Abstract
Background and Aims
Potassium transporters belonging to the KT/KUP/HAK family are important for various aspects of plant life including mineral nutrition and the regulation of development. Genes encoding these transporters are present in the genomes of all plants, but have not been found in the genomes of Protista or Animalia. The aim of this Botanical Briefing is to analyse the function of KT/KUP/HAK transporters from evolutionary, molecular and physiological perspectives.
Scope
This Briefing covers the phylogeny and evolution of KT/KUP/HAK transporters, the role of transporters in plant mineral nutrition and potassium homeostasis, and the role of KT/KUP/HAK transporters in plant development.
Key words: Potassium transporters, mineral nutrition, K+ transport, plant development, homeostasis, salt stress, root-hairs, cell expansion
INTRODUCTION
Potassium is an essential macronutrient, which is important for various aspects of plant life. In contrast to other metals, the concentration of potassium in living cells is very high and in plants, particularly, may reach up to 8 % of the dry weight (Evans and Sorger, 1966). The high abundance of this cation in the cell implies its function in maintenance of cellular osmolarity and compensation of negative electrical charges associated with organic molecules. As a major cellular solute, K+ is important for turgor-dependent cell expansion, e.g. in plant tropisms and the regulation of stomatal aperture. At the biochemical level, the potassium-rich environment is essential for activity of various cytosolic enzymes (Evans and Sorger, 1966).
Although potassium is one of the most abundant elements in the Earth's crust, the bulk of it is not readily available to plants because they acquire this mineral from the soil solution only in ionic form. Owing to the fast uptake of the nutrient by plants, the actual concentrations of K+ at the surface of a root are even lower than in bulk soil solution and often fall down to the μm range (for a recent review, see Ashley et al., 2006). In order to maintain an adequate potassium status under conditions of potassium deficiency, plants have developed a sophisticated mechanism of potassium acquisition, which enables them to transport the cation against >1000-fold concentration gradients.
In classical experiments on potassium acquisition in barley, Epstein demonstrated for the first time that potassium uptake in plants is mediated by at least two systems, characterized by low and high affinity for K+ (Epstein et al., 1961). Because K+ concentrations in soil are quite variable (Ashley et al., 2006), the existence of multiple K+ uptake systems in roots is vital for the efficient mineral nutrition in a dynamic environment.
Channels and carrier-type transporters, which in the recent Transporter Classification (TC) System (Busch, 2002) are attributed to the Class 1 (channels/porins) and Class 2 (electrochemical potential-driven transporters), respectively, constitute two major pathways for K+ acquisition in plants. Porters (TC: 2·A) are the largest group of the Class 2 transporters and incorporate uniporters, symporters and antiporters. An important feature of antiporter- and symporter-mediated processes is that they can be energized by the proton- or sodium-motive force and therefore some Class 2 transporters are capable of transporting a substrate against steep concentration gradients. This property of the Class 2 transporters implies their role in K+ acquisition at low external [K+]. Indeed it has been demonstrated that K+ uptake in Arabidopsis thaliana roots must be energized if external [K+] are <1 mm (Maathuis and Sanders, 1993). The required extra energy for K+ uptake can be provided either through coupled transport mediated by a carrier-type transporter or through a hypothetical K+-motive ATPase (Kochian and Lucas, 1988). The existence of K+ ATPase in the plant plasma membrane has not been confirmed yet and therefore carrier-type (Class 2) transporters have been suggested to facilitate K+ uptake in the high-affinity range of concentrations. Low-affinity K+ transport is conventionally attributed to channels (Maathuis and Sanders, 1997). Recent findings suggest, however, that channels also contribute to high-affinity K+ uptake. Disruption of AKT1 potassium channels in the akt1-1 Arabidopsis mutant, for instance, is associated with reduced ability of plants to grow in low (10 µm) external [K+] (Hirsch et al., 1998). This potassium-dependent growth defect was also accompanied by reduced permeability of the plasma membrane to K+. Effects of the akt1-1 mutation on plant growth, however, were observed only in the presence of NH4+ in the nutrient medium (Hirsch et al., 1998). The conditional redundancy of the AKT1 channel strongly indicates that its function in high-affinity K+ uptake can be complemented by a different system, which is sensitive to NH4+. It has also been demonstrated that the K+ transport capacity of this second system is stimulated by increased concentrations of H+ and Na+ in the medium (Spalding et al., 1999). Because of this, it is quite likely that this system comprises the carrier-type (Class 2) transporters, in which the energization of K+ uptake is achieved through coupling with H+ or Na+ transport.
In plants, there are four multi-gene families of Class 2 transporters (Mäser et al., 2001) which may contribute to K+ transport: Trk/HKT (TC: 2·A·38), KEA (K+/H+ antiporters, TC: 2·A·37), CHX (cation/H+ antiporters, TC: 2·A·37) and KT/KUP/HAK (TC: 2·A·72). The HKT1 transporter from wheat (Triticum aestivum), belonging to the Trk/HKT family, has been shown to mediate K+/Na+ co-transport in heterologous expression systems (Rubio et al., 1995). In arabidopsis, however, the HKT1 orthologue does not contribute to potassium nutrition and is solely involved in the long-distance transport of sodium (Berthomieu et al., 2003). HKT transporters from barley (Hordeum vulgare) and rice (Oryza sativa) have also been shown to function just as sodium uniporters (Garciadeblas et al., 2003; Haro et al., 2005). The CHX gene family in arabidopsis consists of 28 members, which are homologous to mammalian and bacterial Cation/H+ eXchangers. Most of these transporters facilitate Na+ transport, but one member of the family, AtCHX17, is involved in K+ acquisition and homeostasis in arabidopsis (Cellier et al., 2004). KEA transporters have been identified in the arabidopsis genome through their homology to bacterial K+/H+ transporters. So far, there is no physiological evidence that KEA transporters play a role in potassium transport in plants, but activation of KEA5 by potassium deficiency (Shin and Schachtman, 2004) may imply its involvement in K+ homeostasis.
Plant transporters homologous to bacterial kup (K+ uptake) were first identified and cloned from arabidopsis and barley (Quintero and Blatt, 1997; Santa-Maria et al., 1997). Different research groups have used different acronyms for kup orthologues in plants (Quintero and Blatt, 1997; Santa-Maria et al., 1997; Fu and Luan, 1998; Kim et al., 1998) and these transporters are commonly referred as the KT/KUP/HAK gene family. This review is focused primarily on this family of potassium transporters because of their importance for a variety of essential physiological processes in plants including nutrient acquisition and regulation of plant development.
PHYLOGENY AND EVOLUTION OF KT/KUP/HAK TRANSPORTERS
Bacterial Kup transporters
The Kup (TrkD) potassium uptake system in bacteria was identified by Epstein and Kim (1971) through mutagenesis of K+-dependent (Kdp) Escherichia coli strains carrying the kdpABC5 mutation. Introduction of the trkD1 mutation into a strain with mutated trkA and kdp K+ transport systems further exacerbated growth in low [K+] media (Epstein and Kim, 1971). Half-maximal growth rate was observed at 16 mm K+ in the strain TK401 (kdpABC5 trkA401 trkD1), in which all three K+ uptake systems, Kdp, TrkA and TrkD, were mutated as compared with 2·3 mm in the strain TK133 (kdpABC5 trkA133) with functional TrkD (Rhoads et al., 1976). An ability to grow in low [K+] was only moderately affected by the trkD1 mutation in a kdpABC5 background (Rhoads et al., 1976). The latter fact suggests that the Kup (TrkD) transporters may be functionally redundant in E. coli at standard experimental conditions and their role in K+ uptake is masked by the more active TrkA system. K+ uptake through the Kup (TrkD) was approx. 2-fold stimulated when pH was reduced from 7·0 to 5·6, while the TrkA system was inhibited under these conditions by 50 % (Rhoads et al., 1976). In agreement with these data it was subsequently demonstrated that the bacterial Kup transporter is crucial for K+ uptake by cells under stress conditions such as hyper-osmolarity at low pH (Trchounian and Kobayashi, 1999). Stimulation of K+ uptake by low external pH suggests that this transporter may function as a K+/H+ symporter.
Occurrence of KT/KUP/HAK transporter genes in prokaryotic and eukaryotic genomes
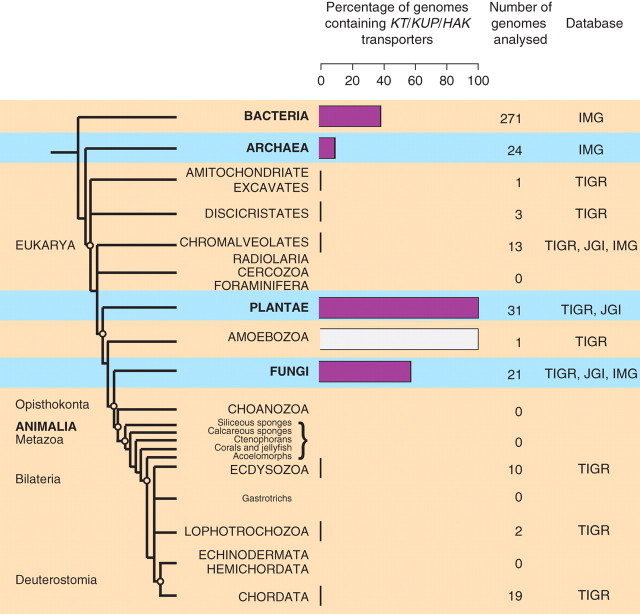
Although KT/KUP/HAK transporters were first identified in bacterial cells, their genes are not ubiquitously present in prokaryotic genomes. As based on the analysis of the Integrated Microbial Genomes (IMG) database (http://img.jgi.doe.gov) only 38 % of species in Bacteria contain these transporters (Fig. 1). This percentage is even lower in Archaea (IMG, http://img.jgi.doe.gov), where only two species out of the 24 analysed contain the kup gene (Fig. 1). The results of genomic analysis are consistent with physiological data, indicating that Kup transporters may play a crucial role in some habitats (Trchounian and Kobayashi, 1999), but are functionally redundant in others (Epstein and Kim, 1971; Rhoads et al., 1976). Analysis of the Gene Indices database at the Institute for Genomic Research (TIGR; http://www.tigr.org/tdb/tgi/index.shtml) and the eukaryotic database at the DOE Joint Genome Institute (JGI; http://genome.jgi-psf.org/euk_home.html) shows that among eukaryotes, Kup orthologues are found in Plantae, Fungi and probably Amoebozoa. Interestingly, all plant genomes analysed contain genes encoding KT/KUP/HAK transporters, while in Fungi these genes are found only in 12 species out of 21 [plant indices containing <5000 unique sequences (Cocoa and Petunia) were excluded from the analysis]. Some ESTs homologous to the 5′ end of KT/KUP/HAK transporter genes were generated from Dictyostelium discoideum transcripts (http://www.tigr.org/tdb/tgi/protist.shtml).
Fig. 1.
Genomic distribution of KT/KUP/HAK transporters. KT/KUP/HAK transporter genes are present in all plant genomes studied so far (see text for details), but were not found in Protista and Animalia. The presence of these transporters in Amoebozoa requires further confirmation. The evolutionary tree is adapted from Pennisi (2003).
Correlation between the genomic occurrence of KT/KUP/HAK transporters and the predominant mode of nutrition in a taxonomic group
As shown in Fig. 1, KT/KUP/HAK transporter genes are found primarily in organisms that acquire nutrients through absorption. The concentration of nutrients like K+ is often very low in surrounding media and therefore an absorptive mode of nutrition requires a more sophisticated system of membrane transporters as compared with ingestion. Organisms obtaining nutrients through ingestion forage on potassium-rich organic matter and probably do not require KT/KUP/HAK transporters for their potassium homeostasis. Apart from D. discoideum, which in the feeding stage obtains nutrients through ingestion (phagocytosis), no KT/KUP/HAK transporters have been found in the genomes of such organisms.
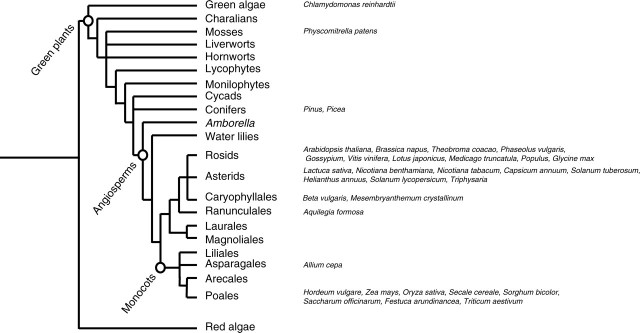
In plants, KT/KUP/HAK transporter genes have been found in evolutionarily diverse organisms ranging from green algae to angiosperms (Fig. 2). The ubiquitous presence of these genes in plants implies that they play an important role in nutrient acquisition and in the ability of plants to survive in potassium-poor environments. The evolution of such an ability was a prerequisite for the colonization of land by plants, because the concentration of potassium in soil may be up to 100-fold lower than in ocean water (Adams, 1971; Garciadeblas et al., 2002). The presence of KT/KUP/HAK transporter genes in ancestral plant genomes was probably crucial for development of terrestrial plants.
Fig. 2.
KT/KUP/HAK transporter genes in plant genomes. Data were obtained through the mining of TIGR and JGI genomic databases. The evolutionary tree is adapted from Pennisi (2003).
The Fungi is another major Kingdom in the Eukaryota, in which KT/KUP/HAK transporter genes are found. Although fungi, as well as plants, are sessile and obtain nutrients by absorption, they are chemoheterotrophs and feed on either living organisms or dead organic matter. These sources are rich in minerals and adequate potassium supply in some fungi can probably be achieved without KT/KUP/HAK transporters. This may explain why only 57 % of fungal genomes analysed contain these genes.
ROLE OF KT/KUP/HAK TRANSPORTERS IN MINERAL NUTRITION AND POTASSIUM HOMEOSTASIS IN PLANTS AND FUNGI
HAK1 transporter from Schwanniomyces occidentalis is crucial for high affinity potassium uptake
Although KT/KUP/HAK transporter genes are not ubiquitous in fungi, they are crucial for fungal nutrition in some habitats. It is not surprising therefore that the first eukaryotic KT/KUP/HAK transporter gene, HAK1 (High Affinity K+ transporter), was cloned from Schwanniomyces occidentalis, an ascomycete yeast which is able to grow in a nutrient-poor environment (Banuelos et al., 1995). Particularly remarkable is the ability of S. occidentalis to accumulate K+ from solutions in which the concentration of this nutrient is as low as 0·03 µm. If the cytosol contains around 100 mm K+, this organism is able to transport the cation against a >3000 000-fold concentration gradient! The extraordinarily high K+ concentrative capacity of S. occidentalis is largely due to the activity of the HAK1 potassium transporter (Banuelos et al., 1995). This remarkable finding provided the first experimental evidence that KT/KUP/HAK transporters might constitute a major high-affinity potassium acquisition system in eukaryotes.
Phylogeny of plant KT/KUP/HAK transporters
In plants the transporters are present as multi-gene families and in arabidopsis, for instance, there are 13 KT/KUP/HAK genes, while the rice genome contains at least 27 members of the family. As proposed recently (Rubio et al., 2000; Banuelos et al., 2002) all KT/KUP/HAK transporters can be grouped into four distinct clusters. This idea is confirmed by the phylogenetic analysis of the KT/KUP/HAK transporter genes for which full-length sequences are available. The most studied members of the KT/KUP/HAK family belong to the two largest groups, which were named Clusters I and II (Banuelos et al., 2002). Current analysis indicates that all plants have transporters homologous to members of Cluster I or Cluster II. To date, genes belonging to Cluster III are found only in arabidopsis and rice. The smallest cluster is number IV, which comprises only four rice genes.
The role of Cluster I transporters in high affinity K+ uptake
All transporters of Cluster I characterized so far display high affinity for the substrate and therefore they are likely to play a key role in potassium acquisition, particularly when K+ availability is low (Banuelos et al., 2002; Rodriguez-Navarro and Rubio, 2006). The HvHAK1 transporter from barley is likely to represent Epstein's high-affinity uptake system because its Km (27 µm; Santa-Maria et al., 1997) is remarkably close to the Km of System I (21 µm; Epstein et al., 1961). In agreement with the proposed function of HvHAK1 as the main high-affinity potassium uptake system in barley roots, its expression is induced by potassium starvation (Santa-Maria et al., 1997). Similarly, expression of HvHAK1 orthologues in tomato (Solanum lycopersicum syn. Lycopersicon esculentum; LeHAK5) and arabidopsis (AtHAK5) is activated by low external [K+] (Wang et al., 2002; Ahn et al., 2004; Hampton et al., 2004; Gierth et al., 2005). Genetic mapping of natural variations in [K+] in arabidopsis also indicated that AtHAK5 is associated with potassium accumulation (Harada and Leigh, 2006). In line with its function in potassium acquisition, AtHAK5 is expressed in the epidermis of main and lateral roots (Gierth et al., 2005). The role of AtHAK5 in K+ uptake was further verified through analysis of 86Rb+ fluxes in athak5 mutant plants (Gierth et al., 2005).
How do plants sense potassium deficiency?
Induction of AtHAK5 expression by K+ deficiency raises an important question: how is this response triggered? Do plants sense external K+ or are conditions of mineral deficiency recognized through internal signalling associated with low plant potassium status?
There are several lines of evidence indicating that internal signals regulate AtHAK5 expression (Gierth et al., 2005) and K+ uptake in general (Siddiqi and Glass, 1987). This idea is also supported by the finding that expression of AtHAK5 is enhanced in mutants in which the K+ channel AtAKT1 is disrupted (Hampton, 2005). Disruption of AtAKT1 obviously affects potassium supply (Hirsch et al., 1998) and alterations in plant potassium status may trigger AtHAK5 expression. The signal transduction cascade that regulates responses to low [K+] has yet to be identified, but there is strong evidence that jasmonate, ethylene and reactive oxygen species pathways may be involved (for recent reviews, see Amtmann et al., 2005; Ashley et al., 2006).
Some studies suggest, however, that the sensing of external [K+] may also be involved in the regulation of potassium homeostasis (Maathuis and Sanders, 1997) and K+ transporters are prime candidates for this function. Gating of the GORK channel, for instance, is regulated by external [K+] and therefore this potassium channel may serve as a sensor of the cation (Ivashikina et al., 2001). The AtKUP4/TRH1 potassium transporter may also be involved in the sensing of external [K+] and regulation of potassium-dependent root development (Vicente-Agullo et al., 2004; Ashley et al., 2006).
K+/H+ co-transport energizes high-affinity K+ uptake
In plant cells, the negative electrical potential across the plasma membrane is generated primarily through activity of the H+-ATPase, which transports H+ from the cell to the external medium. This potential constitutes a component of the driving force for the uptake of cations. At high- and mid-ranges of external concentrations, K+ transport can be driven entirely by transmembrane electrical potential, but K+ acquisition from K+-deprived soil requires an additional source of energy (Banuelos et al., 1995). The proton-electrochemical gradient, which normally is inwardly directed, may energize potassium uptake, and indeed it is utilized by some KT/KUP/HAK transporters, which co-transport K+ and H+ (Rodriguez-Navarro, 2000).
The role of Cluster II transporters in ionic homeostasis
The physiological functions of Cluster II transporters are probably quite diverse and their role in potassium nutrition is not well defined. Most of these transporters are likely to facilitate the low-affinity K+ transport complementing potassium channels (Senn et al., 2001; Garciadeblas et al., 2002). Some transporters of Cluster II are probably localized to the tonoplast similarly to OsHAK10 (Garciadeblas et al., 2002). The putative function of the tonoplast KT/KUP/HAK transporters is to facilitate K+ efflux from the vacuole. The efflux process is particularly important for the maintenance of K+ homeostasis in K+-deprived plants. Under such conditions, the K+ content of vacuoles is quite low and transport of the cation from the organelle to the cytosol must be energized (Walker et al., 1996; Garciadeblas et al., 2002). In contrast to K+ channels, the KT/KUP/HAK transporters can facilitate such transport by utilizing the energy of the trans-tonoplast H+ gradient. It has been shown that under conditions of K+ deprivation, export of K+ from the vacuole can be mediated by a K+/H+ symporter with a 1 : 1 stoichiometry (Walker et al., 1996).
In arabidopsis, the Cluster II genes AtHAK6 and AtHAK2, alongside AtHAK11 of Cluster III, may be involved in plant responses to salinity because their expression is affected by increased salt concentrations (Maathuis, 2006). AtHAK6 and AtHAK11, however, are up-regulated while the amount of the AtKUP2 transcript is reduced under the stress conditions. Owing to physical and chemical similarities between the two alkali cations, sodium interference under conditions of salt stress affects K+ acquisition and the activity of potassium-dependent enzymes, including transporters of Cluster II (Banuelos et al., 1995). Higher expression of AtHAK6 and ATHAK11 may help to achieve the required rate of potassium transport and to restore Na+/K+ balance in plants at high salt concentrations. AtKUP2 is known to regulate cell size (Elumalai et al., 2002; see below) and therefore the reduced expression of this transporter could be important for developmental rather than physiological responses to salt stress.
Comparative studies of KT/HAK/KUP transporters from barley and the sea grass Cymodocea nodosa indicate a remarkable correlation between their characteristics and plant habitat. The transport activity of the HvHAK2 is extremely sensitive to the presence of Na+, whereas CnHAK1 from C. nodosa is practically unaffected by salt (Garciadeblas et al., 2002).
POTASSIUM TRANSPORTERS AND PLANT DEVELOPMENT
Regulation of the cell size
Some KT/KUP/HAK transporters have been found to play important roles in plant development. The mutation shy3-1 in the AtKUP2 gene, for instance, causes a reduction in the size of arabidopsis shoot cells (Elumalai et al., 2002). Because K+ is a major cellular solute, impairment in potassium homeostasis may weaken cell turgor and thus restrict rates of cell expansion. However, [K+] in the shy3-1 mutant is only marginally lower than in wild-type plants. Puzzlingly, this mutation had little effect on the ability of the protein to transport K+ when produced in a heterologous expression system, and the atkup2 null mutant did not display a morphological phenotype (Elumalai et al., 2002). Currently, no clear explanation of the observed effects is available, but partial dominance of shy3-1 alongside other observations suggests that the mutation may cause gain of function and/or alteration of protein properties (Elumalai et al., 2002). It remains obscure, however, whether the affected function is the K+ transport. Alternatively it has been suggested that the shy3-1 mutation may affect activities of other proteins, which normally interact with AtKUP2 and are involved in regulation of the cell size (Elumalai et al., 2002).
The role of the KT/KUP/HAK transporters in turgor-dependent growth has been demonstrated in rapidly expanding cotton fibres (Gossypium hirsutum), where expression of the GhKT1 member of the gene family correlates positively with build-up of turgor pressure (Ruan et al., 2001). In growing grapevine fruits (Vitis vinifera) expression of VvKUP1 and VvKUP2 potassium transporter genes is also strongly dependent on developmental stage. It is likely that these transporters are required for the potassium-driven cell expansion in young grape berries (Davies et al., 2006).
Root-hair development and gravitropic responses
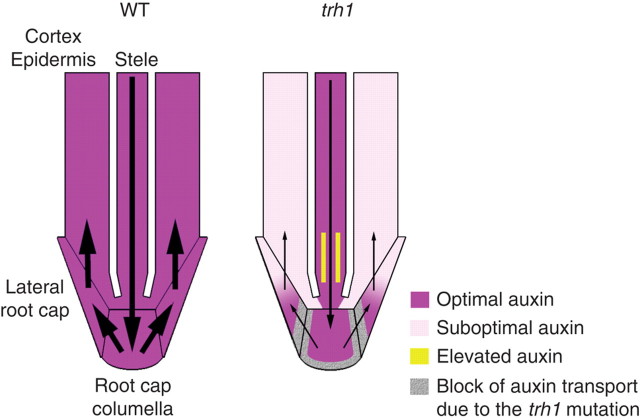
The trh1 (tiny root-hair 1) mutant, in which the AtKUP4/TRH1 gene is disrupted, was identified through phenotyping of a T-DNA-tagged arabidopsis population (Rigas et al., 2001). Root-hairs are initiated in this mutant, but they fail to elongate and appear as small bulges on the surface of epidermal cells (Rigas et al., 2001). Apart from this defect, trh1 roots display agravitropic behaviour when grown on vertical agar plates (Vicente-Agullo et al., 2004; Grabov et al., 2005). Surprisingly, none of the morphological defects observed in trh1 is rescued by increased external [K+]. Therefore, similarly to shy3-1, trh1 phenotypes are unlikely to be due to a K+ deficit per se. This idea was further supported by the observation that the highest level of expression of the TRH1 gene was observed in the root-cap and not in epidermal cells, the development of which is affected by the trh1 mutation (Vicente-Agullo et al., 2004). The fact that both root-hair elongation and gravitropic responses in trh1 are complemented by exogenous auxin points to a role for the TRH1 potassium transporter in auxin signalling and transport. Indeed, experiments with the trh1 mutant expressing an auxin-sensitive DR5–GUS construct demonstrate that disruption of the TRH1 transporter is associated with distortion of auxin profiling in the arabidopsis root (Vicente-Agullo et al., 2004). The DR5–GUS expression pattern in the trh1 root provides evidence that TRH1 is required for the facilitation of polar auxin transport in the root-cap. Figure 3 illustrates auxin distribution in wild-type and trh1 plants. According to this scheme, disruption of the TRH1 transporter partially blocks auxin flow through the root-cap, resulting in sub-optimal concentrations of the phytohormone in the cortex and epidermis. In accordance with the scheme in Fig. 3, blockage of auxin transport in the root-cap should also cause build-up of the phytohormone in the central cylinder, which indeed was observed in experiments with trh1 plants expressing the DR5–GUS construct.
Fig. 3.
Auxin fluxes in the root-tip of wild-type (WT) and trh1 plants. The trh1 mutation partially blocks auxin transport through the root-cap. This blockage causes a reduction in overall acropetal auxin transport through the stele and decreases in the auxin concentration in the cortex and epidermis. Defects in root-hair development and gravitropic behaviour in the trh1 mutant are due to sub-optimal concentrations of auxin in the epidermis. Reproduced from Vicente-Agullo et al. (2004) with permission.
Although root-hair growth is independent of external [K+], the trh1 phenotype is rescued by phosphate deficiency (Muller and Schmidt, 2004). This fact again supports the idea that the trh1 phenotype is due to a defect in signalling rather than in potassium supply for turgor-dependent growth.
It is not clear yet how the K+ transporter facilitates auxin transport. TRH1 could either transport auxin directly or create the electrochemical gradients that drive phytohormone transport across the plasma membrane. A role for TRH1 in the regulation of trans-plasma membrane H+ gradients, for instance, is quite plausible because many KT/KUP/HAK transporters are known to co-transport K+ and H+ (Rodriguez-Navarro, 2000). Alterations in H+ transport may affect auxin transport in the trh1 mutant, but this hypothesis has yet to be tested experimentally.
CONCLUDING REMARKS
The various functions of plant KT/KUP/HAK transporter genes range from mineral nutrition to the regulation of cell growth and development. Although all the transporters studied so far do transport potassium and some of them are important for potassium uptake, some morphological defects in kt/kup/hak knock-outs are not directly linked to impairment in potassium acquisition and/or homeostasis.
ACKNOWLEDGEMENTS
This work was supported by the BBSRC, the Royal Society and the Central Research Fund of the University of London. The author is grateful to John Mansfield and Alonso Rodriguez-Navarro for critical reading of the manuscript.
LITERATURE CITED
- Adams F. Soil solution. In: Carson EW, editor. The plant root and its environment. Charlottesville, VA: University Press of Virginia; 1971. pp. 441–481. [Google Scholar]
- Ahn SJ, Shin R, Schachtman DP. Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiology. 2004;134:1135–1145. doi: 10.1104/pp.103.034660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Hammond JP, Armengaud P, White PJ. Nutrient sensing and signalling in plants: potassium and phosphorus. Advances in Botanical Research. 2005;43:209–257. [Google Scholar]
- Ashley MK, Grant M, Grabov A. Plant responses to potassium deficiencies: a role for potassium transport proteins. Journal of Experimental Botany. 2006;57:425–436. doi: 10.1093/jxb/erj034. [DOI] [PubMed] [Google Scholar]
- Banuelos MA, Klein RD, Alexander-Bowman SJ, Rodriguez-Navarro A. A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO Journal. 1995;14:3021–3027. doi: 10.1002/j.1460-2075.1995.tb07304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuelos MA, Garciadeblas B, Cubero B, Rodriguez-Navarro A. Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiology. 2002;130:784–795. doi: 10.1104/pp.007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthomieu P, Conejero G, Nublat A, Brackenbury WJ, Lambert C, et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance. EMBO Journal. 2003;22:2004–2014. doi: 10.1093/emboj/cdg207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W. The Transporter Classification (TC) System, 2002. Critical Reviews in Biochemistry & Molecular Biology. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- Cellier F, Conejero G, Ricaud L, Luu DT, Lepetit M, Gosti F, Casse F. Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. The Plant Journal. 2004;39:834–846. doi: 10.1111/j.1365-313X.2004.02177.x. [DOI] [PubMed] [Google Scholar]
- Davies C, Shin R, Liu W, Thomas MR, Schachtman DP. Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. Journal of Experimental Botany. 2006;57:3209–3216. doi: 10.1093/jxb/erl091. [DOI] [PubMed] [Google Scholar]
- Elumalai RP, Nagpal P, Reed JW. A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. The Plant Cell. 2002;14:119–131. doi: 10.1105/tpc.010322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein E, Rains DV, Elzam OE. Resolution of dual mechanisms of potassium absorption by barley roots. Proceedings of the National Academy of Sciences of the USA. 1961;49:684–692. doi: 10.1073/pnas.49.5.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein W, Kim BS. Potassium transport loci in Escherichia coli K-12. Journal of Bacteriology. 1971;108:639–644. doi: 10.1128/jb.108.2.639-644.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans HJ, Sorger GJ. Role of mineral elements with emphasis on the univalent cations. Annual Review of Plant Physiology. 1966;17:47–76. [Google Scholar]
- Fu HH, Luan S. AtKUP1: a dual-affinity K+ transporter from Arabidopsis. The Plant Cell. 1998;10:63–73. doi: 10.1105/tpc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciadeblas B, Benito B, Rodriguez-Navarro A. Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa. Plant Molecular Biology. 2002;50:623–633. doi: 10.1023/a:1019951023362. [DOI] [PubMed] [Google Scholar]
- Garciadeblas B, Senn ME, Banuelos MA, Rodriguez-Navarro A. Sodium transport and HKT transporters: the rice model. The Plant Journal. 2003;34:788–801. doi: 10.1046/j.1365-313x.2003.01764.x. [DOI] [PubMed] [Google Scholar]
- Gierth M, Maser P, Schroeder JI. The potassium transporter AtHAK5 functions in K+ deprivation-induced high-affinity K+ uptake and AKT1 K+ channel contribution to K+ uptake kinetics in Arabidopsis roots. Plant Physiology. 2005;137:1105–1114. doi: 10.1104/pp.104.057216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabov A, Ashley MK, Rigas S, Hatzopoulos P, Dolan L, Vicente-Agullo F. Morphometric analysis of root shape. New Phytologist. 2005;165:641–652. doi: 10.1111/j.1469-8137.2004.01258.x. [DOI] [PubMed] [Google Scholar]
- Hampton CR. University of Birmingham; 2005. Caesium uptake and accumulation by Arabidopsis thaliana. PhD Thesis. [Google Scholar]
- Hampton CR, Bowen HC, Broadley MR, Hammond JP, Mead A, Payne KA, et al. Cesium toxicity in Arabidopsis. Plant Physiology. 2004;136:3824–3837. doi: 10.1104/pp.104.046672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Leigh RA. Genetic mapping of natural variation in potassium concentrations in shoots of Arabidopsis thaliana. Journal of Experimental Botany. 2006;57:953–960. doi: 10.1093/jxb/erj081. [DOI] [PubMed] [Google Scholar]
- Haro R, Banuelos MA, Senn ME, Barrero-Gil J, Rodriguez-Navarro A. HKT1 mediates sodium uniport in roots: pitfalls in the expression of HKT1 in yeast. Plant Physiology. 2005;139:1495–1506. doi: 10.1104/pp.105.067553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- Ivashikina N, Becker D, Ache P, Meyerhoff O, Felle HH, Hedrich R. K+ channel profile and electrical properties of Arabidopsis root hairs. FEBS Letters. 2001;508:463–469. doi: 10.1016/s0014-5793(01)03114-3. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI. AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. The Plant Cell. 1998;10:51–62. doi: 10.1105/tpc.10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian LV, Lucas WJ. Potassium transport in roots. Advances in Botanical Research. 1988;15:93–178. [Google Scholar]
- Maathuis FJM. The role of monovalent cation transporters in plant responses to salinity. Journal of Experimental Botany. 2006;57:1137–1147. doi: 10.1093/jxb/erj001. [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Sanders D. Energization of potassium uptake in Arabidopsis thaliana. Planta. 1993;191:302–307. [Google Scholar]
- Maathuis FJM, Sanders D. Regulation of K+ absorption in plant root cells by external K+: Interplay of different plasma membrane K+ transporters. Journal of Experimental Botany. 1997;48:451–458. doi: 10.1093/jxb/48.Special_Issue.451. [DOI] [PubMed] [Google Scholar]
- Maser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiology. 2001;126:1646–1667. doi: 10.1104/pp.126.4.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M, Schmidt W. Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiology. 2004;134:409–419. doi: 10.1104/pp.103.029066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. Drafting a tree. Science. 2003;300:1694. doi: 10.1126/science.300.5626.1694. [DOI] [PubMed] [Google Scholar]
- Quintero FJ, Blatt MR. A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Letters. 1997;415:206–211. doi: 10.1016/s0014-5793(97)01125-3. [DOI] [PubMed] [Google Scholar]
- Rhoads DB, Waters FB, Epstein W. Cation transport in Escherichia coli. VIII. Potassium transport mutants. Journal of General Physiology. 1976;67:325–341. doi: 10.1085/jgp.67.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Desbrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, et al. TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. The Plant Cell. 2001;13:139–151. doi: 10.1105/tpc.13.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro A. Potassium transport in fungi and plants. Biochimica et Biophysica Acta — Reviews on Biomembranes. 2000;1469:1–30. doi: 10.1016/s0304-4157(99)00013-1. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro A, Rubio F. High-affinity potassium and sodium transport systems in plants. Journal of Experimental Botany. 2006;57:1149–1160. doi: 10.1093/jxb/erj068. [DOI] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Furbank RT. The control of single-celled cotton fiber elongation by developmentally reversible gating of plasmodesmata and coordinated expression of sucrose and K+ transporters and expansin. The Plant Cell. 2001;13:47–60. doi: 10.1105/tpc.13.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-Maria GE, Rodriguez-Navarro A. Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiologia Plantarum. 2000;109:34–43. [Google Scholar]
- Santa-Maria GE, Rubio F, Dubcovsky J, Rodriguez-Navarro A. The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. The Plant Cell. 1997;9:2281–2289. doi: 10.1105/tpc.9.12.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn ME, Rubio F, Banuelos MA, Rodriguez-Navarro A. Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. Journal of Biological Chemistry. 2001;276:44563–44569. doi: 10.1074/jbc.M108129200. [DOI] [PubMed] [Google Scholar]
- Shin R, Schachtman DP. Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proceedings of the National Academy of Sciences of the USA. 2004;101:8827–8832. doi: 10.1073/pnas.0401707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi MY, Glass ADM. Regulation of K+ influx in barley – evidence for a direct control of influx by K+ concentration of root cells. Journal of Experimental Botany. 1987;38:935–947. [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD. Potassium uptake supporting plant growth in the absence of AKT1 channel activity – inhibition by ammonium and stimulation by sodium. Journal of General Physiology. 1999;113:909–918. doi: 10.1085/jgp.113.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trchounian A, Kobayashi H. Kup is the major K+ uptake system in Escherichia coli upon hyper-osmotic stress at a low pH. FEBS Letters. 1999;447:144–148. doi: 10.1016/s0014-5793(99)00288-4. [DOI] [PubMed] [Google Scholar]
- Vicente-Agullo F, Rigas S, Desbrosses G, Dolan L, Hatzopoulos P, Grabov A. Potassium carrier TRH1 is required for auxin transport in Arabidopsis roots. The Plant Journal. 2004;40:523–535. doi: 10.1111/j.1365-313X.2004.02230.x. [DOI] [PubMed] [Google Scholar]
- Walker DJ, Leigh RA, Miller AJ. Potassium homeostasis in vacuolated plant cells. Proceedings of the National Academy of Sciences of the USA. 1996;93:10510–10514. doi: 10.1073/pnas.93.19.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Garvin DF, Kochian LV. Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots: evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiology. 2002;130:1361–1370. doi: 10.1104/pp.008854. [DOI] [PMC free article] [PubMed] [Google Scholar]