. H., Goodfellow M. (1985) in Chemical Methods in Bacterial Systematics (Goodfellow M., Minnikin D. E. eds.) pp. 237–265, Academic Press, Inc., London [Google Scholar]
- 27. Smith C. A., Want E. J., O'Maille G., Abagyan R., Siuzdak G. (2006) Anal. Chem. 78, 779–787 [DOI] [PubMed] [Google Scholar]
- 28. Inhoffen H. H., Quinkert G., Schutz S., Friedrich G., Tober E. (1958) Chem. Ber. 91, 781–791 [Google Scholar]
- 29. Inhoffen H. H., Bruckner K., Grundel R. (1954) Chem. Ber. 87, 1–13 [Google Scholar]
- 30. Mincione E., Bovicelli P., Forcellese M. L. (1989) Synth. Commun. 19, 723–735 [Google Scholar]
- 31. Leppik R. A. (1982) Biochem. J. 202, 747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Slomp G., Jr., Johnson J. L. (1958) J. Am. Chem. Soc. 80, 915–921 [Google Scholar]
- 33. Lehman T. C., Hale D. E., Bhala A., Thorpe C. (1990) Anal. Biochem. 186, 280–284 [DOI] [PubMed] [Google Scholar]
- 34. Lehman T. C., Thorpe C. (1990) Biochemistry 29, 10594–10602 [DOI] [PubMed] [Google Scholar]
- 35. Isler O., Ruegg R., Wursch J., Gey K. F., Pletscher A. (1957) Helv. Chim. Acta 40, 2369–2373 [Google Scholar]
- 36. Bloch K. (1965) Science 150, 19–28 [DOI] [PubMed] [Google Scholar]
- 37. Popják G., Cornforth J. W. (1966) Biochem. J. 101, 553–568 [PMC free article] [PubMed] [Google Scholar]
- 38. Popják G., Goodman W. S., Cornforth J. W., Cornforth R. H., Ryhage R. (1961) J. Biol. Chem. 236, 1934–1947 [PubMed] [Google Scholar]
- 39. Popják G., Edmond J., Anet F. A., Easton N. R., Jr. (1977) J. Am. Chem. Soc. 99, 931–935 [DOI] [PubMed] [Google Scholar]
- 40. Horinouchi M., Hayashi T., Koshino H., Kurita T., Kudo T. (2005) Appl. Environ. Microbiol. 71, 5275–5281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hayakawa S., Kanematsu Y., Fujiwara T. (1967) Nature 214, 520–521 [DOI] [PubMed] [Google Scholar]
- 42. Hashimoto S., Hayakawa S. (1977) Biochem. J. 164, 715–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakamatsu T., Beppu T., Arima K. (1980) Agric. Biol. Chem. 44, 1469–1474 [Google Scholar]
- 44. Kieslich K. (1985) J. Basic Microbiol. 25, 461–474 [DOI] [PubMed] [Google Scholar]
- 45. van der Geize R., Grommen A. W., Hessels G. I., Jacobs A. A., Dijkhuizen L. (2011) PLoS Pathog. 7, e1002181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Capyk J. K., Casabon I., Gruninger R., Strynadka N. C., Eltis L. D. (2011) J. Biol. Chem. 286, 40717–40724 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.