Abstract
Exposure of metazoan organisms to hypoxia engages a metabolic switch orchestrated by the hypoxia-inducible factor 1 (HIF-1). HIF-1 mediates induction of glycolysis and active repression of mitochondrial respiration that reduces oxygen consumption and inhibits the production of potentially harmful reactive oxygen species (ROS). Here, we show that FoxO3A is activated in hypoxia downstream of HIF-1 and mediates the hypoxic repression of a set of nuclear-encoded mitochondrial genes. FoxO3A is required for hypoxic suppression of mitochondrial mass, oxygen consumption, and ROS production and promotes cell survival in hypoxia. FoxO3A is recruited to the promoters of nuclear-encoded mitochondrial genes where it directly antagonizes c-Myc function via a mechanism that does not require binding to the consensus FoxO recognition element. Furthermore, we show that FoxO3A is activated in human hypoxic tumour tissue in vivo and that FoxO3A short-hairpin RNA (shRNA)-expressing xenograft tumours are decreased in size and metabolically changed. Our findings define a novel mechanism by which FoxO3A promotes metabolic adaptation and stress resistance in hypoxia.
Keywords: FoxO3A, hypoxia, metabolism, mitochondrion, Myc
Introduction
In order for metazoan organisms to survive under conditions of low oxygen, a metabolic switch is engaged resulting in the increased conversion of glucose to lactate. This phenomenon, known as the Pasteur effect, was until recently thought to result from increased glycolytic flux with mitochondrial respiration passively decreasing due to lack of oxygen. Recent reports have, however, demonstrated that metabolic adaptation to hypoxia requires an active suppression of mitochondrial function in order to control oxygen consumption and prevent excessive generation of potentially harmful reactive oxygen species (ROS; Kim et al, 2006; Papandreou et al, 2006; Fukuda et al, 2007; Zhang et al, 2008). Hypoxia-inducible factor 1 (HIF-1) is the master transcription factor orchestrating the cellular hypoxic response, and thus is deeply involved in the metabolic adaptation to hypoxia. HIF-1 is a heterodimer comprising the constitutively expressed HIF-1β subunit as well as the oxygen-regulated HIF-1α subunit (Wang et al, 1995). In the presence of oxygen, HIF-1α is, upon modification by a family of prolyl hydroxylase domain proteins (Bruick and McKnight, 2001; Epstein et al, 2001; Ivan et al, 2001), bound by the VHL E3 ubiquitin ligase complex and degraded via the ubiquitin/proteasome pathway (Kallio et al, 1999; Maxwell et al, 1999; Kamura et al, 2000). Since oxygen is a limiting factor for proline hydroxylation, HIF-1α accumulates in hypoxia and increases glycolytic flux through direct transcriptional induction of glucose transporters and glycolytic enzymes (Iyer et al, 1998; Seagroves et al, 2001). Moreover, recent lines of evidence have shown that additional HIF-dependent mechanisms balance energy and redox homeostasis. Specifically, this involves a HIF-1-mediated switch in the subunit composition of cytochrome c oxidase from COX4-1 to COX4-2 (Fukuda et al, 2007) and the direct transcriptional activation of pyruvate dehydrogenase kinase 1 (PDK1). PDK1 inactivates pyruvate dehydrogenase, thereby inhibiting the conversion of pyruvate to acetyl-CoA and thus shunting it away from entry into the mitochondrion and instead towards lactate production (Kim et al, 2006; Papandreou et al, 2006). Long-term exposure to hypoxia leads to HIF-1-dependent induction of mitochondrial autophagy through upregulation of BNIP3 (Zhang et al, 2008). In addition, HIF-1 has been shown to suppress mitochondrial biogenesis by inhibiting the growth-promoting transcription factor c-Myc (hereafter termed Myc) through transcriptional induction of the Myc antagonist Mxi1 (Zhang et al, 2007). Importantly, interference with these adaptation mechanisms leads to increased oxygen consumption, elevated ROS production, and cell death (Kim et al, 2006; Papandreou et al, 2006; Zhang et al, 2008).
The evolutionary conserved FoxO family of transcription factors is involved in the control of a vast array of biological processes including cell-cycle control, apoptosis, glucose metabolism, DNA damage repair, angiogenesis, and resistance to oxidative stress (Arden, 2008; Dansen and Burgering, 2008). Four different members have been identified in mammalian cells: FoxO1, FoxO3A, FoxO4, and the largely uncharacterized FoxO6. The phosphorylation of FoxO factors by the serine-threonine kinase Akt/PKB and their subsequent inactivation by nuclear exclusion is accepted as their canonical mode of regulation. However, many signalling pathways converge at the level of FoxO factors; for instance, the negative regulation by Akt can be overridden by activating signals emanating from stress-activated pathways (Brunet et al, 2004; Essers et al, 2004; Lehtinen et al, 2006). In Drosophila, FoxO directs a substantial fraction of the transcriptional changes in response to nutrient availability (Gershman et al, 2007; Teleman et al, 2008). In C. elegans, the FoxO homologue DAF-16 promotes longevity and resistance to hypoxic stress (Scott et al, 2002; Mabon et al, 2009). These examples emphasize the diverse roles of FoxO factors as signal integrators, and along these lines FoxO3A was recently found to be transcriptionally induced by hypoxic stress downstream of HIF-1 (Bakker et al, 2007).
In the present study, we investigated the role of FoxO3A in the transcriptional response to hypoxia. We found that FoxO3A contributes significantly to the regulation of both hypoxia-induced and -repressed genes and specifically suppresses a number of nuclear-encoded mitochondrial genes by directly antagonizing Myc at the promoters of these genes. FoxO3A short-hairpin RNA (shRNA)-expressing cells are thus dysfunctional in downregulating mitochondrial mass, oxygen consumption, and ROS in hypoxia, and exhibit increased sensitivity to hypoxia-induced apoptosis. Our findings in tissue culture experiments are supported by in vivo data, which show that FoxO3A is activated in hypoxic human tumour tissue and also demonstrates that the growth of FoxO3A shRNA-expressing xenografts is markedly impaired and correlates with a decreased glucose uptake.
Results
FoxO3A is a target gene of HIF-1α
To investigate the expression of FoxO factors in hypoxia, we analysed protein levels in HeLa, CaKi, MCF-7 cells, and in primary MEFs in normoxia, hypoxia and upon treatment with deferoxamine (DFO), a chemical hypoxia mimetic. While FoxO1 and FoxO4 were not regulated in a consistent way by hypoxic treatment or DFO, we found FoxO3A is induced in all tested cell lines and MEFs in hypoxia as well as by DFO (Supplementary Figure S1A). Moreover, the induction of FoxO3A protein (Supplementary Figure S1B) and mRNA (Supplementary Figure S1C) as determined by qPCR was abrogated by siRNA-mediated knockdown of HIF-1α and to a lesser extent HIF-2α, suggesting that both isoforms contribute to the hypoxic induction. Indeed, knockdown of HIF-1α and HIF-2α in combination further abrogated the hypoxic induction of FoxO3A protein (Supplementary Figure S1D). Furthermore, cell fractionation experiments demonstrated that the observed induction of FoxO3A protein in hypoxia was reflective of increases in both the cytosolic and the nuclear compartments (Supplementary Figure S1E).
Upon inspection of the proximal region of the human FoxO3A promoter, we found that it contains three putative hypoxia-response elements (HREs), which are conserved between human and mouse. Reporter gene assays performed with a 181-bp fragment encompassing these HREs demonstrated that it was responsive to hypoxia and/or co-transfection of HIF-1α or HIF-2α. Mutation of either of the HREs severely decreased the responsiveness of the reporter and mutation of all HREs in combination led to a near complete abrogation of the inducibility of the reporter by HIFα isoforms (Supplementary Figure S2A and B).
These results strongly suggest that FoxO3A is a direct target of HIF-1 and HIF-2. This was confirmed for HIF-1 by chromatin immunoprecipitation (ChIP) with an antibody against HIF-1α showing enriched hypoxic binding of endogenous HIF-1α in proximity of the proposed HREs in HeLa cells (Supplementary Figure S2C). Taken together, these results clearly demonstrate that FoxO3A is induced by hypoxia as a direct target gene of HIF-1, confirming published data (Bakker et al, 2007).
FoxO3A is required for hypoxic repression of mitochondrial genes
In order to study a potential biological role for FoxO3A in the hypoxic response, we utilized shRNA technology. We established stable FoxO3A knockdown HeLa cell clones that expressed two different hairpin sequences. The efficiency of the knockdowns in normoxia and hypoxia was validated by immunoblotting, which also indicated that knockdown of FoxO3A had no effect on the levels of HIF-1α protein in hypoxia (Figure 1A). We performed three parallel experiments in which Ctrl (empty vector) and FoxO3A knockdown cells were incubated in normoxia and hypoxia (0.5% O2) for 16 h and performed DNA microarray analysis on the RNA extracted from each sample.
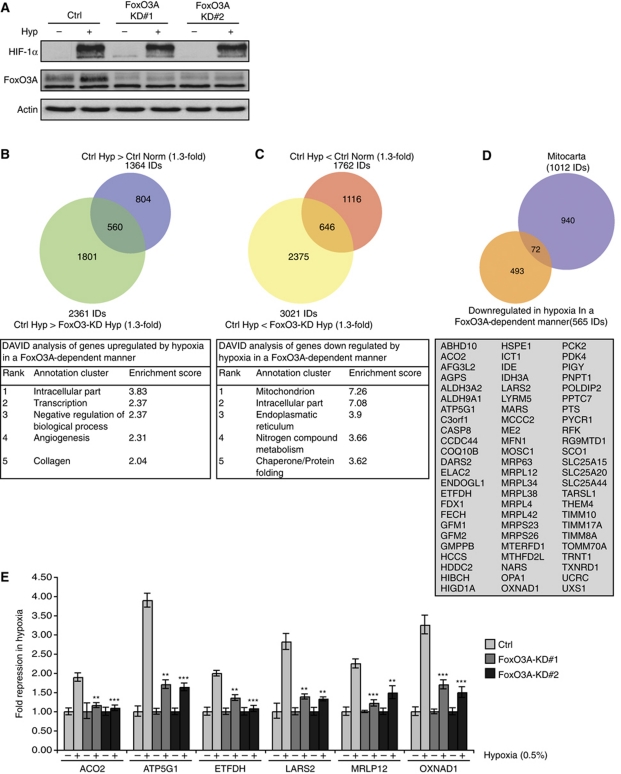
Figure 1.
FoxO3A contributes to the transcriptional regulation of the hypoxic response and is required for hypoxic repression of mitochondrial genes. (A) HeLa cell clones stably expressing FoxO3A shRNA or empty knockdown vector (Ctrl) were incubated for 16 h in normoxia or hypoxia (0.5% O2) and analysed by immunoblotting. (B, C) RNA from three experiments performed with Ctrl and FoxO3A-KD#1 as in (A) was extracted, labelled, and hybridized to Affymetrix HG-U133 Plus 2.0 microarrays. Statistical analysis was applied to identify probe sets upregulated (B) or downregulated (C) by hypoxia in a FoxO3A-dependent manner. DAVID analysis for each group is shown below, respectively. (D) Overlay of Mitocarta gene list with the list of hypoxia-repressed FoxO3A-dependent genes. The list of genes in the overlap is shown below. (E) Validation of six FoxO3A-dependent hypoxia-repressed mitochondrial genes by qPCR. The experiment was performed as in (A). The results are normalized to 18S rRNA and are shown as fold repression in hypoxia. Error bars indicate mean±s.d. of three independent experiments, **P<0.01, ***P<0.001 using Student's t-test.
Statistical analysis of the DNA microarray data showed that of the 1364 probe sets induced by hypoxia in control cells (>1.3-fold difference, P<0.02), 560 were lower expressed in hypoxic FoxO3A-KD compared with the expression levels in hypoxic control cells (>1.3-fold difference, P<0.02) (Figure 1B; see also Supplementary Table S1). This result identifies FoxO3A as an important contributor to the transcriptional induction of a significant fraction of hypoxia-induced genes. Interestingly, hypoxia-repressed probe sets were also dependent on FoxO3A: of the 1762 probe sets repressed by hypoxia in control cells (>1.3-fold difference, P<0.02), 646 were higher expressed in hypoxic FoxO3A-KD compared with the expression levels of hypoxic control cells (>1.3-fold difference, P<0.02) (Figure 1C; see also Supplementary Table S2). In addition, we noted that a substantial number of probe sets were already affected by the knockdown of FoxO3A in normoxic conditions. A total of 1861 probe sets were thus downregulated (>1.3-fold difference, P<0.02) by knockdown of FoxO3A in normoxia while 2145 were upregulated (>1.3-fold difference, P<0.02), demonstrating that FoxO3A affects the expression of many genes under normoxic conditions (see Supplementary Tables S3 and S4). Interestingly, gene ontology analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) bioinformatics tool (Dennis et al, 2003) showed that for the genes repressed by hypoxia in a FoxO3A-dependent manner, the most highly enriched GO term was ‘mitochondrion’ (Figure 1C, lower panel). Further analysis of the FoxO3A-dependent hypoxia-induced or -repressed genes using the Molecular Signatures Database (MSigDB) (Subramanian et al, 2005) showed that in addition to the expected hypoxia-related gene sets, the two curated gene sets ‘MOOTHA_HUMAN_MITODB_6_2002’ and ‘MOOTHA_MITOCHONDRIA’, comprising lists of 440 and 455 mitochondrial genes, respectively (Mootha et al, 2003), were highly significantly (P<10−8) overlapping with the list of FoxO3A-dependent hypoxia-repressed probe sets (Supplementary Tables S5 and S6).
In order to identify the mitochondrial genes in the list of 646 FoxO3A-dependent hypoxia-repressed probe sets, we converted this list to unique gene IDs (565 gene IDs) and subsequently overlaid it with the Mitocarta gene list (Figure 1D), which comprises 1013 human genes encoding proteins with known mitochondrial localization (Pagliarini et al, 2008). We found that a total of 72 of the 565 FoxO3A-dependent gene IDs encoded mitochondrial proteins (Figure 1D, lower box). Of these, six genes were selected for further analysis. Aconitase 2 (ACO2), mitochondrial ATP synthase subunit 9 isoform 1 (ATP5G1), electron transfer flavoprotein dehydrogenase (ETFDH), and oxidoreductase NAD-binding domain containing 1 (OXNAD1) are involved in mitochondrial metabolism or electron transport, and mitochondrial ribosomal protein L12 (MRPL12) and mitochondrial leucyl-tRNA synthetase 2 (LARS2) are part of the mitochondrial protein synthesis machinery. The selection is thus representative of several different aspects of mitochondrial function. In order to verify the expression changes observed in the microarray experiments, we analysed the expression of these six genes by qPCR in both FoxO3A knockdown cell clones. Our results did indeed confirm that the hypoxic repression of these genes was strongly compromised in both FoxO3A knockdown cell clones (Figure 1E). Consistent with our finding that FoxO3A is induced downstream of HIF-1 in hypoxia, we found that knockdown of HIF-1α also compromised the hypoxic repression of these genes (Supplementary Figure S3).
FoxO3A suppresses mitochondrial function and promotes cell survival in hypoxia
We next asked whether the FoxO3A knockdown cells would display a functional phenotype reflecting the observed changes in hypoxic gene expression patterns. Determination of mitochondrial mass by staining with the cardiolipin-binding dye nonyl acridine orange (NAO) followed by flow cytometry showed that both FoxO3A knockdown cell clones had more mitochondrial mass than control cells. Importantly, this difference was enhanced by hypoxia treatment (0.5% O2) for 24 h, due to the impaired ability of the FoxO3A knockdown cells to downregulate their mitochondrial mass in hypoxia (Figure 2A and B). The oxygen consumption of control and FoxO3A knockdown cells incubated in normoxia or hypoxia (0.5% O2) for 20 h was determined in Oxyograph chambers. When allowed to respire in full media, the FoxO3A knockdown cells consumed 13% more oxygen per cell per minute than their control counterparts in normoxia. After hypoxic treatment this difference was, however, further augmented as the FoxO3A knockdown cells failed to appropriately downregulate oxygen consumption, resulting in an oxygen consumption that was 84% higher than that of the control cells (Figure 2C), thus reflecting the results obtained from the determination of mitochondrial mass. Oxygen consumption measurements performed in glutamine-free media showed that the enhanced oxygen consumption of the FoxO3A knockdown cells in hypoxia is in part due to increased glutaminolysis. Depriving the cells of glucose resulted in a strong increase in oxygen consumption for all samples, presumably due to the Crabtree effect, but again highlighted the increased capacity of the FoxO3A knockdown cells for oxygen consumption (Figure 2C).
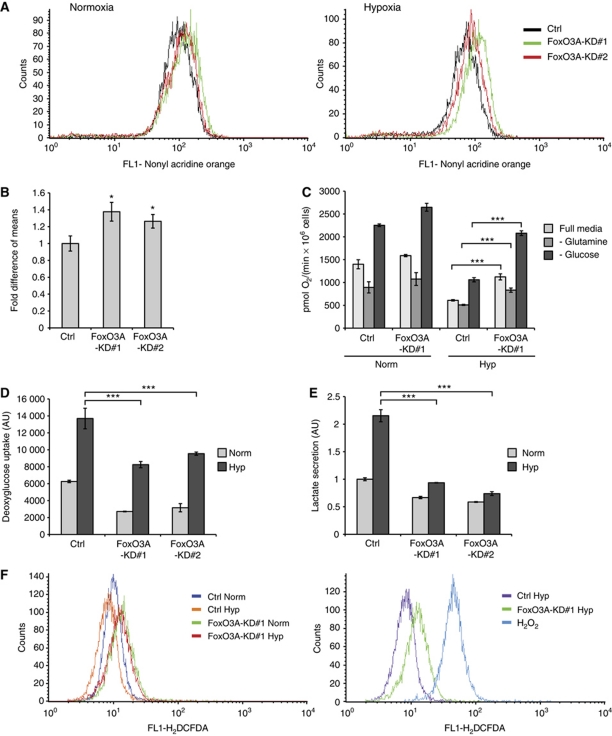
Figure 2.
FoxO3A knockdown cell clones have increased mitochondrial mass and are impaired in metabolic adaptation to hypoxia. (A) HeLa Ctrl and FoxO3A knockdown cell clones were incubated for 24 h in normoxia or hypoxia (0.5% O2) and analysed for mitochondrial mass by NAO staining followed by flow cytometry. (B) Bar diagram summarizing the results for hypoxia-treated samples of three experiments performed as in (A). Error bars indicate mean±s.d. of three independent experiments, *P<0.05 using Student's t-test. (C) Ctrl and FoxO3A knockdown cell clones were exposed to normoxia or hypoxia (0.5% O2) for 20 h and analysed for oxygen consumption in full media, glucose-free media, or glutamine-free media using an Oxyograph oxygen electrode. Error bars indicate mean±s.d. of three independently performed experiments, ***P<0.001 using Student's t-test. (D) Ctrl and FoxO3A knockdown cell clones were analysed for glucose uptake in normoxia and hypoxia (0.5%, 20 h) by use of radioactively labelled deoxyglucose. The glucose uptake was normalized to protein content. Error bars indicate mean±s.d. of three independently performed experiments, ***P<0.001 using Student's t-test. (E) Lactate secretion was determined in Ctrl and FoxO3A knockdown cell clones after 20 h incubation in either normoxia or hypoxia, normalized to protein content and presented as fold of normoxic Ctrl. Error bars indicate mean±s.d. of three independently performed experiments, ***P<0.001 using Student's t-test. (F) ROS levels in HeLa Ctrl and FoxO3A knockdown cell clones incubated in normoxia or hypoxia (0.5% O2) for 24 h were determined by DCFDA staining followed by flow cytometry (left panel). HeLa cells treated with 100 μM tert-butylperoxide for 15 min were used as a positive control (right panel).
In order to investigate whether the observed changes in mitochondrial mass and oxygen consumption would be accompanied by changes in glycolytic flux, we performed deoxyglucose uptake and lactate production measurements. Indeed, both FoxO3A knockdown cell clones displayed significantly lower deoxyglucose uptake and lactate production consistent with a less glycolytic phenotype (Figure 2D and E). Furthermore, the ROS levels in control and FoxO3A knockdown cells were determined by DCFDA (5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate) staining followed by flow cytometry. This analysis showed that FoxO3A knockdown cells have increased ROS levels when compared with control cells and, notably, fail to efficiently downregulate these under prolonged hypoxia (Figure 2F). Taken together, these results indicate that FoxO3A knockdown cells have increased mitochondrial mass and, importantly, in hypoxia fail to properly adapt their mitochondrial metabolism and to control ROS levels.
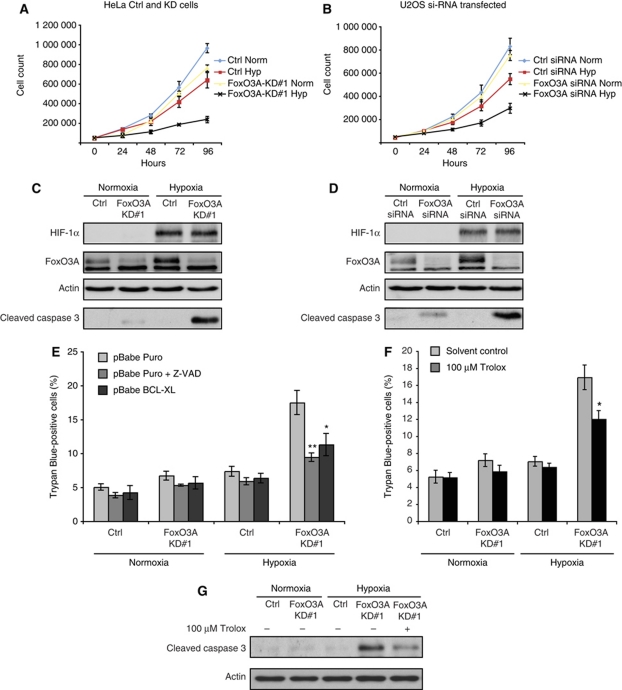
In light of this inability, we asked whether a knockdown of FoxO3A would also impact cell proliferation and survival in hypoxia. Growth curves of stable (knockdown and control) HeLa cell clones (Figure 3A) and of U2OS cells transiently transfected with Ctrl or FoxO3A siRNA (Figure 3B) showed that knockdown of FoxO3A resulted in decreased cell numbers in hypoxia. In order to investigate whether the reduced cell counts in hypoxia upon depletion of FoxO3A could be the result of increased sensitivity to apoptosis, we examined levels of the apoptotic marker cleaved caspase 3 by immunoblotting. Both HeLa stable FoxO3A knockdown cell clones and transiently FoxO3A siRNA transfected U2OS cells showed an increase in cleaved caspase 3 after 24 h incubation in hypoxia (Figure 3C and D), indicating increased apoptosis. Trypan blue staining showed a clear increase of dead cells in hypoxia-treated HeLa FoxO3A knockdown cell clones, which, importantly, could be suppressed by ectopic expression of the antiapoptotic factor BCL-XL or treatment with the caspase inhibitor Z-VAD (Figure 3E). In addition, pretreatment of cells with the antioxidant Trolox partially rescued the FoxO3A knockdown cells from cell death in hypoxia (Figure 3F), correlating with a decrease in cleaved caspase 3 (Figure 3G). This indicates that the elevated ROS levels in FoxO3A knockdown cells can sensitize them to hypoxia-induced apoptosis.
Figure 3.
FoxO3A knockdown cells show increased sensitivity to hypoxia-induced apoptosis. (A, B) HeLa Ctrl and FoxO3A-KD#1 cell clones (A) and U2OS cells transiently transfected with siRNA oligos (B) were plated in triplicate and 24 h later incubated in normoxia or hypoxia (0.2% O2). Every 24 h, samples were trypsinized, stained with trypan blue and viable cells were counted. Error bars indicate mean±s.d. of three independent experiments. (C, D) HeLa Ctrl and FoxO3A-KD#1 cell clones (C) and U2OS cells transiently transfected with siRNA oligos (D) were incubated for 24 h in normoxia or hypoxia (0.2% O2) and subsequently analysed by immunoblotting. (E) HeLa Ctrl and FoxO3A-KD#1 cell clones were transfected with pBabe Puro or pBabe BCL-XL expression plasmid and subsequently incubated for 48 h in normoxia or hypoxia (0.2% O2) in the presence or absence of Z-VAD. All cells were collected, stained with trypan blue, and live/dead cell numbers were determined. Error bars indicate mean±s.d. of three independent experiments, *P<0.05, **P<0.01 using Student's t-test. (F) HeLa Ctrl and FoxO3A-KD#1 cell clones were incubated for 48 h in normoxia or hypoxia (0.2% O2) in the presence or absence of 100 μM antioxidant Trolox or solvent control. All cells were collected, stained with trypan blue, and live/dead cell numbers were determined. Error bars indicate mean±s.d. of three independent experiments, *P<0.05 using Student's t-test. (G) HeLa Ctrl and FoxO3A-KD#1 cell clones preincubated in the absence or presence of 100 μM antioxidant Trolox were incubated for 24 h in normoxia or hypoxia (0.2% O2) and subsequently analysed by immunoblotting.
FoxO3A counteracts Myc to suppress mitochondrial genes in hypoxia
Having characterized the impact of FoxO3A deficiency on hypoxic metabolism and cell survival, we aimed to gain mechanistic insights into how FoxO3A might suppress expression of nuclear-encoded mitochondrial genes in hypoxia. Results from the MSigDB analysis not only showed overlaps with mitochondrial gene sets but, interestingly, several curated gene sets relating to Myc (Supplementary Table S6). The gene sets ‘WEI_MYCN_TARGETS_WITH_E_BOX’, ‘DANG_BOUND_BY_MYC’, and ‘SCHUHMACHER_MYC_TARGETS_UP’ (Schuhmacher et al, 2001; Zeller et al, 2003; Wei et al, 2008) were all significantly overlapping (P<10−6) with the list of FoxO3A-dependent hypoxia-repressed probe sets. The well-established role for Myc in regulating mitochondrial function (Li et al, 2005; Kim et al, 2008) prompted us to investigate this further.
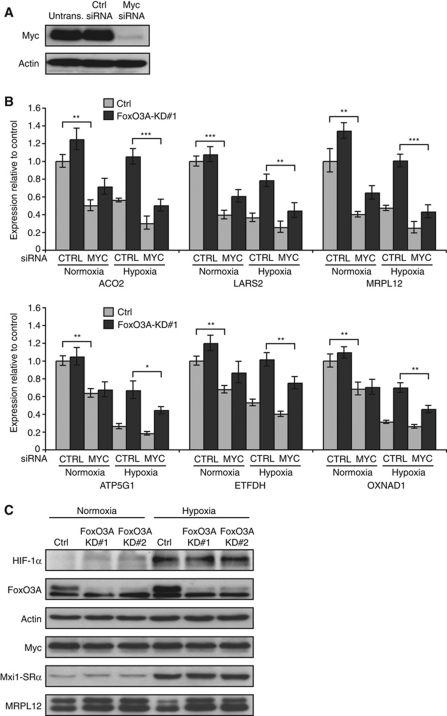
In order to investigate if the hypoxia-repressed FoxO3A-dependent mitochondrial genes could be co-regulated by Myc, we utilized Myc siRNA (Figure 4A). Interestingly, knockdown of Myc not only reduced the mRNA levels of all six model genes in Ctrl HeLa cell clones but, importantly, also partially restored the hypoxic repression in the FoxO3A knockdown cell clones (Figure 4B), indicating that they may be target genes of Myc and that FoxO3A may exert its repressive effect on these genes by interfering with Myc function. Results from analogous experiments performed with transiently transfected FoxO3A and Myc siRNA oligos in WI-38 (Supplementary Figure S4A and B) and TIG-3 (Supplementary Figure S5A and B) fibroblasts confirmed this pattern. FoxO3A was previously shown to counteract Myc by upregulating the expression of Mxi1-SRα (Delpuech et al, 2007), which competes with Myc for heterodimerization with Max and, more recently, FoxO3A was shown to induce expression of the Myc-targeting microRNAs miR-34b/c and miR-145 (Gan et al, 2010; Kress et al, 2011). However, our microarray data did not indicate any significant change in Mxi1-SRα mRNA levels between hypoxic Ctrl and FoxO3A knockdown cell clones. In agreement with this, results from qPCR (Supplementary Figure S6) and immunoblots (Figure 4C) showed that the induction of Mxi1-SRα mRNA and protein in hypoxia was unaffected by the knockdown of FoxO3A. Furthermore, knockdown of Mxi1-SRα had no impact on the hypoxic repression of the FoxO3A-dependent mitochondrial genes (Supplementary Figure S7A and B). Knockdown of FoxO3A did not result in any notable difference in the mRNA levels of Myc, Max, or Mad1 (Supplementary Figure S6). Immunoblots showed that neither hypoxia nor knockdown of FoxO3A had any major effect on the protein levels of Myc, making it unlikely that FoxO3A could be affecting Myc activity in hypoxia through a microRNA-dependent effect (Figure 4C). Finally, immunoblots confirmed that the deregulation previously observed for the mRNA levels of the mitochondrial ribosomal subunit MRPL12 upon knockdown of FoxO3A was translated into comparable changes in protein levels (Figure 4C, bottom).
Figure 4.
FoxO3A-dependent hypoxia-repressed mitochondrial genes are Myc responsive. (A) Verification of Myc knockdown. HeLa cells were transfected with Ctrl or Myc siRNA or left untransfected. Cells were lysed 24 h after transfection and analysed by immunoblotting. (B) Hela Ctrl and FoxO3A knockdown cell clones were transfected with Ctrl siRNA or siRNA directed against Myc and 24 h later incubated in normoxia or hypoxia (0.5% O2) for 20 h. RNA was extracted and analysed by qPCR. The results are normalized to 18S rRNA and are shown as fold of normoxic Ctrl. Error bars indicate mean±s.d. of three independent experiments, *P<0.05, **P<0.01, ***P<0.001 using Student's t-test. (C) Hela Ctrl and FoxO3A knockdown cell clones were incubated for 20 h in normoxia or hypoxia (0.5% O2). Protein was extracted and analysed by immunoblotting.
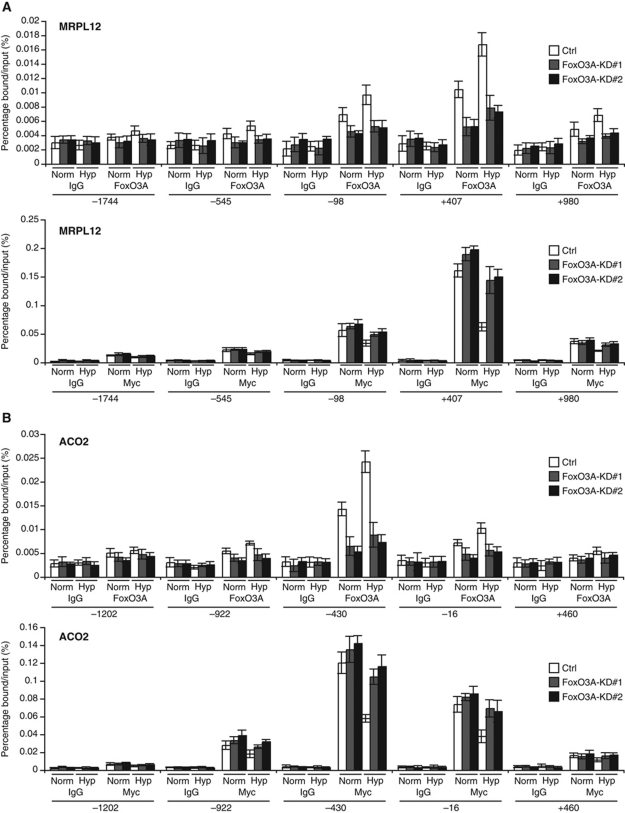
Having found no evidence for an indirect mode of regulation of the mitochondrial genes by FoxO3A in hypoxia through a modulation of Myc or Mxi1-SRα protein levels, we were prompted to consider that FoxO3A could be directly repressing these genes. To investigate this possibility, we took advantage of an inducible FoxO3A allele in the form of U2OS cells stably expressing a FoxO3A-ER fusion, in which three Akt-dependent phosphorylation sites were mutated to alanine (FoxO3A(A3)-ER). Activation of FoxO3A(A3)-ER by addition of 4-hydroxy-tamoxifen (4-OHT) for 10 h resulted in a clear repression of the mRNA levels of all model genes apart from ETFDH, which may be differentially regulated in U2OS cells compared with HeLa cells (Supplementary Figure S8). Importantly, inhibition of translation by pretreatment of the cells with cycloheximide prior to addition of 4-OHT did not abrogate the repressive function of FoxO3A(A3)-ER (Supplementary Figure S8), indicating that the repression does not require de novo protein synthesis and thus likely occurs by direct action of FoxO3A on the promoters of the repressed genes. To test this hypothesis, we performed ChIP on chromatin from Ctrl and FoxO3A knockdown cells incubated in normoxia and hypoxia (0.5% O2) for 16 h. We scanned the promoters (−2000 to +1000) of MRPL12, ACO2, LARS2, and OXNAD1 for binding of endogenous FoxO3A and Myc by designing primers at ∼500 bp intervals where DNA sequence allowed. Binding of endogenous FoxO3A was observed at distinct peaks for both MRPL12 and ACO2 promoters and was further enriched upon hypoxic treatment (Figure 5A and B, upper panels). As expected, the enrichment of FoxO3A binding was clearly decreased in the FoxO3A knockdown cell clones in normoxia as well as in hypoxia. Interestingly, ChIP results for Myc showed strong binding of Myc to the promoters with peak binding at positions detected by the same primer sets as for FoxO3A (Figure 5A and B, lower panels). Binding of Myc to the promoters was decreased in Ctrl cells upon incubation in hypoxia consistent with the ability of the cells to downregulate expression of these genes in hypoxia. However, Myc binding in the FoxO3A knockdown cell clones did not decrease to the same levels as observed for the Ctrl cells in hypoxia. This suggests a model in which FoxO3A directly antagonizes Myc in hypoxia and facilitates its displacement from the promoters of this group of hypoxia-repressed nuclear-encoded mitochondrial genes. Similar binding patterns and regulations were also found for FoxO3A and Myc at the OXNAD1 and LARS2 promoters (Supplementary Figure S9A and B). Closer inspection of the DNA sequence in proximity of the regions where Myc and FoxO3A exhibited peak enrichment revealed the existence of canonical and non-canonical E-boxes in MRPL12, ACO2, and OXNAD1 at positions +627, −141, and −418 relative to the start sites of transcription, respectively. For LARS2, no E-box element could be found in close proximity of the observed peak binding. However, ChIP and high-throughput sequencing data obtained for Myc have recently revealed that 36% of Myc-bound promoters in vivo lack any known E-box sequence (Kim et al, 2008).
Figure 5.
FoxO3A directly antagonizes Myc at the promoters of hypoxia-repressed mitochondrial genes. (A, B) HeLa Ctrl, FoxO3A-KD#1, and FoxO3A-KD#2 cell clones were incubated in normoxia or hypoxia (0.5% O2) for 16 h before crosslinking. ChIP was performed using anti-FoxO3A (upper panel), anti-Myc (lower panel), and IgG (negative control) antibodies. Primers scanning the MRPL12 (A) and ACO2 (B) loci were designed. Results were generated by qPCR and are presented as percentage bound/input. Error bars indicate mean±s.d.
The ability of FoxO3A to directly counteract Myc at these promoters prompted us to consider whether this could be a general phenomenon. In order to investigate this, we examined the mRNA expression of a selection of the well-established Myc targets CAD, ODC, and nucleolin (Bello-Fernandez et al, 1993; Boyd and Farnham, 1999; Greasley et al, 2000). We found no defect in the ability of FoxO3A knockdown cell clones to repress these genes in hypoxia when compared with Ctrl cells (Supplementary Figure S10A). In accordance with this, ChIP assays revealed that Myc displacement from the E-boxes of CAD and ODC in hypoxia was not impaired in FoxO3A knockdown cells (Supplementary Figure S10B, lower panel). Importantly, no binding of FoxO3A was found at these loci (Supplementary Figure S10B, upper panel). Taken together, our findings suggest that the direct action of FoxO3A on the promoters of hypoxia-repressed nuclear-encoded mitochondrial genes applies to a specific subclass of Myc target genes.
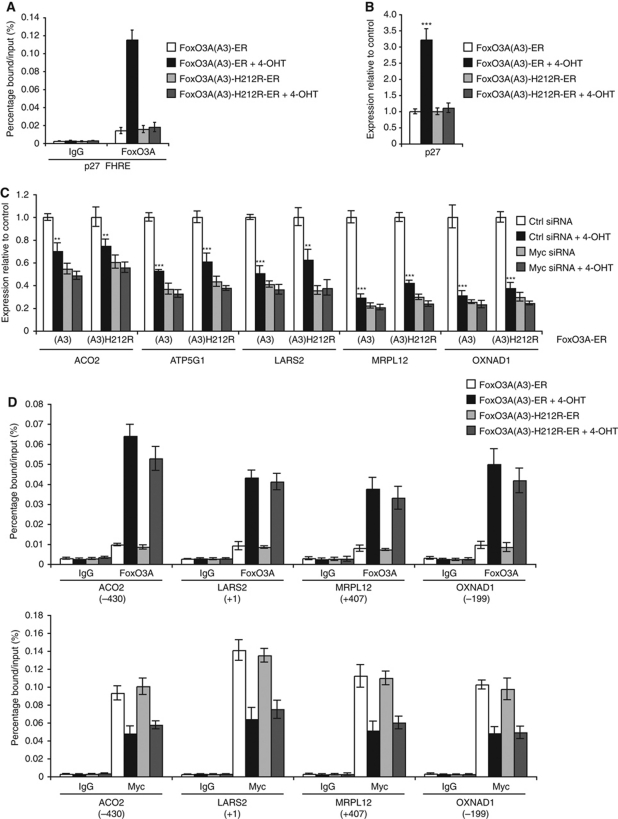
In order to gain further mechanistic insight into the FoxO3A-mediated repression of these Myc-dependent mitochondrial genes, we took advantage of a previously characterized FoxO3A point mutant FoxO3A(A3)-H212R that is incapable of binding to the consensus FoxO DNA recognition element (FHRE) (Ramaswamy et al, 2002; Czymai et al, 2010). Whereas FoxO3A(A3)-ER bound an established FHRE in the p27 promoter (Hu et al, 2004) and significantly upregulated the mRNA expression of p27 upon stimulation with 4-OHT, FoxO3A(A3)-H212R-ER was unable to bind the FHRE and to induce the expression of p27 mRNA (Figure 6A and B). However, both FoxO3A(A3)-ER and FoxO3A(A3)-H212R-ER were capable of repressing the selected mitochondrial genes. As observed for the other cell types, knockdown of Myc in the FoxO3A-ER expressing U2OS cell clones led to a clear decrease in expression of the selected mitochondrial genes. Notably, the activation of FoxO3A(A3)-ER or FoxO3A(A3)-H212R-ER by 4-OHT after knockdown of Myc had no significant additional repressive effect (Figure 6C). ChIP results demonstrated a clear association of both FoxO3A(A3)-ER and FoxO3A(A3)-H212R-ER upon 4-OHT stimulation to the loci where peak binding was previously observed for endogenous FoxO3A in hypoxia (Figure 6D, upper panel). Moreover, the association of FoxO3A(A3)-ER and FoxO3A(A3)-H212R-ER was inversely correlated with Myc binding as previously observed for endogenous FoxO3A and Myc in hypoxia (Figure 6D, lower panel; see Supplementary Figures S11 and S12 for extended ChIP coverage of the MRPL12, ACO2, OXNAD1, and LARS2 loci). Taken together, these results demonstrate that FoxO3A-mediated repression of the selected mitochondrial genes does not require FHRE binding and appears to function by facilitating the displacement of Myc from a subset of its target gene promoters. However, although a direct interaction between FoxO3A and Myc has been reported (Chandramohan et al, 2008), we were unable to detect any interaction between ectopically expressed HA-FoxO3A(A3) and Myc in normoxia or hypoxia (Supplementary Figure S13).
Figure 6.
Repression of hypoxia-repressed mitochondrial genes by an inducible FoxO3A allele is independent of binding to FHRE. (A) FoxO3A(A3)-ER or FoxO3A(A3)-H212R-ER stably expressing U2OS cell clones were incubated in the absence or presence of 250 nM 4-OHT for 2 h before crosslinking. ChIP was performed using anti-FoxO3A and IgG (negative control) antibodies. Binding of FoxO3A to an FHRE in the p27 promoter was analysed by qPCR and is presented as percentage bound/input. Error bars indicate mean±s.d. (B) FoxO3A(A3)-ER or FoxO3A(A3)-H212R-ER stably expressing U2OS cell clones were incubated in the absence or presence of 250 nM 4-OHT for 10 h. RNA was extracted and analysed by qPCR. The results are normalized to 18S rRNA and are shown as fold of normoxic Ctrl. Error bars indicate mean±s.d. of three experiments, ***P<0.001 using Student's t-test. (C) FoxO3A(A3)-ER or FoxO3A(A3)-H212R-ER stably expressing U2OS cell clones were transfected with Ctrl siRNA or siRNA against Myc 24 h prior to incubation in the absence or presence of 250 nM 4-OHT for 10 h. RNA was extracted and analysed by qPCR. The results are normalized to 18S rRNA and are shown as fold of Ctrl. Error bars indicate mean±s.d. of three experiments, **P<0.01, ***P<0.001 using Student's t-test. (D) The experiment was preformed as in (A). ChIP was performed using anti-FoxO3A, anti-Myc, and IgG (negative control) antibodies. Binding of FoxO3A (upper panel) and Myc (lower panel) to the peak areas of ACO2, LARS2, MRPL12, and OXNAD1 was analysed by qPCR and is presented as percentage bound/input. Error bars indicate mean±s.d.
FoxO3A is activated in hypoxic tumour tissue in vivo
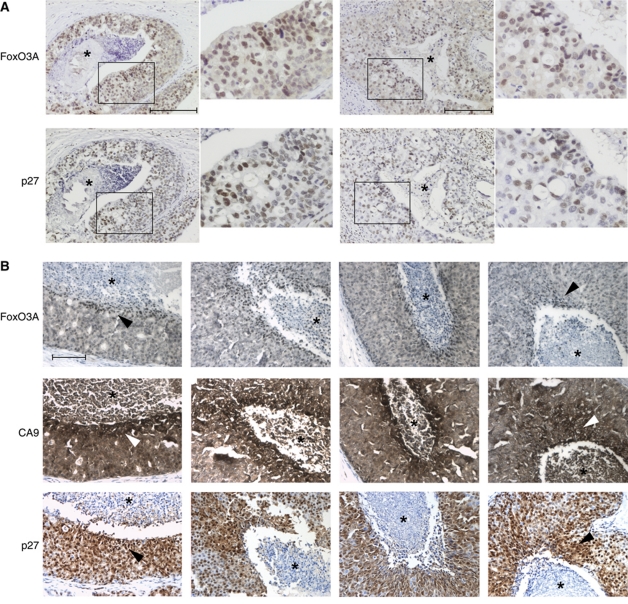
Having found evidence from tissue culture experiments for an important role for FoxO3A in orchestrating metabolic adaptation and promoting survival in hypoxic cancer cells, we next asked whether FoxO3A was activated in vivo in hypoxic tumour tissue. To address this question, we performed immunohistochemical stainings on sections of ductal breast carcinoma in situ (DCIS), a tumour type known to contain hypoxic areas (Helczynska et al, 2003). Stainings for FoxO3A revealed a clear tendency towards a strong, nuclear-localized staining in the perinecrotic area of the tumours (Figure 7A). Indeed, when adjacent sections of the same tumours were co-stained for FoxO3A, the HIF target carbonic anhydrase 9 (CA9), and the FoxO3A target p27, the staining pattern indicated elevated expression of these proteins in overlapping areas (Figure 7B). A statistical analysis performed on 86 DCIS lesions from 9 patients confirmed a significant correlation between increased CA9 and FoxO3A staining towards the necrotic core of the tumours (Supplementary Table S7; Supplementary Figure S14).
Figure 7.
FoxO3A is activated in hypoxic human tumour tissue. (A) Human samples of carcinoma in situ of the breast were immunohistochemically stained using a FoxO3A antibody. Two different tumours are shown with a magnified view to the right of each. Scale bars indicate 100 μM. (B) Human samples for carcinoma in situ of the breast were analysed for the immunohistochemical localization of FoxO3A, CA9, and p27 in adjacent slides. The necrotic cores of the tumours are highlighted by an *. Scale bars indicate 100 μm.
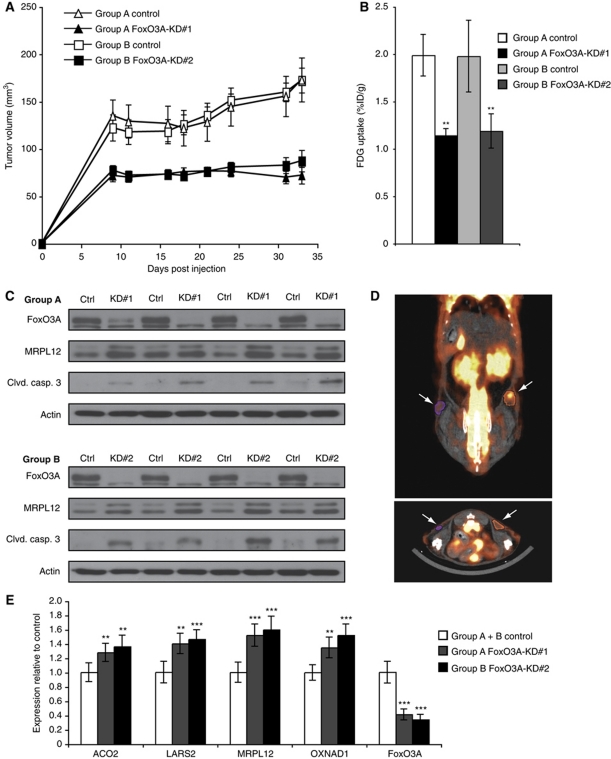
In order to test whether the inability of FoxO3A knockdown cells to successfully adapt metabolism and promote survival in hypoxia would also impact the growth of xenograft tumours in vivo, Ctrl and FoxO3A knockdown cell clones were inoculated in nude mice and tumour growth was monitored over 33 days. The growth of the HeLa xenografts was significantly impaired by the knockdown of FoxO3A (P<0.01 for all time points post injection; Figure 8A). We investigated whether this was associated with physiological alterations in vivo by assessing the glucose uptake. On the final day of the experiment, animals were injected with the glucose analogue fluorodeoxyglucose (FDG) labelled with 18F. Non-invasive positron emission tomography (PET) was performed to assess the FDG accumulation in the tumours. We found that the accumulation of FDG relative to the volume of the tumour was significantly impaired for both FoxO3A-KD#1 and FoxO3A-KD#2 xenografts compared with their control counterparts (Figure 8B), which could likely be a consequence of their inability to appropriately downregulate mitochondrial activity in hypoxia and shift the balance in favour of glycolysis. An example of a fused PET and computer tomography (CT) image of a FoxO3A knockdown and a control tumour is depicted in Figure 8D. Immunoblots on protein extracts from the xenograft tumours confirmed the knockdown of FoxO3A and demonstrated elevated protein levels of MRPL12 as well as of cleaved caspase 3 in the knockdown tumours (Figure 8C). Consistent with this, stainings of xenograft tumour sections for cleaved caspase 3 by immunofluorescence showed an increase in positive staining foci in the FoxO3A knockdown tumours compared with control tumours (Supplementary Figure S15A and B). Moreover, RNA extracted from the tumours confirmed significantly increased mRNA expression of the selected mitochondrial genes in both FoxO3A-KD#1 and FoxO3A-KD#2 tumours compared with their control counterparts (Figure 8E).
Figure 8.
FoxO3A knockdown xenograft tumours are metabolically changed and exhibit impaired growth and increased apoptosis. (A) HeLa Ctrl and FoxO3A knockdown cell clones were injected subcutaneously into the left and right flanks of nude mice, respectively. Growth of the tumour xenografts was monitored using a caliper. Error bars indicate s.e.m. (n=12). (B) At the final day of the experiment, F-18-FDG accumulation in FoxO3A knockdown tumours and control tumours was assessed by PET. Error bars indicate s.e.m. (n=12), **P<0.01 using Student's t-test. (C) Protein was extracted from four Ctrl and FoxO3A knockdown tumours of comparable size within each experimental group and the extracts were analysed by immunoblotting. (D) Example of a PET/CT fusion image showing glucose uptake of Ctrl (right arrow) and FoxO3A knockdown (left arrow) tumours. (E) RNA was extracted from eight xenograft tumours originating from Ctrl, FoxO3A-KD#1, and FoxO3A-KD#2 cells and analysed by qPCR. The results are normalized to 18S rRNA and are shown as fold of Ctrl. Error bars indicate mean±s.d., **P<0.01, ***P<0.001 using Student's t-test.
Discussion
In this study, we have shown that FoxO3A plays an important role in the transcriptional program that facilitates metabolic adaptation in hypoxia. Not only does FoxO3A contribute markedly to the upregulation of many hypoxia-induced genes but, importantly, also plays a crucial role in the hypoxic repression of a substantial number of nuclear-encoded mitochondrial genes. Previous studies have elucidated a number of different, specific mechanisms by which HIF-1 can adapt metabolism to hypoxic conditions. This includes inhibition of acetyl-CoA synthesis by induction of PDK-1, COX4 subunit switching, and promotion of mitochondrial autophagy through BNIP3 (Kim et al, 2006; Papandreou et al, 2006; Fukuda et al, 2007; Zhang et al, 2007, 2008). The metabolic phenotype of FoxO3A knockdown cells bears a resemblance to the phenotypes demonstrated in several of the above-mentioned studies upon interference with these adaptation mechanisms. Such noteworthy parallels include increased mitochondrial mass, oxygen consumption, and ROS production, correlating with an increased sensitivity to apoptotic cell death in hypoxia. The beneficial effects of suppressing mitochondrial oxygen consumption in hypoxia are, however, not limited to the control of ROS production. This adaptation also serves to preserve oxygen for non-energy-producing cellular functions that require oxygen such as enzymatic reactions catalysed by oxidases, hydroxylases, and histone demethylases. Moreover, when considering a hypoxic tumour in vivo, reducing oxygen consumption helps to extend the diffusion limit of oxygen and thus prevents the exposure of cells to anoxic conditions that can induce cell death (Denko, 2008; Wheaton and Chandel, 2010).
Our finding that knockdown of FoxO3A in both HeLa and U2OS cells leads to increased cell death in hypoxia is consistent with findings made by others in MCF-7 cells. The authors propose that this is due to disruption of a negative feedback loop from FoxO3A to HIF-1 via induction of CITED2, leading to increased HIF-1-dependent induction of proapoptotic genes RTP801 and NIX in hypoxia (Bakker et al, 2007). We, however, did not find these genes differentially induced in FoxO3A knockdown cells when compared with Ctrl. Indeed, our microarray data indicate that 560 out of 2361 hypoxia-induced probe sets are dependent on FoxO3A for their induction, suggesting that, at least in the conditions applied in this study, FoxO3A is a positive contributor to the hypoxia-induced component of the hypoxic response. This notion is consistent with results from another microarray-based study demonstrating that FoxO3−/− neural stem cells show decreased expression of hypoxia-induced genes (Renault et al, 2009).
While the role of FoxO factors and in particular FoxO3A in promoting stress resistance and controlling ROS is well established (Miyamoto et al, 2007; Tothova et al, 2007), this has so far been attributed to transcriptional induction of antioxidant enzymes that control the amount of ROS such as manganese superoxide dismutase, catalase, and peroxiredoxin (Kops et al, 2002; Essers et al, 2004; Chiribau et al, 2008). However, none of the probe sets for these enzymes were induced in HeLa cells by hypoxia in a FoxO3A-dependent manner. Instead our findings in this study define a novel, repressive mechanism by which FoxO3A prevents inappropriate, hypoxic Myc activity by directly antagonizing Myc at the promoters of mitochondrial genes (summarized in Figure 9). Although Myc is a proto-oncogene that drives cell proliferation, it is also known to sensitize cells to cell death in hypoxic conditions (Brunelle et al, 2004).
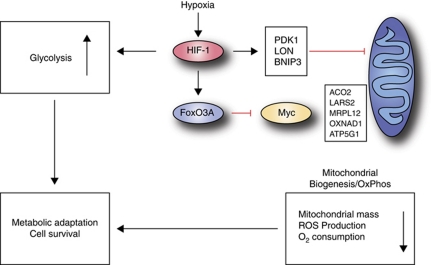
Figure 9.
Model. Hypoxia induces stabilization of HIF-1, which in turn leads to transcriptional regulation of its target genes. This leads to an increase in glycolytic flux through transcriptional upregulation of glucose transporters and glycolytic enzymes. In addition, HIF-1 transactivates target genes involved in inhibiting mitochondrial activity (PDK-1 and BNIP3) and adjusting oxygen consumption (LON protease) to hypoxic conditions. The hypoxic induction of FoxO3A leads to a direct and specific repression of a subset of nuclear-encoded mitochondrial genes that are Myc target genes. The combined effects of these adaptive measures serve to balance oxygen, ROS, and energy homeostasis in hypoxia, which is required for cell survival.
The interplay between FoxO factors and Myc has been the focus of many studies and recent advances have offered insight into the mechanisms by which these transcription factors counteract each other. This includes the induction of Mxi1-SRα by FoxO3A (Delpuech et al, 2007) and, recently identified, the upregulation of two miRNAs targeting Myc (Gan et al, 2010; Kress et al, 2011). In the present study, we cannot find evidence that either these miRNAs or Mxi1-SRα play a significant role in the antagonistic effect of FoxO3A on Myc in hypoxia: Myc protein levels were virtually unchanged by hypoxia or knockdown of FoxO3A and the induction of Mxi1-SRα in hypoxia appears to be largely independent of FoxO3A. Hypoxic expression of Mxi1-SRα is thus likely to be primarily driven by HIF-1 and may be involved in the repression of other Myc target genes. In addition to this, a direct antagonistic effect of HIF-1 on Myc function has been demonstrated for target genes involved in cell-cycle regulation (Koshiji et al, 2004; Gordan et al, 2007).
Interestingly, our experiments using the inducible FoxO3A(A3)-ER allele indicate that an additional hypoxic signal is not required since FoxO3A(A3)-ER activation in normoxia is sufficient to repress the genes tested. This observation suggests that activation of FoxO3A in another context such as growth factor deprivation would lead to suppression of mitochondrial genes. In agreement with this, a study performed in Drosophila showed that a group of genes involved in mitochondrial biogenesis were not only repressed and subsequently activated in vivo upon starvation and re-feeding, respectively, but also repressed by expression of a constitutively nuclear Drosophila FoxO orthologue (dFoxO(A3)) in vitro (Gershman et al, 2007). A potential role for the Drosophila Myc (dMyc) in this regulation was not investigated but, interestingly, the authors noted that the promoter regions of genes repressed in a dFoxO-dependent manner only rarely contained consensus FoxO binding elements as opposed to the genes that were induced by starvation and dFoxO(A3).
Similar to this, we found no consensus FoxO binding elements in proximity of the regions to which our ChIP experiments demonstrated that FoxO3A binds in hypoxia. On the contrary, we observed an inversely correlated binding of Myc to the same regions, which for ACO2, MRPL12, and OXNAD1 correlate with nearby potential binding sites for Myc, and crucially we have demonstrated that a mutant FoxO3A unable to bind the FHRE is still capable of mediating the repression of these genes. We, therefore, propose that FoxO3A displaces Myc from the promoters of this group of nuclear-encoded mitochondrial genes in hypoxia via a mechanism that does not require direct binding to a FHRE. The ability of FoxO DNA binding mutants to repress transcription has previously been demonstrated (Ramaswamy et al, 2002; Czymai et al, 2010) and a model similar to the one put forward in this study has been proposed for the interplay between FoxO3A and Myc in the regulation of the cyclin D2 promoter (Bouchard et al, 2004). The exact mechanism of recruitment does, however, remain elusive and awaits investigation by genome-wide approaches.
Our findings in this study indicate that the inability of cancer cells to downregulate oxygen consumption and ROS production in hypoxia upon FoxO3A knockdown not only correlates with decreased cell proliferation in hypoxia in tissue culture experiments but also with in vivo xenograft experiments. Interestingly, a recent study demonstrates that shRNA-mediated downregulation of the mitochondrial ribosomal proteins MRPL12 or MRPL28 lead to clear increases in the growth of xenograft tumours as well as decreased oxygen consumption and increased glucose uptake (Chen et al, 2009). Considering these results, the inability of FoxO3A knockdown cells to appropriately downregulate MRPL12 in hypoxia likely confers a significant adaptive disadvantage and, taken together, highlights regulation of mitochondrial ribosomal function as a potential key to metabolic adaptation in hypoxia. The observed decrease in glucose uptake of FoxO3A knockdown xenografts when compared with control tumours underlines this deficit of metabolic adaptation. Although the experiments performed in cell culture show an important correlation between dysregulation of metabolism, ROS production, and cell death, it should be stressed that we cannot exclude that other hypoxic gene expression changes caused by knockdown of FoxO3A, such as the reduced activation of many hypoxia-induced genes, might contribute to the observed cell and tumour growth phenotypes.
While several studies have demonstrated a tumour suppressor function for FoxO transcription factors (Bouchard et al, 2007; Paik et al, 2007), the data presented in the present study define an important role for FoxO3A in promoting metabolic adaptation and cell survival in hypoxic cancer cells. Our xenograft data are in agreement with results from a recent study showing that knockdown of FoxO3A in MDA-MB-231 breast cancer cells injected into the fat pads of nude mice led to a significant decrease in tumour volume (Storz et al, 2009).
Moreover, our in situ immunohistochemical stainings for FoxO3A in human ductal breast carcinoma show that FoxO3A is indeed activated in hypoxic regions of human tumour tissue. This observation is particularly interesting when considering that another recent study has shown a significant association between strong nuclear FoxO3A staining and poor overall survival of breast cancer patients (Chen et al, 2010).
The role of FoxO3A in tumourigenesis is thus likely to be context dependent. While the inactivation of FoxO factors in early tumour formation by increased signalling through growth factor cascades offers a proliferative advantage, it is conceivable that at later stages, limitations in the access to serum factors in combination with intratumoural hypoxia can reactivate FoxO3A allowing metabolic adaptation and thus promoting tumour cell survival.
Materials and methods
Cell culture
All cell lines were grown in high-glucose Dulbecco's modified Eagle's medium (Invitrogen) containing Glutamax supplemented with 10% fetal calf serum, penicillin, and streptomycin. The cells were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air (20% O2). For incubations in hypoxia, the cells were kept in an atmosphere containing 0.5 or 0.2% oxygen and 5% CO2 at 37°C in a hypoxia incubator for the indicated time periods. The stable cell clones were maintained in 1.5 μg/ml puromycin at all times.
shRNA stable cell clones
HeLa stable knockdown cell clones were established by calcium phosphate-mediated transfection of HeLa cells with two different pLKO.1 shRNA constructs from Open Biosystems targeting FoxO3A. Product numbers were FoxO3A-KD#1 (RHS3979-9630810) and FoxO3A-KD#2 (RHS3979-9607487). pLKO.1 empty vector was used for control cells. The cells were selected in full media containing 1.5 μg/ml puromycin.
siRNA transfections
U2OS cells were transfected with oligonucleotides at a final concentration of 50 nM using Oligofectamine (Invitrogen) according to the manufacturer's protocol. HeLa, WI-38, and TIG-3 cells were transfected at a final concentration of 20 nM using RNAiMAX (Invitrogen) according to the manufacturers instructions. For control siRNA transfections, Allstars negative control (Qiagen) was used and the following duplex specific for FoxO3A: sense 5′-GCUCUUGGUGGAUCAUCAAdTdT-3′ and antisense 5′-UUGAUGAUCCACCAAGAGCdTdT-3′ custom synthesized by Qiagen. For HIF-1α: forward 5′-CUGAUGACCAGCAACUUGAdTdT-3′ and reverse 5′-UCAAGUUGCUGGUCAUCAGdTdT-3′ custom synthesized by Qiagen. For HIF-2α: forward 5′-CAGCAUCUUUGAUAGCAGUdTdT-3′ and reverse 5′-ACUGCUAUCAAAGAUGCUGdTdT-3′ custom synthesized by Qiagen. For Mxi1-SRα, we used Stealth RNAi (Invitrogen, #1299003). For Myc, we used MYC siGenome SMART pool (Dharmacon).
Quantitative PCR
Total RNA was isolated from cells using an RNeasy mini kit (Qiagen) and treated with DNase I. cDNA was synthesized with a Taq-Man Reverse Transcription kit (Applied Biosystems). Quantitative PCR was performed with SYBR Green 2 × PCR master mix (Applied Biosystems) in an ABI Prism 7300 Real Time PCR system (Applied Biosystems). Melting temperature profiles of final products were used to ensure amplicon specificity. The relative fold change in expression of each mRNA was calculated using the ΔΔCt method relative to 18S rRNA or RPLP0. See Supplementary Table S8 for primer details.
Chromatin immunoprecipitation
ChIP analysis was performed as previously described (Beyer et al, 2008). Analysis of the samples was performed by qPCR as described above. The antibodies used for ChIP were anti-HIF-1α (Abcam, ab2185), anti-FoxO3A (Santa Cruz Biotechnology, sc11351), anti-Myc (Santa Cruz Biotechnology, sc-764), anti-HA (Santa Cruz Biotechnology, sc-805), and rabbit IgG (Sigma). See Supplementary Table S8 for primer details.
Luciferase reporter assays
Reporter assays were performed as described previously (Beyer et al, 2008). The mutated reporters Δ1, Δ2, and Δ1Δ2 were made by site-directed mutagenesis using the Quick Change kit (Stratagene) with the following primers: Δ1: 5′-CTCGAGGCGCGCGAGCTCACAGATCTGCACTCACACACGCACGCC-3′; Δ2: 5′-CTCGAGGCGCGCGAGCTCACACACGCGCACTCACATATGCCCGCC-3′; Δ1Δ2: 5′-CTCGAGGCGCGCGAGCTCACAGATCTGCACTCACATATGCCCGCC-3′. All constructs were verified by sequencing.
Supplementary Material
Acknowledgments
We thank Signe Breum for general technical assistance and Anna Fossum for assistance with flow cytometry. This study was supported by grants from The Danish Cancer Society and The Danish Cancer Research Foundation.
Author contributions: KSJ performed all biochemical and cell biological studies. TB and KTJ performed xenograft tumour studies in mice. TB and AK performed FDG uptake measurements and calculations. IT and BQ performed oxygen consumption measurements. RB performed the DNA microarray analysis. EN, HM, and GL performed the immunohistochemical stainings. CB created the FoxO3A(A3)-ER-expressing U2OS cell clones. PS conceived the hypothesis, advised on experiments, and supervised the project. KSJ and PS wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arden KC (2008) FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene 27: 2345–2350 [DOI] [PubMed] [Google Scholar]
- Bakker WJ, Harris IS, Mak TW (2007) FOXO3a is activated in response to hypoxic stress and inhibits HIF1-induced apoptosis via regulation of CITED2. Mol Cell 28: 941–953 [DOI] [PubMed] [Google Scholar]
- Bello-Fernandez C, Packham G, Cleveland JL (1993) The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci USA 90: 7804–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer S, Kristensen MM, Jensen KS, Johansen JV, Staller P (2008) The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem 283: 36542–36552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA (2007) FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev 21: 2775–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Marquardt J, Bras A, Medema RH, Eilers M (2004) Myc-induced proliferation and transformation require Akt-mediated phosphorylation of FoxO proteins. EMBO J 23: 2830–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Farnham PJ (1999) Coexamination of site-specific transcription factor binding and promoter activity in living cells. Mol Cell Biol 19: 8393–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Brunelle JK, Santore MT, Budinger GR, Tang Y, Barrett TA, Zong WX, Kandel E, Keith B, Simon MC, Thompson CB, Hay N, Chandel NS (2004) c-Myc sensitization to oxygen deprivation-induced cell death is dependent on Bax/Bak, but is independent of p53 and hypoxia-inducible factor-1. J Biol Chem 279: 4305–4312 [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME (2004) Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303: 2011–2015 [DOI] [PubMed] [Google Scholar]
- Chandramohan V, Mineva ND, Burke B, Jeay S, Wu M, Shen J, Yang W, Hann SR, Sonenshein GE (2008) c-Myc represses FOXO3a-mediated transcription of the gene encoding the p27(Kip1) cyclin dependent kinase inhibitor. J Cell Biochem 104: 2091–2106 [DOI] [PubMed] [Google Scholar]
- Chen J, Gomes AR, Monteiro LJ, Wong SY, Wu LH, Ng TT, Karadedou CT, Millour J, Ip YC, Cheung YN, Sunters A, Chan KY, Lam EW, Khoo US (2010) Constitutively nuclear FOXO3a localization predicts poor survival and promotes Akt phosphorylation in breast cancer. PLoS One 5: e12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cairns R, Papandreou I, Koong A, Denko NC (2009) Oxygen consumption can regulate the growth of tumors, a new perspective on the Warburg effect. PLoS One 4: e7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiribau CB, Cheng L, Cucoranu IC, Yu YS, Clempus RE, Sorescu D (2008) FOXO3A regulates peroxiredoxin III expression in human cardiac fibroblasts. J Biol Chem 283: 8211–8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czymai T, Viemann D, Sticht C, Molema G, Goebeler M, Schmidt M (2010) FOXO3 modulates endothelial gene expression and function by classical and alternative mechanisms. J Biol Chem 285: 10163–10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansen TB, Burgering BM (2008) Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol 18: 421–429 [DOI] [PubMed] [Google Scholar]
- Delpuech O, Griffiths B, East P, Essafi A, Lam EW, Burgering B, Downward J, Schulze A (2007) Induction of Mxi1-SR alpha by FOXO3a contributes to repression of Myc-dependent gene expression. Mol Cell Biol 27: 4917–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denko NC (2008) Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat Rev Cancer 8: 705–713 [DOI] [PubMed] [Google Scholar]
- Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4: P3. [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, Burgering BM (2004) FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J 23: 4802–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, Semenza GL (2007) HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129: 111–122 [DOI] [PubMed] [Google Scholar]
- Gan B, Lim C, Chu G, Hua S, Ding Z, Collins M, Hu J, Jiang S, Fletcher-Sananikone E, Zhuang L, Chang M, Zheng H, Wang YA, Kwiatkowski DJ, Kaelin WG Jr, Signoretti S, DePinho RA (2010) FoxOs enforce a progression checkpoint to constrain mTORC1-activated renal tumorigenesis. Cancer Cell 18: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershman B, Puig O, Hang L, Peitzsch RM, Tatar M, Garofalo RS (2007) High-resolution dynamics of the transcriptional response to nutrition in Drosophila: a key role for dFOXO. Physiol Genomics 29: 24–34 [DOI] [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC (2007) HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 11: 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greasley PJ, Bonnard C, Amati B (2000) Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res 28: 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helczynska K, Kronblad A, Jogi A, Nilsson E, Beckman S, Landberg G, Pahlman S (2003) Hypoxia promotes a dedifferentiated phenotype in ductal breast carcinoma in situ. Cancer Res 63: 1441–1444 [PubMed] [Google Scholar]
- Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC (2004) IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117: 225–237 [DOI] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG Jr (2001) HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292: 464–468 [DOI] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 12: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio PJ, Wilson WJ, O’Brien S, Makino Y, Poellinger L (1999) Regulation of the hypoxia-inducible transcription factor 1alpha by the ubiquitin-proteasome pathway. J Biol Chem 274: 6519–6525 [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW (2000) Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci USA 97: 10430–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee JH, Iyer VR (2008) Global identification of Myc target genes reveals its direct role in mitochondrial biogenesis and its E-box usage in vivo. PLoS One 3: e1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Tchernyshyov I, Semenza GL, Dang CV (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3: 177–185 [DOI] [PubMed] [Google Scholar]
- Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM (2002) Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 419: 316–321 [DOI] [PubMed] [Google Scholar]
- Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE (2004) HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. EMBO J 23: 1949–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress TR, Cannell IG, Brenkman AB, Samans B, Gaestel M, Roepman P, Burgering BM, Bushell M, Rosenwald A, Eilers M (2011) The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol Cell 41: 445–457 [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A (2006) A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 125: 987–1001 [DOI] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV (2005) Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol 25: 6225–6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabon ME, Scott BA, Crowder CM (2009) Divergent mechanisms controlling hypoxic sensitivity and lifespan by the DAF-2/insulin/IGF-receptor pathway. PLoS One 4: e7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ (1999) The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399: 271–275 [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, Chen C, Hosokawa K, Nakauchi H, Nakayama K, Nakayama KI, Harada M, Motoyama N, Suda T, Hirao A (2007) Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell 1: 101–112 [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D et al. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134: 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA (2007) FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 128: 309–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC (2006) HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3: 187–197 [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR (2002) A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2: 81–91 [DOI] [PubMed] [Google Scholar]
- Renault VM, Rafalski VA, Morgan AA, Salih DA, Brett JO, Webb AE, Villeda SA, Thekkat PU, Guillerey C, Denko NC, Palmer TD, Butte AJ, Brunet A (2009) FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5: 527–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, Eick D (2001) The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res 29: 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott BA, Avidan MS, Crowder CM (2002) Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science 296: 2388–2391 [DOI] [PubMed] [Google Scholar]
- Seagroves TN, Ryan HE, Lu H, Wouters BG, Knapp M, Thibault P, Laderoute K, Johnson RS (2001) Transcription factor HIF-1 is a necessary mediator of the pasteur effect in mammalian cells. Mol Cell Biol 21: 3436–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz P, Doppler H, Copland JA, Simpson KJ, Toker A (2009) FOXO3a promotes tumor cell invasion through the induction of matrix metalloproteinases. Mol Cell Biol 29: 4906–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Hietakangas V, Sayadian AC, Cohen SM (2008) Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab 7: 21–32 [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegue E, DePinho RA, Gilliland DG (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339 [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA 92: 5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei JS, Song YK, Durinck S, Chen QR, Cheuk AT, Tsang P, Zhang Q, Thiele CJ, Slack A, Shohet J, Khan J (2008) The MYCN oncogene is a direct target of miR-34a. Oncogene 27: 5204–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheaton WW, Chandel NS (2010) Hypoxia regulates cell metabolism. Am J Physiol Cell Physiol 300: C385–C393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O’Donnell KA, Dang CV (2003) An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 4: R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL (2008) Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 283: 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang CV, Semenza GL (2007) HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell 11: 407–420 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.