Abstract
Obesity is a life-threatening factor and is often associated with dysregulation of gene expression. Here, we show that the CNOT3 subunit of the CCR4–NOT deadenylase complex is critical to metabolic regulation. Cnot3+/− mice are lean with hepatic and adipose tissues containing reduced levels of lipids, and show increased metabolic rates and enhanced glucose tolerance. Cnot3+/− mice remain lean and sensitive to insulin even on a high-fat diet. Furthermore, introduction of Cnot3 haplodeficiency in ob/ob mice ameliorated the obese phenotype. Hepatic expression of most mRNAs is not altered in Cnot3+/− vis-à-vis wild-type mice. However, the levels of specific mRNAs, such as those coding for energy metabolism-related PDK4 and IGFBP1, are increased in Cnot3+/− hepatocytes, having poly(A) tails that are longer than those seen in control cells. We provide evidence that CNOT3 is involved in recruitment of the CCR4–NOT deadenylase to the 3′ end of specific mRNAs. Finally, as CNOT3 levels in the liver and white adipose tissues decrease upon fasting, we propose that CNOT3 responds to feeding conditions to regulate deadenylation-specific mRNAs and energy metabolism.
Keywords: deadenylase, energy metabolism, insulin resistance, mRNA decay, obesity
Introduction
Posttranscriptional mechanisms are important for various biological events, and their dysregulation is linked to a variety of disorders, including cancer, diabetes, and neuronal defects. Among the posttranscriptional controls of gene expression, regulation of mRNA stability is vitally important, as it determines the availability of mRNAs for translation. Indeed, recent microarray analyses show that nearly half of the changes in gene expression in response to cellular signalling occur at the level of mRNA decay (Fan et al, 2002; Cheadle et al, 2005).
Most mRNAs have a poly(A) tail at their 3′ ends, which plays important roles in the regulation of translation and degradation of mRNAs. Once poly(A) tail shortening takes place, being catalysed by deadenylases, mRNA decay from either the 5′ or the 3′ end proceeds (Garneau et al, 2007). In addition, the 3′ untranslated region (3′UTR) of mRNAs has been implicated in the regulation of mRNA decay. RNA-binding proteins that interact with sequences at the 3′UTR, such as AU-rich element (ARE) and the microRNA-binding sites (Garneau et al, 2007; Filipowicz et al, 2008), interact with the CCR4–NOT deadenylase complex (Lykke-Andersen and Wagner, 2005; Belloc and Mendez, 2008; Fabian et al, 2009), suggesting that the proteins in the CCR4–NOT complex is important in controlling gene expression and thus various biological activities.
The CCR4–NOT complex is a large (>2 MDa) multi-subunit protein complex conserved from yeast to humans and serves as a major deadenylase (Collart and Timmers, 2004). In yeast, two components of the complex, Ccr4p and Caf1p, possess deadenylase activity (Tucker et al, 2001). The mammalian orthologues of Ccr4p are CNOT6 and CNOT6L, and those of Caf1p are CNOT7 and CNOT8 (Dupressoir et al, 2001; Yamashita et al, 2005; Morita et al, 2007; Aslam et al, 2009). Recent structural analyses of the CNOT6L complexed with nucleotides revealed a deadenylase mechanism involving a pentacovalent phosphate transition (Wang et al, 2010). In contrast to the enzymatic subunits, the function of the non-deadenylase subunits, CNOT1–3, CNOT9, and CNOT10, is elusive. Some of them are implied to be involved in the control of deadenylase activity (Tucker et al, 2002; Temme et al, 2010). In Drosophila, miRNA-dependent deadenylation is suppressed by CNOT1 depletion (Behm-Ansmant et al, 2006) and CNOT2 depletion affects the length of mRNA poly(A) tails (Temme et al, 2004). Slight poly(A) tail lengthening is seen in Not3 mutants (Tucker et al, 2002). Furthermore, Drosophila NOT3 recruits the CCR4–NOT deadenylase to its target mRNA (Chicoine et al, 2007).
In yeast, the CCR4–NOT complex plays important roles in cell growth, glucose metabolism, and DNA damage response (Collart, 2003). The mammalian CCR4–NOT complex is also suggested to be relevant to biological functions. Knockdown of the expression of the enzymatic subunit, CNOT6, CNOT6L, CNOT7, or CNOT8, reduces cell growth (Morita et al, 2007; Aslam et al, 2009; Mittal et al, 2011). Knockdown of CNOT2 induces apoptotic cell death (Ito et al, 2011). CNOT3 depletion in embryonic stem cell results in differentiation into trophectoderm lineage (Hu et al, 2009). Cnot7-knockout mice are viable, but defective in spermatogenesis, resulting in male sterility (Berthet et al, 2004; Nakamura et al, 2004). CNOT7-knockout mice also have bone-mass increases that are due to enhanced bone formation (Washio-Oikawa et al, 2007). These intriguing findings provide a glimpse into the physiological importance of the CCR4–NOT deadenylase and direct evidence for the involvement of the 3′UTR and CCR4–NOT-mediated deadenylation in these biological phenomena is to be provided.
In this study, we addressed the biological significance of CNOT3 and found that mice haplodeficient in Cnot3 are lean due to poor fat accumulation. We provide evidence that CNOT3 is involved in the regulation of the CCR4–NOT-mediated deadenylation of some specific, but not all, mRNAs that are involved in energy metabolism. We also found that the expression of the CNOT3 protein, but not other subunits of the CCR4–NOT complex, is lowered in the liver and white adipose tissues of fasted mice compared with that in the fed mice. Taken together, we propose that CNOT3 could function in sensing nutrients and alter the deadenylase activity of the CCR4–NOT complex to control the length of the poly(A) tails and eventually expression of mRNAs coding for proteins relevant to the energy metabolism.
Results
CNOT3 reduction leads to leanness and diminishes liver and adipose tissue weight
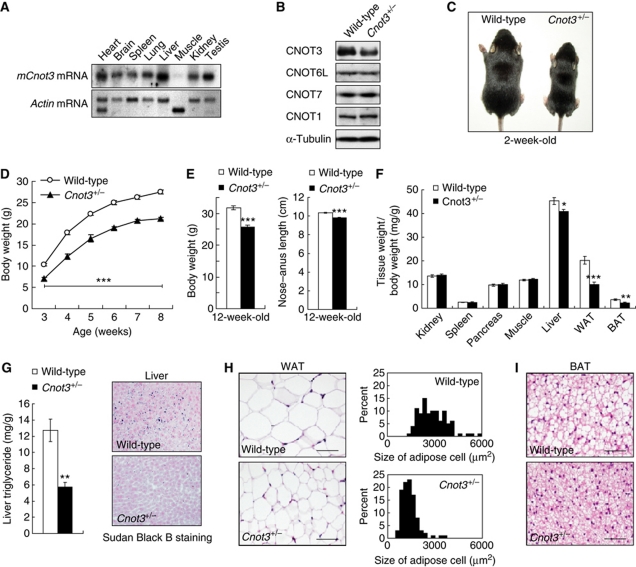
Northern blot analyses showed that Cnot3 was expressed well in the various tissues examined except that its expression was very low in muscle (Figure 1A). The data suggest that CNOT3 plays roles in various tissues. To dissect the physiological roles of CNOT3, we produced mice lacking the Cnot3 gene. The success of the procedure was confirmed by Southern blot and PCR analysis (Supplementary Figure S1). As described recently by others (Neely et al, 2010), Cnot3−/− mice did not develop past embryonic day 6.5 (Supplementary Table SI). Cnot3+/− mice were alive and fertile, and the expression levels of CNOT3 in Cnot3+/− mice were half of that in wild-type mice (Figure 1B). This decrease did not affect the expression levels of the other components, such as CNOT1, 6L, and 7. Cnot3+/− mice were smaller than their wild-type littermates. The difference in body weight between wild-type and Cnot3+/− mice was apparent in the newborn mice (Figure 1C) and remained throughout development (Figure 1D). At 12 weeks of age, Cnot3+/− mice weighed ∼20% less than wild-type mice (Figure 1E, left). The nose–anus length of Cnot3+/− mice was also reduced by about 5% at 12 weeks of age compared with wild-type mice (Figure 1E, right). Dissection of Cnot3+/− mice revealed a reduction in the size of almost all organs (Supplementary Figure S2); however, when the organ weights were normalized to body weight, the differences disappeared in most organs except for the liver and adipose tissues (Figure 1F). In Cnot3+/− mice, hepatic lipids accumulated poorly (Figure 1G) and the adipocytes in white and brown adipose tissues (WAT and BAT, respectively) were smaller than those in wild-type mice (Figure 1H and I). Histological analysis revealed that almost all tissues, including the thyroid, pituitary, adrenal gland, growth plate, and salivary gland, were normal (unpublished observation). Therefore, CNOT3 might have a specific function in the liver and adipose tissues.
Figure 1.
Leanness and reduced lipid content in Cnot3+/− mice. (A) Expression of Cnot3 mRNA in mouse tissue. A membrane filter containing mRNAs from multiple mouse tissues (Clontech) was hybridized with a probe specific for Cnot3. Actin-specific probe was used as a loading control. (B) Immunoblotting of CNOT3, CNOT6L, CNOT7, and CNOT1 protein in the Cnot3+/− livers. (C) Gross appearance of 2-week-old Cnot3+/− mice and wild-type littermates. (D) Growth curve of wild-type and Cnot3+/− mice from 3 to 8 weeks after birth. n=10 for each genotype. (E) Comparison of body weight (left) and body length (right) of 12-week-old Cnot3+/− mice and wild-type littermates. (F) The relative weight of the indicated organs. The weight of the organ was normalized to body weight. n=8–10 for each genotype. (G) Lipid levels in the liver. (Left panel) Liver triglyceride levels of 12-week-old wild-type and Cnot3+/− mice. Triglycerides in the homogenized liver were extracted with 2-propanol and measured with a Triglyceride E-Test Kit. n=5 for each genotype. (Right panel) Sudan Black B staining of liver sections from 12-week-old wild-type and Cnot3+/− mice. (H) Histological analysis of and cell size distribution in epididymal WATs of 12-week-old wild-type and Cnot3+/− mice. (I) Histological analysis of BATs of 12-week-old wild-type and Cnot3+/− mice. All values represent mean±s.e.m. *P<0.05; **P<0.01 and ***P<0.001.
Metabolic balance is disordered in Cnot3+/− mice
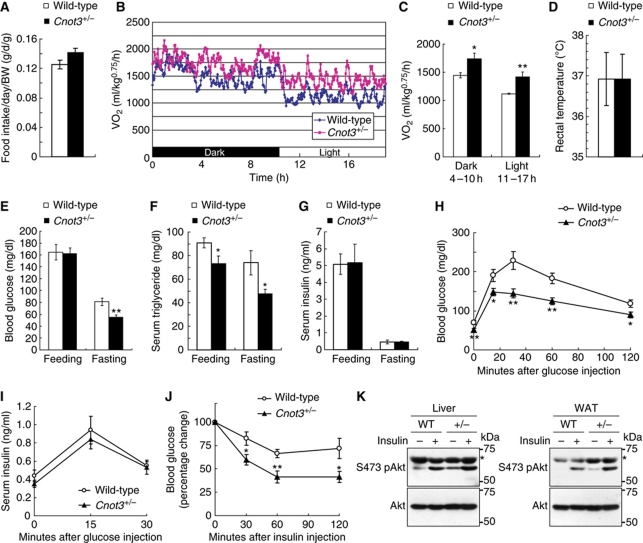
The rate of food intake per day appeared to be slightly higher in Cnot3+/− mice than in wild-type mice, but the difference was not significant (Figure 2A). Therefore, it appears that nutrients are burned more efficiently in Cnot3+/− mice than in wild-type mice. Consistent with this notion, whole-body oxygen consumption was higher in Cnot3+/− mice during dark and light periods than in wild-type mice (Figure 2B and C). In all, 24 h oxygen consumption rates were 20% higher in Cnot3+/− mice than in wild-type mice (1.611±0.088 l/kg0.75/h and 1.347±0.035 l/kg0.75/h, respectively). There was no significant difference in rectal temperature (Figure 2D). These results suggest that the leanness of Cnot3+/− mice results from an enhanced metabolic rate.
Figure 2.
Increased glucose homeostasis, insulin sensitivity, and metabolic rates in Cnot3+/− mice. (A) Average daily food intake normalized to body weight. Daily food intake per mouse was measured over 7 days. n=10 for each genotype. (B, C) Oxygen consumption (VO2) over 24 h (B) and average VO2 (C) of wild-type and Cnot3+/− mice. The data were normalized to body weight0.75. n=5 for each genotype. (D) Rectal temperatures of wild-type and Cnot3+/− mice. n=5 for each genotype. (E–G) Blood tests. Blood glucose levels (E), serum triglyceride concentrations (F), and serum insulin concentrations (G) in fed or fasted wild-type and Cnot3+/− mice. n=6–13 for each genotype. (H, I) Glucose tolerance tests. Mice were deprived of food for 16 h before the experiment. Blood glucose levels (H) and serum insulin levels (I) in wild-type and Cnot3+/− mice were measured at the indicated times following intraperitoneal injection of glucose. n=8–10 for each genotype. (J) Insulin tolerance tests. Blood glucose levels in wild-type and Cnot3+/− mice were measured at the indicated times following intraperitoneal injection of insulin. n=12 for each genotype. (K) Immunoblotting of phospho (Ser-473) and total Akt protein in the liver and WAT of wild-type and Cnot3+/− mice, with or without insulin stimulation. *Unspecific signals. All values represent mean±s.e.m. *P<0.05 and **P<0.01.
We then examined glucose and lipid metabolism in Cnot3+/− mice and found significant decreases in blood glucose under fasting conditions (Figure 2E) and serum triglyceride levels under feeding and fasting conditions (Figure 2F), in comparison with wild-type mice. By contrast, no significant differences were detected in the serum insulin levels of wild-type and Cnot3+/− mice under the same conditions (Figure 2G). A glucose tolerance test revealed that the blood glucose levels of Cnot3+/− mice remained significantly lower than those of wild-type mice after glucose administration (Figure 2H). The insulin response to glucose was virtually the same between wild-type and Cnot3+/− mice (Figure 2I), indicating that the control of insulin levels was normal in Cnot3+/− mice. These data suggest that insulin sensitivity is increased in Cnot3+/− mice. Indeed, an insulin tolerance test revealed a greater decrease in blood glucose levels in Cnot3+/− mice than in wild-type mice in response to insulin (Figure 2J). Moreover, insulin-stimulated Akt phosphorylation in the liver and WAT of Cnot3+/− mice was increased relative to wild-type mice (Figure 2K). Thus, we conclude that Cnot3+/− mice exhibit enhanced glucose tolerance and that signalling downstream of insulin receptor is enhanced.
Cnot3+/− mice are resistant to high-fat diet-induced obesity
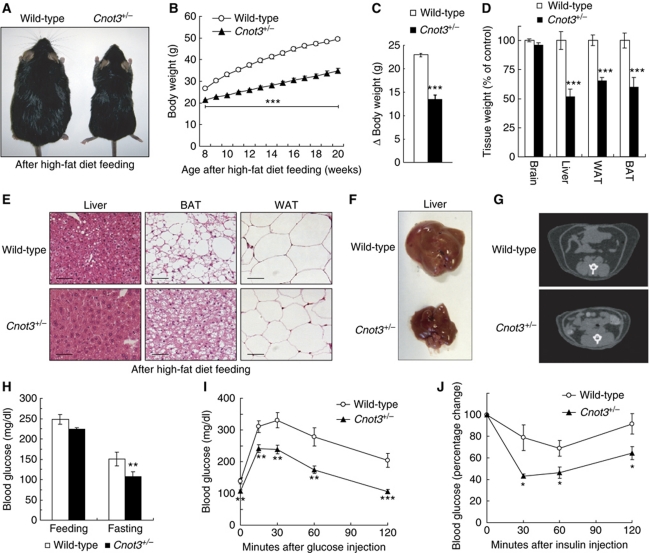
As the above data suggest that Cnot3+/− mice are protected against diet-induced obesity, we challenged Cnot3+/− mice with a high-fat (32% wt/wt fat) diet for 12 weeks. As shown in Figure 3, Cnot3+/− mice were resistant to high-fat diet (HFD)-induced obesity; they were less obese than wild-type mice (Figure 3A) and the weight accumulation of Cnot3+/− mice was significantly reduced compared with that of wild-type mice (Figure 3B) after the period of HFD feeding. The net weight gain of wild-type and Cnot3+/− mice was 23.0±0.4 and 13.5±1.0 g, respectively (Figure 3C). Notably, the weight of the liver, WAT, and BAT of the Cnot3+/− mice was less than that of wild-type mice (Figure 3D). Poor fat accumulation in the liver, white adipose tissue, and brown adipose tissue was observed in Cnot3+/− mice fed a HFD (Figure 3E). Moreover, the development of fatty livers was much less significant in Cnot3+/− mice than in wild-type mice (Figure 3F). Macroscopic and computed tomographic analyses showed that both visceral and subcutaneous fat depots were greatly decreased in Cnot3+/− mice relative to wild-type mice fed HFDs (Figure 3G). In addition, blood glucose levels in Cnot3+/− mice were lower than those in wild-type mice on a HFD (Figure 3H). Blood glucose levels remained lower in Cnot3+/− mice than in wild-type mice in both glucose tolerance tests (Figure 3I) and insulin tolerance tests (Figure 3J), suggesting that Cnot3+/− mice are insulin sensitive even on a HFD.
Figure 3.
Resistance to diet-induced obesity and related metabolic disorder in Cnot3+/− mice. (A) Gross appearance of 20-week-old Cnot3+/− mice and their wild-type littermates after HFD feeding. (B) Growth curves of wild-type and Cnot3+/− mice during HFD feeding. n=16–20 for each genotype. HFD feeding started at 8 weeks of age. (C) Changes in body weight of wild-type and Cnot3+/− mice. n=16–20 for each genotype. (D) The relative weights of the indicated organs from wild-type and Cnot3+/− mice. n=6 for each genotype. (E) Histological analysis of the liver, epididymal WATs, and BATs of wild-type and Cnot3+/− mice. (F) Liver morphology of wild-type and Cnot3+/− mice at the end of HFD feeing. (G) CT scan analysis of wild-type and Cnot3+/− mice. (H) Blood glucose levels of wild-type and Cnot3+/− mice after 16 h of HFD feeding or fasting at the end of the HFD feeding. n=9–10 for each genotype. (I, J) Glucose and insulin tolerance tests. Blood glucose levels in wild-type and Cnot3+/− mice were measured at each indicated time point following intraperitoneal injection of glucose or insulin. n=8–10 for each genotype. All values represent mean±s.e.m. *P<0.05; **P<0.01 and **P<0.01.
The obese phenotype of ob/ob mice is ameliorated by the reduction of CNOT3
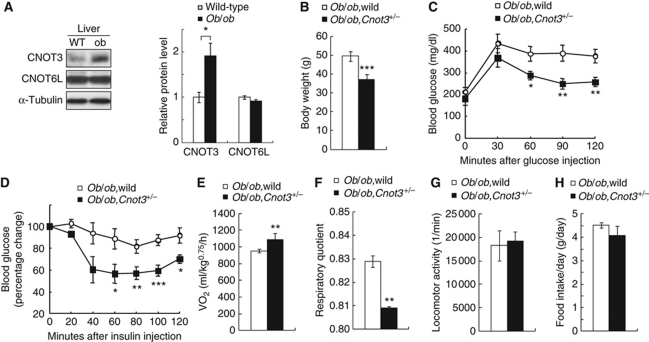
Given that suppressed CNOT3 expression had an anti-obesity effect, we addressed whether introduction of CNOT3 haplodeficiency (Cnot3+/−) into ob/ob mice could improve the obese phenotype. Note that ob/ob mice showed stronger hepatic expression of CNOT3 compared with wild-type mice (Figure 4A). As expected, ob/ob,Cnot3+/− mice were less obese than ob/ob mice: body weight, glucose tolerance, and insulin sensitivity were all ameliorated in ob/ob,Cnot3+/− mice (Figure 4B–D) compared with ob/ob mice. In ob/ob,Cnot3+/− mice, oxygen consumption rate was increased (Figure 4E) and respiratory quotient was lower (Figure 4F) compared with ob/ob mice, indicating greater utilization of fat versus carbohydrates as an energy source. Little difference was observed in locomotor activity (Figure 4G) or food intake (Figure 4H) between ob/ob and ob/ob,Cnot3+/− mice. Thus, we conclude that reduction of the CNOT3 expression in ob/ob mice ameliorates their obese phenotype.
Figure 4.
Improvement of obesity and insulin resistance in ob/ob;Cnot3+/− mice. (A) Increased expression of CNOT3 in the liver of ob/ob mice. Immunoblotting of CNOT3 and CNOT6L in wild-type and ob/ob mice (left), and quantification of the data (right). Levels were normalized to α-tubulin. n=3 for each genotype. (B) Decreased body weight in 12-week-old ob/ob,Cnot3+/− mice. n=10 for ob/ob mice. n=4 for ob/ob,Cnot3+/− mice. (C, D) Glucose (C) and insulin (D) tolerance tests. Blood glucose levels were measured at each indicated time point following intraperitoneal glucose or insulin injection. n=6 for ob/ob mice. n=4 for ob/ob,Cnot3+/− mice. (E–H) Comparison of average VO2 (E), respiratory quotient (F), locomotor activity (G), and average daily food intake (H) between ob/ob and ob/ob,Cnot3+/− mice. VO2 were normalized to body weight0.75. Respiratory quotient was calculated by carbon dioxide production/oxygen consumption. Daily food intake per mouse was measured over 7 days. All values represent mean±s.e.m. *P<0.05; **P<0.01 and ***P<0.001.
Reduction of CNOT3 correlates with up-regulation of genes involved in metabolism
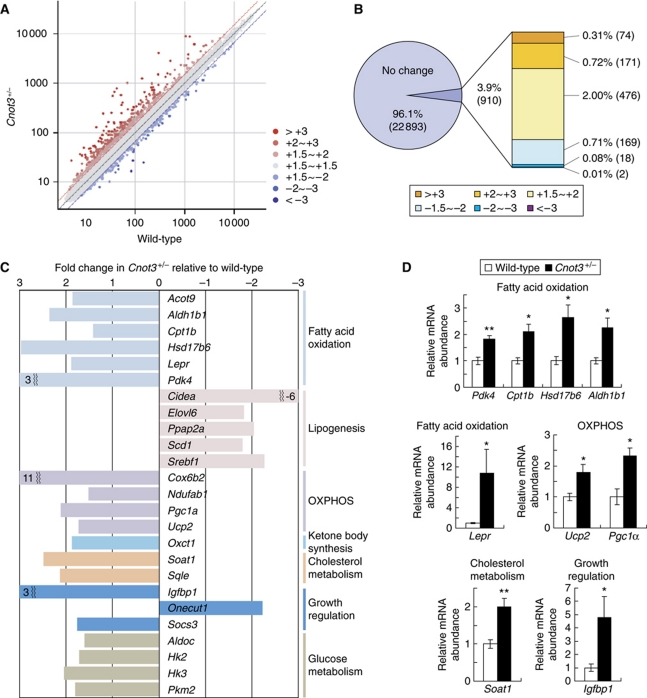
To examine the underlying mechanisms by which CNOT3 is involved in the control of metabolic balance, we compared the gene expression profiles of wild-type and Cnot3+/− mice using Affymetrix microarray technology. We chose to analyse the liver not only because the liver is the essential metabolic organ, playing a major role in glucose and lipid metabolism, but also because metabolism-related function in the liver was affected in Cnot3+/− mice as shown above. We assumed that CNOT3 could affect the abundance of mRNAs by regulating the CCR4–NOT deadenylase activity. In 12-week-old Cnot3+/− mice, most of the mRNAs (96%) showed no significant differences in abundance, and only a small fraction of mRNAs (∼1%) showed a difference of >2.0-fold compared with those in wild-type mice (Figure 5A and B). Thus, CNOT3 appeared to regulate the CCR4–NOT deadenylase activity for a fraction, but not all, of the liver mRNAs. Of ∼23 000 mRNA transcripts, ∼250 were up-regulated >2.0-fold while 20 were down-regulated >2.0-fold in the liver of Cnot3+/− mice compared with that of wild-type mice (Figure 5B). The genes whose expression was altered are listed in Supplementary Table SII.
Figure 5.
Deadenylase regulates genes involved in metabolism in the liver. (A) A scatter plot of mRNA expression values in the livers isolated from 12-week-old wild-type (x axis) and Cnot3+/− (y axis) mice. n=2 for each genotype. (B) A proportion of each group was categorized as a fraction of the fold change in Cnot3+/− livers relative to wild-type livers. Genes displaying a fold change of −1.5 to +1.5 were considered within the normal range. (C) Fold change in expression values of genes related to fatty acid oxidation, lipogenesis, oxidative phosphorylation (OXPHOS), ketone body synthesis, cholesterol metabolism, growth regulation, and glucose metabolism from the microarray data shown in (A) and listed in Supplementary Figure S2. (D) Real-time PCR analysis of selected genes involved in fatty acid oxidation, oxidative phosphorylation, cholesterol metabolism, and growth regulation in the livers of wild-type and Cnot3+/− mice. Hprt mRNA levels were used for normalization. n=3–5 for each genotype. All values represent mean±s.e.m. *P<0.05 and **P<0.01.
KEGG (Kyoto Encyclopedia of Genes and Genomes) Pathway analysis revealed that the largest proportion of altered genes was involved in metabolic processes, and genes involved in lipid metabolism were the most enriched (19 genes, P (raw)=0.04). As shown in Figure 5C, the genes up-regulated in Cnot3+/− mice include lipid catabolism-related genes such as Acot9 (acyl-CoA thioesterase 9), Aldh1b1 (aldehyde dehydrogenase 1 family, member B1), Cpt1b (carnitine palmitoyltransferase 1b), Hsd17b6 (17-β hydroxysteroid dehydrogenase 6), Lepr (leptin receptor), and Pdk4 (pyruvate dehydrogenase kinase 4). By contrast, lipogenic genes such as Elovl6 (elongation of long chain fatty acids family member 6), Scd1 (sterol O-acyltransferase 1), and Srebf1 (sterol regulatory element binding transcription factor 1) were down-regulated in Cnot3+/− mice. These results indicate that increased fat oxidation and decreased lipogenesis contribute to poor fat accumulation in Cnot3+/− mice. Glycolytic genes such as Aldoc (aldolase C) and Hk2/3 (hexokinase 2/3) were also up-regulated in Cnot3+/− mice (Figure 5C). Furthermore, energy consumption-related genes, such as Cox6b2 (cytochrome c oxidase subunit VIb polypeptide 2), Pgc1α (peroxisome proliferative activated receptor, γ, coactivator 1 α), and Ucp2 (uncoupling protein 2), as well as the metabolism and growth regulatory gene Igfbp1 (insulin-like growth factor binding protein 1) were up-regulated in Cnot3+/− mice (Figure 5C). We then validated the data by quantitative RT–PCR analysis and confirmed that the expression levels of Pdk4, Cpt1b, Hsd17b6, Aldh1b1, Pgc1α, Ucp2, Lepr, Igfbp1, and Soat1 were significantly increased in the livers of Cnot3+/− mice relative to control mice (Figure 5D). We tentatively concluded that altered expression of these genes underlies the leanness of Cnot3+/− mice.
Control of poly(A) tail length by CNOT3 involves the 3′UTR of target mRNAs
To determine whether up-regulation of the mRNAs in Cnot3+/− mice is caused by malfunction of the CCR4–NOT deadenylase, we measured the length of the poly(A) tails of mRNAs whose hepatic expression was augmented in Cnot3+/− mice. Among the up-regulated mRNAs, we focused on two mRNA species: Pdk4 and Igfbp1 mRNAs. PDK4 (pyruvate dehydrogenase kinase 4) inhibits pyruvate dehydrogenase activity, which is often correlated with enhanced utilization of fatty acids (Sugden and Holness, 2003). IGFBP1 is involved in the control of mitogenic and metabolic actions (Siddals et al, 2002), and elevated expression of IGFBP1 produces a lean phenotype (Rajkumar et al, 1995). The increase in Pdk4 and Igfbp1 mRNA levels in the livers of Cnot3+/− mice was confirmed by northern blot (Figure 6A). To demonstrate that down-regulation of CNOT3 led the inhibition of the poly(A) tail shortening of the Igfbp1 and Pdk4 mRNAs, we prepared their 3′ poly(A)-containing portions by utilizing RNase H and short stretches of nucleotide sequences that were complementary to the mRNAs' sequences (Figure 6B, left). Northern blot analyses of the enriched samples revealed that the poly(A) tails of the two mRNA species from Cnot3+/− mice were longer than those from wild-type mice (Figure 6B, right). After removal of the poly(A) tails, the lengths of the Igfbp1 or Pdk4 mRNA samples from Cnot3+/− mice were virtually the same as that from wild-type mice. As a control, we tested Gapdh mRNA, a transcript whose expression is not affected by the Cnot3 haplodeficiency, and found that there was virtually no difference in the length of poly(A) tail between wild-type and Cnot3+/− mice (Figure 6B).
Figure 6.
CNOT3 reduction affects the poly(A) tail length of mRNAs involved in lipid metabolism and growth. (A) Northern blot of Igfbp1 and Pdk4 mRNAs from the livers of wild-type and Cnot3+/− mice. (B) Comparison of the poly(A) tail lengths of the Pdk4, Igfbp1, and Gapdh mRNAs between wild-type and Cnot3+/− mice. RNAs were prepared from the livers. The experimental procedure (left) and northern blot data (right) are presented. Completely deadenylated mRNAs were prepared by RNase H treatment in the presence of oligo(dT). Estimated length of the poly(A) tails is indicated on the right of each panel. (C) Luciferase assay with reporter plasmids harbouring the 3′UTRs of Pdk4, Igfbp1, or Lpl mRNA in wild-type and Cnot3+/− hepatocytes. n=3 for each genotype. (D) Half-life of the reporter mRNA. Wild-type and Cnot3+/− hepatocytes transfected with a reporter plasmid harbouring the 3′UTR of Pdk4 mRNA were incubated with actinomycin D (5 μg/ml) for the indicated time length. The level of the reporter mRNA was determined by the qPCR method and normalized to that of Hprt mRNA, and plotted semilogarithmically. (E) Luciferase assay (top) with reporter plasmids (bottom) harbouring the various segments of Pdk4 3′UTR in wild-type and Cnot3+/− hepatocytes. ARE, GRE, and possible miRNA-binding sites were indicated. miRanda-mirSVR algorithm provided by microRNA.org was used for the possible miRNA-binding site. (F) Association of CNOT3 with the 3′UTR of Pdk4 mRNA. Cell lysates from HepG2 cells transfected with the reporter plasmids harbouring the 3′UTR of Pdk4 were immunoprecipitated with anti-CNOT3 and control-IgG antibodies. The immunoprecipitates were analysed by RT–PCR using primers specific for the Firefly luciferase mRNA. (G) Association of CNOT6L with Pdk4 mRNA in a CNOT3-dependent manner. Proteins in the hepatocyte lysates prepared from wild-type and Cnot3+/− mice were immunoprecipitated with the monoclonal anti-CNOT6L and control-IgG antibodies. RT–PCR analysis of the immunoprecipitates was performed with primers specific to Pdk4 mRNA. All values represent mean±s.e.m. *P<0.05.
To investigate the mechanism by which the CNOT3 subunit of the CCR4–NOT complex regulates Igfbp1 and Pdk4 mRNA deadenylation, the 3′UTR of the Pdk4, Igfbp1, or Lpl mRNA was inserted into a luciferase reporter plasmid (Gopfert et al, 2003). The luciferase reporter assay in wild-type and Cnot3+/− hepatocytes showed that the 3′UTRs of Pdk4 and Igfbp1, but not Lpl, were responsible for CNOT3-mediated suppression of the reporter genes (Figure 6C). The data argued that the 3′UTRs of Pdk4 and Igfbp1 mRNAs are important for determining the availability of these mRNAs for translation in a manner dependent on CNOT3. To clarify this issue, we measured the half-life of the luciferase reporter mRNA in wild-type and Cnot3+/− cells. The cells transfected with the reporter were treated with actinomycin D to inhibit de novo transcription, and subsequently the time course of the mRNA level was analysed. We found that the rate of decline in the level of Pdk4 3′UTR-containing reporter mRNA was lower in Cnot3+/− hepatocytes than in wild-type hepatocytes (Figure 6D). This difference in rates suggested that CNOT3 is involved in the 3′UTR-mediated mechanism of Pdk4 mRNA decay. Further examination of the activity of luciferase reporters linked to various segments of the Pdk4 3′UTR revealed that sequences present in segments spanning basepairs 1–460 and 921–1643 were sensitive to the amount of CNOT3 (Figure 6E). The 921–1643 segment contains two possible AREs, with the first ARE (ARE1) conserved between humans and mice. As the conserved ARE is likely to be relevant to deadenylation-dependent decay (Barreau et al, 2006; Garneau et al, 2007; Marchese et al, 2010), we examined the luciferase activity of the ARE1-depleted reporter construct (Figure 6E). However, ARE1 was not sensitive to CNOT3, leaving open the possibility of the involvement of other regulatory elements (discussed below).
RT–PCR analysis of anti-CNOT3-immunoprecipitated RNAs showed that CNOT3 physically interacted with a sequence within the 3′UTR of the Pdk4 mRNA (Figure 6F). The data suggested that CNOT3 contributes to the decay of target mRNAs by recruiting the deadenylase complex to their 3′ ends. To substantiate this notion, we assessed whether CNOT6L, a catalytic subunit of the CCR4–NOT deadenylase complex, is associated with Pdk4 mRNA in a CNOT3-dependent manner. We carried out RNA–immunoprecipitation microarray (RIP-CHIP) analysis using the anti-CNOT6L antibody and the hepatocyte lysates. Pdk4 mRNA was identified in the CNOT6L immunoprecipitates and the association of Pdk4 mRNA with CNOT6L was confirmed by immunoprecipitation–RT–qPCR analysis (Figure 6G). The association between Pdk4 mRNA and CNOT6L was decreased in Cnot3+/− hepatocytes compared with that in wild-type hepatocytes. These results suggest that CNOT3 is involved in the control of Pdk4 mRNA stability and in the association of the CNOT6L-containing CCR4–NOT deadenylase complex with the Pdk4 mRNA.
A possible involvement of CNOT3 in a nutrient-sensing mechanism
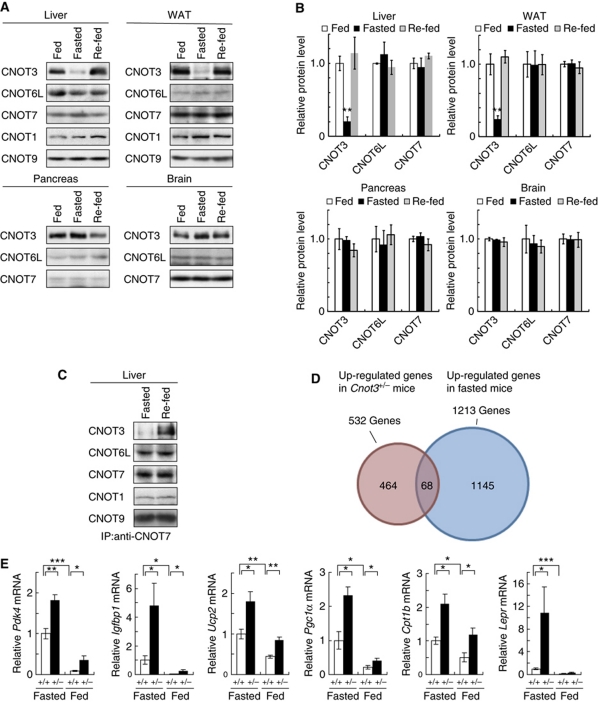
Analysis of gene expression in Cnot3+/− hepatocytes revealed that expression of Igfbp1 and Pdk4 was in inverse relationship with the CNOT3 level. As Igfbp1 and Pdk4 are up-regulated in various tissues upon starvation (Cotterill et al, 1993; Jeoung et al, 2006), we addressed if the CNOT3 expression is other way around. Examination of the CNOT3 expression under different feeding conditions revealed that the level of CNOT3 in the liver and adipose tissues was greatly decreased after 24 h fasting and returned to nearly control levels upon re-feeding (Figure 7A and B). The data imply that the CNOT3 expression is altered responding to the nutrients in these tissues. CNOT3 expression was little altered in the brain, pancreas, and testis in different feeding conditions. The decrease in CNOT3 protein levels in the fasted mice did not parallel its mRNA level (Supplementary Figure S3), suggesting that the level of CNOT3 was regulated posttranscriptionally. By contrast, the levels of other CCR4–NOT components, including CNOT6L and CNOT7 deadenylases, remained virtually the same under both conditions (Figure 7A and B). The amount of CNOT3 in anti-CNOT7 immunoprecipitates from the livers of fasted mice was less than that detected in the fed mice (Figure 7C). Therefore, the CCR4–NOT complex in the liver of fasted mice associates poorly with the CNOT3 protein, which resembles that in Cnot3+/− mice.
Figure 7.
Alternation of CNOT3 expression level by responding to nutrient status and identification of CNOT3-regulated mRNAs. (A) Reduced expression of CNOT3 in the liver and WAT of fasted mice. Western blots of CNOT3 and other CCR4–NOT subunits expression in the liver, WAT, brain, and pancreas of mice fed ad libitum, 24 h fasted, and 24 h re-fed. (B) Quantification of the data is shown in (A). Ad libitum-fed, open columns; 24 h fasted, filled columns; and 24 h re-fed mice, semifilled columns. Levels were normalized to α-tubulin. n=3 for each condition. All values are mean±s.e.m. *P<0.01 versus fed mice. (C) Levels of CNOTs in the CCR4–NOT complex. Immunoblotting of the anti-CNOT7 immunoprecipitates prepared from the liver of mice fed ad libitum, 24 h fasted and 24 h re-fed. (D) Identification of genes regulated by CNOT3. Global gene expression profiling of liver RNAs was assessed for 8-week-old mice. Note that the data shown in Figure 5 were from the analysis of 12-week-old mice. Comparisons were made between wild-type and Cnot3+/− mice (circled red) and between feeding and fasted wild-type mice (circled blue). (E) Real-time PCR analysis of selected genes in the livers of 8-week-old wild-type and Cnot3+/− mice under fasting or feeding conditions (n=3–5 for each genotype). Data represent mean values±s.e.m. *P<0.05; **P<0.01 and ***P<0.001.
Then we examined whether expression of hepatic mRNAs that are elevated in the Cnot3+/− liver is higher in fasted than fed mice by microarray analysis. In the livers of 8-week-old wild-type mice, ∼1200 mRNA transcripts were up-regulated upon fasting versus feeding. Of these mRNAs, 68 corresponded to the genes up-regulated in the livers of 8-week-old Cnot3+/− mice (Figure 7D; Supplementary Table SIII). Among the 68 mRNA species, expression of Pdk4, Pgc1a, Cpt1b, Ucp2, Lepr, Igfbp1, and Irs2 was validated by quantitative real-time RT–PCR. The data confirmed elevated expression of all of these mRNAs in the fasted mice as well as in Cnot3+/− mice (Figure 7E). Therefore, we propose that CNOT3 plays a part in controlling the expression of the metabolism-related genes by responding to nutrients. It may be noted that 36 mRNAs out of the 68 mRNA species have the ARE core sequence (AUUUA) in their 3′UTRs (Supplementary Table SIV). We also found that some consensus sequence comprised the 7-nucleotide match to the seed sequence of miRNAs. For example, miR-325 and miR-298 were identified among them (Supplementary Table SIV).
Discussion
Accumulating evidence shows that leanness is caused by multiple factors and results from either the suppression or the overexpression of various genes (Reitman, 2002). To date, various types of lean mice have been generated by disrupting genes such as those coding for regulators of gene expression, which include transcription factors C/EBPβ (Tanaka et al, 1997; Liu et al, 1999) and PGC-1α (Lin et al, 2004) and translation regulators 4E-BP1 (Tsukiyama-Kohara et al, 2001) and S6K1 (Um et al, 2004). In the present study, we provide evidence that impairment of the deadenylase, another regulator of gene expression, induces lean phenotype. We have shown that mice heterozygous for Cnot3, a gene encoding a subunit of the CCR4–NOT deadenylase complex, are lean and resistant to diet-induced hepatic steatosis and obesity. We further provide evidence that CNOT3 targets metabolism-related mRNAs, such as Pdk4 and Igfbp1 mRNAs, so that CCR4–NOT can act to deadenylate these mRNAs.
In lean mice, the intake of nutrition from the intestine is decreased and/or catabolism is accelerated compared with wild-type mice (Reitman, 2002). As the food intake of Cnot3+/− mice was virtually the same as that of wild-type mice, catabolism of nutrients was supposedly increased in Cnot3+/− mice compared with wild-type littermates. Supportingly, whole-body oxygen consumption rates were significantly increased in Cnot3+/− mice, which suggests that increased burning of energy that would otherwise be stored is occurring somewhere in the body. As increased muscle oxidation is linked to the lean phenotype of genetically modified mice (Li et al, 2000), muscle oxidation might be increased in Cnot3+/− mice. However, this is less likely because CNOT3 expression in muscle was very low in wild-type mice to begin with. Instead, we provided evidence that increased oxidation of nutrients occurred in the liver, which is one of the most important organs for energy metabolism. Indeed, Cnot3+/− mice demonstrated fewer circulating glucose and triglycerides and poorer lipid accumulation in the liver than wild-type mice. Consistently, the level of catabolism-related mRNAs is increased in the liver of Cnot3+/− mice as compared with that of wild-type mice. As Cnot3 was expressed in various organs including the liver, adipose tissues, heart, and brain, burning of energy or energy expenditure could potentially be increased in the other tissues of Cnot3+/− mice as well.
The CCR4–NOT deadenylases could act to any poly(A)-containing mRNA species as is evidenced by the in vitro deadenylating activity of the catalytic subunits (Morita et al, 2007; Temme et al, 2010). In yeast, although the Ccr4–Not complex could potentially affect the expression of most of the genes, depletion of CCR4 and CAF1 affected the expression of distinct sets of genes under different growth conditions (Azzouz et al, 2009). In Drosophila, depletion of Caf1/Not7 and Not1 was reported to inhibit general deadenylation (Temme et al, 2004; Behm-Ansmant et al, 2006), but knockdown of Caf1/Not7 and Not1 affects the levels of only a subset of mRNAs (Eulalio et al, 2009). In NIH3T3 cells, Cnot6L depletion induces poly(A) tail shortening of p27Kip mRNA but not p21Cip mRNA, indicating that CNOT6L differentially targets distinct mRNAs (Morita et al, 2007). It should be noted that proper selection of target mRNAs by the CCR4–NOT complex may involve the function of microRNA, because CCR4–NOT is implicated in microRNA-mediated mRNA decay (Behm-Ansmant et al, 2006; Wu et al, 2006; Wakiyama et al, 2007; Fabian et al, 2009). Therefore, although CCR4–NOT deadenylase can potentially shorten poly(A) tails of any mRNA species, its use and/or activity would be regulated in a manner that depends on cell types and/or growth conditions. This would explain why expression of only a subset of mRNAs is affected in the liver of Cnot3+/− mice (discussed more below).
Accelerated expression of mRNAs for energy-metabolizing reactions in the livers of Cnot3+/− mice suggests that CNOT3 is responsible for mRNA metabolism by controlling the CCR4–NOT deadenylase. There is one preceding report showing that deadenylase activity is involved in the regulation of metabolism, with the finding that a loss of the circadian deadenylase Nocturine (NOC) confers diet-induced obesity (Green et al, 2007). NOC is thought to control specific circadian pathways related to lipid uptake and/or utilization probably by targeting as yet unidentified mRNAs. Although the mRNAs with altered expression in Cnot3+/− mice compared with wild-type mice are candidate CCR4–NOT targets, expression of some of these mRNAs may be altered as a result of secondary effects. Provided that CNOT3 positively control the CCR4–NOT deadenylase activity, the mRNAs directly targeted by the CCR4–NOT deadenylase should have longer poly(A) tails in Cnot3+/− compared with wild-type mice. We showed here that the Pdk4 and Igfbp1 mRNAs clarified the criteria. Efforts to identify other direct targets of the CCR4–NOT deadenylase under the control of CNOT3 in the liver are in progress.
Yeast NOT3 depletion suppresses, albeit slightly, cellular deadenylase activity, resulting in mRNA stabilization (Tucker et al, 2002). Although reduction of CNOT3 expression in Cnot3+/− mice did not affect the levels of other components of the CCR4–NOT complex, it may have caused the conformational change of the whole complex, resulting in the alteration of the deadenylation activity. In addition, as only a subset of mRNAs was affected by Cnot3 haplodeficiency, CNOT3 may mediate recruitment of the CCR4–NOT deadenylase to the 3′ end of particular mRNAs. Indeed, RNA–immunoprecipitation–RT–qPCR analysis using the monoclonal anti-CNOT6L antibody and liver lysates from wild-type and Cnot3+/− mice suggested involvement of CNOT3 in the recruitment. There is a report that the Drosophila CCR4–NOT complex is recruited to the Bic-C mRNA through an interaction between CNOT3 and the Bic-C mRNA-binding protein (Chicoine et al, 2007). Likewise, mammalian CNOT3 may interact with RNA-binding proteins to regulate the recruitment of CCR4–NOT to target mRNAs. The 3′UTRs of Pdk4 and Igfbp1 mRNAs contain various cis-acting elements such as AREs that function as docking platforms for RNA-binding proteins, which in turn recruit the mRNA decay machinery (Barreau et al, 2006). Intriguingly, CCR4–NOT interacts with ARE-binding proteins such as TTP (tristetraprolin) (Lykke-Andersen and Wagner, 2005), suggesting that some ARE-binding proteins participate in the deadenylation of Pdk4 and Igfbp1 mRNAs. However, ARE1 in the 3′UTR of Pdk4 mRNA (Figure 6E) is not responsible by itself for the CCR4–NOT-dependent degradation. It may be worth mentioning that the human CNOT8 deadenylase of the complex interacts with the RNA-binding protein PUM1 (Goldstrohm et al, 2006). The PUM1-binding motif was also found in the 3′UTRs of both the Pdk4 and the Igfbp1 mRNAs. Furthermore, recent studies show that GREs influence decay of mRNAs (Lee et al, 2010), and that GW182 protein binds to the CCR4–NOT complex to promote the miRNA-dependent mRNA decay (Fabian et al, 2009). The 921–1643 region of the 3′UTR of Pdk4 mRNA contains one GRE and binding sites for some conserved miRNAs (such as miR-23, miR-324, miR-429) in addition to ARE1, suggesting that these sequences cooperate to control the stability of the Pdk4 mRNA. In addition, the stability of Pdk4 mRNA is also controlled by the CNOT3-responding element within the 1–460 region of the 3′UTR that contains GRE and possibly other elements. Further studies should clarify the precise mechanism by which CCR4–NOT is recruited to its target mRNAs.
Cnot3+/− mice are not only leaner but also smaller in size and weigh less than wild-type mice. This phenotype of Cnot3+/− mice resembles that observed in Igfbp1 transgenic mice (Rajkumar et al, 1995). Our present data showed that Igfbp1 mRNA was elevated more than threefold in Cnot3+/− mice compared with wild-type mice. Consequently, the level of the IGFBP1 protein was elevated in the livers of Cnot3+/− mice (Supplementary Figure S4). IGFBP1 inhibits IGF-mediated mitogenic activity (McGuire et al, 1992) and differentiation of preadipocytes in vitro (Siddals et al, 2002). On the contrary, diet-induced obesity is prevented in Igfbp1 transgenic mice (Rajkumar et al, 1999). Therefore, elevated expression of Igfbp1 mRNA may contribute to the anti-obese and growth retardation phenotypes of Cnot3+/− mice to a certain extent.
As the CNOT3 expression in the liver of wild-type mice dynamically changes depending on the dietary condition, we assume that CNOT3 is involved in nutrient-sensing mechanism. This function is important for animals to adapt to the availability of nutrients. PDK4 and IGFBP1 are known to be induced in various tissues upon starvation (Cotterill et al, 1993; Jeoung et al, 2006). PDK4 regulates pyruvate dehydrogenase activity and acetyl-CoA production by balancing between fatty acid oxidization and glucose oxidization (Sugden and Holness, 2003). IGFBP1 regulates mitogenic and metabolizing activity through its interaction with IGF (Jones and Clemmons, 1995). Their up-regulation has been shown to be mediated by several transcription factors, such as PPARα (Sugden and Holness, 2003), FOXO (Kwon et al, 2004), and ATF4 (Averous et al, 2005). Our present data suggest that starvation-induced up-regulation of PDK4 and IGFBP1 could be at least in part due to stabilization of their mRNAs. Consistently, recent evidence showed involvement of translational regulation in amino-acid starvation-induced IGFBP1: namely the Igfbp1 mRNA is stabilized by a mechanism that involves the 3′UTR of Igfbp1 mRNA (Averous et al, 2005).
Obesity, characterized by excess body fat, is a prevalent public health concern and is caused by genetic abnormality and/or related to lifestyle. Therefore, the obesity is one of the central medical concerns and the methodologies of improving obesity have been developed. A recent genetic study shows significant linkage of an extreme-obesity phenotype with chromosome 19q13.33–q13.43 (Bell et al, 2005) where the CNOT3 gene is mapped. As reduction of CNOT3 expression effectively decreases the amount of body fat, which for example is evidenced by amelioration of the obese phenotype of ob/ob mice by lowering the level of Cnot3, CNOT3 could be a novel molecular target for anti-obesity. Yet, it should be noted that complete loss of CNOT3 is fatal (Supplementary Table SI; Neely et al, 2010), affecting cell proliferation (Hu et al, 2009). We also have to be aware of the increment of the stress-induced heart failure symptoms in the Cnot3 haplodeficient mice (Neely et al, 2010). The symptoms may be mediated by the transcriptional regulation. Therefore, proper targeting to deadenylation activity of the CCR4–NOT complex would be necessary to develop materials that are effectively anti-obese. In addition, a recent genetic study shows significant linkage of an extreme-obesity phenotype with chromosome 19q13.33–q13.43 (Bell et al, 2004). As the CNOT3 gene maps to 19q13, further genetic studies are needed to clarify the link between CNOT3 and metabolic disorders.
In conclusion, a reduction in CNOT3 levels affects the expression of mRNAs that encode proteins important for lipid metabolism, glucose metabolism, oxidative phosphorylation, and growth regulation. Furthermore, we propose here that CNOT3 signals to the CCR4–NOT deadenylases in response to nutrients to regulate decay of mRNAs important for energy metabolism.
Materials and methods
Mice
Generation of Cnot3-deficient mice is described in Supplementary materials and methods. Mice were housed in cages and maintained on a 12-h light–dark cycle, and they had access to water ad libitum. Mice were fed a normal chow diet (NCD) (CA-1, CLEA Japan Inc.) or a HFD (HFD32, CLEA Japan Inc.). For fasting analysis, mice were housed individually and deprived of food for 24 h. ob/+ mice were purchased from Charles River Japan. To generate ob/ob,Cnot3+/− double-mutant mice, ob/+,Cnot3+/− mice were mated with ob/+ mice. For all analyses, we used 8-week-old male mice unless otherwise noted. Experiments were conducted according to the guidelines for animal use issued by the Committee of Animal Experiments, Institute of Medical Science, University of Tokyo.
Blood analysis
For glucose tolerance tests, mice were fasted for 16 h. NCD and HFD mice were given an intraperitoneal injection of glucose (0.75 mg/g body weight and 0.5 mg/g body weight, respectively). For insulin tolerance tests, NCD mice and HFD mice were given human insulin (0.75 mU/g body weight and 1.5 mU/g body weight, respectively). Blood glucose was measured from tail blood using the glucose oxidase method (Sanwa Kagaku). Levels of serum triglycerides and insulin were determined using a Triglyceride E-Test (Wako) and a Mouse Insulin ELISA Kit (Morinaga), respectively.
Metabolic studies
Oxygen consumption (VO2) was determined with an O2/CO2 metabolic measuring system (Model MK-500, Muromachikikai) at 24°C, as described (Oike et al, 2005). VO2 is expressed as the volume of O2 consumed per kg0.75 weight per hour. VO2, carbon dioxide production, and the respiratory quotient were analysed during a 48-h time period. Continuous measurements were obtained over a 24-h time period. Consumption of foods was measured for 7 consecutive days. Rectal temperature was monitored using an electronic thermistor (Model BAT-12) equipped with a rectal probe (RET-3, Physitemp), as described (Oike et al, 2005).
Histological analysis of tissue and CT scan analysis
After dissection, all tissues were fixed in 10% formaldehyde overnight. Paraffin-embedded sections were analysed by haematoxylin and eosin staining. Morphometric analysis of adipose tissues from 500 cells per genotype was performed with Photoshop. For Sudan III staining, the liver was frozen embedded. The adiposity of mice was examined using a CT scanner (LaTheta, ALOKA) as described (Oike et al, 2005). CT scanning from the diaphragm to the bottom of the abdominal cavity was performed at 2-mm intervals.
Antibodies and reagents
Rabbit polyclonal antibodies against CNOT1, CNOT3, CNOT6L, CNOT7, and CNOT9 were as described (Morita et al, 2007). Mouse monoclonal antibodies against CNOT6L and CNOT7 were generated using recombinant human CNOT6L (amino acid 155–555) and full-length human CNOT7, respectively. Their specificities were verified by immunoprecipitation and immunoblotting of proteins from cells depleted expression of corresponding proteins as well as from control cells. Anti-Akt (#9272) and anti-phospho-Akt (Ser-473) (#9271) antibodies were purchased from Cell Signaling. Anti-GFP (598) antibodies were from UBI. Human insulin was from Eli Lilly and Co.
Immunoprecipitation and immunoblotting
Tissues were homogenized in lysis buffer (1% NP-40, 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM NaF, 1 mM Na3VO4) using a glass homogenizer and centrifuged for 10 min at 4°C. Proteins in the lysates were immunoprecipitated and immunoblotted as described (Suzuki et al, 2002).
Microarray analysis
Total RNAs were extracted with ISOGEN according to the manufacturer's protocol (Nippon Gene) and purified with an RNeasy Kit (Qiagen). cDNA and biotin-labelled cRNA were synthesized according to the protocols from Affymetrix. After fragmentation of cRNA, 20 μg biotin-labelled cRNA was hybridized to the GeneChip Mouse Genome 430 2.0 Array. To determine the average difference for each probe set, a global normalization method (Robust Multi-array Average) was used. Selected probe set IDs were converted to the manufacture's annotation. Enrichment of pathway was analysed by Fisher's exact test followed by Bonferonni's correction. The complete data set reported herein has been submitted to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) and can be obtained under accession numbers GSE18924 and GSE18925.
RNA analysis
For real-time PCR, total RNAs (0.5 μg) were used for reverse transcription with oligo(dT)12–18 primer (Invitrogen) using the SuperScript III First-Strand Synthesis System (Invitrogen). Real-time quantitative PCR reactions were carried out using SYBR Premix Ex Taq (Takara) and the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Hprt mRNA levels were used for normalization. Primers used for PCR reactions are listed in Supplementary Table SV. Northern blot analyses were carried out as previously described (Yoshida et al, 2000). PCR-amplified DNA fragments of mouse Cnot3 (nucleotides 1–500), mouse Pdk4 (nucleotides 2989–3430), mouse Igfbp1 (nucleotides 981–1489), and mouse Gapdh (nucleotides 921–1220) cDNAs were used as hybridization probes. An RNase H-Poly(A) treatment assay was performed as described (Mishima et al, 2006), using total RNA extracted and mixed with 25 pmol oligoDNA complementary to the 3′UTRs of the indicated mRNAs (Igfbp1, 5′-TGGTGTGCTCCAGAGTATAAATATACTATA-3′; Pdk4, 5′-CAAAACAACTATACATCAGATTACCCAAAT-3′; Gapdh, 5′-CAAAGTTGTCATTGAGAGCAARGCCAGCCC-3′). The 3′ fragments of the indicated mRNAs were detected by northern blotting. Low range RNA ladder marker (Thermo Scientific) was used to estimate the length of poly(A) tail. To measure mRNA stability, cells were treated with actinomycin D (5 μg/ml), and total RNAs were extracted at the indicated time points and subjected to qPCR analysis. The amount of reporter mRNAs was normalized to that of Hprt mRNA. For RNA–immunoprecipitation–RT–PCR analysis, proteins in the cell lysates were immunoprecipitated with anti-CNOT3, anti-CNOT6L, and control-IgG antibodies. The RNAs in the immunoprecipitates were purified using ISOGEN. The following primers were used for PCR analysis: Firefly luciferase, 5′-TGGAAGACGCCAAAAACATA-3′ and 5′-GTATTCAGCCCATATCGTTTCAT-3′; Pdk4 mRNA, 5′-TTTTGCATTGTAGATGTTGTCCTT-3′ and 5′-TCAACCAATGTGGGAGTCCA-3′. To predict miRNA-binding sequence, miRanda-mirSVR algorithm provided by microRNA.org was used.
Luciferase assays
The 3′UTRs of Pdk4, Igfbp1, Lpl mRNAs were inserted downstream of the luciferase coding region to produce Luc plus 3′UTR constructs (Figure 6C and D). pRL-TK was used as a control. The plasmid DNA was transfected into mouse primary hepatocytes (Hashita et al, 2008), and the luciferase assays were carried out as described (Morita et al, 2007).
Statistical analyses
All values represent mean±s.e.m. Differences between groups were examined for statistical significance using Student's t-test (two-tailed distribution with two-sample equal variance). We considered a P-value of <0.05 statistically significant.
Supplementary Material
Acknowledgments
We thank J Miyazaki for the CAG-Cre mice; R Sakamoto for the generation of the Cnot3-knockout mice; and O Gavrilova, RF Whittier, M Ohsugi, T Nakazawa, and K Yokoyama for valuable discussions. This work was supported by a grant-in-aid from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT), Japan, by a research fund from the Uehara Memorial Foundation, and by the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms), MEXT, Japan. MM was supported by grant-in-aid from Japan Society for the Promotion of Science (JSPS), an Ajinomoto Scholarship, and Uehara Memorial Foundation.
Author contributions: MM and TY designed this study and wrote the manuscript. MM performed the experiments and analysed the data. YO, MT, and TK measured oxygen consumption rate, respiratory quotient, and rectal temperature. TN analysed the microarray data. NY helped with the generation of Cnot3-deficient mice. MM and TY communicated with TS, TN, and MO about data interpretation and writing of the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aslam A, Mittal S, Koch F, Andrau JC, Winkler GS (2009) The Ccr4-Not deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell 20: 3840–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averous J, Maurin AC, Bruhat A, Jousse C, Arliguie C, Fafournoux P (2005) Induction of IGFBP-1 expression by amino acid deprivation of HepG2 human hepatoma cells involves both a transcriptional activation and an mRNA stabilization due to its 3′UTR. FEBS Lett 579: 2609–2614 [DOI] [PubMed] [Google Scholar]
- Azzouz N, Panasenko OO, Deluen C, Hsieh J, Theiler G, Collart MA (2009) Specific roles for the CCR4-NOT complex subunits in expression of the genome. RNA 15: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne H (2006) AU-rich elements and associated factors: are there unifying principles. Nucleic Acid Res 33: 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CG, Benzinou M, Siddiq A, Lecoeur C, Dina C, Lemainque A, Clément K, Basdevant A, Guy-Grand B, Mein CA, Meyre D, Froguel P (2004) Genome-wide linkage analysis for severe obesity in French Caucasians finds significant susceptibility locus on chromosome 19q. Diabetes 53: 1857–1865 [DOI] [PubMed] [Google Scholar]
- Bell CG, Walley AJ, Froguel P (2005) The genetics of human obesity. Nat Rev Genet 6: 221–234 [DOI] [PubMed] [Google Scholar]
- Belloc E, Mendez R (2008) A deadenylation negative feedback mechanism governs meiotic metaphase arrest. Nature 452: 1017–1021 [DOI] [PubMed] [Google Scholar]
- Berthet C, Morera AM, Asensio MJ, Chauvin MA, Morel AP, Dijoud F, Magaut JP, Durand P, Rouault JP (2004) CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol Cell Biol 24: 5808–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG (2005) Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics 6: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicoine J, Benoit P, Gamberi C, Paliouras M, Simonelig M, Lasko P (2007) Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev Cell 13: 691–704 [DOI] [PubMed] [Google Scholar]
- Collart MA (2003) Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313: 1–16 [DOI] [PubMed] [Google Scholar]
- Collart MA, Timmers HT (2004) The eukaryotic Ccr4-not complex: a regulatory platform integrating mRNA metabolism with cellular signaling pathways? Prog Nucleic Acid Res Mol Biol 77: 289–322 [DOI] [PubMed] [Google Scholar]
- Cotterill AM, Holly JMP, Wass JAH (1993) The regulation of insulin-like growth factor binding protein (IGFBP)-1 during prolonged fasting. Clin Endocri 39: 357–362 [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Morel AG, Barbot W, Loirreau P, Corbo L, Heidmann T (2001) Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2: 9S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E (2009) Deadenylation is a widespread effect of miRNA regulation. RNA 15: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, Chen CY, Shyu AB, Yates JR III, Hannon GJ, Filipowicz W, Duchaine TF, Sonenberg N (2009) Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 24: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Yang X, Wang W, Wood WH III, Becker KG, Gorospe M (2002) Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Nat Acad Sci USA 99: 10611–10616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M (2006) PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 13: 533–539 [DOI] [PubMed] [Google Scholar]
- Gopfert U, Kullmann M, Hengst L (2003) Cell cycle-dependent translation of p27 involves a responsive element in its 5′-UTR that overlaps with a uORF. Hum Mol Genet 12: 1767–1779 [DOI] [PubMed] [Google Scholar]
- Green CB, Douris N, Kojima S, Strayer CA, Fogerty J, Lourim D, Keller SR, Besharse JC (2007) Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc Natl Acad Sci USA 104: 9888–9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashita T, Sakuma T, Akada M, Nakajima A, Yamahara H, Ito S, Takesako H, Nemoto N (2008) Forkhead box A2-mediated regulation of female-predominant expression of the mouse Cyp2b9 gene. Drug Metab Dispos 36: 1080–1087 [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ (2009) A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 23: 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Inoue T, Yokoyama K, Morita M, Suzuki T, Yamamoto T (2011) CNOT2 depletion disrupts and inhibits the CCR4-NOT deadenylase complex and induces apoptotic cell death. Genes Cells 16: 368–379 [DOI] [PubMed] [Google Scholar]
- Jeoung NH, Wu P, Joshi MA, Jaskiewics J, Bock CB, DePaoli-Roach AA, Harris RA (2006) Role of pyruvate dehydrogenase kinase isoenzyme 4 (PDHK4) in glucose homeostasis during starvation. Biochem J 397: 417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JI, Clemmons DR (1995) Insulin-like growth factors and their binding proteins: biological actions. Endocrine Rev 16: 3–34 [DOI] [PubMed] [Google Scholar]
- Kwon HS, Huang B, Unterman TG, Harris RA (2004) Protein kinase B-alpha inhibits human pyruvate dehydrogenase kinase-4 gene induction by dexamethasone through inactivation of FOXO transcription factors. Diabetes 53: 899–910 [DOI] [PubMed] [Google Scholar]
- Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ (2010) Systematic analysis of cis-elements in unstable mRNA demonstrates that GUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One 5: e11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Nolte LA, Ju JS, Han DH, Coleman T, Holloszy JO, Semenkovich CF (2000) Skeletal muscle respiratory uncoupling prevents diet-induced obesity and insulin resistance in mice. Nat Med 6: 1115–1120 [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI et al. (2004) Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119: 121–135 [DOI] [PubMed] [Google Scholar]
- Liu S, Croniger C, Arizmendi C, Harada-Shiba M, Ren J, Poli V, Hanson RW, Friedman JE (1999) Hypoglycemia and impaired hepatic glucose production in mice with a deletion of the C/EBPbeta gene. J Clin Invest 103: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Wagner E (2005) Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev 19: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese FP, Aubareda A, Tudor C, Saklatvala J, Clark AR, Dean JLE (2010) MAPKAP kinase blocks Tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem 285: 27590–27600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire WL Jr, Jackson JG, Figuerroa JA, Shimasaki S, Powell DR, Yee D (1992) Regulation of insulin-like growth factor-binding protein (IGFBP) expression by breast cancer cells: use of IGFBP-1 as an inhibitor of insulin-like growth factor action. J Natl Cancer Inst 84: 1336–1341 [DOI] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K (2006) Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol 16: 2135–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal S, Aslam A, Doidge R, Medica R, Winkler SG (2011) The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4-Not complex contribute to the prevention of cell death senescence. Mol Biol Cell 22: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Suzuki T, Nakamura T, Yokoyama K, Miyasaka T, Yamamoto T (2007) Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol Cell Biol 27: 4980–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Yao R, Ogawa T, Suzuki T, Ito C, Tsunekawa N, Inoue K, Ajima R, Miyasaka T, Yoshida Y, Ogura A, Toshimori K, Noce T, Yamamoto T, Noda T (2004) Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat Genet 36: 528–533 [DOI] [PubMed] [Google Scholar]
- Neely GG, Kuba K, Cammarato A, Isobe K, Amann S, Zhang L, Murata M, Elmen L, Gupta V, Arora S, Sarangi R, Dan D, Fujisawa S, Usami T, Xia CP, Keene AC, Alayari NN, Yamakawa H, Elling U, Berger C et al. (2010) A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell 141: 142–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oike Y, Akao M, Yasunaga K, Yamauchi T, Morisada T, Ito Y, Urano T, Kimura Y, Kubota Y, Maekawa H, Miyamoto T, Miyata K, Matsumoto S, Sakai J, Nakagata N, Takeya M, Koseki H, Ogawa Y, Kadowaki T, Suda T (2005) Angiopoietin-related growth factor antagonizes obesity and insulin resistance. Nat Med 11: 400–408 [DOI] [PubMed] [Google Scholar]
- Rajkumar K, Barron D, Lewitt MS, Murphy LJ (1995) Growth retardation and hyperglycemia in insulin-like growth factor binding protein-1 transgenic mice. Endocrinology 136: 4029–4034 [DOI] [PubMed] [Google Scholar]
- Rajkumar K, Modric T, Murphy LJ (1999) Impaired adipogenesis in insulin-like growth factor binding protein-1 transgenic mice. J Endocrinol 162: 457–465 [DOI] [PubMed] [Google Scholar]
- Reitman ML (2002) Metabolic lessons from genetically lean mice. Annu Rev Nutr 22: 459–482 [DOI] [PubMed] [Google Scholar]
- Siddals KW, Westwood M, Gibson JM, White A (2002) IGF-binding protein-1 inhibits IGF effects on adipocyte function: implications for insulin-like actions at the adipocyte. J Endocrinol 174: 289–297 [DOI] [PubMed] [Google Scholar]
- Sugden MC, Holness MJ (2003) Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab 284: 855–862 [DOI] [PubMed] [Google Scholar]
- Suzuki T, K-Tsuzuku J, Ajima R, Nakamura T, Yoshida Y, Yamamoto T (2002) Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev 16: 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Yoshida N, Kishimoto T, Akira S (1997) Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J 16: 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E (2004) A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 23: 2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, Wahle E (2010) Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 16: 1356–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, Harper ME, Tremblay ML, Sonenberg N (2001) Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med 7: 1128–1132 [DOI] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R (2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G (2004) Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature 431: 200–205 [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S (2007) Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev 21: 1857–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Morita M, Yang X, Suzuki T, Yang W, Wang J, Ito K, Wang Q, Zhao C, Bartlam M, Yamamoto T, Rao Z (2010) Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J 29: 2566–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio-Oikawa K, Nakamura T, Usui M, Yoneda M, Ezura Y, Ishikawa I, Nakashima K, Noda T, Yamamoto T, Noda M (2007) Cnot7-null mice exhibit high bone mass phenotype and modulation of BMP actions. J Bone Miner Res 22: 1217–1223 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103: 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB (2005) Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Tanaka S, Umemori H, Minowa O, Usui M, Ikematsu N, Hosoda E, Imamura T, Kuno J, Yamashita T, Miyazono K, Noda M, Noda T, Yamamoto T (2000) Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103: 1085–1097 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.