Background: The vital interaction between components of the TIM23 import complex and presequences is poorly characterized.
Results: A direct interaction between presequences and components of TIM23 was demonstrated and characterized.
Conclusions: Trans binding sites exhibit stronger interaction than cis sites.
Significance: Stronger binding to the trans side of the TIM23 complex provides an additional driving force for mitochondrial protein import.
Keywords: Biophysics, Membrane Proteins, Mitochondria, Mitochondrial Transport, Protein Cross-linking
Abstract
Precursor proteins that are imported from the cytosol into the matrix of mitochondria carry positively charged amphipathic presequences and cross the inner membrane with the help of vital components of the TIM23 complex. It is currently unclear which subunits of the TIM23 complex recognize and directly bind to presequences. Here we analyzed the binding of presequence peptides to purified components of the TIM23 complex. The interaction of three different presequences with purified soluble domains of yeast Tim50 (Tim50IMS), Tim23 (Tim23IMS), and full-length Tim44 was examined. Using chemical cross-linking and surface plasmon resonance we demonstrate, for the first time, the ability of purified Tim50IMS and Tim44 to interact directly with the yeast Hsp60 presequence. We also analyzed their interaction with presequences derived from precursors of yeast mitochondrial 70-kDa heat shock protein (mHsp70) and of bovine cytochrome P450SCC. Moreover, we characterized the nature of the interactions and determined their KDs. On the basis of our results, we suggest a mechanism of translocation where stronger interactions of the presequences on the trans side of the channel support the import of precursor proteins through TIM23 into the matrix.
Introduction
The majority of mitochondrial proteins are translocated from the cytosol with the assistance of several multimeric translocases that are present in the various mitochondrial compartments (1–4). In the outer membrane, the TOM3 (translocase of the outer mitochondrial membrane) complex serves both as a receptor for recognition of mitochondrial precursor proteins and as the main portal of protein entry into mitochondria (5, 6). The TOM complex is composed of the primary receptors Tom20 and Tom70 and the core subunits Tom40, Tom22, Tom7, Tom6, and Tom5. Tom40 forms the protein-conducting channel, providing a route for precursor proteins to cross the outer membrane. On their way to the matrix, proteins that contain cleavable amino-terminal targeting signals are transferred from the TOM complex to the TIM23 (translocase of the inner mitochondrial membrane) translocase. The latter is a multisubunit complex consisting of at least ten subunits. Its core is composed of two multispanning integral inner membrane proteins, Tim17 and Tim23. These proteins form a channel that allows proteins to cross, or integrate into the inner mitochondrial membrane. An additional subunit, Tim50, serves as a receptor in the mitochondrial intermembrane space (IMS) and has a role in maintaining the permeability barrier of the channel (7–11). Together, these three proteins constitute the cis side of the inner membrane channel. It was shown that the soluble domains of Tim23 and Tim50 can form a complex that is essential for translocation through the TIM23 translocase (12, 13). The final step of protein import into the matrix requires the ATP-dependent action of the mitochondrial import motor of the TIM23 complex, also referred to as a presequence translocase-associated protein import motor (PAM). The import motor is predominantly located on the trans side of the TIM23 channel. Its central components are the ATP-hydrolyzing 70 kDa heat shock protein (mHsp70) and Tim44. Tim44 is a peripheral inner membrane protein that binds simultaneously to mHsp70 and to the translocation channel of the TIM23 complex, anchoring the soluble mHsp70 to the translocation channel. However, additional roles in the regulation and coordination of the motor have also been attributed to this protein (4, 14). mHsp70 promotes translocation by undergoing conformational changes that are controlled by ATP hydrolysis. Several cycles of binding and release from the chaperone facilitate inward movement of the precursor protein into the matrix (6, 15, 16). Additional components of the motor are suggested to play essential regulatory roles in either the function or stability of the motor. These include the accessory proteins Mge1, Tim14/Pam18, Tim16/Pam16, and Pam17 (4, 17–21).
On their way to the matrix, precursor proteins bind to various components of the TIM23 complex that are located on either the cis or trans sides. The three components studied in this work were shown to be in close proximity to imported precursor proteins using cross-linking and immunoprecipitation in vivo. On the cis side of the membrane, Tim50 is the first component to encounter precursor proteins. Tim50 could be cross-linked to various preproteins transiting the import channel, and therefore the function of a receptor was attributed to this subunit (7, 9–11). Another component on the cis side, the soluble domain of Tim23, was suggested to receive precursor proteins from Tim50 and to transfer them further into the channel (7, 22–24). On the trans side of the membrane, Tim44 could be cross-linked to precursor proteins (25–29). Subsequent transfer of the imported precursors from Tim44 to mHsp70 drives the translocation process to completion. Notably, an in vitro study using the soluble domain of Tim23 showed that it indeed interacts with presequences but with only very weak affinity (KD = 0.47 mm) (23). In contrast, a direct interaction between Tim50 or Tim44 with presequences has not been demonstrated thus far. In fact, previous studies failed to show evidence of interaction between Tim44 and the P5 peptide, which serves as a model peptide for mitochondrial targeting signals (30, 31), thereby casting doubt on the ability of Tim44 to bind presequences. In this study, we analyzed the interaction of three matrix targeting signals with purified mature Tim44 and with the purified soluble domains of Tim50 and Tim23. We demonstrate for the first time that Tim44 and Tim50 are capable of binding presequences in vitro. We also show that the affinity of the cis side to the presequence is much weaker than that of the trans side. The significance of our results is discussed in light of what is known about the mechanism of protein translocation across the mitochondrial inner membrane.
EXPERIMENTAL PROCEDURES
Protein Purification
Purification of yeast Tim23IMS, Tim50IMS, Tim44, and Ssc1 (mHsp70) was carried out as described previously (12, 32).
Peptides
All peptides were purchased from Sigma. pHsp60 (MLRSSVVRSRATLRPLLRRAYS) represents the entire presequence of yeast mitochondrial Hsp60. pHsp70 represents the entire presequence of yeast mitochondrial Hsp70 (MLAAKNILNRSSLSSSFRIATRL). pP450SCC-WT represents the cleaved functional targeting sequence of bovine mitochondrial cytochrome P450SCC, in which Cys-16 was changed to Thr (MLARGLPLRSAVLKATPPIYC). In pP450SCC-Mut, three basic amino acid residues of pP450SCC-WT were replaced by Ser or Thr. In addition, Thr-14 was changed to Ser, and Cys-16 was changed to Thr (MLASGLPLSSAVLSATPPIYC). P5 is part of the presequence of aspartate aminotransferase from chicken (CALLLSAPRR). For the SPR experiments, a single Cys residue was added to the carboxyl or amino terminus of pHsp60 and pHsp70, respectively.
Cross-linking Experiments
Cross-linking of the various proteins was carried out with the entire presequence of Hsp60 labeled in its C terminus with biotin. For Tim50IMS and Tim23IMS, cross-linking was performed with 50 μm disuccinimidyl suberate (DSS) at room temperature for 30 min in 20 mm Na-HEPES (pH 7.4), 5 mm MgCl2, 50 mm KCl, 200 mm NaCl, 5% glycerol, and 0.01% Triton X-100. The cross-linking reaction was stopped by the addition of SDS-containing sample buffer followed by boiling for 3 min. The cross-linked products were analyzed by SDS-PAGE with a 12% or 14% acrylamide gel and subjected to Western blotting with streptavidin-HRP according to standard procedures. For experiments with Tim44 or mHsp70, the cross-linking conditions were essentially the same except that the buffer contained 20 mm Na-HEPES (pH 7.4), 10 mm MgCl2, 100 mm KCl, 300 mm NaCl, and 0.01% Triton X-100.
SPR Experiments
Binding experiments were performed with a ProteOn XPR36 instrument (Bio-Rad) in running buffer containing 20 mm Na-HEPES (pH 7.5), 5 mm MgCl2, 50 mm KCl, 200 mm NaCl, 5% glycerol and 0.005% Tween 20 for Tim50IMS and Tim23IMS, or 20 mm Na-HEPES (pH 8), 400 mm NaCl, 5% glycerol, and 0.005% Tween 20 for Tim44. All peptides were immobilized through a single cysteine residue on a general layer capacity (GLC) sensor chip to a level of approximately 1000 resonance units (RU) as described previously (33). Briefly, the chip was activated twice for 160 s with a mixture of 0.1 m 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (EDC) and 0.025 m sulfo-N-hydroxy-sulfo-succinimide (sulfo-NHS) (TS-Pierce). Next, 0.5 m cysteine (in running buffer) was injected for 360 s followed by an injection of 25 mm Ellman's reagent (5,5′-dithio-bis-(2-nitrobenzoic acid) in PBSX10 for 600 s. A reference channel underwent the same procedure except for pHsp60 immobilization. After immobilization of pHsp60 (0.0375 mg/ml in running buffer), blocking was performed with 50 mm cysteine (in 10 mm acetate (pH 4.5), 1 m NaCl) for 200 s followed by a 200 s injection of 1 m ethanolamine (pH 8.5). The chip was rotated 90°, and five different concentrations of the various proteins were simultaneously injected at a flow rate of 30 μl/min.
RESULTS
The Soluble Domain of Tim50 Interacts with Mitochondrial Targeting Presequences
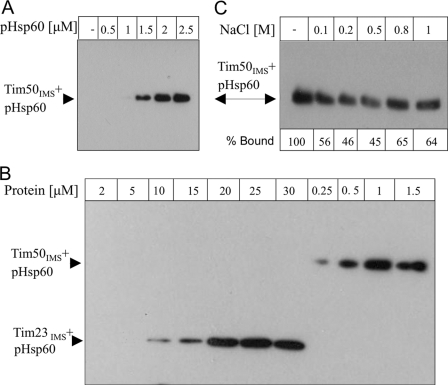
We established two methods for following the interaction between the purified soluble domain of Tim50 (Tim50IMS) and presequence peptides. For this purpose, we first chose the entire presequence of yeast Hsp60 (pHsp60). In the first method, biotinylated pHsp60 was incubated with Tim50IMS to allow complex formation, followed by mild cross-linking with a low concentration of DSS, resolution of the cross-linking products by SDS-PAGE, and detection with streptavidin conjugated to horseradish peroxidase. We examined binding at increasing concentrations of pHsp60 (Fig. 1A). Concentration-dependent binding was observed, and at 1–1.5 μm of pHsp60, a significant binding was detected. A previous study has shown that the soluble domain of Tim23 (Tim23IMS) can bind presequences (pALDH and pCoxIV) with a very low affinity of about 0.47 mm (23). Accordingly, we examined whether Tim23IMS also shows weak binding to biotin-pHsp60. In this experiment, we kept the concentration of pHsp60 constant (2 μm) and examined its interaction with increasing concentrations of either Tim50IMS or Tim23IMS. The interaction with Tim50IMS was maximal at a concentration of approximately 1.5 μm, whereas a complex between Tim23IMS and pHsp60 could only be observed at 10 μm protein and was maximal at 20 μm (Fig. 1B). The latter result confirms that Tim23IMS binds presequences with very weak affinity compared with Tim50IMS.
FIGURE 1.
Direct binding of Tim50IMS to pHsp60. A complex was formed between biotin-labeled pHsp60 and Tim50IMS by incubating the proteins for 20 min. The complex was stabilized by addition of 50 μm DSS for 30 min at room temperature. Cross-linked products were analyzed by SDS-PAGE followed by Western blotting with streptavidin-HRP. A, interaction of Tim50IMS (1.5 μm) with increasing concentrations of pHsp60. B, interaction of 2 μm pHsp60 with increasing concentrations of Tim50IMS or Tim23IMS. C, interaction of 1.5 μm Tim50IMS with 2 μm pHsp60 in the presence of increasing NaCl concentrations.
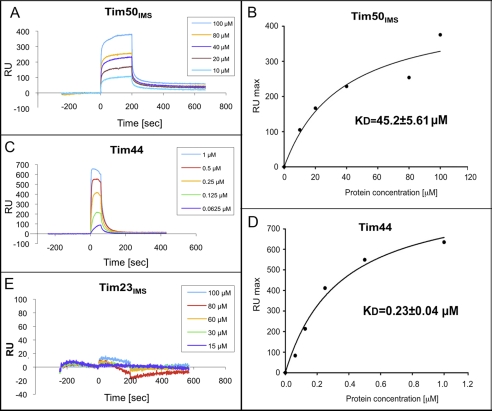
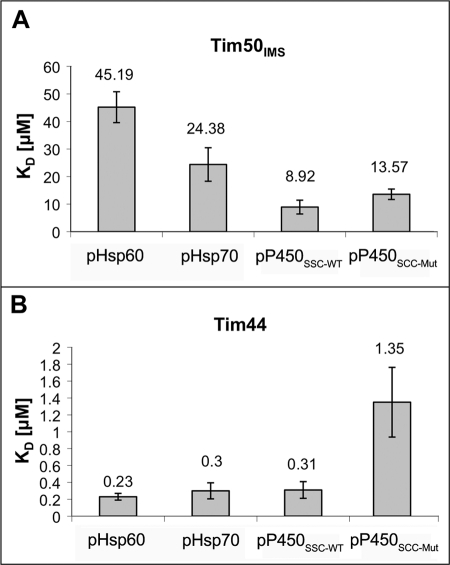
To obtain further support for our results, we analyzed the binding characteristics of pHsp60 by an additional method, SPR. The peptide was immobilized on the SPR sensor chip via a single cysteine residue that was added to its carboxyl terminus. A typical sensogram is presented in Fig. 2A. Binding between the immobilized peptide and Tim50IMS took place in a concentration-dependent manner. Using equilibrium analysis, we estimated the KD between Tim50IMS and pHsp60 to be approximately 45 μm (Fig. 2B). Two results indicate that the binding observed is specific. 1) When we examined the interaction between Tim23IMS and pHsp60 using SPR, no binding was seen, even at a protein concentration of 100 μm (Fig. 2E). This result is in agreement with the cross-linking results, which showed that an approximately 20-fold concentration of Tim23IMS over Tim50IMS was required to obtain maximal binding. 2) When the model peptide, P5, was immobilized on the sensor chip, it was not able to bind to Tim50IMS, although an interaction with the bacterial chaperone DnaK could be detected (supplemental Fig. S1). To show that the observed results are not unique for pHs60 but reflect a general mechanism, we also examined the binding of two additional presequences to Tim50IMS: the entire presequence of yeast mitochondrial Hsp70 (pHsp70) and the cleaved functional targeting sequence of bovine mitochondrial cytochrome pP450SCC (pP450SCC-WT) (34). Both presequences were found to associate with Tim50IMS, with a Kd of approximately 25 μm and approximately 9 μm, respectively (Fig. 3A). To summarize this part, we were able to demonstrate a direct binding in vitro between Tim50IMS and the presequences of several precursor proteins. Notably, when examined by SPR, Tim23IMS did not bind to any of the peptides examined in this study (supplemental Fig. S2).
FIGURE 2.
Binding between pHsp60 and components of the TIM23 complex was examined using SPR analysis. Cys-pHsp60 was immobilized on the GLC sensor chip at a density of approximately 1000 RU via a single Cys residue added at its C terminus (see “Experimental Procedures”). The chip was rotated 90°, and Tim50IMS, Tim23IMS, or Tim44 were injected at a flow rate of 30 μl/min at the five indicated concentrations. Typical sensograms of the interaction of Tim50IMS (A), Tim44 (C), or Tim23IMS (E) with pHsp60 are presented. For Tim50IMS and Tim23IMS, the 200 s association phase was followed by 400 s or 300 s of buffer injection, respectively, to allow complex dissociation. For Tim44, a 60 s association phase was followed by a 300 s dissociation phase. B and D, representative plots for the equilibrium analysis of the KD. Indicated are the mean KD values ± S.E. on the basis of at least eight experiments. The values were calculated using the ProteOn Manager software (Bio-Rad).
FIGURE 3.
Summary of SPR equilibrium analysis of Tim50IMS and Tim44 with the various presequences. Column diagrams summarizing the equilibrium KD values of Tim50IMS (A) and Tim44 (B) with the various presequences. The average KD of at least five measurements is indicated, and the error bars represent mean ± S.E.
In the next step, we sought to determine the nature of the forces that mediate the interaction between presequences and Tim50IMS. To this end, we created a complex between Tim50IMS and pHsp60 in the presence of increasing concentrations of salt. The addition of low salt concentrations led to a partial dissociation of the complex. However, complete dissociation was not observed, even at a concentration as high as 1 m NaCl (Fig. 1C). Therefore, although electrostatic interactions may play some role in the stability of the pHsp60-Tim50IMS complex, the fact that the majority of the complex remains intact at 1 m NaCl (approximately 65%) suggests that hydrophobic forces play a major role in complex formation, as reported recently on the basis of data from in vivo studies (35).
The Tim50IMS-Tim23IMS Complex Can Bind the Presequence Peptide pHsp60
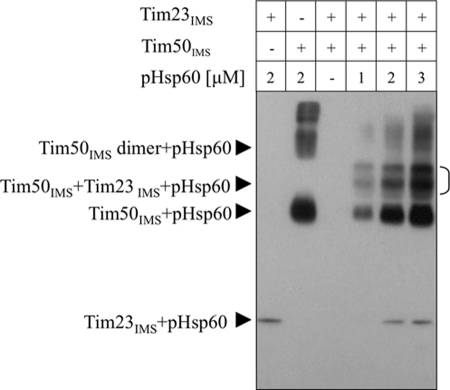
Because Tim50 and Tim23 interact via their IMS domains and because both could bind pHsp60, albeit with different affinities, we examined whether a complex of these domains is also able to bind a presequence. We first formed a complex between Tim23IMS (30 μm) and Tim50IMS (10 μm) and then added increasing concentrations of pHsp60. Complex formation was detected by cross-linking as described in Fig. 1. As expected, Tim50IMS alone showed very strong binding to the peptide, whereasTim23IMS displayed a much weaker affinity (Fig. 4). Importantly, a strong binding of pHsp60 was also observed with the Tim23IMS-Tim50IMS complex. Previous studies showed that the Tim50IMS-Tim23IMS interaction has a KD of approximately 60 μm (12), which is much lower than that reported for Tim23IMS with presequences. Taken together, our results imply that in the intermembrane space, binding of presequences to Tim50 alone or in complex with Tim23 comprises the first stage of protein import. In either case, the presequence will end up bound to the Tim50-Tim23 complex for further translocation through the channel.
FIGURE 4.
Interaction between the complex of Tim50IMS-Tim23IMS and pHsp60. Tim50IMS (10 μm) and Tim23IMS (30 μm) were incubated together for 10 min to allow complex formation and subjected to cross-linking with biotinylated pHsp60 as described for Fig. 1A, except that the experiment was carried out with 1 mm DSS. The brackets indicate higher cross-linking products of the Tim23IMS-Tim50IMS-pHsp60 complex.
Tim44 Interacts with Model Mitochondrial Presequences
Previous studies carried out with purified Tim44 failed to detect binding to P5. This hydrophobic decapeptide represents only a part of the mitochondrial targeting sequence of aspartate aminotransferase from chicken. Because this peptide was able to interact with mitochondrial Hsp70, it was initially assumed that it would also bind to Tim44 (36), a prediction that turned out to be not viable (30, 31). However, the possibility that P5 does not contain all the elements required for association with Tim44 was not excluded. To test this possibility, we examined the association of pHsp60 with Tim44 using cross-linking as carried out for Tim50IMS. Interestingly, Tim44 was able to interact with pHsp60 in a peptide concentration-dependent manner, exhibiting a substantial signal at 1–1.5 μm (Fig. 5A). To obtain a precise estimation of the affinity between Tim44 and presequences, we conducted SPR experiments similar to those described for Tim50IMS. Surprisingly, the interaction of Tim44 with the three presequences pHsp60, pHsp70, and pP450SCC-WT was the strongest among the proteins analyzed in this study, with a KD of 0.23 μm, 0.30 μm, 0.31 μm, respectively (Figs. 2, C and D, and 3B). In agreement with previous results, Tim44 could not bind to P5 in SPR, again confirming the specificity of our system (supplemental Fig. S1D).
FIGURE 5.
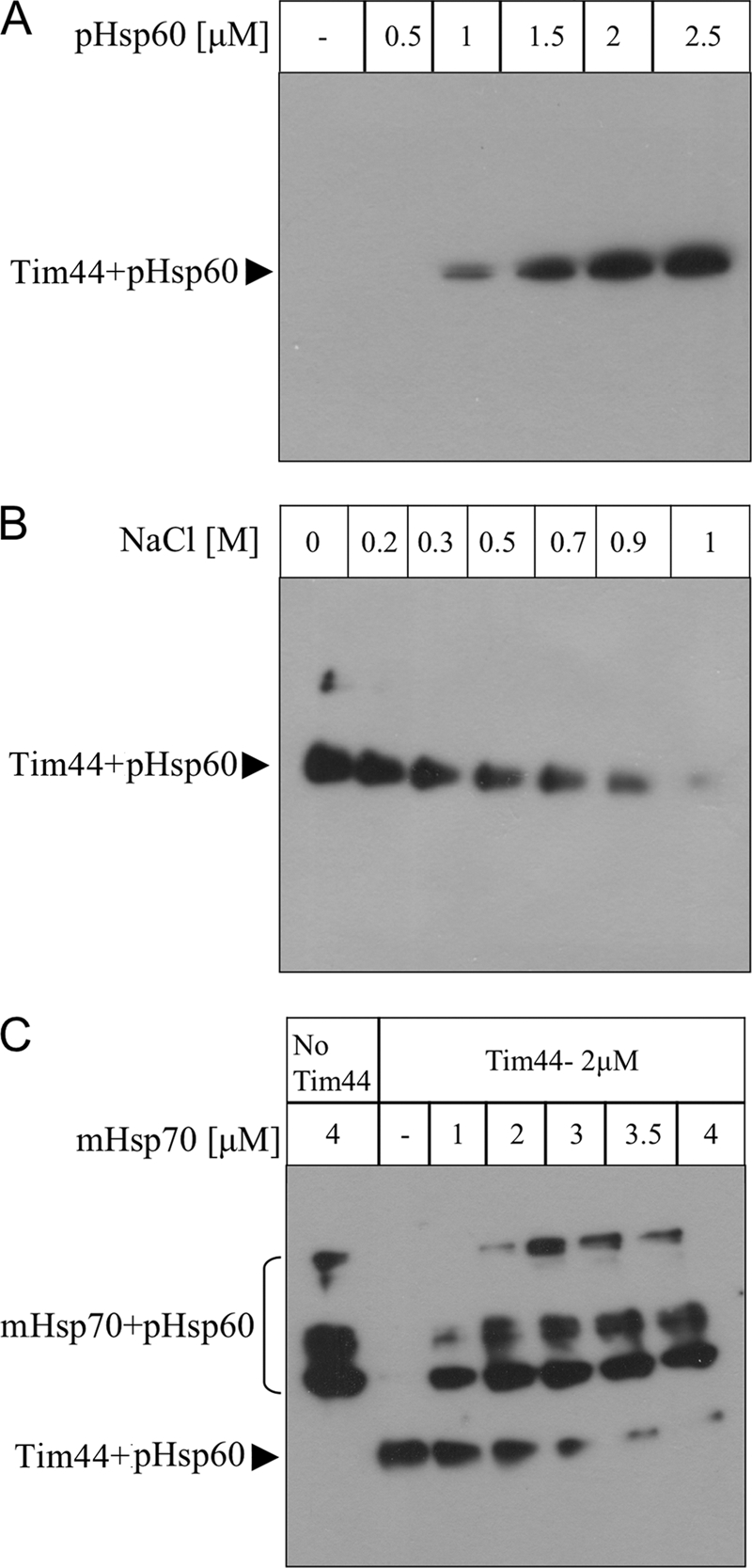
Direct binding of Tim44 to pHsp60. The interaction between Tim44 and pHsp60 was analyzed using cross-linking with DSS essentially as described in Fig. 1. A, the interaction of Tim44 (1.5 μm) with increasing concentrations of pHsp60. B, the interaction of 1.5 μm Tim44 with 1.5 μm pHsp60 in the presence of increasing NaCl concentrations. C, the interaction of 2 μm Tim44 with 0.9 μm pHsp60 in the presence of 1 mm ADP and increasing concentrations of mHsp70. In this experiment, the DSS concentration was 0.2 mm.
Next, to identify the forces that contribute to the interaction between presequences and Tim44, we examined the effect of increasing concentrations of salt on complex formation with pHsp60. Increasing the NaCl concentration led to a decrease in the formation of the pHsp60-Tim44 complex (Fig. 5B). This complex was almost completely dissociated upon addition of 1 m NaCl. Thus, we suggest that the pHsp60-Tim44 interaction is driven mainly by electrostatic forces, in contrast to the hydrophobic interaction observed for Tim50IMS.
To gain further support for the differences between Tim50IMS and Tim44 in regard to forces that govern their interaction with presequences (hydrophobic versus hydrophilic), we analyzed an additional peptide, pP450SCC-Mut. In this peptide, three basic amino acid residues of pP450SCC-WT were replaced by serine or threonine residues, and it was shown that this peptide does not constitute a functional presequence (34). Indeed, this peptide showed an approximately 4-fold increase in the KD value for Tim44 compared with pP450SCC-WT, whereas for Tim50IMS there was no significant difference (Fig. 3). This is consistent with our findings that Tim50IMS interacts with presequences mainly through hydrophobic forces, whereas positively charged residues play a key role in Tim44 binding.
The Presequence Can Be Detected Either Bound to Tim44 or mHsp70
A fundamental step in the translocation process is the transfer of the precursor from Tim44 to mHsp70 to promote its entry into the matrix. The addition of P5 to a preformed mHsp70-Tim44 complex has been shown to lead to its dissociation (30–32). Thus, it was concluded that mHsp70 can bind either Tim44 or the presequence, but not both simultaneously. Because the P5 peptide does not bind to Tim44 it was not possible to demonstrate a direct transfer of this peptide from Tim44 to mHsp70 (30–32). Hence, in the following experiment, we examined the effect of increasing the concentration of mHsp70 on the Tim44-pHsp60 complex. Increasing the mHsp70 concentration led to dissociation of pHsp60 from Tim44, enabling its transfer to the chaperone (Fig. 5C). This result should be taken in perspective of the in vivo concentrations of Tim44 and mHsp70 (30, 37, 38), where the latter is present in a large excess over Tim44, a fact that ensures that such a transfer will be favorable in vivo as well.
DISCUSSION
Binding of precursor proteins to various components belonging to the TIM23 translocase represents essential steps during their import into the matrix. Proteins participating in such interactions may include channel forming subunits, receptors, and other constituents that modulate and regulate the translocation process. Previous in vivo studies identified several proteins (including Tim23, Tim50, Tim44, and mHsp70) that are found in close proximity to a translocating precursor. However, a direct interaction was only demonstrated with the soluble domain of Tim23 in vitro. This left the question unresolved as to whether Tim50 and Tim44 can bind precursors directly. In this work, we used cross-linking and SPR to demonstrate for the first time the specific binding of Tim50IMS and Tim44 to model presequences in vitro. In addition, we confirm that Tim23IMS displays a weak affinity for presequences, as reported previously (23), which supports the validity of our approach. Interestingly we found that components that are located on the cis side of the membrane (Tim50 and Tim23) bind presequences with a relatively low affinity, whereas at the trans side, Tim44 shows strong binding (Fig. 2). Moreover, we observed that the essential complex formed between Tim23 and Tim50 can bind pHsp60, suggesting that the integrity of this complex is not affected upon presequence binding. Finally, we show that despite the strong interaction between Tim44 and pHsp60, it can be transferred further to mHsp70.
On the basis of our results, we suggest the following model for the translocation of precursor proteins into the matrix after emerging from the TOM complex. In the IMS, the precursor protein binds to Tim50 and Tim23, each with a low affinity. This step is followed by the formation of a ternary complex of Tim50-Tim23-precursor that was demonstrated to be essential for maintaining the interaction of precursor proteins with these components in vivo (7). At this stage, opening of the channel exposes the imported presequence to the pulling effect of the membrane potential. Significant directionality of presequence movement is imbued by Tim44, which binds the presequence with much higher affinity than at the cis side of the inner membrane. Further transfer of the presequence to mHsp70 allows for completion of its import into the matrix. This work did not address the effect of the actual channel on the translocating precursor. Further work on this topic will increase our understanding of additional forces that contribute to the directional movement of precursor proteins into the matrix.
Supplementary Material
Acknowledgments
We thank Gideon Schreiber and Ori Cohavi for invaluable advice with the SPR experiments and Ohad Iosefson for useful ideas.
This work was supported by the German-Israeli Foundation for Scientific Research and Development Grant GIF-1012/08, SFB594 and Israel Science Foundation Grant 452/09. This work was also supported by the Argentinean Friends of Tel-Aviv University and by the Ori Foundation in Memory of Ori Levi (to D. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- TOM
- translocase of the outer mitochondrial membrane
- TIM
- translocase of the inner mitochondrial membrane
- IMS
- intermembrane space
- SPR
- surface plasmon resonance
- DSS
- disuccinimidyl suberate
- sulfo-NHS
- N-hydroxy-sulfo-succinimide
- GLC
- general layer capacity
- RU
- resonance unit
- EDC
- 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride.
REFERENCES
- 1. Schmidt O., Pfanner N., Meisinger C. (2010) Nat. Rev. Mol. Cell Biol. 11, 655–667 [DOI] [PubMed] [Google Scholar]
- 2. Endo T., Yamano K. (2009) Biol. Chem. 390, 723–730 [DOI] [PubMed] [Google Scholar]
- 3. Mokranjac D., Neupert W. (2010) Biochim. Biophys. Acta 1797, 1045–1054 [DOI] [PubMed] [Google Scholar]
- 4. Marom M., Azem A., Mokranjac D. (2011) Biochim. Biophys. Acta 1808, 990–1001 [DOI] [PubMed] [Google Scholar]
- 5. Rapaport D. (2005) J. Cell Biol. 171, 419–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mokranjac D., Neupert W. (2005) Biochem. Soc. Trans. 33, 1019–1023 [DOI] [PubMed] [Google Scholar]
- 7. Mokranjac D., Sichting M., Popov-Celeketi D., Mapa K., Gevorkyan-Airapetov L., Zohary K., Hell K., Azem A., Neupert W. (2009) Mol. Biol. Cell 20, 1400–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Meinecke M., Wagner R., Kovermann P., Guiard B., Mick D. U., Hutu D. P., Voos W., Truscott K. N., Chacinska A., Pfanner N., Rehling P. (2006) Science 312, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto H., Esaki M., Kanamori T., Tamura Y., Nishikawa S., Endo T. (2002) Cell 111, 519–528 [DOI] [PubMed] [Google Scholar]
- 10. Mokranjac D., Paschen S. A., Kozany C., Prokisch H., Hoppins S. C., Nargang F. E., Neupert W., Hell K. (2003) EMBO J. 22, 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Geissler A., Chacinska A., Truscott K. N., Wiedemann N., Brandner K., Sickmann A., Meyer H. E., Meisinger C., Pfanner N., Rehling P. (2002) Cell 111, 507–518 [DOI] [PubMed] [Google Scholar]
- 12. Gevorkyan-Airapetov L., Zohary K., Popov-Celeketic D., Mapa K., Hell K., Neupert W., Azem A., Mokranjac D. (2009) J. Biol. Chem. 284, 4865–4872 [DOI] [PubMed] [Google Scholar]
- 13. Tamura Y., Harada Y., Shiota T., Yamano K., Watanabe K., Yokota M., Yamamoto H., Sesaki H., Endo T. (2009) J. Cell Biol. 184, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marom M., Safonov R., Amram S., Avneon Y., Nachliel E., Gutman M., Zohary K., Azem A., Tsfadia Y. (2009) Biochemistry 48, 11185–11195 [DOI] [PubMed] [Google Scholar]
- 15. van der Laan M., Rissler M., Rehling P. (2006) FEMS Yeast Res. 6, 849–861 [DOI] [PubMed] [Google Scholar]
- 16. Stojanovski D., Pfanner N., Wiedemann N. (2007) Methods Cell Biol. 80, 783–806 [DOI] [PubMed] [Google Scholar]
- 17. Neupert W., Brunner M. (2002) Nat. Rev. Mol. Cell Biol. 3, 555–565 [DOI] [PubMed] [Google Scholar]
- 18. Voos W., Röttgers K. (2002) Biochim. Biophys. Acta 1592, 51–62 [DOI] [PubMed] [Google Scholar]
- 19. Matouschek A., Pfanner N., Voos W. (2000) EMBO Rep. 1, 404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Popov-Celeketi D., Mapa K., Neupert W., Mokranjac D. (2008) EMBO J. 27, 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van der Laan M., Chacinska A., Lind M., Perschil I., Sickmann A., Meyer H. E., Guiard B., Meisinger C., Pfanner N., Rehling P. (2005) Mol. Cell Biol. 25, 7449–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bauer M. F., Sirrenberg C., Neupert W., Brunner M. (1996) Cell 87, 33–41 [DOI] [PubMed] [Google Scholar]
- 23. de la Cruz L., Bajaj R., Becker S., Zweckstetter M. (2010) Protein Sci. 19, 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alder N. N., Sutherland J., Buhring A. I., Jensen R. E., Johnson A. E. (2008) Mol. Biol. Cell 19, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blom J., Kübrich M., Rassow J., Voos W., Dekker P. J., Maarse A. C., Meijer M., Pfanner N. (1993) Mol. Cell Biol. 13, 7364–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scherer P. E., Manning-Krieg U. C., Jenö P., Schatz G., Horst M. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11930–11934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanamori T., Nishikawa S., Shin I., Schultz P. G., Endo T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gruhler A., Arnold I., Seytter T., Guiard B., Schwarz E., Neupert W., Stuart R. A. (1997) J. Biol. Chem. 272, 17410–17415 [DOI] [PubMed] [Google Scholar]
- 29. Merlin A., Voos W., Maarse A. C., Meijer M., Pfanner N., Rassow J. (1999) J. Cell Biol. 145, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Q., D'Silva P., Walter W., Marszalek J., Craig E. A. (2003) Science 300, 139–141 [DOI] [PubMed] [Google Scholar]
- 31. D'Silva P., Liu Q., Walter W., Craig E. A. (2004) Nat. Struct. Mol. Biol. 11, 1084–1091 [DOI] [PubMed] [Google Scholar]
- 32. Slutsky-Leiderman O., Marom M., Iosefson O., Levy R., Maoz S., Azem A. (2007) J. Biol. Chem. 282, 33935–33942 [DOI] [PubMed] [Google Scholar]
- 33. Yosef E., Politi R., Choi M. H., Shifman J. M. (2009) J. Mol. Biol. 385, 1470–1480 [DOI] [PubMed] [Google Scholar]
- 34. Komiya T., Rospert S., Schatz G., Mihara K. (1997) EMBO J. 16, 4267–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamamoto H., Itoh N., Kawano S., Yatsukawa Y., Momose T., Makio T., Matsunaga M., Yokota M., Esaki M., Shodai T., Kohda D., Hobbs A. E., Jensen R. E., Endo T. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Azem A., Oppliger W., Lustig A., Jenö P., Feifel B., Schatz G., Horst M. (1997) J. Biol. Chem. 272, 20901–20906 [DOI] [PubMed] [Google Scholar]
- 37. Moro F., Sirrenberg C., Schneider H. C., Neupert W., Brunner M. (1999) EMBO J. 18, 3667–3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rassow J., Maarse A. C., Krainer E., Kübrich M., Müller H., Meijer M., Craig E. A., Pfanner N. (1994) J. Cell Biol. 127, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.