Abstract
Background and Aims
Hybridization is an important evolutionary phenomenon, and therefore a detailed understanding of the dynamics of interspecific gene flow and resulting morphological and genetic patterns is of widespread interest. Here hybridization between the polyploids Cardamine pratensis and C. raphanifolia at four localities is explored. Using different types of data, the aim is to provide simultaneous and direct comparisons between genotype and phenotype variation patterns in the studied hybrid populations.
Methods
Evidence of hybridization has been gathered from morphology, molecular markers (amplified fragment length polymorphism and chloroplast DNA sequences), pollen viability, karyology and nuclear DNA content.
Key Results
All data support extensive gene flow occurring in the hybrid populations. A wide range of morphological and genetic variation is observed, which includes both parental and intermediate types. Unbalanced pollen fertility and several ploidy levels are recorded.
Conclusions
Incongruence reported between genotype and phenotype suggests that parental phenotypes are affected by introgression, and intermediate hybrid phenotypes can be genetically closer to one of the parents. Thus, it is evident that morphology, when used alone, can be misleading for interpreting hybridization, and critical evaluation of other data is needed.
Key words: AFLP, Brassicaceae, Cardamine pratensis, Cardamine raphanifolia, flow cytometry, hybridization, morphometrics, polyploidy, Spain
INTRODUCTION
Interspecific hybridization and polyploidy are sources of genetic variation and novel genetic combinations in plant populations. Furthermore, hybridization and polyploidy can have important evolutionary consequences, inducing plant diversification and speciation (Arnold, 1992; Rieseberg, 1998; Buerkle et al., 2000; Hegarty and Hiscock, 2005). Hybridization is a common feature of vascular plants, considering the estimation that at least 25 % of extant plant species are involved in hybridization and introgression with related species (Mallet, 2005) and that most flowering plants have polyploidy in their history (Soltis et al., 2004). Nevertheless, spontaneous hybridization is not distributed evenly over plant genera and families, as surveyed by Ellstrand et al. (1996), but seems to be most common among outcrossing species with favourable reproductive strategies (vegetative reproduction and agamospermy) and among polyploids. Examples of hybridization, introgression and hybrid speciation have also been reported from the Brassicaceae, indicating that this is a significant evolutionary force in the family. Some Brassicaceae genera have been subjected to thorough studies that provide examples of well-explored hybrid systems and hybrid speciation at both the diploid and polyploid levels (reviewed by Marhold and Lihová, 2006).
As in many other genera, interspecific hybrids have been described in the genus Cardamine L. Early studies of hybridization were based almost exclusively on observations of gross morphology (reviewed by Lihová and Marhold, 2006). However, closer examination of such putative hybrids in some cases led to the rejection of the hybrid status [see the examples mentioned by Lövkvist (1956) and Marhold et al. (2002)]. Morphological intermediacy is indeed expected in natural hybrids, but parental and transgressive characters can also appear (Rieseberg and Ellstrand, 1993; Rieseberg and Carney, 1998). Moreover, intermediacy can be observed in species of non-hybrid origins (Wilson, 1992). Thus, morphological characters alone are of limited value in detecting hybridization, and additional data are needed to prove hybrid origin. More recently, several studies have employed molecular markers to document hybridization events in Cardamine. In addition to the well-studied case of C. × insueta Urbanska and C. schulzii Urbanska, a hybrid speciation event associated with human-induced habitat disturbance (Urbanska, 1987; Urbanska et al., 1997), hybridization was documented to occur between two Pyrenean endemic diploids C. amara subsp. pyrenaea Sennen and C. crassifolia Pourr. (C. × enriquei Marhold, Lihová and Perný; Marhold et al., 2002), and hybrid status was also proved for the pentaploid C. × ferrarii Burnat (Cardamine asarifolia L. × C. amara L.; Lihová et al., 2006). For the latter, polytopic and reciprocal hybridization was inferred from chloroplast DNA (cpDNA) sequence data.
The present study reports on spontaneous hybridization occurring between two highly polyploid taxa (C. pratensis L. s.s. and C. raphanifolia Pourr. s.s.) on several sites in north-western Spain. The genus Cardamine comprises several polyploid complexes, among which the C. pratensis and C. raphanifolia groups are the most prominent in Europe (Lihová et al., 2004; Marhold et al., 2004). While the former is widely distributed in Eurasia, the latter is restricted to the European Mediterranean. Recent molecular studies elucidated relationships within and among taxa from these groups, and indicated that the C. raphanifolia group does not represent a monophyletic lineage (Lihová et al., 2004; Marhold et al., 2004; Lihová and Marhold, 2006). In Spain, the C. pratensis group is represented by two diploid endemics (C. castellana Lihová and Marhold and C. crassifolia Pourr.) and C. pratensis s.s., which occurs in diploid to heptaploid populations distributed from the central to northern parts of the Iberian Peninsula (Lihová et al., 2003). From the C. raphanifolia group, two closely related endemic taxa are represented: hexa- and octoploid C. raphanifolia s.s., and tetra- and hexaploid C. gallaecica (M.Laínz) Rivas. Mart. and Izco (Perný et al., 2005). Cardamine raphanifolia s.s. occurs in the northern Iberian Peninsula from the Pyrenees to western Cordillera Cantabrica and does not display any geographic pattern in cytotype distribution (Perný et al., 2005: Fig. 1). In contrast, some trends have been observed for cytotypes of C. pratensis s.s. (Lihová et al., 2003: Fig. 1). Both polyploid species exhibit similar habitat preferences and come into contact in several regions of the northern Iberian Peninsula, where populations of different ploidy levels are also found. During field research, sympatric occurrence of C. pratensis s.s. and C. raphanifolia s.s. has been observed at several localities. Morphologically peculiar, putatively hybrid populations were found in the Cordillera Cantabrica Mountains in north-western Spain.
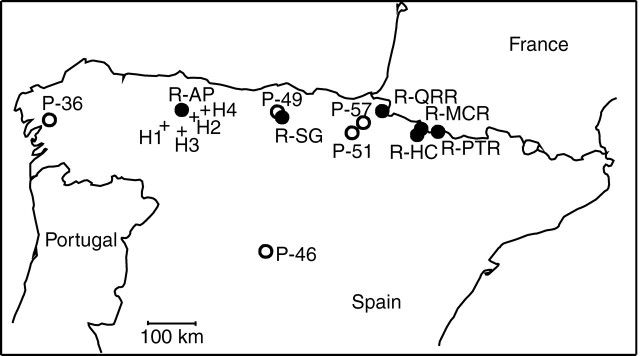
Fig. 1.
Map showing sample sites of the pure populations of C. pratensis s.s. (open circles), C. raphanifolia s.s. (filled circles) and the studied hybrid populations (crosses, H1–H4) in the Cordillera Cantabrica Mountains in north-western Spain. Population abbreviations follow Table 1.
In the present study, putative hybrids are characterized, using data from morphology, pollen viability, genetic markers [AFLPs (amplified fragment length polymorphisms) and cpDNA sequences], karyology and nuclear DNA content. The AFLP technique has been successfully used to study hybridization events and reticulations, because it produces a high number of polymorphic characters that can characterize and discriminate between closely related parental genomes (e.g. Bleeker, 2003; Guo et al., 2005; Salmon et al., 2005; Paun et al., 2006). cpDNA markers, in turn, reveal maternal lineages and can elucidate the direction of interspecific gene flow and indicate introgression that might not be detected by other nuclear markers (see, for example, Bleeker and Hurka, 2001). Morphological intermediacy, pollen sterility and intermediate chromosome numbers have been considered as straightforward indications for hybridization since early studies (Rieseberg and Ellstrand, 1993; Levin, 2002), and are also examined in the present study. A more critical evaluation of hybrid morphology, however, is currently stressed, due to complex character expression expected in hybrids (see above). Here different types of data are combined to allow simultaneous and direct comparisons between genotype and phenotype variation patterns in the putative hybrid populations. The morphological and genetic patterns observed are compared with those of the related diploid hybrid C. × enriquei and, furthermore, inferences on evolutionary history of the parental species are made.
MATERIALS AND METHODS
Plant material
Population samples were collected from four localities in the Cordillera Cantabrica Mountains (Table 1, Fig. 1), where C. pratensis s.s., C. raphanifolia s.s. and putative hybrids were found intermingled or growing in close proximity (hereafter called ‘hybrid populations’, referring to both parental and putative hybrid individuals). More detailed sampling was undertaken at one of these localities (H3, located at Puerto de Pajares). Parental species were also sampled from the area where they form pure and spatially well-separated populations (Fig. 1, Table 1), in order to assess their variation not affected by hybridization or introgression (hereafter called ‘pure populations’). These latter populations were selected from previous studies (Lihová et al., 2003; Perný et al., 2005) to cover species' distribution ranges, and morphological, genetic and karyological variation present in the Iberian Peninsula. For morphometric studies, the number of sampled individuals per locality ranged between 17 and 40 for pure populations, and between 29 and 109 for hybrid populations (Table 1). Where flower buds were retained, pollen fertility was estimated from the maximum number of specimens included in morphometric analyses (see Table 1). For AFLPs, a selection of 85 individuals was made (with the focus on hybrid populations), collected as fresh leaf samples taken from the labelled plants directly in the field before the plants were pressed for specimens used in the morphometric and pollen analyses. This means that although a smaller number of individuals were analysed for AFLPs, each of the corresponding voucher specimens was included in the morphometric analyses, allowing direct individual-level comparison between morphological and genetic variation patterns (see Results). Based on the AFLP variation pattern, 29 individuals (from both the hybrid and pure populations) that spanned the whole AFLP variation range were selected and sequenced to ascertain their trnL-trnF cpDNA haplotype. The trnL-trnF intergenic spacer was previously sequenced in both C. pratensis and C. raphanifolia, for which two distinct haplotypes were resolved, respectively (Lihová et al., 2004). In addition, from each of the hybrid populations, living plants were collected in the field and transferred to the experimental garden of the Institute of Botany, Slovak Academy of Sciences, Bratislava. The cultivated material was analysed for chromosome numbers (14 individuals) and used in flow cytometric measurements of nuclear DNA content (114 individuals). Chromosome numbers of the pure populations were published in previous studies (Lihová et al., 2003; Perný et al., 2005). Voucher specimens of the analysed material are deposited in the herbarium SAV.
Table 1.
Origin of the analysed Cardamine populations, information on chromosome numbers (2n) and numbers of individuals (n) used for flow cytometry, AFLPs, morphometrics and pollen viability
| Population abbreviation | Locality (all in Spain) and collection data | 2n | FCM (n) | AFLP (n) | Morphometrics (n) | Pollen (n) | trnL-trnF GenBank accession no. |
|---|---|---|---|---|---|---|---|
| Hybrid populations | |||||||
| H1 | Prov. León, road 623, crossroad to San Emiliano, 1165 m, 42°56·97′N, 6°0·14′W, 26 May 2003, J.L. and M.P. | 46, 56 | 8 | 8 | 29 | 22 | EF067935 EF067947 |
| H2 | Prov. Asturias, Puerto de Vegarada, above Redipuertas, 1336 m, 43°1·14′N, 5°26·63′W, 22 May 2003, J.L. and M.P. | 48, approx. 88 (two individuals) | 18 | 10 | 64 | 63 | EF067936 EF067937 EF067946 |
| H3 | Prov. León, Puerto de Pajares, 1380–1440 m, 42°59·92′–42°59·56′ N, 5°45·42′–5°45·54′ W, 20 May 2003, J.L. and M.P. | 48, 56, 64, 72 (three individuals), approx. 80 | 58 | 39 | 109 | 57 | EF0679400–45 EF067949 EF067950 EF067954 EF067957 |
| H4 | Prov. León, NW of Isoba, between Isoba and Puerto de San Isidro, 1270–1527 m, 43°2·17′–43°4·02′N, 5°18·95′–5°23·04′W, 24 May 2003, J.L. and M.P. | 44, 64 | 30 | 11 | 53 | 50 | EF067938–39 EF067948 EF067956 EF067958 |
| Cardamine pratensis L.– pure populations | |||||||
| P-46 | Prov. Madrid, Sierra Guadarrama, El Paular, Monasterio de El Paular, 1160 m, 12 May 2001, J.L. | 561 | – | 2 | 26 | 20 | EF067955 |
| P-36 | Prov. A Coruña, Portomuoro, river Tambre, 160 m, 6 Apr 2000, J.L. and I. Pulgar | 301 | 2 | 28 | 26 | EF067951 | |
| P-49 | Prov. Cantabria, between Campillo and S. Roque de Riomiera, 750 m, 7 May 2001, J.L. and M. Herrera | 321 | – | 1 | 29 | 24 | |
| P-57 | Prov. Navarra, Larraín, Sierra de Aralar, 1 km of Baraibar, 600 m, 6 May 2001, J.L. and I. Biurrun | 461 | – | 2 | 25 | EF067952 | |
| P-51 | Prov. Álava, Urcabustaiz, Beluntza, 640 m, 6 May 2001, JL and I. Biurrun | 481 | – | 2 | 26 | 27 | EF067953 |
| Cardamine raphanifolia Pourr.– pure populations | |||||||
| R-AP | Prov. Asturias, Pajares, below Puerto de Pajares, 1860 m, 24 May 2001, M.P. and F. Llamas | 642 | – | 1 | 20 | 12 | EF067930 |
| R-SG | Prov. Guipuzcoa, Otzaurte, 600 m, 22 May 2001, M.P. and I. Biurrun | 48, 642 | – | 1 | 17 | 6 | |
| R-QRR | Prov. Navarra, Quinto Real, 2·2 km S of Irurito, 700 m, 8 May 2001, J.L. and I. Biurrun | 642 | – | 1 | 20 | 20 | EF067931 |
| R-PTR | Prov. Huesca, Puerto de Portalet, SW of the road, near the frontier Spain–France, 1790 m, 1 June 2001, M.P. and G. Sanz | 482 | – | 1 | 26 | 27 | |
| R-MCR | Prov. Huesca, Selva de Oza, N slope of Monte Campanil, 30 May 2001, M.P. and L. Villar | 482 | – | 1 | 40 | 21 | |
| R-HC | Prov. Huesca, Selva de Oza (N of Hecho), 1150 m, 30 May 2001, M.P. and L. Villar | 482 | – | 3 | 29 | 28 | EF067932–34 |
Denotation of pure populations of C. pratensis and C. raphanifolia follows the codes as used in Lihová et al. (2003) and Perný et al. (2005), respectively. Chromosome numbers with superscripts were previously published in 1Lihová et al. (2003), and 2Perný et al. (2005).
Pollen viability estimation
As a measure of male fertility, pollen viability was assessed using acetocarmine staining (Marks, 1954). Numerous specimens from both the hybrid and pure populations were analysed (Table 1). Anthers were removed from a single flower bud per individual and chopped in a drop of acetocarmine jelly on a microscope slide to release pollen grains. At least 100 grains per flower were observed, and viable (well-stained) and non-viable (shrunken and unstained) grains were recorded. Pollen quality was expressed as the percentage of viable grains. Specimens from the hybrid populations were subsequently sorted into five classes, according to the pollen viability values: 0–20, 21–40, 41–60, 61–80 and 81–100 %, and used in morphometric analyses (see below).
Morphometric analyses
In order to evaluate morphological variation and differentiation between C. pratensis, C. raphanifolia and their putative hybrids, 17 quantitative morphological characters were measured and 12 ratios derived (Table 2). Out of the 17 characters, 12 characters were vegetative, measured on herbarium material of field-collected specimens. For measurements of the five floral characters, floral parts of one well-developed flower per plant were attached by adhesive tape to paper, dried and then scanned on a Microtek 9800XL scanner. Floral parts were measured with the program Carnoy (Schols et al., 2002). Eight characters relating to the size of leaves and stem (see Table 2) were used for computing ratios only. Two data sets were assembled, one including measurements from 11 pure populations (286 specimens × 21 characters), and another one consisting of both the pure and hybrid populations (572 specimens × 21 characters). First, principal component analysis (PCA) based on the former data set was performed to show the overall morphological variation and relationships among individuals from the pure populations. Secondly, PCA of individuals from both the pure and hybrid populations was run, with the latter marked according to their assignment to the pollen viability class. Individuals without values on pollen viability were excluded. The aim of this second analysis was to display the positions of individuals from hybrid populations with respect to the pure populations, as well as to correlate morphological patterns (parental and intermediate phenotypes) with the pollen viability values. In both cases, an R type of PCA based on a correlation matrix was used (Sneath and Sokal, 1973; Krzanowski, 1990).
Table 2.
List of characters used in the morphometric analyses of the studied Cardamine populations, including eigenvectors (column PCA, prin1) expressing correlations of the measured characters with the first principal component of PCA (see Fig. 3), and total canonical structure (column CDA, can 1) expressing correlations of the measured characters with the canonical axis of CDA (see Fig. 4)
| List of morphological characters | PCA | CDA |
|---|---|---|
| Vegetative characters | prin1 | can1 |
| Width of stem (WIS) | 0·325 | 0·774 |
| Number of stem branches (NB) | 0·201 | 0·538 |
| Height of stem* (LSL)† | – | – |
| Height of stem‡ (LSI)† | – | – |
| Number of stem leaves (NL) | 0·110 | 0·393 |
| Length of middle stem leaf§ (LC2)† | – | – |
| Length of the uppermost stem leaf (LC3)† | – | – |
| Maximum number of pairs of lateral leaflets on the middle stem leaf (NFS) | –0·296 | –0·829 |
| Length of terminal leaflet on the middle stem leaf (LTS)† | – | – |
| Width of terminal leaflet on the middle stem leaf (WTS)† | – | – |
| Length of first lateral leaflets on the middle stem leaf (LLS)† | – | – |
| Width of first lateral leaflets on the middle stem leaf (WLS)† | – | – |
| Floral characters | ||
| Average length of sepals (LS) | 0·266 | 0·281 |
| Average length of petals (LP) | 0·129 | −0·203 |
| Average width of petals (WP) | 0·207 | −0·054 |
| Average length of longer filaments (LFL) | 0·346 | 0·648 |
| Average length of shorter filaments (LFS) | 0·360 | 0·671 |
| Character ratios | ||
| WIS/LSL | 0·201 | 0·460 |
| LSL/LSI | 0·141 | 0·501 |
| NL/LSL | −0·095 | −0·222 |
| LC2/LSL | 0·076 | −0·071 |
| LC3/LSL | 0·053 | −0·129 |
| LC3/LC2 | −0·004 | −0·150 |
| WTS/LTS | 0·334 | 0·903 |
| WLS/LLS | 0·327 | 0·891 |
| LLS/LTS | −0·051 | −0·430 |
| WLS/WTS | −0·068 | −0·207 |
| WP/LP | 0·144 | 0·110 |
| LFS/LFL | 0·229 | 0·324 |
* Height of stem from the stem base to the base of the uppermost stem leaf.
† Characters not included in the morphometric analyses, used only to compute ratios.
‡ Height of stem from the stem base to the base of the peduncle of the lowermost flower/fruit.
§ The leaf closest to the LSL/2 point.
Thirdly, canonical discriminant analysis (CDA; Klecka, 1980) was performed based on the data set of the pure populations. This method, in contrast to PCA, maximizes between-group differences and thus expresses the extent of morphological differentiation between the defined groups (i.e. C. pratensis and C. raphanifolia from the pure populations). The resulting discriminant function was applied to individuals from the hybrid populations, and these were accordingly depicted on the histogram. This procedure was carried out to find an individual's position in morphological space with respect to the pure populations. According to the position expressed by the canonical score, individuals from the hybrid populations were sorted into three classes: morphologically corresponding to C. pratensis (score less than − 2; see Results); morphologically corresponding to C. raphanifolia (score > 1); and morphologically intermediate (score − 2 to + 1). Although distributions of some characters deviated to some extent from the normal distribution, both PCA and CDA have proven to be sufficiently robust against such violations (Legendre and Legendre, 1998). Results of the tests of significance determining the importance and minimum number of canonical axes, which are valid only for the data with the normal distribution (Klecka, 1980), were not taken into consideration. All morphometric analyses were run using the SAS statistical package (SAS Institute, 2000).
Molecular analyses (AFLP and cpDNA sequences)
Genomic DNA was extracted from silica gel-dried leaves using the cetyltrimethyl ammonium bromide (CTAB) procedure (Doyle and Doyle, 1987) with modifications according to Schönswetter et al. (2002). The AFLP procedure (Vos et al., 1995) followed the general protocol (Applied Biosystems, 1996) with minor modifications. Restriction–ligation, pre-selective and selective amplifications were performed as described in detail by Kučera et al. (2006). Three primer combinations giving clear and reproducible bands, applied before in previous Cardamine studies (Lihová et al., 2003; Kučera et al., 2006), were used for selective amplifications: EcoRI-AAG-(HEX) + MseI-CTG, EcoRI-ATC-(6-FAM) + MseI-CAG and EcoRI-AGC-(NED) + MseI-CTG (VBC Genomics, Vienna, Austria). The AFLP fragments were loaded on 4·5 % polyacrylamide gels with an internal size standard GeneScan 500 ROX (Applied Biosystems) and electrophoresed on an automated sequencer (ABI 377). Raw data were analysed in GeneScan 3·1·2. (Applied Biosystems) and then in the Genographer program (version 1·6·0, Montana State University, 1999; http://hordeum.msu.montana.edu/genographer/). The presence or absence of each AFLP fragment within each individual was scored (only well scorable and unambiguous fragments were analysed) and translated into a binary data matrix. Reproducibility of the obtained fragments was tested by repeating AFLP procedures for a few selected individuals, and running the products on different gels.
The AFLP data matrix was first inspected for variation patterns present among the analysed individuals. The total number of AFLP fragments, the number of polymorphic fragments, the proportion of group-specific fragments (pure C. pratensis, pure C. raphanifolia and hybrid populations) and patterns of fragment sharing were recorded. Principal coordinate analysis (PCoA) was then performed to obtain the general view on the AFLP variation pattern. This was based on the secondary matrix computed using Jaccard's coefficient, and the analysis was run with SYN-TAX 2000 (Podani, 2001). In the resulting ordination graph, individuals from the pure and hybrid populations were marked by different symbols to allow illustration of their genetic similarities. In addition, individuals from the hybrid populations were labelled according to their canonical score expressing the three main groups of morphological phenotypes (see above), in order to relate their morphological and genetic positions.
The trnL-trnF region of cpDNA was amplified in a standard polymerase chain reaction (PCR) procedure, using the universal primers e and f of Taberlet et al. (1991). Amplifications were performed in a 40 µL reaction volume with 1·4 mm MgCl2, 0·2 mm dNTPs, 0·2 µm each primer, 10 × PCR buffer and 1 U of Taq polymerase (Fermentas, St Leon-Rot, Germany), and run in a Mastercycler ep Gradient S (Eppendorf, Germany). The PCR cycle profile was 94 °C for 5 min, 35 cycles at 94 °C (1 min), 54 °C (1 min), 72 °C (45 s), then final extension at 72 °C for 10 min, and cooling to 4 °C. Amplification products were checked on a 1·5 % agarose gel and purified using the Spinprep PCR clean-up kit (Calbiochem, Darmstadt, Germany) following the manufacturer's protocol. Sequencing, using the original PCR primers, was performed at the BITCET Consortium at the Department of Molecular Biology, Comenius University, Bratislava. The region was sequenced mostly in the forward direction only (after a pilot study that involved sequencing of both strands). Sequences were aligned manually using BioEdit (version 7·0·4·1; Hall, 1999). Due to multiple trnF gene duplications and subsequent pseudogene formation (Koch et al., 2005), ambiguous regions at the 3′ end of the spacer were eliminated from the alignment, and only 394 aligned positions from the 5′ end downstream were considered. The sequences obtained from the pure populations were compared with the two haplotypes previously identified for C. pratensis and C. raphanifolia, respectively (Lihová et al., 2004), and those from the hybrid populations were assigned to either of these two groups. Haplotypes were mapped onto the ordination graph of PCoA of AFLP data.
Chromosome numbers and flow cytometry
To assess chromosome number variation in the hybrid populations, chromosome numbers were directly counted in a few selected plants from each locality, and relative nuclear DNA contents were screened using flow cytometry in 114 plants (Table 1). Chromosome numbers were counted from mitotic metaphase plates of root tips obtained from cultivated plants. The protocol for root tip treatment and preparation of microscopic slides can be found in Marhold et al. (2002).
Flow cytometry was used to estimate nuclear DNA content and to relate it to ploidy levels based on the comparison of values obtained from the plants of known chromosome numbers (Doležel, 1997). Multiple individuals per population, representing both parental and intermediate phenotypes, were analysed (the number of individuals per locality is given in Table 1). Relative nuclear DNA content was assessed on the basis of fluorescence intensity of nuclei stained using the AT-specific DAPI (4′,6-diamidino-2-phenylindole) fluorochrome (Doležel, 1997). Two internal standards were needed to span the range of DNA contents of the measured plants. For most measurements, Glycine max ‘Polanka’ (2C = 2·50 pg; Doležel et al., 1994) was used as a standard, but in a few cases where the peaks overlapped with those of the measured plant, Bellis perennis, with a higher DNA content (2C = 3·38 pg; Schönswetter et al., 2007), was used. Tissue from a fresh young leaf of the analysed individual was co-chopped with the internal standard in 0·5 mL of ice-cold Otto I buffer (0·1 m citric acid monohydrate, 0·5 % Tween-20; Otto, 1990) using a razor blade. The obtained suspension was filtered through a 42 µm nylon mesh. After a short incubation, 1 mL of Otto II buffer (0·4 M Na2HPO4.12H2O) supplemented with β-mercaptoethanol (2 µL mL−1) and DAPI (4 µg mL−1) was added to the flow-through containing the suspension of nuclei. The fluorescence intensity of the stained nuclei was measured using a PA I flow cytometer with an HBO-100 mercury arc lamp (Partec GmbH, Germany). The relative nuclear DNA content (expressed in arbitrary units) was determined from the ratio of G1 peak positions of the analysed sample/standard. For each individual, three independent measurements on different days were carried out. For each measurement, the coefficient of variation (CV) for both the internal standard and the analysed sample was calculated. Differences in nuclear DNA content values measured for the same individual in the three separate measurements were calculated as the standard errors of the means, and expressed as a percentage of the mean value.
RESULTS
Pollen viability estimation
Male fertility, indicated by pollen stainability, was relatively high in all examined pure populations of C. pratensis and C. raphanifolia (Table 3, Fig. 2). The mean population values in C. pratensis ranged from 76·8 to 94·7 % pollen viability, and in C. raphanifolia from 70·8 to 92·8 %. In contrast, the mean population values of the hybrid populations varied from 25·8 % in H1 to 63·8 % in H3, and considerably unbalanced values were observed for individual plants (ranging from 0 to 100 %; Table 3, Fig. 2).
Table 3.
Pollen viability observed in the studied Cardamine populations, expressed as minimum, maximum and mean values ± s.d.
| C. pratensis | P-46 | P-36 | P-49 | P-57 | P-51 | |
| Min–max | 63–100 % | 64–94 % | 20–97 % | 34–100 % | 39–95 % | |
| Mean ± s.d. | 94·7 ± 8·2 % | 78·2 ± 9·0 % | 82·1 ± 18·1 % | 79·9 ± 19·7 % | 76·8 ± 15·8 % | |
| C. raphanifolia | R-AP | R-SG | R-QRR | R-PTR | R-MCR | R-HC |
| Min–max | 56–90 % | 85–90 % | 61–84 % | 50–88 % | 63–99 % | 75–99 % |
| Mean ± s.d. | 79·9 ± 8·9 % | 87·3 ± 2·0 % | 70·8 ± 6·1 % | 75·3 ± 8·0 % | 85·8 ± 9·7 % | 92·8 ± 6·0 % |
| Hybrid populations | H1 | H2 | H3 | H4 | ||
| Min–max | 0–67 % | 5–100 % | 0–94 % | 0–99 % | ||
| Mean ± s.d. | 25·8 ± 22·1 % | 56 ± 28 % | 63·8 ± 23·2 % | 60 ± 29 % |
Population abbreviations and the number of individuals are specified in Table 1.
Fig. 2.
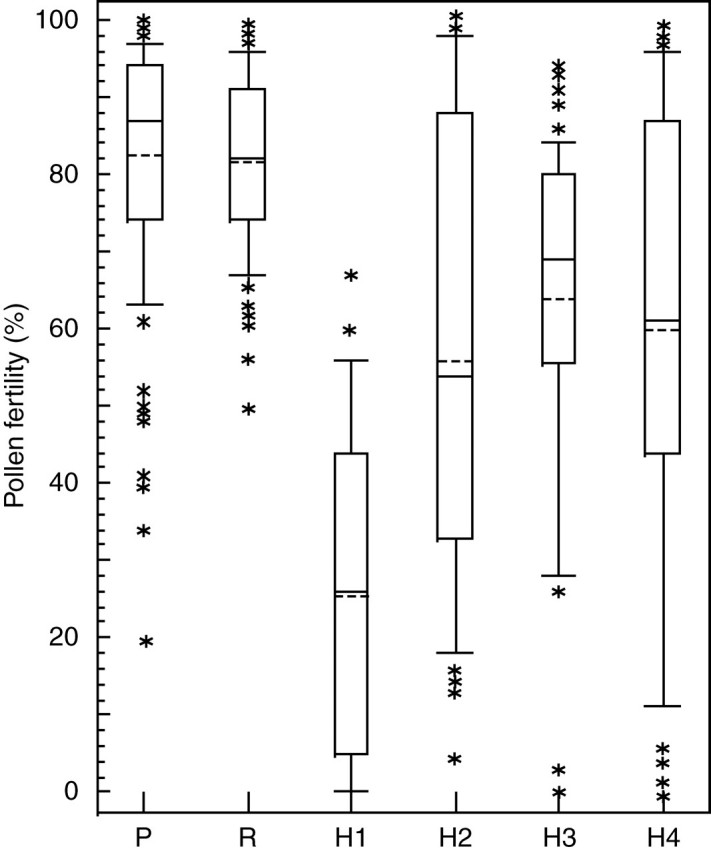
Variation in pollen viability observed in C. pratensis from five pure populations (P), C. raphanifolia from six pure populations (R) and in the four studied hybrid populations (H1–H4, see Table 1). Rectangles define 25th and 75th percentiles; horizontal full lines show the median; horizontal dashed lines show the mean; whiskers are from the 10th to the 90th percentiles; and asterisks show extreme values.
Morphometric analyses
PCA based on 21 measured characters in individual plants from the pure populations resulted in two groups clearly separated along the first axis, corresponding to C. pratensis and C. raphanifolia (figure not shown). As can be inferred from the eigenvectors, expressing correlations of the characters with the first axis, several characters are responsible for this differentiation: average length of filaments, width of stem at base, number of pairs of lateral leaflets/segments and shapes of lateral and terminal leaflets/segments on the middle stem leaf (expressed as ratios, see Table 2). It should be noted that only quantitative characters were included in the PCA, but C. pratensis and C. raphanifolia are also distinct in respect of the morphology of the rhizome (shortened and erect in the former vs. elongated, prostrate or ascending in the latter), formation of the rosette of basal leaves (true rosette in the former vs. no basal rosette in the latter) and morphology of the stem leaves (at least upper stem leaves pinnatisect in the former vs. all leaves pinnate in the latter; see Lihová et al., 2003; Perný et al., 2005).
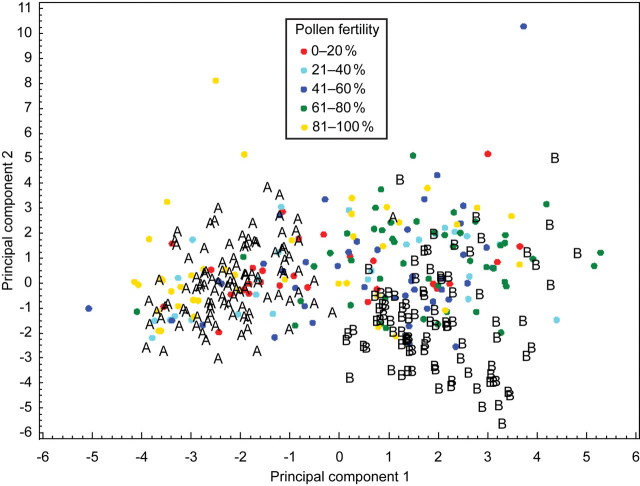
PCA based on individual plants from both the pure and hybrid populations (Fig. 3) showed the same pattern in respect of the clear differentiation between pure C. pratensis and C. raphanifolia. Individuals from the hybrid populations were placed interspersed in the ordination space, i.e. covering the whole variation range of both species, as well as the space between the individuals of the parental species. Although this was not included in the PCA, many individuals from hybrid populations showed equivocal and intermediate character states in rhizome morphology, formation of the basal rosette and morphology of stem leaves (pinnate or pinnatisect), supporting the PCA results based on quantitative characters. When focusing on the pollen viability of plants from the hybrid populations and their positions in the ordination space, it becomes apparent that there is no correlation between the sterility and morphological intermediacy. Sterile plants (0–20 % pollen viability) recorded in the hybrid populations were found not only in intermediate positions, but also among plants from the pure populations exhibiting typical parental phenotypes. Similarly, highly fertile plants (81–100 %) from the hybrid populations were found across the whole ordination space, including intermediate positions and thus clearly intermediate phenotypes (Fig. 3).
Fig. 3.
Principal component analysis of individuals of C. pratensis from the pure populations (A), C. raphanifolia from the pure populations (B) and individuals from the hybrid populations (coloured circles) based on 21 morphological characters. The first two axes explain 25·7 and 19·1 % of the total variation. Colours denote pollen stainability classes as indicated on the diagram.
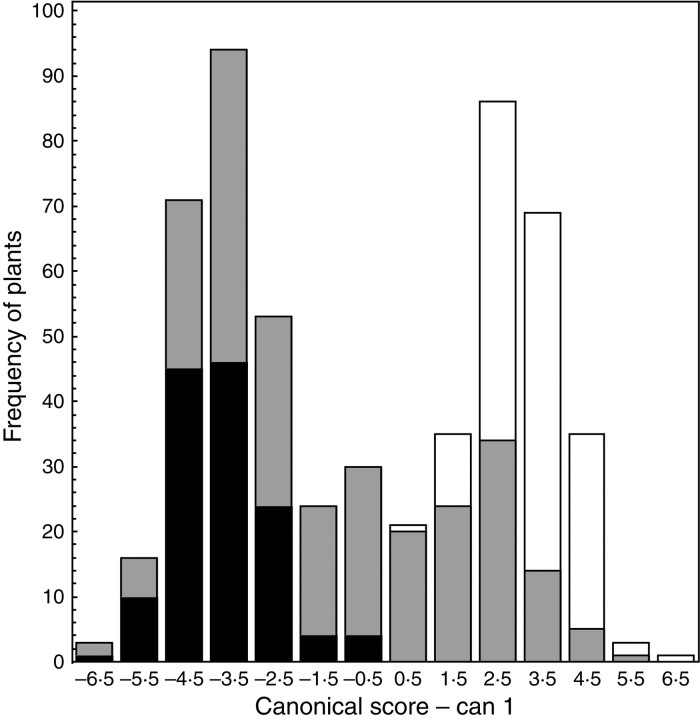
In accordance with PCA results, CDA based on individuals from the pure populations of C. pratensis and C. raphanifolia with these two taxa defined as groups revealed two morphologically differentiated entities with no overlap at all (Fig. 4). Characters showing the highest correlation with the canonical axis, and thus best separating these two taxa, were the same as in PCA (Table 2). The discriminant function resulting from this analysis was used for computing corresponding canonical scores of individuals from the hybrid populations. In the histogram depicting their positions it is seen that both typical parental phenotypes are present, and that many specimens occupied intermediate positions, corresponding to the phenotypes not found (or only very rarely) in the pure populations (Fig. 4).
Fig. 4.
Canonical discriminant analysis of C. pratensis (white) and C. raphanifolia (black) originating from the pure populations, based on 21 morphological characters. Individuals from the hybrid populations (grey) were additionally depicted on the resulting histogram according to their canonical scores.
Molecular analyses (AFLP and cpDNA sequences)
The total number of the scored AFLP fragments using three primer combinations was 164; 150 of them were polymorphic. In the hybrid populations, a total of 121 fragments were recorded; 105 of them were polymorphic. The hybrid populations shared 33 fragments with C. pratensis from the pure populations, which were absent in C. raphanifolia; 21 fragments with C. raphanifolia, which were absent in C. pratensis; and 42 fragments simultaneously with both C. pratensis and C. raphanifolia; 25 fragments were specific to the hybrid populations and were not detected among individuals from the pure populations (most probably due to the significantly smaller sampling within the pure populations). In the few individuals of C. raphanifolia from the pure populations, 87 fragments were recorded; 69 of them were polymorphic. In C. pratensis from the pure populations, 94 fragments were resolved; 60 of them polymorphic. Approximately half of the fragments resolved within each species was species specific (absent in the other species; 52 for C. pratensis, 45 for C. raphanifolia), while the other half of fragments were shared between the species (42 fragments). In C. raphanifolia, eight of those 45 specific fragments were species-specific fixed (present in each individual, absent from C. pratensis pure populations).
In the PCoA ordination diagram, individuals from the pure populations were placed in two distinctly separated groupings, corresponding to C. pratensis and C. raphanifolia, respectively (Fig. 5A). This pattern clearly documents that two differentiated groups of AFLP genotypes can be delimited. Individuals from the hybrid populations were resolved in the respective groups of parental genotypes as well as in the space between them, spanning a large variation range. It is apparent that both typical parental genotypes as well as a number of intermediate genotypes can be found in the hybrid populations.
Fig. 5.
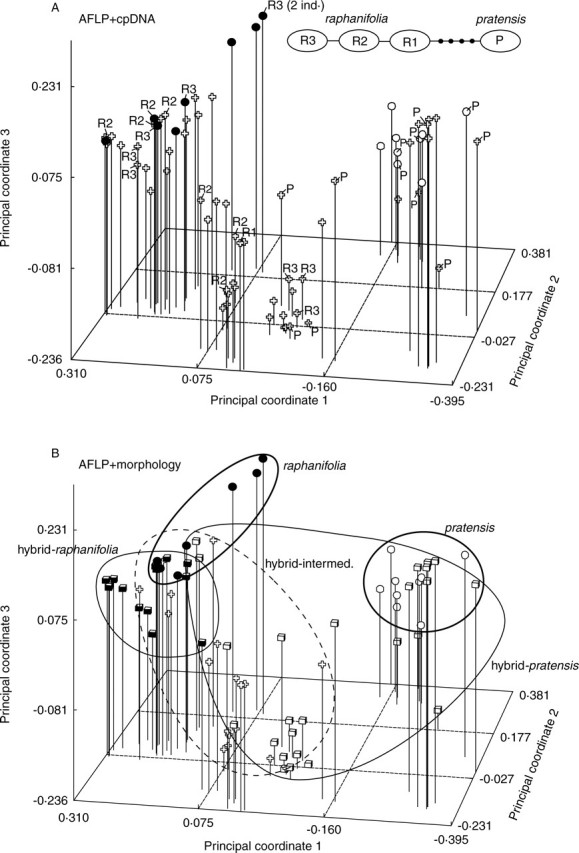
Principal coordinate analysis of AFLP data of 85 individuals from both pure and hybrid populations. The first three axes explain 18·8, 11·5 and 7·9 % of the total variation. (A) Individuals from the pure populations of C. pratensis are marked by open circles, those of C. raphanifolia by filled circles, and individuals from the hybrid populations by crosses. The unrooted haplotype parsimony network based on the trnL-trnF intergenic spacer is additionally depicted, and the corresponding haplotypes determined in 29 individuals are mapped onto the ordination graph (R1–R3, P). In the haplotype network, circles represent haplotypes, lines are single mutational steps, and dots are non-detected haplotypes. (B) Individuals from the pure populations of C. pratensis are marked by open circles (encircled by a bold line), those of C. raphanifolia by filled circles (encircled by a bold line) and individuals from the hybrid populations are sorted into three classes according to their morphology (expressed by the canonical score): morphologically corresponding to C. pratensis (open cubes; encircled by a normal line), to C. raphanifolia (filled cubes; encircled by a normal line) and morphologically intermediate (crosses; encircled by a dashed line).
GenBank accession numbers of the trnL-trnF sequences are given in Table 1. Out of 29 individuals analysed for trnL-trnF variation, nine were sampled from the pure populations. The sequences obtained were consistent with the pattern resolved in the previous study by Lihová et al. (2004), and allowed unequivocal distinction between the haplotypes of C. pratensis and C. raphanifolia (see Fig. 5A). Among the 20 individuals sampled from the hybrid populations, eight showed haplotypes of C. pratensis and 12 of C. raphanifolia. The haplotypes detected are in congruence with the AFLP variation pattern, as can be seen in Fig. 5A. Genetically intermediate genotypes, as assessed from AFLPs, possess either C. pratensis- or C. raphanifolia-specific haplotypes, while individuals with one of the parental AFLP genotypes are characterized by the presence of the respective haplotypes of the parental species.
Individuals from the hybrid populations were further assigned to the three classes of morphological phenotypes, following their canonical scores in CDA: morphologically corresponding to C. pratensis, to C. raphanifolia and to morphological intermediates. When mapping this information onto the PCoA graph of AFLP data, the following pattern can be seen (Fig. 5B). Individuals morphologically corresponding to C. pratensis exhibit a wide range of AFLP genotypes, not only those typical for C. pratensis from pure populations, but also intermediate ones, as well as those approaching the AFLP genotypes found in C. raphanifolia from the pure populations. Both C. pratensis- and C. raphanifolia-specific haplotypes were found in these plants. Individuals morphologically corresponding to C. raphanifolia, on the other hand, mostly possessed AFLP genotypes similar to those found in the pure populations of this species, and only a few of them showed signs of introgression, shifted towards the genetically intermediate genotypes. Only C. raphanifolia-specific haplotypes were found here. Morphologically intermediate individuals exhibited either clearly intermediate AFLP genotypes or genotypes close to those in C. raphanifolia from the pure populations, and showed only haplotypes specific to C. raphanifolia.
Chromosome numbers and flow cytometry
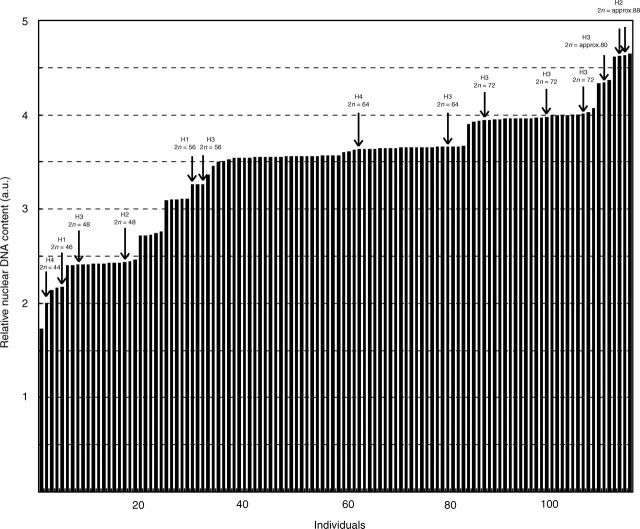
Flow cytometric measurements produced histograms with reasonably low CVs, being on average 3·54 for standards and 4·52 for the measured samples. The standard errors of the means expressed as the percentage of mean values did not exceed 0·7 %, indicating that the data are precise and reliable. The DNA content values screened in 114 individuals from the hybrid populations are presented in Fig. 6. Data on chromosome numbers counted are given in Table 1 and also included in Fig. 6. As can be seen, several ploidy levels and chromosome numbers were found, ranging from 2n = 44 to 2n = approx. 88. These include regular numbers known for pure populations of C. pratensis in Spain (2n = 16, 30, 32, 46, 48, 56; see Lihová et al., 2003) and for C. raphanifolia (2n = 48, 64; see Perný et al., 2005), as well as some other, unusual chromosome numbers, in some cases even extending the known ploidy levels of the parental species such as 2n = 44, 72, approx. 80, approx. 88. The wide range of chromosome numbers and ploidy levels indicates extensive gene flow in the hybrid populations, with apparently various meiotic and zygotic chromosomal constitutions.
Fig. 6.
Relative nuclear DNA content measured in 114 individuals from the hybrid populations (H1–H4, see Table 1) expressed in arbitrary units (a.u.) and shown in increasing order. Individuals with the counted chromosome numbers are indicated.
DISCUSSION
Molecular evidence for hybridization: AFLP and cpDNA variation patterns
Hybridization is an important evolutionary phenomenon, and therefore a detailed understanding of the processes involved in hybridization is of widespread interest. Frequently addressed questions include the identity of parental species, the directionality and frequency of hybridization events, hybrid-zone structure, hybrid fitness, genetic variability of hybrids and evolutionary potential (Rieseberg and Carney, 1998). Most recently, advanced molecular tools provide the opportunity to gain insights into the consequences of hybridization at the level of the genome and transcriptome (Hegarty and Hiscock, 2005). These investigations are focused mainly on well-explored genera such as Arabidopsis, Brassica, Gossypium, Helianthus or Spartina, and allow fundamental questions about hybrid speciation to be addressed (reviewed by Hegarty and Hiscock, 2005).
In the present study, suspected hybridization was explored in four Cardamine populations that turned out to represent very dynamic and complex hybrid swarms worthy of future investigations. Evidence for hybridization has been gathered from multiple data sources, and the present sampling design allowed direct, individual-based comparison between genetic and morphological variation patterns. DNA fingerprinting methods, such as AFLPs employed in the present study, have been successfully used to document hybridization events and to confirm hybrid origins in many previous studies (e.g. Gobert et al., 2002; Bleeker, 2003; Guo et al., 2005; van Droogenbroeck et al., 2006; Paun et al., 2006). Despite the drawback of the dominant inheritance and anonymity of generated AFLP fragments, this technique produces a high number of polymorphic and reproducible markers that can distinguish between closely related taxa. If species-specific AFLP fragments are available, i.e. if there are fixed genetic differences between the putative parental taxa, these can be screened in potential hybrid individuals. Additive banding patterns of species-specific AFLP fragments have been expected particularly in early generation hybrids, and this approach has been successfully followed in several studies exploring hybridization events and hybrid speciation (e.g. Bleeker, 2003; Salmon et al., 2005; Kadereit et al., 2006). However, as is the case here, it can be problematic to find species-specific markers, especially when closely related and highly polyploid taxa of unknown origin are involved. While eight specific and fixed fragments were recorded among selected individuals of C. raphanifolia, no such fragments were found in C. pratensis, precluding exploration of AFLP additive patterns in putative hybrids. These individuals were drawn from previously analysed geographically distant and genetically differentiated populations to provide a reasonable representation of AFLP genotypes for each of the species (Lihová et al., 2003; Perný et al., 2005). The diagram of PCoA presented here showed that the generated AFLP fragments (and their frequency) discriminated well between the two species, a result in accordance with previous studies with more extensive sampling and encompassing several other related species (see Lihová and Marhold, 2006). In the present study, the main focus, however, was on pattern variations within the putative hybrid populations. Indeed, considerable genetic variation in respect of AFLP genotypes was observed; except for the parental genotypes, numerous individuals in intermediate positions were also found, unequivocally pointing to genetic admixture and, thus, their hybrid origin (Fig. 5). Except for the PCoA presented here, being a distance-based method, some other powerful methods for the analysis of multilocus molecular data that allow assignment to parental species and estimation of admixture proportion are available. The Bayesian model-based analyses (Corander and Marttinen, 2006) have recently gained much interest, as they may be more informative for the estimation of gene flow and admixture proportions than a largely graphical analysis such as PCoA. Similarly, the maximum likelihood hybrid index analysis can be used to quantify the genetic contribution of hybridizing taxa to putative hybrid individuals (Buerkle, 2005). However, the complex ploidy level variation observed in the present study precludes utilization of a simple, model-based approach (assuming Hardy–Weinberg proportions for diploids). If ploidy levels were simultaneously estimated for each individual included in the AFLP analysis, a fairly complex model should be specified for a complete admixture analysis. Therefore, PCoA appears to be most powerful in this case, expressing and displaying overall pattern variations in polymorphic AFLP markers, and thus intermediate AFLP genotypes of hybrid individuals that combine parental ones can also be detected in this way. Patterns of fragment sharing between the hybrid populations and individuals from the pure populations are also in favour of the genetic admixture in the hybrid populations as displayed by PCoA.
Distribution of the species-specific cpDNA haplotypes in the hybrid populations, which is in accordance with the AFLP pattern, provided further evidence for hybridization. While C. pratensis-specific haplotypes were identified in individuals with AFLP genotypes corresponding to this species, and vice versa as for C. raphanifolia, both haplotypes were present among apparent hybrid individuals. It can be inferred from this picture that hybridization clearly proceeds in both directions (cpDNA is maternally inherited in the Brassicaceae; Harris and Ingram, 1991).
Morphometric approach; discordance between morphological and molecular patterns
In earlier studies, morphological intermediacy was taken as the main evidence of hybridity, and different approaches have been suggested to express the intermediacy as well as to distinguish hybrid phenotypes from those resulting from divergence. Hybrid indices, ordination methods (PCA, PCoA or scatter diagrams) as well as ‘character count procedure’ have been used to document intermediate phenotypes that putatively arose by hybridization (Wilson, 1992). Although the character intermediacy still remains the most important and first indication of hybrid origin from the phenotype perspective, it is also clear that character expression in hybrids is rather complex and to a certain extent also unpredictable. Even F1 and early generation hybrids have been observed to be a mosaic of parental, intermediate and transgressive characters (Rieseberg and Ellstrand, 1993; Rieseberg and Carney, 1998; Schwarzbach et al., 2001). Employment of molecular markers yields independent data and independent indication for hybrid origins, and subsequently allows direct comparison with morphology. Several studies have efficiently combined data from morphology and molecular markers to detect and explore hybridization (Hardig et al., 2000; Marhold et al., 2002; Tovar-Sánchez and Oyama, 2004).
In the present study, four sympatric populations of C. pratensis and C. raphanifolia were sampled, which, when growing in separate stands, can be unequivocally distinguished by several qualitative and quantitative characters. In four populations, denoted here as hybrid, however, a phenotype continuum was observed that completely blurred morphological differences between the two species. For these two reasons (morphological continuum and complex character expression in hybrids), no attempt was made to classify the plants as parental or hybrid prior to morphological and molecular analyses. A different approach was used to express the observed morphological variation and to relate it to genetic variation patterns. To define and delimit the parental morphological range, pure populations of both species were sampled and used as reference in two morphometric analyses, PCA and CDA. The former provided an unequivocal illustration that both typical parental phenotypes as well as an array of those which display intermediate character states can be found in hybrid populations; a few individuals exhibited apparently non-parental values and appeared in somewhat distant positions in the ordination space. In CDA, a canonical function was derived in order to differentiate between the parental phenotypes and to obtain a standardized classification criterion for parental and non-parental hybrid phenotypes. When this morphology-based classification was superimposed on the AFLP and cpDNA pattern, a partial discordance between the data sets was observed. Plants, which from morphology would be identified as C. pratensis, in fact may represent hybrids or introgressed individuals, as assessed from their genetic patterns. Some morphologically intermediate specimens correspond genetically to C. raphanifolia. On the other hand, morphology attributable to C. raphanifolia appears to be a rather good indicator of AFLP variation pattern (see Fig. 5B). Generally, however, it is apparent that morphology does not allow explanation or reliable prediction of the AFLP variation pattern and cpDNA haplotype, and vice versa. Morphology, when used alone, can be misleading for interpreting hybridization, and critical evaluation of other data is needed. Parental phenotypes may in fact be more or less affected by hybridization and introgression, while intermediate genotypes may genetically approach either of the parents.
Extensive interspecific hybridization observed between C. pratensis and C. raphanifolia, resulting in the formation of dynamic hybrid swarms, may also be significant for unravelling phylogenetic relatedness and polyploid evolutionary history of the parental species. Previous studies addressing phylogenetic relationships among taxa from the polyploid complexes of C. raphanifolia, C. pratensis and C. amara suggested extensive reticulate evolution in the past, and that the origin of taxa from the C. raphanifolia group should be traced among taxa from all three complexes (Lihová et al., 2004; Marhold et al., 2004; see also Lihová and Marhold, 2006). In this context, it can be assumed that the lack of species-specific fixed AFLP fragments in C. pratensis in respect to C. raphanifolia, but not vice versa, might reflect the polyploid origin of the latter, where C. pratensis or its progenitors may have been involved. If this was true, then we could speculate about the causes of the morphological and genetic discordance (as shown in Fig. 5B). The genome of C. pratensis included in C. raphanifolia might be the reason why hybrids revealed by genetic data morphologically resemble C. pratensis rather than having intermediate morphology; the genome of C. pratensis would prevail in the genetic constitution of the hybrids. An alternative explanation for the discordance, however, might be the overall pattern of introgression, where backcrossing may occur asymmetrically and hybrids hybridize more frequently with C. pratensis than with C. raphanifolia individuals.
Dynamics of interspecific gene flow in the studied hybrid populations
One of the most straightforward indications for hybridization used since early studies is the reduced fertility in hybrids that results from genetic differences between the parental species and the attendant meiotic disturbances (Levin, 2002). The reduction in fertility undoubtedly represents a reproduction barrier and restricts mating and backcrossing. If interbreeding among F1 hybrids and backcrossing occurs, even a limited amount, it may lead either to the constitution of balanced and novel genotypes with restored fertility that become reproductively isolated, or to a dynamic hybrid swarm in an overlap zone between the two species, characterized by extensive gene flow, backcrosses and introgression (Buerkle et al., 2000; Mallet, 2005). In C. pratensis and C. raphanifolia, the latter scenario was inferred from both molecular and morphological data, as well as a full range of pollen fertility values and chromosomal variation found in individuals sampled from hybrid populations. In this context, the unusual chromosome number reports of 2n = 40, 44, 46, 50, 72 originally attributed to C. raphanifolia by Landolt in Perný et al. (2005) and Lövkvist (1956) should also be highlighted, which, based on the present study, most probably refer to hybrid individuals.
Clearly different patterns resulting from hybridization events were observed in two recently examined Cardamine hybrids, Pyrenean diploid C. × enriquei (C. amara ssp. pyrenaea × C. crassifolia, Marhold et al., 2002) and the Swiss/Italian pentaploid C. × ferrarii (hexaploid C. asarifolia × diploid C. amara ssp. amara; Lihová et al., 2006). In both cases, morphological distinction between the parental taxa and the hybrids was clear and unequivocal, as was the high pollen sterility in intermediate plants, which was in contrast to largely fertile pollen in the parents. No continuous variation in the pollen fertility was observed, as in the case of the hybrid populations studied here. Although the possibility for introgression and backcrossing was also invoked for populations with the occurrence of C. × enriquei, the hybrids appeared morphologically and genetically much more homogeneous, and backcrossing and introgression, if they occur, seem to be rather limited there (Marhold et al., 2002; Lihová et al., 2006). It might be hypothesized that isolation barriers due to genetic incompatibilities occurring between (a) the two diploids or (b) the diploid and the hexaploid parent (see above) are much stronger than can be expected when the two hybridizing species are both high polyploids with assumed close evolutionary history. This might explain the contrasting patterns observed in the previous two reports on hybridization in Cardamine, and the present study. Detailed investigations on gamete formation and pathways of gene flow were performed in the case of the thoroughly studied triploid hybrid C. × insueta from Switzerland (Urbanska et al., 1997). Based on experimental crosses as well as investigation of natural stands, unusual meiotic behaviour during both micro- and macrosporogenesis was documented in the triploid hybrid, resulting either in polarized haploid and diploid gametes or in unreduced triploid gametes. Thus, the heterogamy allows reproduction of the hybrid and its recurrent formation, as well as backcrosses with one of the parents (Urbanska et al., 1997). In the context of the present study, it would also be interesting to explore gamete formation and reproduction in the polyploid hybrid populations studied here, where, from the large variation in the relative nuclear DNA content and chromosome numbers detected, irregular meiotic processes are expected. Complex chromosomal variation has been observed even in the parental taxa, but has not been studied at the level of meiosis and gametes so far.
Ecological aspects of the hybrid formation and establishment
From the ecological point of view, at three localities (H1, H2 and H4) all representing wet meadows and pastures, plants of both parental phenotypes and hybrid ones were found intermingled, without any ecological or spatial pattern apparent at first sight. At the H3 locality (Puerto de Pajares), however, a different pattern was observed. Whereas plants of parental phenotypes together with the scarce occurrence of intermingled hybrid phenotypes grew along brooks on a pasture, providing a relatively stable and less disturbed habitat, a dense population consisting of individuals of exclusively intermediate phenotype (and intermediate genotypes as resolved by AFLPs) was spreading in a drainage ditch along the road and on an opposite mound (see Lihová and Marhold, 2006: Fig. 4). The role of environmental disturbance for creating new niches or open habitat space available for hybrids is widely recognized (Urbanska, 1987; Rieseberg and Carney, 1998), and apparently was also crucial for the establishment of the dense hybrid population at this particular locality. Similarly, the successful establishment of the above-mentioned hybrid C. × insueta is associated with the colonization of man-made drained, fenced and fertilized hay meadows (Urbanska, 1987; Urbanska et al., 1997). Several other examples can also be mentioned, among them Rorippa × armoracioides (Tausch) Fuss growing at ruderal sites and on abandoned agricultural fields (Bleeker, 2003).
The above-mentioned examples illustrate recent and still ongoing interspecific hybridizations, where the hybrids have remained localized in the vicinity, or sympatry with parental species, and have not been observed to spread beyond their distribution range so far. Interspecific hybridization and subsequent recombination in hybrids can, however, generate novel genotypes that may colonize new areas, exhibit elevated rates of response to selection and finally result in adaptive radiation (Seehausen, 2004).
ACKNOWLEDGEMENTS
This research was supported by the Grant Agency VEGA, Slovak Republic (project no. 6055/26 to J.L.) and the Ministry of Education, Youth and Sports of the Czech Republic (grant no. 0021620828 to K.M.). AFLP data were generated at the Department of Systematic and Evolutionary Botany, Faculty Center Botany of the University of Vienna, Austria; thanks are due to T.F. Stuessy, K. Tremetsberger and G. Kadlec for their support and supervision. We thank P. Trávníček (Institute of Botany, Czech Academy of Sciences, Průhonice, Czech Republic) for assistance with flow cytometry, F. Llamas, A. Fernandez and J. Ignacio (University of León, Spain) for their help in the field, and M. Beilstein (Harvard University, Cambridge, MA, USA) for improvement to the English and useful comments on the manuscript.
LITERATURE CITED
- Applied Biosystems. AFLP™ plant mapping protocol. Foster City, CA: PE Applied Biosystems; 1996. [Google Scholar]
- Arnold MJ. Natural hybridization as an evolutionary process. Annual Review of Ecology and Systematics. 1992;23:237–261. [Google Scholar]
- Bleeker W. Hybridization and Rorippa austriaca (Brassicaceae) invasion in Germany. Molecular Ecology. 2003;12:1831–1841. doi: 10.1046/j.1365-294x.2003.01854.x. [DOI] [PubMed] [Google Scholar]
- Bleeker W, Hurka H. Introgressive hybridization in Rorippa (Brassicaceae): gene flow and its consequences in natural and anthropogenic habitats. Molecular Ecology. 2001;10:2013–2022. doi: 10.1046/j.1365-294x.2001.01341.x. [DOI] [PubMed] [Google Scholar]
- Buerkle CA. Maximum-likelihood estimation of a hybrid index based on molecular markers. Molecular Ecology. 2005;5:684–687. [Google Scholar]
- Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. The likelihood of homoploid hybrid speciation. Heredity. 2000;84:441–451. doi: 10.1046/j.1365-2540.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- Corander J, Marttinen P. Bayesian identification of admixture events using multi-locus molecular markers. Molecular Ecology. 2006;15:2833–2843. doi: 10.1111/j.1365-294X.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- Doležel J. Application of flow cytometry for the study of plant genomes. Journal of Applied Genetics. 1997;38:285–302. [Google Scholar]
- Doležel J, Doleželová M, Novák FJ. Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana) Biologia Plantarum. 1994;36:351–357. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin of the Botanical Society of America. 1987;19:11–15. [Google Scholar]
- van Droogenbroeck B, Kyndt T, Romeijn-Peeters E, van Thuyne W, Goetghebeur P, Romero-Motochi JP, Gheysen G. Evidence of natural hybridization and introgression between Vasconcellea species (Caricaceae) from Southern Ecuador revealed by chloroplast, mitochondrial and nuclear DNA markers. Annals of Botany. 2006;97:793–805. doi: 10.1093/aob/mcl038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellstrand NC, Whitkus R, Rieseberg LH. Distribution of spontaneous plant hybrids. Proceedings of the National Academy of Sciences of the USA. 1996;93:5090–5093. doi: 10.1073/pnas.93.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert V, Moja S, Colson M, Taberlet P. Hybridization in the section Mentha (Lamiaceae) inferred from AFLP markers. American Journal of Botany. 2002;89:2017–2023. doi: 10.3732/ajb.89.12.2017. [DOI] [PubMed] [Google Scholar]
- Guo Y-P, Saukel J, Mittermayr R, Ehrendorfer F. AFLP analyses demonstrate genetic divergence, hybridization, and multiple polyploidization in the evolution of Achillea (Asteraceae-Anthemideae) New Phytologist. 2005;166:273–290. doi: 10.1111/j.1469-8137.2005.01315.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hardig TM, Brunsfeld SJ, Fritz RS, Morgan M, Orians CM. Morphological and molecular evidence for hybridization and introgression in a willow (Salix) hybrid zone. Molecular Ecology. 2000;9:9–24. doi: 10.1046/j.1365-294x.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- Harris SA, Ingram R. Chloroplast DNA and biosystematics: the effect of intraspecific diversity and plastid transmission. Taxon. 1991;40:393–412. [Google Scholar]
- Hegarty MJ, Hiscock SJ. Hybrid speciation in plants: new insights from molecular studies. New Phytologist. 2005;165:411–423. doi: 10.1111/j.1469-8137.2004.01253.x. [DOI] [PubMed] [Google Scholar]
- Kadereit JW, Uribe-Convers S, Westberg E, Comes HP. Reciprocal hybridization at different times between Senecio flavus and Senecio glaucus gave rise to two polyploid species in north Africa and south-west Asia. New Phytologist. 2006;169:431–441. doi: 10.1111/j.1469-8137.2005.01604.x. [DOI] [PubMed] [Google Scholar]
- Klecka WR. Discriminant analysis. Sage University papers, series: quantitative applications in the social sciences, no. 19. Beverly Hills, CA: Sage Publications; 1980. [Google Scholar]
- Koch MA, Dobeš Ch, Matschinger M, Bleeker W, Vogel J, Kiefer M, Mitchell-Olds T. Evolution of the trnF (GAA) gene in Arabidopsis relatives and the Brassicaceae family: monophyletic origin and subsequent diversification of a plastidic pseudogene. Molecular Biology and Evolution. 2005;22:1032–1043. doi: 10.1093/molbev/msi092. [DOI] [PubMed] [Google Scholar]
- Krzanowski WJ. Principles of multivariate analysis. Oxford: Clarendon Press; 1990. [Google Scholar]
- Kučera J, Lihová J, Marhold K. Taxonomy and phylogeography of Cardamine impatiens and C. pectinata (Brassicaceae) Botanical Journal of the Linnean Society. 2006;152:169–195. [Google Scholar]
- Legendre P, Legendre L. Numerical ecology. 2nd edn. Amsterdam: Elsevier; 1998. [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Lihová J, Marhold K. Phylogenetic and diversity patterns in Cardamine (Brassicaceae) – a genus with conspicuous polyploid and reticulate evolution. In: Sharma AK, Sharma A, editors. Plant genome: biodiversity and evolution, vol. 1C: Phanerogams (angiosperms – dicotyledons) Enfield, NH: Science Publishers, Inc; 2006. pp. 149–186. [Google Scholar]
- Lihová J, Tribsch A, Marhold K. The Cardamine pratensis (Brassicaceae) group in the Iberian Peninsula: taxonomy, polyploidy and distribution. Taxon. 2003;52:783–802. [Google Scholar]
- Lihová J, Fuertes Aguilar J, Marhold K, Nieto Feliner G. Origin of the disjunct tetraploid Cardamine amporitana (Brassicaceae) assessed with nuclear and chloroplast DNA sequence data. American Journal of Botany. 2004;91:1231–1242. doi: 10.3732/ajb.91.8.1231. [DOI] [PubMed] [Google Scholar]
- Lihová J, Shimizu KK, Marhold K. Allopolyploid origin of Cardamine asarifolia (Brassicaceae): incongruence between plastid and nuclear ribosomal DNA sequences solved by a single-copy nuclear gene. Molecular Phylogenetics and Evolution. 2006;39:759–786. doi: 10.1016/j.ympev.2006.01.027. [DOI] [PubMed] [Google Scholar]
- Lövkvist B. The Cardamine pratensis complex. Outline of its cytogenetics and taxonomy. Symbolae Botanicae Upsalienses. 1956;14/2:1–131. [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends in Ecology and Evolution. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Marhold K, Lihová J. Polyploidy, hybridization and reticulate evolution: lessons from the Brassicaceae. Plant Systematics and Evolution. 2006;259:143–174. [Google Scholar]
- Marhold K, Lihová J, Perný M, Grupe R, Neuffer B. Natural hybridization in Cardamine (Brassicaceae) in the Pyrenees: evidence from morphological and molecular data. Botanical Journal of the Linnean Society. 2002;139:275–294. [Google Scholar]
- Marhold K, Lihová J, Perný M, Bleeker W. Comparative ITS and AFLP analysis of diploid Cardamine (Brassicaceae) taxa from closely related polyploid complexes. Annals of Botany. 2004;93:507–520. doi: 10.1093/aob/mch073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks GE. An aceto-carmine glycerol jelly for use in pollen fertility counts. Stain Technology. 1954;29:277. doi: 10.3109/10520295409115483. [DOI] [PubMed] [Google Scholar]
- Otto F. DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. Methods in Cell Biology. 1990;33:105–110. doi: 10.1016/s0091-679x(08)60516-6. [DOI] [PubMed] [Google Scholar]
- Paun O, Stuessy TF, Hörandl E. The role of hybridization, polyploidization and glaciation in the origin and evolution of the apomictic Ranunculus cassubicus complex. New Phytologist. 2006;171:223–236. doi: 10.1111/j.1469-8137.2006.01738.x. [DOI] [PubMed] [Google Scholar]
- Perný M, Tribsch A, Stuessy T, Marhold K. Taxonomy and cytogeography of Cardamine raphanifolia and C. gallaecica (Brassicaceae) in the Iberian Peninsula. Plant Systematics and Evolution. 2005;254:69–91. [Google Scholar]
- Podani J. SYN-TAX 2000. Computer programs for data analysis in ecology and systematics. User's manual. Budapest, Hungary: Scientia Publishing; 2001. [Google Scholar]
- Rieseberg LH. Molecular ecology of hybridization. In: Carvalho GR, editor. Advances in molecular ecology. Amsterdam: IOS Press; 1998. pp. 243–265. [Google Scholar]
- Rieseberg LH, Carney SE. Tansley review no. 102. Plant hybridization. New Phytologist. 1998;140:599–624. doi: 10.1046/j.1469-8137.1998.00315.x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Ellstrand NC. What can molecular and morphological markers tell us about plant hybridisation? Critical Reviews in Plant Sciences. 1993;12:213–241. [Google Scholar]
- Salmon A, Ainouche ML, Wendel J. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae) Molecular Ecology. 2005;14:1163–1175. doi: 10.1111/j.1365-294X.2005.02488.x. [DOI] [PubMed] [Google Scholar]
- SAS Institute. SAS OnlineDoc®, version 8 (available online). Cary, NC: SAS Institute; 2000. [Google Scholar]
- Schols P, Dessein S, D'hondt C, Huysmans S, Smets E. Carnoy: a new digital measurement tool for palynology. Grana. 2002;41:124–126. [Google Scholar]
- Schönswetter P, Tribsch A, Barfuss M, Niklfeld H. Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium Sternb. & Hoppe (Campanulaceae) in the European Alps. Molecular Ecology. 2002;11:2637–2647. doi: 10.1046/j.1365-294x.2002.01651.x. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Suda J, Popp M, Weiss-Schneeweiss H, Brochmann Ch. Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints, cpDNA sequences, nuclear DNA content and chromosome numbers. Molecular Phylogenetics and Evolution. 2007;42:92–103. doi: 10.1016/j.ympev.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Schwarzbach AE, Donovan LA, Rieseberg LH. Transgressive character expression in a hybrid sunflower species. American Journal of Botany. 2001;88:270–277. [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends in Ecology and Evolution. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Sneath PHA, Sokal RR. Numerical yaxonomy. Principles and practice of numerical classification. San Francisco, CA: W. H. Freeman and Company; 1973. [Google Scholar]
- Soltis DE, Soltis PS, Tate JA. Advances in the study of polyploidy since plant speciation. New Phytologist. 2004;161:173–191. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tovar-Sánchez E, Oyama K. Natural hybridization and hybrid zones between Quercus crassifolia and Q. crassipes (Fagaceae) in Mexico: morphological and molecular evidence. American Journal of Botany. 2004;91:1352–1363. doi: 10.3732/ajb.91.9.1352. [DOI] [PubMed] [Google Scholar]
- Urbanska K. Disturbance, hybridization and hybrid speciation. In: van Andel J, Bakker JP, Snaydon RW, editors. Disturbance in grasslands. Dordrecht, The Netherlands: Dr W. Junk Publishers; 1987. pp. 285–301. [Google Scholar]
- Urbanska KM, Hurka H, Landolt E, Neuffer B, Mummenhoff K. Hybridization and evolution in Cardamine (Brassicaceae) at Urnerboden, Central Switzerland: biosystematic and molecular evidence. Plant Systematics and Evolution. 1997;204:233–256. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23::4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P. On inferring hybridity from morphological intermediacy. Taxon. 1992;41:11–23. [Google Scholar]