Abstract
Background and Aims
The species-poor and little-studied genus Verhuellia has often been treated as a synonym of the genus Peperomia, downplaying its significance in the relationships and evolutionary aspects in Piperaceae and Piperales. The lack of knowledge concerning Verhuellia is largely due to its restricted distribution, poorly known collection localities, limited availability in herbaria and absence in botanical gardens and lack of material suitable for molecular phylogenetic studies until recently. Because Verhuellia has some of the most reduced flowers in Piperales, the reconstruction of floral evolution which shows strong trends towards reduction in all lineages needs to be revised.
Methods
Verhuellia is included in a molecular phylogenetic analysis of Piperales (trnT-trnL-trnF and trnK/matK), based on nearly 6000 aligned characters and more than 1400 potentially parsimony-informative sites which were partly generated for the present study. Character states for stamen and carpel number are mapped on the combined molecular tree to reconstruct the ancestral states.
Key Results
The genus Peperomia is generally considered to have the most reduced flowers in Piperales but this study shows that this is only partially true. Verhuellia, with almost equally reduced flowers, is not part of or sister to Peperomia as expected, but is revealed as sister to all other Piperaceae in all analyses, putting character evolution in this family and in the perianthless Piperales in a different light. A robust phylogenetic analysis including all relevant taxa is presented as a framework for inferring patterns and processes of evolution in Piperales and Piperaceae.
Conclusions
Verhuellia is a further example of how a molecular phylogenetic study can elucidate the relationships of an unplaced taxon. When more material becomes available, it will be possible to investigate character evolution in Piperales more thoroughly and to answer some evolutionary questions concerning Piperaceae.
Key words: Verhuellia, Peperomia, Piper, Piperales, Piperaceae, character evolution, morphology, phylogeny, ancestral state reconstruction, stochastic character mapping
INTRODUCTION
Verhuellia, first described by Miquel (1843) in tribe Peperomieae, is a species-poor genus in Piperaceae, known from very few collections and localities on Cuba and Hispaniola (Haiti and Dominican Republic). Besides Peperomia and Verhuellia, three other genera have been included in this tribe (Acrocarpidium, Erasmia and Phyllobryon; Miquel, 1852, 1868; de Candolle, 1869). Based on synapomorphies such as the unicarpellate ovary and the two disporangiate anthers, Acrocarpidium, Erasmia and Phyllobryon have now been included in Peperomia (Samain et al., 2007). This is also substantiated by molecular results (Wanke et al., 2006a).
Miquel (1843) included three species in Verhuellia, V. brasiliensis, V. elegans and V. serpens. As a consequence of the transfer of V. brasiliensis and V. serpens to Peperomia, V. elegans has to be considered as the type of the genus. Saralegui Boza (2004) correctly, but without argumentation, designated this as the type species. In total, nine Verhuellia species names have been published of which only two are still accepted, V. elegans and V. hydrocotylifolia. All other species are now included in Peperomia or treated as synonyms of the remaining species of Verhuellia. A detailed taxonomic discussion will be published elsewhere.
Verhuellia has only been superficially studied. Only one historical literature source describes some morphological characters in more detail (Schmitz, 1872a,b) and thorough anatomical studies are lacking. Tebbs (1993) synonymized Verhuellia with Peperomia without much argumentation. In contrast, Saralegui Boza (2004) considered Verhuellia as a separate genus, although discussion is lacking.
Phylogenetic analyses using molecular data have proved useful in elucidating relationships of unplaced and misplaced taxa, putting evolutionary trends in many plant groups in a different light and often leading to new insights in generally accepted concepts (e.g. APG II, 2003). We use this approach in Piperales to clarify the relationships of Verhuellia. During the last 5 years, several studies dedicated to the phylogeny and evolution of Piperales or Piperaceae have been published (Jaramillo and Manos, 2001; Jaramillo et al., 2004; Neinhuis et al., 2005; Wanke et al., 2006a, 2007), but the relationships of Verhuellia were not discussed because it was not included. Hence, its position remains unclear and needs reinvestigation based on molecular inference and a re-evaluation of morphological characters.
As reported by Jaramillo et al. (2004), the perianthless species of Piperales are being used as a model for examining floral development and evolution, because the simple flowers facilitate ontogenetic studies and inferences about their evolution. With the exception of Manekia and Verhuellia, the floral morphology and ontogeny of all genera in this group have been studied comprehensively (Tucker, 1975, 1976, 1979, 1980, 1981, 1982a, 1982b, 1985; Liang and Tucker, 1989, 1990, 1995; Tucker et al., 1993; Lei and Liang, 1998, 1999).
Piperales are one of the most species-rich and heterogeneous clades in the magnoliids and include much variability in growth form and life history (Wanke et al., 2007) and morphological characters (inflorescence position, presence or absence of perianth, number of stigmas; e.g. Igersheim and Endress, 1998; Doyle and Endress, 2000). For interpretation of character evolution in Piperales, a robust phylogenetic analysis including all relevant taxa is needed. The flowers of Verhuellia are among the most reduced in the perianthless Piperales, although those of Peperomia have been assumed to be the most reduced (Jaramillo et al., 2004). As a consequence, the reconstruction of floral evolution, showing strong trends towards reduction in all lineages of Piperales, needs to be revised.
MATERIALS AND METHODS
Fewer than 30 herbarium collections of Verhuellia are available and most of these have been investigated for the present study (B, BM, C, G, GH, K, NY, MA, S, U, US). Moreover, apart from one recently collected specimen of V. elegans, all of these specimens are at least 60 years old. Due to their considerable age and their preservation, the DNA is likely to be severely fragmented and detailed morphological and anatomical studies are seriously hindered.
DNA isolation followed methods described in Borsch et al. (2003). Specimens used in the phylogenetic study are listed in Table 1. The trnT-trnL sequences were largely generated for the present study, whereas those for the trnL-F and trnK/matK were mostly taken from Neinhuis et al. (2005) and Wanke et al. (2007). The trnK/matK region was generally amplified in two parts with an overlap of 250–400 bp, using the primers listed in Table 2 as described by Wanke et al. (2007). Similarly, the trnT-F region was also amplified in two parts, with a minimal overlap in the 5′ trnL gene as described by Neinhuis et al. (2005) for the trnL-F region. In some species, the trnK/matK and the trnT-F regions were amplified in three parts due to long insertions of AT-rich microsatellites. After gel electrophoresis, the PCR products were purified using a gel extraction kit (Macherey-Nagel). Direct sequencing used the CEQ DTCS Quick Start Kit (Beckman Coulter) with a CEQ 8000 sequencer, following standard protocols for each kit.
Table 1.
Taxa used in the present study, including the origin of the material studied (field or collection), voucher information and the herbarium where the voucher is deposited, as well as GenBank accession numbers (including source), are given
| Family | Species | Origin | Voucher (herbarium) | GenBank accession number | ||
|---|---|---|---|---|---|---|
| trnK/matK | trnT-L | trnL-F | ||||
| Aristolochiaceae | Aristolochia albida Duch. | BG Bonn, 17419 | Neinhuis 92 (DR) | DQ5320644 | EF422813 | AY6891536 |
| Aristolochia arborea Linden | BG Bonn, 02560 | Neinhuis 93 (DR) | DQ5320444 | EF422810 | AY6891756 | |
| Aristolochia eriantha Mart. & Zucc. | BG Bonn, 12952 | Neinhuis 99 (DR) | DQ5320544 | EF422812 | AY6891636 | |
| Aristolochia pistolochia L. | France, Cassis, Calenque d'en Veau | leg. Kreft, Wanke 037 (DR) | DQ5320625 | EF422814 | DQ5320245 | |
| Aristolochia gigantea Mart. & Zucc. | BG Bonn, 02099 | Neinhuis 101 (DR) | DQ8821874 | EF422811 | AY6891656 | |
| Asarum yakusimense Masam. | BG Bonn, 14276 | Neinhuis 91 (DR) | DQ8821971 | EF422808 | AY6891506 | |
| Saruma henryi Oliver | BG Bonn, 02618 | Neinhuis 120 (DR) | DQ5320334 | AY1453407 | AY1453407 | |
| Thottea siliquosa (Lamk.) Ding Hou | India, Kerala (BG Bonn, 09037) | Neinhuis 121 (DR) | DQ5320354 | EF422809 | AY6891516 | |
| Lactoridaceae | Lactoris fernandeziana Phil. | Chile, Masatierra Island (Juan Fernandez) | Crawford & Stuessy 11950 | DQ8821954 | AY1453247 | AY1453426 |
| Piperaceae | Manekia naranjoana (C.DC.) Callejas | Costa Rica | O. Vargas s.n. (DUKE) | DQ8822394 | EF422821 | EF422821 |
| Manekia sydowii (Trel.) Arias, Callejas & Bornstein | Columbia, Antioquia | MAJ038 (DUKE) | DQ8822384 | EF422820 | EF422820 | |
| Peperomia gracillima S. Watson | BG Bonn, 06005 | Wanke 060 (DR) | DQ2127163 | EF422825 | EF422825 | |
| Peperomia graveolens Rauh & Barthlott | Ecuador, El Oro | Rauh & Barthlott 35122 (HEID) | DQ2127223 | EF422826 | EF422826 | |
| Peperomia maypurensis Kunth | BG Bonn, 11132 | Wanke 006 (DR) | DQ2127353 | EF422827 | AY6891466 | |
| Peperomia pitcairnensis C.DC. | BG Bonn, 17744 | Wanke 007 (DR) | DQ2127623 | EF422828 | AY6891456 | |
| Peperomia marmorata Hook. f. | BG Bonn, 17527 | Wanke 064 (DR) | DQ2127253 | EF422829 | EF422829 | |
| Piper angustum#1 Rudge | Miss. Bot. Gard. | Acc. 910150 | – | EF422824 | EF422824 | |
| Piper betle#2 L. | BG Cologne | Neinhuis s.n. (DR) | – | EF422823 | EF422823 | |
| Piper cf. magnificum#2 Hort. ex Gentil | BG Bonn, 05020 | Wanke 069 (DR) | DQ8822094 | |||
| Piper ornatum#1 N.E.Br. | BG Bonn, 18144 | Wanke 005 (DR) | DQ8822114 | – | ||
| Piper sp. | BG Bonn, 00854 | Borsch 3475 (BONN) | DQ8822254 | AY1453467 | AY1453467 | |
| Verhuellia elegans Miq. | Dominican Rep., Sierra de Bahoruco | Jiménez & García 3560 (GENT) coll. 24·12·2003 | EF422831 | EF422819 | EF422819 | |
| Verhuellia sp. | Haiti, Massife du Nord | Ekman 8928 (US, GH), coll. 29·05·1927 | EF422830 | EF422818 | EF422818 | |
| Zippelia begoniifolia Blume | BG Kunming, s.n. | Wanke & Neinhuis s.n. (DR) | DQ8822304 | EF422822 | EF422822 | |
| Saururaceae | Anemopsis californica (Nutt.) Hook. & Arn. | BG Bonn, 06422 | Wanke 002 (DR) | DQ8821984 | EF422817 | AY6891426 |
| Gymnotheca chinensis Decne. | BG Bonn, 17072 | Wanke 004 (DR) | DQ8821994 | EF422816 | AY6891416 | |
| Houttuynia cordata Thunb. | BG Bonn, 08120 | Borsch 3481 (BONN) | DQ2127123 | AY1453447 | AY1453447 | |
| Saururus cernuus L. | USA, Florida | Borsch & Wilde 3108 (VPI, FR) | DQ8822002 | AY1453437 | AY1453437 | |
| Saururus chinensis (Lour.) Baill. | BG Bonn, 00312 | Wanke 001 (DR) | DQ2127133 | EF422815 | AY6891406 | |
| Outgroup | ||||||
| Canellaceae | Canella winterana Gaertn. | BG Bonn, 15293 | Borsch 3466 (BONN) | DQ8822402 | AY1453487 | AY1453487 |
| Winteraceae | Drimys winteri#3 J.R. Forst. & G. Forst. | BG Bonn | Borsch 3479 (BONN) | – | EF422807 | EF422807 |
| Tasmannia lanceolata#3 (Poir.) A.C.Sm. | BG Bonn, 00769 | Borsch 3484 (BONN) | DQ8822412 | – | ||
1 Hilu et al., 2003; 2 Müller et al., 2006; 3 Wanke et al., 2006a; 4 Wanke et al. 2007; 5 Wanke et al., 2006b; 6 Neinhuis et al., 2005; 7 Borsch et al., 2003.
# Certain species have been replaced with closely related species within the two datasets (trnK/matK versus trnT-F), indicated by numbers following the names.
(#1: Piper ornatum has been replaced with P. angustum; #2: Piper cf. magnificum has been replaced with P. betle and #3: Tasmannia lanceolata has been replaced with Drimys winteri)
Table 2.
Primers used in this study for the amplification and sequencing of the newly generated data (from previous publications) and newly designed for this study
| Primer | Direction | Region | Sequence (5′–3′) | Design |
|---|---|---|---|---|
| Ver-matK-3000R | Reverse | trnK/matK | CTC TAA AAA CCC CGA ACC TAA T | This study |
| Ver-matK-1800F | Forward | trnK/matK | TTC AGT CAT TGT AGA AAT TCC | This study |
| MG1 | Reverse | trnK/matK | AAC TAG TCG GAT GGA GTA GAT | Liang and Hilu (1996) |
| MG15 | Forward | trnK/matK | ATC TGG GTT GCT AAC TCA ATG | Liang and Hilu (1996) |
| Pi-matK-730R | Reverse | trnK/matK | ATA GAA ATG GA(CT) TCG TTC AAG | Wanke et al. (2006a) |
| Pi-matK-1060F | Forward | trnK/matK | ACT T(AG)T GGT CTC AAC (CT)G | Wanke et al. (2006a) |
| Pi-matK-1480F | Forward | trnK/matK | TCG TAA ACA (CT)AA AAG TAC | Wanke et al. (2006a) |
| AR-matK-1850R | Reverse | trnK/matK | CCA GGC AAG ATA CTA AT | Wanke et al. (2007) |
| Pi-matK-1820R | Reverse | trnK/matK | ACA CTA ATT GGA AGG AGA ATG G | Wanke et al. (2007) |
| AR-matK-1200F | Forward | trnK/matK | TTC CAA AGT CAA AAG AGC G | Wanke et al. (2007) |
| Pi-matK-2800F | Forward | trnK/matK | AAT CTT TCT CAT TAT TAC AGT GG | Wanke et al. (2007) |
| AR-matK-1510R | Reverse | trnK/matK | TAG ACT CCT GAA A(AG)A GAA GTG G | Wanke et al. (2007) |
| Pe-matK-2500R | Reverse | trnK/matK | TTC GCA ATA AAT GCA AAG AGG | Wanke et al. (2007) |
| trnTF-50F | Forward | trnT-L-F | TAC AAA TGC GAT GCT CTA ACC | This study |
| trnL110R | Reverse | trnT-L-F | GAT TTG GCT CAG GAT TGC CC | Borsch et al. (2003) |
| trnTc | Forward | trnT-L-F | CGA AAT CGG TAG ACG CTA CG | Taberlet et al. (1991) |
| trnTf | Reverse | trnT-L-F | ATT TGA ACT GGT GAC ACG AG | Taberlet et al. (1991) |
| Ver-trnTL-500R | Reverse | trnT-L-F | CGA ATG AAA CCA TAG GTA T | This study |
| Ver-trnTL-480F | Forward | trnT-L-F | GGT TGC AAT TCA AAT AAT AAT | This study |
Analyses were based on manually aligned sequence data, guided by analysis of microstructural changes, as performed by Borsch et al. (2003) or Wanke et al. (2007). An indel matrix was prepared using the ‘simple indel coding’ (SIC) approach (Simmons and Ochoterena, 2000) as implemented in SeqState (Müller, 2005). The alignment and the indel matrix are available from TreeBASE (www.treebase.org).
Phylogenetic hypotheses were generated using maximum parsimony and Bayesian inferences (as a basis for the ancestral state reconstruction). Phylogenetic reconstructions using heuristic searches under maximum parsimony (MP) were performed using PAUP* 4·0b10 (Swofford, 2002) via a ratchet approach (Nixon, 1999) implemented in PRAP (Müller, 2004) for easy handling. The following ratchet settings were employed: ten random addition cycles of 500 iterations each with a 25 % of upweighting of the characters in the iterations. Evaluation of support of the MP tree was performed using the bootstrap approach (Felsenstein, 1985), conducting 1000 replicates and random addition searches with ten iterations per cycle, and with decay values using PRAP in combination with PAUP* and the same options in effect as for the ratchet. All partitions of the datasets were analysed separately and in combination.
For Bayesian inference, the program MrBayes v3·1 (Ronquist and Huelsenbeck, 2003) was used, assuming a general time reversible model (GTR) and rate variation among sites following a gamma distribution. The model GTR + G + I was chosen as the one that best fits the data as determined using Modeltest v3·6 (Posada and Crandall, 1998) employing the interface MTgui (Nuin, 2005). Chains were sampled every ten generations and the resulting trees were written to a tree file. Calculation of the consensus tree and the posterior probability (PP) of clades was based upon the trees sampled after the chains converged (25 %). Only PPs of 0·95 and higher were considered significant (alpha = 0·05). Trees were compiled and drawn using TreeGraph (Müller and Müller, 2004).
Character states for stamen and carpel number were compiled in a dataset using Mesquite v.1·11 (Maddison and Maddison, 2006) and mapped on the combined molecular Bayesian phylogeny with SIMMAP v.1·0 Beta 2·1 (Bollback, 2006), a program implementing stochastic character mapping (Nielsen, 2002; Huelsenbeck et al., 2003). The morphological characters were set with equal priors on the bias parameter [π(0) = 0·5 and π(1) = 0·5] and without priors on the rate parameter, i.e. using branch lengths as a rate for the occurrence of morphological change among taxa (Schultz and Churchill, 1999; Huelsenbeck et al., 2003).
RESULTS
Data sets and tree statistics
Because of uncertain homology, eight mutational hotspots were excluded from the trnK/matK matrix and nine from the trnT-L-F matrix (Table 3), mostly mononucleotide repeats (plastid microsatellites). Within the trnK/matK dataset, seven hotspots were observed within the 5′ trnK intron and one in the 3′ trnK intron. For the trnT-L-F region, four hotspots were excluded from the trnT-L spacer, three from the trnL intron and two from the trnL-F spacer.
Table 3.
Location of mutational hotspots and characterization of trees
| trnT-L-F | trnK/matK | Total evidence | |||
|---|---|---|---|---|---|
| Position in alignment | Uncertain homology due to | Position in alignment | Uncertain homology due to | ||
| H1 | 300–345 | Poly T | 369–544 | Poly AT | – |
| H2 | 587–632 | Poly A | 652–660 | Poly C + poly T | – |
| H3 | 689–1116 | Poly T | 815–828 | Poly A | – |
| H4 | 1301–1315 | Poly A | 961–968 | Poly A | – |
| H5 | 1993–2055 | Poly A + poly T | 977–994 | Poly A | – |
| H6 | 2085–2095 | Poly A | 1071–1087 | Poly T | – |
| H7 | 2390–2401 | AT repeats | 1102–1164 | Variability | – |
| H8 | 2816–2827 | Variability | 3115–3170 | Variability | – |
| H9 | 2877–2993 | AT repeats | – | – | – |
| No of trees (MP) | 4 (1) | 3 (6) | 1 (1) | ||
| Length | 1582 (2377) | 2626 (2965) | 4170 (5307) | ||
| Total characters | 2892 (3372) | 3057 (3270) | 5949 (6640) | ||
| Variable characters | 932 (1411) | 1252 (1463) | 2176 (2865) | ||
| MP info. characters | 587 (853) | 895 (1011) | 1470 (1850) | ||
| No. of indels | 480 | 213 | 693 | ||
| CI | 0·782 (0·722) | 0·666 (0·661) | 0·712 (0·689) | ||
| RI | 0·868 (0·816) | 0·831 (0·822) | 0·839 (0·814) | ||
| RC | 0·678 (0·589) | 0·553 (0·544) | 0·597 (0·561) | ||
CI = consistency index; RI = retention index; RC = rescaled consistency index.
Values between brackets are calculated based on substitutions and coded length mutations (indels).
For the combined datasets (excluding the hotspots), the calculations were performed on 1470 potentially parsimony-informative characters, based on substitutions only or on 1850 potentially parsimony-informative characters including coded length mutations as additional characters.
Phylogenetic relationships of the genus Verhuellia
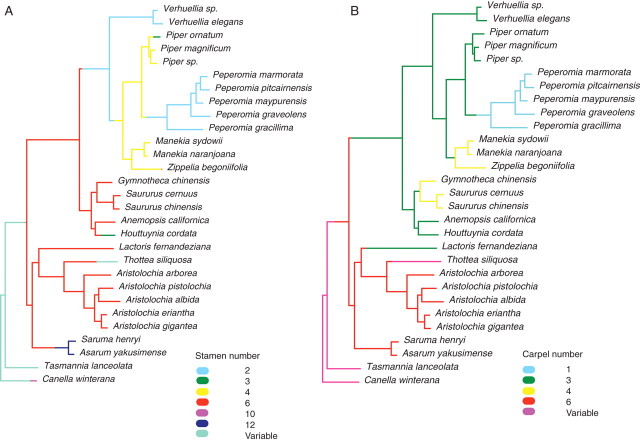
All analyses revealed nearly identical topologies in the strict consensus trees and only a few equally most parsimonious trees were found (Table 3). Incongruence among trees was generally only found within Piper (a polytomy, not shown). Verhuellia is not part of or sister to Peperomia, as expected, but appears as sister to all other Piperaceae (Zippelia + Manekia and Piper + Peperomia) (Fig. 1). This has consequences for the interpretation of character evolution in Piperaceae and Piperales, which can be seen from character mapping based on the ancestral state reconstruction approach (Fig. 2). The root node of perianthless Piperales is characterized by a hexamerous androecium and a trimerous gynoecium, whereas the root node of Piperaceae is characterized by a dimerous androecium and a trimerous gynoecium.
Fig. 1.
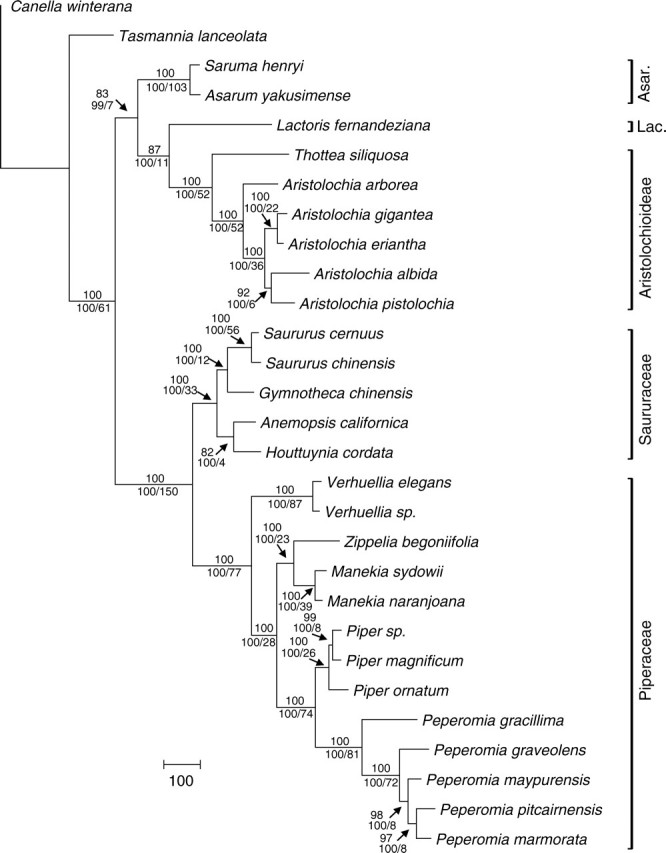
Phylogram of the single most-parsimonious tree obtained with the combined dataset. Independent analyses of the different regions revealed virtually identical topologies to the one obtained from the combined analyses. Support values differ only marginally and are given based on the total evidence above the branches. Below the branches, posterior probabilities (Bayesian analysis) (first), and decay values from the parsimony analysis are given, both calculated from the combined data set. Asar. = Asaroideae; Lac. = Lactoridaceae. Certain species have been replaced by closely related species within the two datasets (trnK/matK versus trnT-F) – see Table 1.
Fig. 2.
Ancestral state reconstruction, based on a Bayesian approach using the combined trnK/matK region and the trnT-L-F region in Piperales showing (A) stamen number and (B) carpel number. Colour changes between nodes indicate transitional states of the given characters (branch lengths are used as a rate for the occurrence of morphological change among taxa). Certain species have been replaced by closely related species within the two datasets (trnK/matK versus trnT-F) – see Table 1.
DISCUSSION
Position of Verhuellia in Piperales
Despite the clarification of the phylogenetic relationships of Verhuellia, living material required for detailed morphological and ontogenetic studies to elucidate character evolution is still not available. The sister group position of Verhuellia to the rest of Piperaceae is unexpected because it shows some superficial similarities to Peperomia. Both the placement near or within Peperomia and the synonymy of Verhuellia with Peperomia were based on characters apparently representing parallel evolution, e.g. the occurrence of only two stamens. However, there are no detailed observations, as thorough studies have not been performed. Authors who have regarded Verhuellia as a separate genus considered it to be closely related to Peperomia as both were placed in the tribe Peperomieae rather than Pipereae (Piper, Zippelia and several taxa now synonymized with Piper; e.g. Miquel, 1843, 1868; de Candolle, 1869). This historical placement of Verhuellia has not previously been questioned.
Morphological affinities to Peperomia and other Piperaceae
In contrast to the species-rich, pantropical genus Peperomia, the species-poor genus Verhuellia has only been the subject of limited studies due to its restricted distribution, limited availability in herbaria, absence in botanic gardens and inaccessibility of material. These genera have been considered as closely related because they are both characterized by some of the most reduced flowers in Piperaceae, superficially similar to each other. Specifically, the similarity in inflorescence morphology and the flowers with a dimerous androecium appear to be strong characters uniting Verhuellia with Peperomia. However, in a clade where all representatives are characterized by marked reduction of floral organs, convergent evolution could disguise true relationships and this could be confounded by the limited observation that has been possible.
Although Tebbs (1993) synonymized Verhuellia with Peperomia on the basis of ‘similar habit, bracts and fruits’, the two genera show a strikingly different habit and differences in floral morphology. The stamens in Verhuellia are tetrasporangiate and show latrorse dehiscence whereas those in Peperomia are disporangiate and show extrorse dehiscence. However, the most conspicuous difference is the number of stigmas (three or four in Verhuellia vs. one in Peperomia, sometimes two-lobed; Dahlstedt, 1900; Yuncker, 1933; Skottsberg, 1947; Sastrapradja, 1968; Remizowa et al., 2005). All these characters provide support for Verhuellia as a distinct genus in Piperaceae, as reported by Saralegui Boza (2004) in the most recent publication in which the genus is mentioned, although the discrepancy with Tebbs (1993) is not discussed.
Reappraising evolution of floral characters
All lineages of Piperales show strong trends towards reduction of floral organs, complicating the use of morphological data in reconstruction of their evolution. This has also been the main reason for different relationships of members of Piperales in classification systems of angiosperms prior to molecular approaches. Clades with similar patterns of reduction of flower organs have also been placed near to or within Piperales, thus providing evidence for parallel evolution of certain traits. A prominent example is the placement of Chloranthaceae within Piperales, mainly based on the reduced flowers (Cronquist, 1988). Similarly, floral details in Saururaceae superficially resemble those of Acoraceae (Igersheim et al., 2001). A trimerous perianth and adaxial prophylls occur in Piperales, monocots and Nymphaeales, suggesting a close relationship and giving rise to the so-called paleoherb hypothesis (Taylor and Hickey, 1990, 1992).
Generally, Peperomia is considered to have the most reduced flowers in Piperales (Jaramillo et al., 2004), but this study shows that this is only partially true. As a consequence of the position of the almost equally reduced genus Verhuellia as sister to all other Piperaceae, character evolution in this family and in the perianthless Piperales needs new attention.
The androecium at the basal node of the perianthless Piperales is hexamerous both with (this study) and without (Jaramillo et al., 2004) inclusion of Verhuellia. However, stamen number at the basal node of Piperaceae differs between the two studies: two with (this study) and four without the inclusion of Verhuellia (Jaramillo et al., 2004).
In contrast to Jaramillo et al. (2004), gynoecial evolution in Piperaceae is not characterized by the reduction from four carpels, as in Zippelia and Manekia ( = Sarcorhachis), to three in Piper, and one in Peperomia. As depicted in Fig. 2B, the gynoecium at the root node of Piperaceae is trimerous and changes to four carpels in the Manekia–Zippelia clade. The same trend is observed in the Gymnotheca–Saururus clade in Saururaceae and could be interpreted as parallel evolution.
Once living material of Verhuellia becomes available, it will be possible to investigate character evolution in the order Piperales more thoroughly. This presents the possibility of answering some evolutionary questions relating to the family Piperaceae.
ACKNOWLEDGEMENTS
Financial support for this study came from the German Science Foundation (DFG NE 681/5-1), the Research Foundation-Flanders (FWO G.0172·07), the Special Research Fund (BOF, Ghent University) the Department of Biology, Ghent University, and the Friends of the Botanic Garden, Ghent. We thank F. Jiménez and R. García (Dominican Republic), Daniel Crawford, Tod Stuessy, Thomas Borsch, O. Vargas, Wilhelm Barthlott, the curator of the herbarium S and the Botanic Gardens of Ghent and Dresden for providing leaf material for DNA extraction.The curators of the herbaria B, BM, C, G, GH, K, NY, MA, S, U and US made their Verhuellia specimens available for study. Finally, we thank Mike Fay and two anonymous reviewers for their extensive suggestions to improve this manuscript and A. M. Muasya and Alexander Vrijdaghs for their support.
LITERATURE CITED
- APG II. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Botanical Journal of the Linnean Society. 2003;141:399–436. [Google Scholar]
- Bollback JP. SIMMAP: stochastic character mapping of discrete traits on phylogenies. BMC Bioinformatics. 2006;7:88. doi: 10.1186/1471-2105-7-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsch T, Hilu KW, Quandt D, Wilde V, Neinhuis C, Barthlott W. Noncoding plastid trnT-trnF sequences reveal a well resolved phylogeny of basal angiosperms. Journal of Evolutionary Biology. 2003;16:558–576. doi: 10.1046/j.1420-9101.2003.00577.x. [DOI] [PubMed] [Google Scholar]
- de Candolle C. Prodromus Systematis Naturalis Regni Vegetabilis. Vol. 16. Paris: Masson: 1869. pp. 235–471. Piperaceae. [Google Scholar]
- Cronquist A. The evolution and classification of flowering plants. New York, NY: New York Botanic Garden; 1988. [Google Scholar]
- Dahlstedt H. Studien über süd- und central-amerikanische Peperomien mit besonderer Berücksichtigung der brasilianischen Sippen. Kongligen Svenska Vetenskaps-Akademiens Handlingar. 1900;33:1–218. [Google Scholar]
- Doyle JA, Endress PK. Morphological phylogenetic analysis of basal angiosperms: comparison and combination with molecular data. International Journal of Plant Sciences. 2000;161:121–151. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Hilu KW, Borsch T, Müller K, Soltis DE, Soltis PS, Savolainen V, et al. Angiosperm phylogeny based on matK sequence information. American Journal of Botany. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Nielsen R, Bollback JP. Stochastic mapping of morphological characters. Systematic Biology. 2003;52:131–158. doi: 10.1080/10635150390192780. [DOI] [PubMed] [Google Scholar]
- Igersheim A, Endress PK. Gynoecium diversity and systematics of paleoherbs. Botanical Journal of the Linnean Society. 1998;127:289–370. [Google Scholar]
- Igersheim A, Buzgo M, Endress PK. Gynoecium diversity and systematics in basal monocots. Botanical Journal of the Linnean Society. 2001;136:1–65. [Google Scholar]
- Jaramillo MA, Manos PS. Phylogeny and patterns of floral diversity in the genus Piper (Piperaceae) American Journal of Botany. 2001;88:706–716. [PubMed] [Google Scholar]
- Jaramillo MA, Manos PS, Zimmer EA. Phylogenetic relationships of the perianthless Piperales: reconstructing the evolution of floral development. International Journal of Plant Sciences. 2004;165:403–416. [Google Scholar]
- Lei LG, Liang HX. Floral development of dioecious species and trends of floral evolution in Piper sensu lato. Botanical Journal of the Linnean Society. 1998;127:225–237. [Google Scholar]
- Lei LG, Liang HX. Variations in floral development in Peperomia (Piperaceae) and their taxonomic implications. Botanical Journal of the Linnean Society. 1999;131:423–431. [Google Scholar]
- Liang H, Hilu KW. Application of the matK gene sequences to grass systematics. Canadian Journal of Botany. 1996;74:125–134. [Google Scholar]
- Liang HX, Tucker SC. Floral development in Gymnotheca chinensis (Saururaceae) American Journal of Botany. 1989;76:806–819. [Google Scholar]
- Liang HX, Tucker SC. Comparative study of floral vasculature in Saururaceae. American Journal of Botany. 1990;77:607–623. [Google Scholar]
- Liang HX, Tucker SC. Floral ontogeny of Zippelia begoniaefolia and its familial affinities: Saururaceae or Piperaceae? American Journal of Botany. 1995;82:681–689. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite version: a modular system for evolutionary analysis. 2006 Version 1·11. http://mesquiteproject.org . [Google Scholar]
- Miquel FAW. Systema Piperacearum. Rotterdam: H.A. Kramers; 1843. [Google Scholar]
- Miquel FAW. Piperaceae. 7–26. In: von Martius CFP, editor. Flora Brasiliensis. Vol. 4. Munich & Leipzig; 1852. [Google Scholar]
- Miquel FAW. De Piperaceis Novae Hollandiae. Verslagen en Mededeelingen der Koninklijke Akademie van Wetenschappen. Afdeeling Natuurkunde ser. 2. 1868;2:53–64. [Google Scholar]
- Müller J, Müller K. TreeGraph: automated drawing of complex tree figures using an extensible tree description format. Molecular Ecology Notes. 2004;4:786–788. [Google Scholar]
- Müller K. PRAP – computation of Bremer support for large datasets. Molecular Phylogenetics and Evolution. 2004;31:780–782. doi: 10.1016/j.ympev.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Müller K. SeqState – primer design and sequence statistics for phylogenetic DNA data sets. Applied Bioinformatics. 2005;4:65–69. doi: 10.2165/00822942-200504010-00008. [DOI] [PubMed] [Google Scholar]
- Müller KF, Borsch T, Hilu KW. Phylogenetic utility of rapidly evolving DNA at high taxonomical levels: contrasting matK, trnT-F, and rbcL in basal angiosperms. Molecular Phylogenetics and Evolution. 2006;41:99–117. doi: 10.1016/j.ympev.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Neinhuis C, Wanke S, Hilu KW, Müller K, Borsch T. Phylogeny of Aristolochiaceae based on parsimony, likelihood, and Bayesian analyses of trnL-trnF sequences. Plant Systematics and Evolution. 2005;250:7–26. [Google Scholar]
- Nielsen R. Mapping mutations on phylogenies. Systematic Biology. 2002;51:729–739. doi: 10.1080/10635150290102393. [DOI] [PubMed] [Google Scholar]
- Nixon KC. The parsimony ratchet, a new method for rapid parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- Nuin PAS. University of Toronto; 2005. MTgui – a simple interface to ModelTest. Program distributed by the author. http://www.genedrift.org/mtgui.php . [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Remizowa M, Rudall PJ, Sokoloff D. Evolutionary transitions among flowers of perianthless Piperales: inferences from inflorescence and flower development in the anomalous species Peperomia fraseri (Piperaceae) International Journal of Plant Sciences. 2005;166:925–943. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Samain MS, Mathieu G, Vanderschaeve L, Wanke S, Neinhuis C, Goetghebeur P. Taxon. Vol. 56. 229–236; 2007. Nomenclature and typification of subdivisional names in the genus Peperomia (Piperaceae) [Google Scholar]
- Saralegui Boza H. Piperaceae. Flora de la República de Cuba. 2004;9:57–60. [Google Scholar]
- Sastrapradja SN. On the morphology of the flower in Peperomia (Piperaceae) species. Annales Bogoriensis. 1968;4:235–244. [Google Scholar]
- Schmitz F. Der morphologische Aufbau von Verhuellia Miq. Flora. 1872a;26:402–415. [Google Scholar]
- Schmitz F. Der morphologische Aufbau von Verhuellia Miq. Flora. 1872b;27:417–424. [Google Scholar]
- Schultz TR, Churchill GA. The role of subjectivity in reconstructing ancestral character states: a Bayesian approach to unknown rates, states, and transformation asymmetries. Systematic Biology. 1999;48:651–664. [Google Scholar]
- Skottsberg C. Peperomia berteroana Miq. and P. tristanensis Christoph., an interesting case of disjunction. Acta Horti Gothoburgensis. 1947;16:251–288. [Google Scholar]
- Simmons MP, Ochoterena H. Gaps as characters in sequence based phylogenetic analyses. Systematic Biology. 2000;49:369–381. [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods) Sunderland, MA: Sinauer; 2002. version 4·0b10. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding chloroplast regions. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Hickey LJ. An Aptian plant with attached leaves and flowers: implications for angiosperm origin. Science. 1990;247:702–704. doi: 10.1126/science.247.4943.702. [DOI] [PubMed] [Google Scholar]
- Taylor DW, Hickey LJ. Phylogenetic evidence for the herbaceous origin of angiosperms. Plant Systemetics and Evolution. 1992;180:137–156. [Google Scholar]
- Tebbs MC. Piperaceae. In: Kubitzki K, Rohwer JG, Bittrich V, editors. The families and genera of vascular plants. Vol. 2. Berlin: Springer-Verlag; 1993. pp. 516–520. [Google Scholar]
- Tucker SC. Floral development in Saururus cernuus (Saururaceae). 1. Floral initiation and stamen development. American Journal of Botany. 1975;62:993–1007. [Google Scholar]
- Tucker SC. Ontogeny of the inflorescence of Saururus cernuus (Saururaceae). 2. Carpel initiation and floral vasculature. American Journal of Botany. 1976;63:289–301. [Google Scholar]
- Tucker SC. Ontogeny of the inflorescence of Saururus cernuus (Saururaceae) American Journal of Botany. 1979;66:227–236. [Google Scholar]
- Tucker SC. Inflorescence and flower development in the Piperaceae. I. Peperomia. American Journal of Botany. 1980;67:686–702. [Google Scholar]
- Tucker SC. Inflorescence and floral development in Houttuynia cordata (Saururaceae) American Journal of Botany. 1981;68:1017–1032. [Google Scholar]
- Tucker SC. Inflorescence and flower development in the Piperaceae. II. Inflorescence development of. Piper. American Journal of Botany. 1982a;69:743–752. [Google Scholar]
- Tucker SC. Inflorescence and flower development in the Piperaceae. III. Floral ontogeny of Piper. American Journal of Botany. 1982b;69:1389–1401. [Google Scholar]
- Tucker SC. Initiation and development of inflorescence and flower in Anemopsis californica (Saururaceae) American Journal of Botany. 1985;72:20–31. [Google Scholar]
- Tucker SC, Douglas AW, Liang HX. Utility of ontogenetic and conventional characters in determining phylogenetic relationships of Saururaceae and Piperaceae (Piperales) Systematic Botany. 1993;18:614–641. [Google Scholar]
- Wanke S, Samain MS, Vanderschaeve L, Mathieu G, Goetghebeur P, Neinhuis C. Phylogeny of the genus Peperomia (Piperaceae) inferred from the trnK/matK region (cpDNA) Plant Biology. 2006a;8:93–102. doi: 10.1055/s-2005-873060. [DOI] [PubMed] [Google Scholar]
- Wanke S, Gonzalez F, Neinhuis C. Systematics of pipevines – combining morphological and fast-evolving molecular characters to investigate the relationships within subfamily Aristolochioideae (Aristolochiaceae) International Journal of Plant Sciences. 2006b;167:1215–1227. [Google Scholar]
- Wanke S, Jaramillo MA, Borsch T, Samain MS, Quandt D, Neinhuis C. Evolution of the Piperales – matK and trnK intron sequence data reveal lineage specific resolution contrast. Molecular Phylogenetics and Evolution. 2007;42:477–497. doi: 10.1016/j.ympev.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Yuncker TG. Revision of the Hawaiian species of Peperomia. Bernice P. Bishop Museum Bulletin. 1933;112:3–131. [Google Scholar]