Abstract
The respiratory chain in the inner mitochondrial membrane contains three large multi-enzyme complexes that together establish the proton gradient for ATP synthesis, and assemble into a supercomplex. A 19-Å 3D map of the 1.7-MDa amphipol-solubilized supercomplex I1III2IV1 from bovine heart obtained by single-particle electron cryo-microscopy reveals an amphipol belt replacing the membrane lipid bilayer. A precise fit of the X-ray structures of complex I, the complex III dimer, and monomeric complex IV indicates distances of 13 nm between the ubiquinol-binding sites of complexes I and III, and of 10–11 nm between the cytochrome c binding sites of complexes III and IV. The arrangement of respiratory chain complexes suggests two possible pathways for efficient electron transfer through the supercomplex, of which the shorter branch through the complex III monomer proximal to complex I may be preferred.
Keywords: amphipols, electron cryo-microscopy, membrane protein complex, respiratory chain, single-particle averaging
Introduction
Mitochondria are the sites of oxidative phosphorylation and produce most of the ATP in animal cells. Their inner membrane contains the five large enzyme complexes of the respiratory chain, namely complex I (NADH dehydrogenase), complex II (succinate dehydrogenase), complex III (cytochrome c reductase/cytochrome bc1 complex), complex IV (cytochrome c oxidase), and complex V (mitochondrial F1Fo ATP synthase). Complexes I, III, and IV use the energy released in electron transfer reactions to pump protons out of the matrix across the inner membrane. The resulting proton gradient powers the synthesis of ATP by complex V (ATP synthase). Electrons are shuttled between the large electron transport complexes by the small, lipid-soluble electron carrier ubiquinol and by cytochrome c, a 12-kDa soluble electron carrier protein.
The mechanisms of the electron and proton transfer reactions in the individual complexes have been studied intensely for decades, and X-ray structures of the three mitochondrial proton pumps have been determined. The 2.8-Å resolution structure of dimeric complex IV from bovine heart mitochondria (Tsukihara et al, 1996) and a 6-Å map of complex I from Yarrowia lipolytica (Hunte et al, 2010) have been reported. Structures of the complex III dimer from chicken (Zhang et al, 1998), bovine heart (Iwata et al, 1998; Huang et al, 2005), and yeast without (Lange et al, 2001) and with (Solmaz and Hunte, 2008) bound cytochrome c at resolutions between 2.0 and 3.7 Å are available. Yet, little is known about how these complexes interact in the cristae membrane to perform their tasks in electron transfer and proton translocation.
Two different models have been proposed. The random collision model (Hackenbrock et al, 1986) states that each complex exists as individual entities that diffuse freely in the lipid bilayer. In this model, transfer of electrons occurs during random and transient collision events. By contrast, the solid-state model (Chance and Williams, 1955) assumes a higher level of organization, such that the electron transport complexes assemble into supercomplexes, where efficient transfer of electrons occurs along predefined pathways. The solid-state model gained support from the discovery of supercomplexes in bovine heart and yeast mitochondria by blue-native polyacrylamide gel electrophoresis (BN-PAGE) (Schägger and Pfeiffer, 2000). A first 3D map of negatively stained supercomplex B from bovine heart, composed of complex I, dimeric complex III and complex IV, provided initial insights into the arrangement of the three complexes in the assembly (Schäfer et al, 2007), but at the limited map resolution of 36 Å they could not be positioned precisely. Moreover, it was not certain to what extent the structure had been distorted by dehydration and shrinkage due to air-drying in negative stain.
Here, we report the 3D structure of mitochondrial supercomplex I1III2IV1, determined at 19 Å resolution by electron cryo-microscopy (cryo-EM). The 3D map of the amphipol-solubilized supercomplex shows a well-defined, unique arrangement of the three component complexes, and indicates the pathways along which the small electron carriers ubiquinone and cytochrome c travel to shuttle electrons between them.
Results
Isolation and purification of amphipol-solubilized mitochondrial supercomplexes
Mitochondria were isolated from bovine heart tissue by standard procedures (Krause et al, 2005) and supercomplexes were solubilized with digitonin at a high detergent-to-protein ratio of 28:1 (Schäfer et al, 2006). To increase the protein concentration for EM, the volume of the solubilized fraction was reduced by a factor of ∼3. This resulted in a monodisperse solution without significant aggregation, but the particles did not partition into the holes of holey carbon films for cryo-EM. To overcome this problem, the detergent was replaced by amphipol A8-35, a polyacrylate-based carbohydrate polymer with carboxylate, octylamine, and isopropylamine sidechains (Popot et al, 2011). The amphipathic polymer interacts with the hydrophobic surface areas of membrane proteins (Zoonens et al, 2005), keeping them soluble in the absence of detergent.
Amphipols have a stabilizing effect on membrane proteins even in detergent solution, and this effect increases significantly when the detergent is removed (Popot et al, 2011). Cyclodextrins can sequester certain detergents from solution (de Grip et al, 1998), and we found that γ-cyclodextrin removed digitonin from solubilized mitochondrial membranes. Figure 1A shows that a 1:1 molar ratio of γ-cyclodextrin to digitonin removed the detergent efficiently. Without amphipols, the protein aggregated and was precipitated by centrifugation. Addition of amphipol A8-35 to a final concentration of 0.2% (w/v) together with γ-cyclodextrin prevented aggregation and the amphipol-solubilized supercomplex remained in solution.
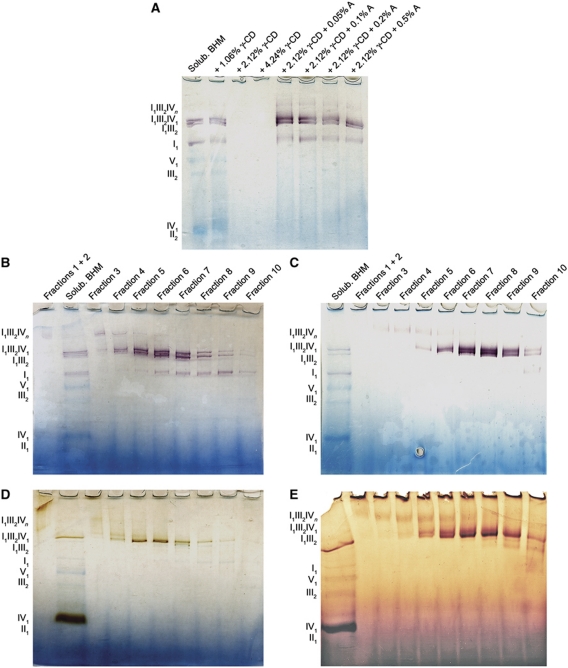
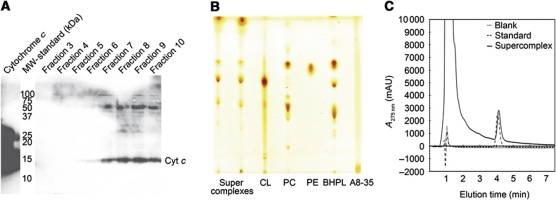
Figure 1.
Purification of active mitochondrial supercomplexes in amphipol A8-35. (A) Digitonin-solubilized mitochondria were incubated with amphipols (A) and γ-cyclodextrin (γ-CD) and the soluble fraction was analysed on 3–10% BN-PAGE. Proteins were stained with NBT for complex I activity. Supercomplexes solubilized with digitonin (B, D) or amphipols (C, E) were separated in linear density gradients. Fractions were collected from bottom to top and analysed by 3–10% BN-PAGE. NBT staining shows complex I activity (A, B); DAB staining shows complex IV activity (D, E). The blue colour is due to Coomassie stain from BN-PAGE.
Supercomplexes were separated by density gradient centrifugation according to a published procedure (Dudkina et al, 2005), with several modifications: digitonin was omitted from the gradient, and the buffer was changed to HEPES pH 7.7, to avoid aggregation of amphipols (Popot et al, 2011). As the apparent mass of the amphipol-solubilized supercomplexes on the gradient was lower than in digitonin, the maximum sugar concentration was reduced to 1.3 M. Protein complexes with molecular masses in the MDa range migrated to positions near the lower end of the gradient, as revealed by BN-PAGE (Figure 1). In-gel activity assays indicated complex I activity in a prominent double band at high molecular weight (Figure 1B and C). In addition, the upper band gave complex IV activity (Figure 1D and E), indicating that it contained supercomplex B (Schäfer et al, 2006). The same enzymatic activities were observed in solution. Overall, gradient purification in amphipols worked better than in digitonin, due to larger differences in apparent molecular mass, which resulted in improved separation in the density gradient.
EM and image processing
Fractions containing amphipol-solubilized supercomplex B were chosen for EM. Samples negatively stained with uranyl acetate indicated a high density of homogenous, well-preserved particles, of which ∼5600 were selected for initial single-particle processing. Class averages showed the characteristic triangular and F-shaped views of supercomplex B in negative stain (Schäfer et al, 2006) (Supplementary Figure S1A). Other class averages were round or oval. As a control, we analysed ∼4700 digitonin-solubilized supercomplexes, which were less uniform but also revealed a number of round or oval class averages (Supplementary Figure S1B). This indicated that the rounded shapes were not primarily due to the amphipols, but more likely to the gentler density gradient purification protocol, whereas samples for the previous negative-stain EM study had been prepared by BN-PAGE and electro elution (Schäfer et al, 2006). As we did not find significant differences between supercomplexes in amphipols and digitonin, we used amphipol-solubilized material exclusively for the 3D reconstruction, considering the generally higher stability of membrane proteins in amphipols, the better separation on density gradients, and the absence of detergents, which facilitates cryo-EM grid preparation.
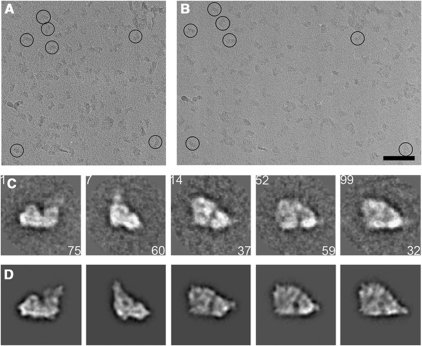
For cryo-EM, samples were vitrified in a thin layer of buffer on continuous carbon support films. This resulted in preferred particle orientations, which facilitated sorting of subpopulations, and enabled us to take advantage of random conical tilt (Radermacher et al, 1987). Two images of each area were recorded at tilt angles of 0° and −45° (Figure 2A and B). Five characteristic views out of the final set of 150 class averages from untilted images are shown in Figure 2C.
Figure 2.
Cryo-EM and single-particle analysis. Amphipol-solubilized supercomplexes in vitrified buffer on continuous carbon support film recorded at a tilt angle of −45° (A) or 0° (B). Scale bar, 100 nm. (C) 5 out of 150 selected class averages of untilted images filtered to 50 Å after the final round of multi-reference alignment and MSA classification (class number in upper left corner, number of particles in lower right corner). (D) Reprojections of the final 19-Å reconstruction corresponding to the class averages shown in (C).
A 3D map was generated and refined by projection matching to a resolution of 19 Å as determined by Fourier shell correlation at FSC=0.143 (Rosenthal and Henderson, 2003) (Supplementary Figure S2A). Angular space is evenly sampled, apart from preferred orientations of the supercomplexes on the carbon support film, which are more densely populated (Supplementary Figure S2B). Dimensions of the complex in the membrane plane were ∼28 nm by 24 nm. Side views of the volume were F-shaped, and top views were roughly triangular, as in negative stain (Schäfer et al, 2007). Reprojections of the final 3D map are compared with the corresponding class averages in Figure 2D.
Docking of X-ray structures
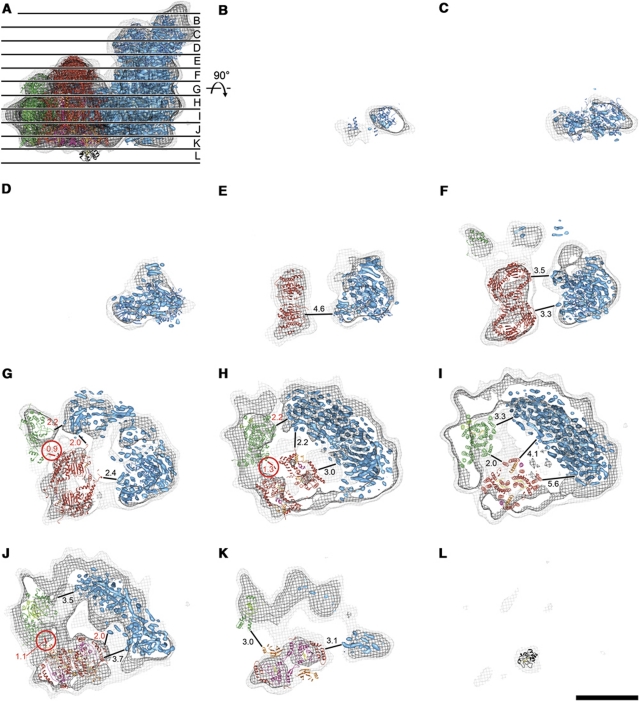
In the 3D map of the supercomplex, the hydrophilic regions of the three component complexes I, III, and IV were clearly resolved at contour levels of 1.3 σ or above, as shown by the map slices in Figure 3. Complex I was identified by the characteristic shape of its matrix arm, which extends 15 nm out of the membrane (Efremov et al, 2010; Hunte et al, 2010) (Figure 3A–G). Another broad 7-nm protrusion on this side of the complex with a central gap and approximate twofold symmetry had the shape and size of the matrix domains of the cytochrome c reductase dimer (Figure 3E–G). A smaller feature on the same side looked like the matrix domain of cytochrome c oxidase (Figure 3G). On the opposite surface of the complex, the volumes of the three complexes protruding into the intermembrane space were also well resolved (Figure 3J and K). By contrast, the transmembrane regions of the three complexes in the two slices between the two membrane surfaces were less well resolved (Figure 3H and I). We attribute this to the lower density of the membrane-embedded parts of the component complexes, which at this resolution would be at an intermediate value between that of soluble protein (1.36 g/ml; Kühlbrandt, 1982) and the hydrophobic membrane interior (estimated at ∼0.9 g/ml). The membrane regions thus scatter electrons less strongly than the extramembraneous regions, and therefore areas occupied by protein or lipid are less distinct in this part of the map.
Figure 3.
Slices through the X-ray structures fitted to the supercomplex map at two different contour levels. X-ray structures of complex I (blue), complex IV (green), complex III (red, with Rieske proteins in orange and c1 in purple), and cytochrome c (black) fitted to the cryo-EM map, drawn at 1.3 σ (light grey) and 4.3 σ (dark grey). (A) Side view along the membrane, showing the position of consecutive 2.2-nm slices in (B–L), starting from the matrix side. Shortest distances (in nm) between complexes are shown in red and distances <1.5 nm are marked with red circles. Scale bar, 10 nm.
The X-ray structures of the three individual respiratory chain complexes were docked manually into the 19-Å map (Figures 3 and 4B and C). As known structures can be fitted to 3D maps to within ∼10% of the map resolution (Rossmann, 2000), we estimate the positional accuracy of our fits to be about 2 Å. Using the gap between the matrix domains of the complex III dimer as a guide (Figure 4D), bovine complex III (PDB 1PP9; Huang et al, 2005) was fitted into the 7-nm protrusion. The twofold axis of the complex III dimer was oriented perpendicular to the membrane plane, and the fit was optimized by moving and rotating the structure along and around this axis. The peripheral subunit 11, which is easily lost during purification, was supplemented from the other structure of the bovine enzyme (PDB 1BGY; Iwata et al, 1998) to obtain a complete set of complex III subunits. This resulted in an excellent overall fit of the complex III structure, with the exception of the flexible Rieske-FeS protein domains (orange in Figures 3K and 7A).
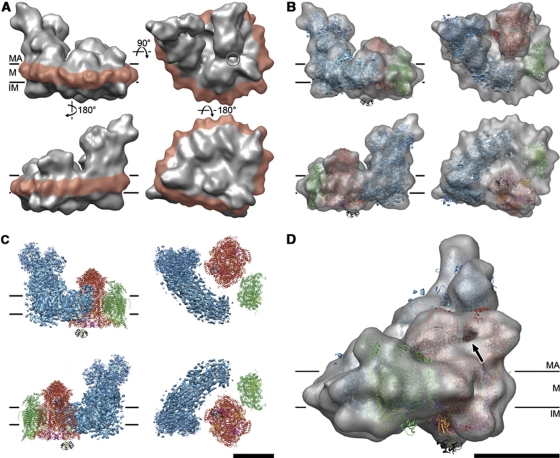
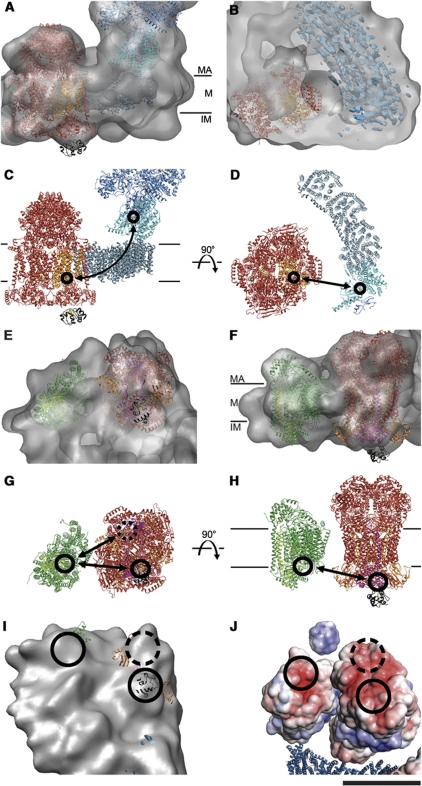
Figure 4.
3D map and fitted X-ray structures. (A) Cryo-EM 3D map as seen from two opposite sides (left), from the matrix (top right), and the intermembrane space (lower right). The amphipol belt is shown in red. The circle marks the gap between complex I and complex III. (B) X-ray structures of component complexes (blue, complex I; red, complex III; green, complex IV) and cytochrome c (black) fitted to the 3D cryo-EM map. (C) Docked X-ray structures without map. (D) Enlarged view with docked X-ray structures. Rieske domains are orange. The arrow points to the gap between the matrix domains of complex III used for positioning the X-ray structure. MA, matrix; M, membrane; IM, intermembrane space. Scale bars, 10 nm.
The fitted complex III dimer served as a starting point for positioning complexes I and IV in the map. The 6-Å map of complex I from Y. lipolytica (Hunte et al, 2010) (provided by Carola Hunte, Universität Freiburg) was fitted to the clear features of the matrix arm and on the intermembrane side in the supercomplex map. This resulted in a very good overall fit but left some peripheral map regions unoccupied (Figure 4B). Presumably, these regions contain protein that is present in the bovine complex, but not in Y. lipolytica. Indeed, the matrix arm in the 3D map of bovine complex I in negative stain (Clason et al, 2010) resembled our cryo-EM map closely (Supplementary Figure S3). By contrast, the higher-resolution X-ray structure of the matrix arm from Thermus thermophilus (PDB 3IAM; Sazanov and Hinchliffe, 2006), which is ∼50% smaller, occupied only about half this volume. The regions not occupied by bacterial complex I included the intermembrane protuberance 2 (IP2) on the intermembrane side of the matrix arm and the distal membrane protuberance (DMP) on the distal end of the membrane arm on the matrix side (Clason et al, 2010) (Figures 4B and 7B). Likewise, the bridge-like structure formed by domains 5 and 6 of the matrix arm and the proximal/central membrane protuberance (PMP/CMP) on the membrane arm (Clason et al, 2010) was present only in the eukaryotic enzyme.
To account for the different angle between matrix and membrane arm in the supercomplex as compared with T. thermophilus (PDB 3M9S; Efremov et al, 2010), the matrix arm from T. thermophilus (PDB 3IAM; Sazanov and Hinchliffe, 2006) and the membrane arm from Escherichia coli (PDB 3M9C; Efremov et al, 2010) were fitted separately. The transmembrane arms of the T. thermophilus and Y. lipolytica complex I were both similarly curved and fitted the supercomplex map well. The membrane arm of E. coli complex I left some of this map region unoccupied.
Complex IV (PDB 1OCC; Tsukihara et al, 1996) was positioned in the supercomplex density adjacent to the complex III dimer and the distal end of the membrane arm of complex I. This space accommodated one complex IV monomer, suggesting that the dimers seen in the X-ray structures might not be required for function. Subunits IV and Va were placed into a protruding density on the matrix side so that the concave side of the protein, which forms the dimer interface in 3D crystals, faced outwards (Figures 3 and 4B). The complex IV monomer in the cryo-EM map is rotated by ∼180° relative to its proposed orientation in the negative-stain reconstruction (Schäfer et al, 2007), where the concave dimer interface faced complex III.
The amphipol belt
The fit of the three respiratory chain complexes left an irregular, belt-shaped region of density around the perimeter of the complex unoccupied. This belt (red in Figure 4A) correlated closely with the hydrophobic membrane regions of complexes I, III, and IV and accordingly was assigned to the amphipols. The amphipol belt was on average ∼2 nm thick, and ∼4 nm wide, similar to a lipid bilayer (le Maire et al, 2000). It showed a number of ∼3-nm protrusions, spaced about 6–10 nm apart, mostly on the curved part of the supercomplex perimeter, suggesting that the amphipathic polymer was not distributed uniformly along the edge of the membrane region. Small-angle neutron scattering has indicated that amphipol A8-35 forms well-defined ∼40 kDa nanoparticles in solution (Gohon et al, 2006). The size of the protrusions suggests that the amphipol may locally form such nanoparticles in the belt. Comparison of supercomplex volumes generated from two equal halves of the data set (Supplementary Figure S4) indicates that the particles are a distinct map feature, implying that they attach to, or form at, the same positions in each supercomplex.
Protein–protein contacts
There were only a few sites where neighbouring complexes came to within ∼1 nm between α-carbons, close enough for ion bridges or hydrogen bonds. Three such sites of potentially strong protein–protein interaction were found between complexes III and IV, two of them on the matrix side, about 2 nm above the membrane surface or at the level of the lipid head groups (Figure 3G and H), and one in the lipid boundary region on the intermembrane side (Figure 3J). The shortest distances between complex I and complexes III and IV appeared longer (∼2 nm) due to the lack of atomic detail in the complex I map but were found at roughly the same levels. No protein–protein contacts were observed within the hydrophobic membrane core (Figure 3I). The hydrophilic protein–protein contacts thus may act as spacers, keeping the complexes in the membrane 2–5 nm apart.
The supercomplex contains cytochrome c, ubiquinol, and cardiolipin
After fitting the structures of the three major membrane protein complexes, further inspection of the 3D map revealed a density on the exterior surface of complex III, below one of its two cytochrome c binding sites. The X-ray structure of the yeast complex III dimer with one bound cytochrome c fitted well to the map of the bovine supercomplex, such that cytochrome c occupied this density and only protruded slightly beyond the map surface (Figures 3L and 4D). This suggested an at least partial occupancy of one of the two cytochrome c binding sites in the supercomplex. The other cytochrome c binding site in the complex III dimer appeared to be empty, as in the X-ray structure (Solmaz and Hunte, 2008). Indeed, analysis of the isolated supercomplex by SDS–PAGE and western blot (Figure 5A) revealed cytochrome c in fractions containing most of the supercomplex I1III2IV1, indicating that it remained bound throughout the purification procedure, and was consequently present in the supercomplex imaged by cryo-EM.
Figure 5.
The supercomplex contains cytochrome c, ubiquinol, and cardiolipin. (A) Western blot showing the presence of cytochrome c in the supercomplex. Amphipol-solubilized supercomplex was purified by density gradient centrifugation and fractions were analysed by 15% SDS–PAGE and western blot with an anti-cytochrome c antibody. Cytochrome c runs at ∼15 kDa. (B) Lipid extracts from two different supercomplex preparations (lanes 1 and 2) and purified lipid standards (lanes 3–5). The supercomplex contains phosphatidyl choline (PC), phosphatidyl ethanolamine (PE), and cardiolipin (CL), which is enriched compared with bovine heart polar lipid extract (BHPL, lane 6). (C) Ubiquinol was quantified by HPLC and comparison to Q10 standards. Each supercomplex contains at least 1 molecule of ubiquinol.
Thin-layer chromatography of an organic extract of gradient purified material revealed significant amounts of bound phospholipids. Cardiolipin was enriched in the supercomplex, compared with bovine heart total lipid (Figure 5B). HPLC of this extract indicated that each supercomplex contained at least one molecule of ubiquinol (Figure 5C).
Position and orientation of substrate binding sites
The ubiquinol-binding site of complex I is located in a pocket formed by the PSST and 49-kDa subunits at the hinge between the matrix and the membrane arms, close to the first FeS-cluster ∼20–25 Å above the membrane surface (Efremov et al, 2010) (Figure 6). The complex III dimer has two ubiquinol-binding sites per monomer in the transmembrane region of cytochrome b (Huang et al, 2005). The site near the matrix side is thought to participate mainly in the Q cycle, while the other is thought to bind reduced ubiquinol (Trumpower, 1990). One of the complex III monomers faces the lipid bilayer, while the other is surrounded by complex I. The shortest distance between the site binding reduced ubiquinol and the site of ubiquinol reduction on complex I was 11.6 nm. Given that this shortest connection would run partly through the aqueous medium, a ∼13-nm trajectory through the membrane as shown in Figure 6C is more likely.
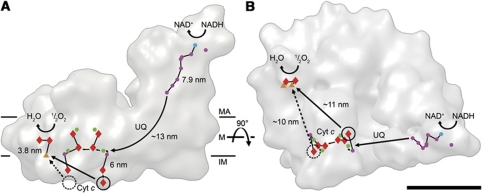
Figure 6.
Substrate binding sites. X-ray structures of complex I (blue ribbon model and electron density as in Figure 3 and 4), bovine complex III (red), IV (green), and cytochrome c (black) docked into the supercomplex map. Ubiquinol-binding sites are located between the 49-kDa and the PSST subunits near the first FeS-cluster above the membrane in complex I and the cytochrome b subunit in complex III (orange). Views from the membrane (A, C) and from the matrix side (B, D) with and without map. Cytochrome c binding sites are located on the intermembrane side below the c1 subunit of complex III (purple) and near two Cu atoms (circle) in subunit II of complex IV (light green). View from the intermembrane space (E, G) and side views (F, H), with and without map. Cytochrome c binding sites are circled and the shortest cytochrome c trajectories are marked with arrows. Dashed circles mark the unoccupied distal cytochrome c binding site. Surface of the supercomplex (I) and surface charge distribution (J) on the intermembrane side of complexes III and IV (red, negative; blue, positive; white, neutral). For comparison, the positively charged surface of cytochrome c is shown above. The blue ribbon diagram shows part of the complex I membrane arm for orientation. MA, matrix; M, membrane; IM, intermembrane space. Scale bar, 10 nm.
The two cytochrome c binding sites on the complex III dimer are located on the c1 subunits in the intermembrane space (Solmaz and Hunte, 2008) (Figure 6). In the supercomplex, one of these sites is occupied (Figures 3L and 6A and I). The presumed cytochrome c binding site on complex IV is on the exterior membrane surface near the two Cu atoms coordinated by subunit II (Tsukihara et al, 1996). The distances from this site to the two sites on complex III are ∼10 nm and ∼11 nm, respectively. The cytochrome c binding site on complex IV is a shallow cavity lined by negative charges, facing the similarly shaped binding sites on complex III, which are also predominantly negatively charged (Figure 6J). This arrangement would make it easy for the small, globular, partly positively charged cytochrome c to pass from one complex to the other like a ball between two cupped hands.
Discussion
Structure determination of membrane protein supercomplexes is a major challenge. With the small quantities that can typically be isolated, 2D or 3D crystallization is not an option. We therefore determined the structure of mitochondrial supercomplex I1III2IV1 by single-particle cryo-EM, using samples kept in solution by amphipols instead of detergent.
Amphipol-solubilized supercomplexes
Amphipols offer a number of advantages for membrane protein studies. As polymeric amphiphiles, their critical micellar concentration is negligible, and their potential to denature sensitive membrane proteins is minimal (Popot et al, 2011). Amphipols bind tightly to membrane proteins, covering hydrophobic surface areas in a 1.5–2.0 nm layer (Zoonens et al, 2005; Gohon et al, 2008), similar to detergents (le Maire et al, 2000). They are able to replace detergents completely, while stabilizing the membrane proteins and keeping them active and in solution so that they behave like soluble proteins (Popot et al, 2011). Amphipols may restrict large-scale movements of transmembrane helices, for example in the membrane arm of complex I, and lock the assembly in a unique conformation, which would be an advantage for single-particle cryo-EM studies. At the same time, they replace the detergent, which often creates problems for specimen preparation, even at low concentration. An earlier study of complex I (Flötenmeyer et al, 2007) had shown that amphipols are suitable for cryo-EM work of membrane protein complexes.
The density gradients indicate that the supercomplexes bind less amphipols than digitonin, and indeed the structural features of the amphipol-solubilized complexes were clear and uniform in negative stain (Supplementary Figure S1A). We decided to prepare the supercomplexes on a thin continuous carbon support film. This is not often done in single-particle cryo-EM, because the ice thickness is more difficult to control than on holey carbon film, and the support film adds background noise. On the other hand, the preferential orientations of the complexes on the support film help in particle selection and classification, and it is then also easier to use the random conical tilt method. This approach has the additional advantage that angular relationships between the particle images are known, providing important constraints in the initial rounds of reconstruction.
Comparison to the 3D map in negative stain
The volume of our map at a contour level of 2.5 σ, which just covers the hydrophilic protein domains, is 3.4 × 106 Å3. The combined protein mass of the bovine heart respiratory chain complexes in the supercomplex is almost exactly 1700 kDa, corresponding to a volume of ∼2.1 × 106 Å3. Therefore, the protein accounts for about 60% of the map volume at this contour level, with lipids and amphipols taking up the rest. By comparison, the volume of the supercomplex in negative stain (Schäfer et al, 2007) was given as representing a mass of 1700 kDa. Accordingly, the volume of the negative-stain map is about 2.1 × 106 Å3, or roughly 60% of the cryo-EM map. The fact that the volume of the negative-stain map coincides with that of the protein in the cryo-EM map must be an accidental result of shrinkage and air-drying, which happened to cancel out the volume contributions of the lipid and detergent.
Lipid content
With only a few points of direct contact and average distances of >2 nm between complexes I, III, and IV (Figure 3), it is likely that the supercomplex is held together at least partly by lipid–protein interactions. From 2D crystals of bacteriorhodopsin (Grigorieff et al, 1995) and aquaporin (Hite et al, 2010), it is known that lipids can mediate strong interactions between membrane proteins. Likewise, at least some of the subunit contacts in the photosystem I structure (Ben-Shem et al, 2003), which are not crystal contacts, must be lipid mediated. It is likely that structural lipid also plays a major role in the mitochondrial supercomplexes. This is consistent with the observation that supercomplexes can be isolated in amounts required for structural and functional studies only after solubilization with the very mild detergent digitonin (Schägger and Pfeiffer, 2000). Presumably, other, more aggressive detergents would remove the structural lipid and thus cause the supercomplexes to dissociate.
If we assume that the respiratory chain supercomplex binds 0.11 g amphipol per gram protein, as observed with complex III (Popot et al, 2011), the total amount of bound amphipol would be 190 kDa. With an amphipol density of 0.74 Da/Å3 (Gohon et al, 2004), this corresponds to almost 10% of the total volume a contour level of 2.5 σ. The remaining 30% of the volume (about 106 Å3) is most likely occupied by lipids. Assuming a lipid density of ∼0.66 Da/Å3, the complex would contain 660 kDa of lipid. The average mass of a membrane lipid is ∼750 Da, or 1500 Da for cardiolipin, so that the supercomplex could contain up to 880 lipid molecules with two fatty acid tails, or up to 440 molecules of cardiolipin. Obviously, this number is strongly dependent on the σ level chosen for calculating the map volume. For example, at a contour level of 3.3 σ, which still encompasses most of the protein density, the map volume is 2.85 × 106 Å3. The protein and amphipol contributions would be unchanged, but the lipid volume would be less than half, accommodating roughly 400 or 200 lipid or cardiolipin molecules, respectively. The total mass for the amphipol-solubilized lipid–protein supercomplex would then be in the range of 2200–2500 kDa.
Our biochemical analysis indicated that the supercomplex is enriched in cardiolipin (Figure 5B). It is known that bovine complexes III and IV depend on cardiolipin for catalytic activity (Gomez Jr and Robinson, 1999; Sedlak and Robinson, 1999), and supercomplex III2IV2 of Saccharomyces cerevisiae requires this lipid for stability (Pfeiffer et al, 2003). Patients suffering from Barth syndrome, who are deficient in cardiolipin synthesis, have unstable respiratory chain supercomplexes (McKenzie et al, 2006). A gap filled with membrane lipid, such as cardiolipin, would facilitate the diffusion of ubiquinol between complex I and III. Assuming a surface area per average lipid of ∼70 Å2 (Demel and De Kruyff, 1976), we estimate that roughly 300 lipid molecules would fit between complexes I, III, and IV. Presumably, the remaining lipid is trapped by the amphipol (Gohon et al, 2008) in an annulus around the perimeter of the supercomplex.
An interesting consequence of the high lipid content of the supercomplex is the lower map density in the membrane region at 19 Å resolution (Figure 3H and I). The same effect is apparent in the cryo-EM maps of a V-type ATPase (Muench et al, 2009), and of the photosystem II supercomplex (Nield et al, 2000) at similar resolution.
Conformational changes
The fit of the X-ray structures to the supercomplex map revealed several rearrangements of surface loops or short helices in all three component complexes (Figure 7). Many of these conformational changes are likely due to crystal contacts, but some of them may be functionally relevant.
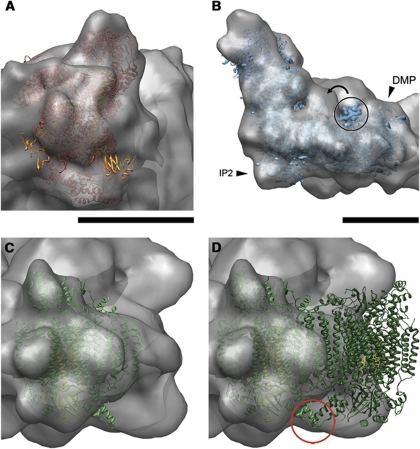
Figure 7.
Conformational changes. (A) Both Rieske domains (orange) of bovine complex III protrude from the cryo-EM map. (B) View with fitted complex I (ribbon model of bacterial complexes and 6-Å electron density of Y. lipolytica 6-complex I). Arrowheads point to the DMP and IP2 domains, which are absent in the bacterial complex. The circle marks the PMP, which is also absent in the bacterial complex and appears to have moved from its position in the X-ray structure (curved arrow). Fit of bovine complex IV monomer (C) and dimer (D). A helix of subunit VIb, which forms part of the dimer interface in the crystal structure, protrudes from the cryo-EM map (red circle). Scale bars, 10 nm.
In complex III, the main differences between the X-ray structure and the supercomplex were found on the intermembrane side at the two Rieske-FeS domains, which both appear to protrude from the cryo-EM map (Figure 7A). This is consistent with the known flexibility of the Rieske proteins, which have to change position and orientation in order to transfer electrons from the ubiquinol at cytochrome b to the heme in the c1 subunit (Zhang et al, 1998). The fitted structure is the stigmatellin-bound form, but the Rieske domains also protruded from the map when the uninhibited form was used instead (not shown). The cryo-EM map is thus an average of several different conformations, in which the density of the Rieske proteins is spread over a larger volume, and therefore they appear to stick out of the map.
In the supercomplex, the PMP/CMP domain of the Y. lipolytica map projects from the cryo-EM map, indicating a different position compared with the X-ray structure (Figure 7B). It will be interesting to see whether this conspicuous domain movement is functionally relevant. The bridge-like connection formed by the complex I domains 5/6 on the matrix arm and the PMP/CMP on the membrane arm appears thicker in the supercomplex than in the isolated complex I (Clason et al, 2010) (Figure 7B; Supplementary Figure S3). It has been suggested that conformational changes of a horizontal helix in the membrane arm of complex I might, by a yet unknown mechanism, couple electron transfer and proton translocation (Efremov et al, 2010). Clason et al (2010) had speculated that the connection between domains 5/6 and PMP/CMP, even though not essential for functional coupling, could explain kinetic differences between the enzymes from Bos taurus and Y. lipolytica. The more robust connection between domains 5/6 and PMP/CMP in the bovine supercomplex may indicate a particularly tight coupling between matrix and membrane arm and thus the two functions accomplished by them in the supercomplex. Likewise, the structure of the matrix arm appears to be more rigid compared with complex I in isolation, because extensive subclassification to account for different angular orientations of the matrix arm, as in negatively stained complex I (Radermacher et al, 2006), was not necessary. This may be an effect of the amphipol locking the supercomplex in a unique conformation.
Another clear difference in conformation was found on the outward-facing surface of complex IV, where a helix of subunit VIb sticks out of the supercomplex density (Figure 7C) but is readily accommodated in an adjacent unoccupied, roughly equal map volume. This helix is involved in dimer contacts in the crystal structures of cytochrome c oxidase (Tsukihara et al, 1996) (Figure 7D), which all show a dimer, whereas in the single-particle EM maps the oxidase is clearly monomeric. The dimer interface is oriented towards the exterior and would thus be available for interaction with other membrane proteins, or with other copies of complex IV. Larger assemblies with up to four copies of complex IV have been found by gel electrophoresis (Schägger and Pfeiffer, 2000).
Electron transfer pathway
The fit of the three component complexes I, III2, and IV allows us to draw conclusions on their functional interaction in the supercomplex. Biochemical analysis of supercomplexes from mouse mitochondria has shown that they contain both ubiquinol and cytochrome c (Acin-Perez et al, 2008). Indeed, both are present in our supercomplex (Figure 5A and C) and one cytochrome c is found attached to its binding site on the complex III monomer next to complex I.
In the supercomplex, the binding sites for the mobile electron carriers are in close proximity and face each other. Of the two complex III monomers, only one is close to the membrane arm of complex I (Figure 3H) and its ubiquinol-binding sites are well placed for efficient electron transfer over the short distance of ∼13 nm (Figure 6C and D). Interestingly, the complex III monomer proximal to complex I is the one that binds cytochrome c (Figures 3L and 6A). It has been shown that in the complex III dimer only one of the two sites is active at a given time (Castellani et al, 2010). The supercomplex structure thus suggests that the proximal complex III monomer may be more active in ubiquinol oxidation, while the distal monomer may be needed to transfer the electron to cytochrome c via its flexible Rieske domain.
Overall, the arrangement of the three component complexes, the short distances between binding sites for the small mobile electron carriers ubiquinol and cytochrome c and the mutual orientation of these binding sites suggest that in the supercomplex, electrons are channelled over short distances along predefined pathways, as postulated by the solid-state model for the mitochondrial electron transfer chain (Chance and Williams, 1955). The binding sites for the soluble electron carrier, cytochrome c, on complexes III and IV are only ∼10 or ∼11 nm apart. Cytochrome c has a diameter of 3.4 nm, which means that in passing from one complex to the other in the supercomplex, it needs to cross a gap of only ∼3 times its size (Figure 6G and J). The protein domains protruding into the intermembrane space might limit the lateral diffusion of cytochrome c along the membrane surface (Figure 6I). Additionally, negatively charged cardiolipin head groups might help to attract cytochrome c to the supercomplex.
The 19-Å 3D cryo-EM map shows how electrons may be shuttled along these pathways over a distance of about 40 nm from their entry site in the matrix arm of complex I to the site where they are finally transferred to molecular oxygen in complex IV (Figure 8). For about half of this distance, the electrons travel through the respiratory chain complexes. For the remaining distance, they are carried by ubiquinol in the membrane or by cytochrome c in the intermembrane space. Although the distance over which the electron is carried by cytochrome c from the proximal binding site to complex IV is longer by ∼1 nm compared with the distal site (dashed in Figure 8), the overall distance from NADH to O2 along this pathway is shorter and involves fewer electron-transfer steps. Therefore the shorter, proximal branch of the complex III dimer may be preferred for electron transport. The supercomplex is stable in detergent and has a well-defined structure, which is likely to be the same as in the mitochondrial membrane. The cryo-EM map of mitochondrial supercomplex I1III2IV1 thus indicates that the three electron transport complexes of the mitochondrial respiratory chain, just like the mitochondrial ATP synthase (Strauss et al, 2008; Davies et al, 2011), are organized into macromolecular assemblies in the mitochondrial inner membrane for efficient energy conversion.
Figure 8.
Electron transfer pathways in the supercomplex. Outline of the supercomplex with cofactors active in electron transport marked in blue (FMN), purple (iron–sulphur-clusters), green (quinols/stigmatellins), red (hemes), and orange (copper atoms), seen from the membrane (A) and from the matrix (B). Electron trajectories are marked in black. The dashed circle marks the distal cytochrome c binding site, which is unoccupied in the supercomplex. Straight arrows in A indicate the shortest distances from the cytochrome c binding sites on complex III to the site of cytochrome c oxidation in complex IV. The shorter, proximal branch may be preferred for electron transport. MA, matrix; M, membrane; IM, intermembrane space; UQ, ubiquinol; Cyt c, cytochrome c. Scale bar, 10 nm.
Materials and methods
Isolation and purification of supercomplexes from bovine heart mitochondria
Bovine heart mitochondria were prepared by differential centrifugation as described (Krause et al, 2005). Mitochondria were solubilized with 1% (w/v) digitonin (Calbiochem, high purity) at a detergent-to-protein ratio of 28:1 (Schäfer et al, 2006). The volume of the solubilized fraction was reduced in concentration devices with a 100-kDa cutoff (Millipore) by a factor of ∼2.8 before purification. For detergent exchange, amphipol A8-35 was added together with γ-cyclodextrin (Fluka) and incubated at 4 °C for 30 min. Precipitated material was removed by centrifugation at 21.500 g for 10 min at 4 °C in a tabletop centrifuge. Linear sugar gradients in 10 mM KCl, 15 mM HEPES, pH 7.7 were prepared on a BioComp Gradient Master (Coombs and Watts, 1985). In all, 2 ml of solubilized mitochondria (∼2 mg mitochondrial protein) was separated by ultracentrifugation at 150 000 g for 21 h at 4 °C in a SW40 rotor (Beckman) according to a protocol modified from Dudkina et al (2005). Gradients were fractionated from bottom to top and investigated for protein content by BN-PAGE in linear gradient gels with 3–10% polyacrylamide (Wittig et al, 2006).
Enzymatic analysis
Functionally active supercomplexes were detected by in-gel assays for activity of NADH dehydrogenase and cytochrome c oxidase (Kuonen et al, 1986; Grandier-Vazeille and Guerin, 1996). Briefly, for complex I, gels were incubated in buffer containing 100 mM Tris, pH 7.4, 768 mM glycine, 0.4% (w/v) NBT (p-nitrotetraazolium blue), and 100 μM β-NADH for 10–15 min. For complex III, gels were incubated for 12 h in 50 mM NaH2PO4, pH 7.4, 219 mM sucrose, 0.5 mg/ml DAB (3,3′diaminobenzidine tetrahydrochloride), 0.5 mg/ml equine heart cytochrome c, and 20 U/ml bovine liver catalase.
SDS–PAGE and western blot
Proteins were separated in 15% SDS–PAGE (Laemmli, 1970) and proteins were transferred to PVDF membranes (Millipore) (Towbin et al, 1979). After blocking the membranes with 5% non-fat dry milk in Tris-buffered saline (10 mM Tris, 150 mM NaCl, 1 mM NaN3, pH 7.5), proteins were probed with a primary antibody directed against cytochrome c (Invitrogen #456100) and a secondary antibody coupled with horseradish peroxidase. After each antibody the membrane was washed with TBS. The chemiluminescence reaction of the antibody-conjugated horseradish peroxidase to detect cytochrome c was started by transferring the membrane into SuperSignal West Pico 1:1 peroxide and luminal solution (Thermo Scientific) and the signal was detected on Amersham Hyperfilm ECL (GE Healthcare).
Thin-layer chromatography
Lipids were extracted from 100 μg of purified supercomplex by adding 7.5 volumes of methanol–chloroform (2:1), followed by vigorous shaking for 2 min. After 10 min incubation at room temperature, another 2.5 volumes of chloroform were added and shaken for 30 s, followed by 2.5 volumes of H2O and shaking for 30 s. After centrifugation at 20 000 g in a tabletop centrifuge, the organic phase was recovered and dried under a stream of nitrogen. The dried extract was dissolved in chloroform and applied to a silica gel 60 F 254 precoated HPTLC plate (Merck). 20 μg of purified lipid standards (Avanti Polar Lipids) was loaded for comparison. The plates were developed in chloroform/methanol/water (65/25/4) in a saturated atmosphere and lipids were stained with iodine vapour.
HPLC analysis
The dried organic extract was dissolved in 60 μl ethanol and 20 μl were loaded on a C30 column (3 μm, 100 × 2.1 mm2, YMC). The sample was eluted under isocratic conditions with diisopropyl ether/methanol (1/4, v/v) at 0.5 ml/min. Absorption was monitored at 275 nm and compared with Q10 standards.
Electron microscopy
For EM, samples were centrifuged through ZEBA desalt columns (Thermo Scientific) to remove the sugar. For initial analysis, samples were adsorbed for 30 s onto glow-discharged 400 mesh copper grids coated with Formvar and carbon and negatively stained with 1% (w/v) uranyl acetate (pH ∼4). Film images were recorded in a FEI Tecnai G2 Spirit electron microscope at 120 kV under low-dose conditions at a calibrated magnification of × 42 600. For cryo-EM, 3 μl samples of a 0.1–0.3 mg/ml solution of amphipol-solubilized supercomplex were adsorbed for 30 s onto glow-discharged 400 mesh copper grids coated with a continuous carbon film and plunge-frozen in liquid ethane after blotting excess liquid. Electron micrographs were collected on a FEI F30 Polara at 200 kV under low-dose conditions at a calibrated magnification of × 58 800 on Kodak SO-163 electron image film. For random conical tilt, two images of the same area were taken, first at −45° or −50° and then at 0° tilt. The nominal defocus ranged from ∼500 to ∼3000 nm on the tilt axis. Some images were recorded at ∼6000 nm defocus. Negatives were developed according to the standard protocol and digitized on a SCAI PhotoScan scanner (Z/I Imaging, Aalen, Germany) on an 8-bit scale at a pixel size of 7 μm, corresponding to 1.19 Å on the specimen. Adjacent pixels were subsequently averaged to 2.38 Å/pixel in IMAGIC5.
Data processing
Particles from negatively stained samples were picked in 128 pixel boxes with Boxer from the EMAN1 software package (Ludtke et al, 1999) and analysed with IMAGIC5 (van Heel et al, 1996) (Image Science Software GmbH).
After binning, digitized cryo-EM images were converted from RAW format, normalized, and inverted in SPIDER (version 18.03) (Frank et al, 1996). CTF parameters were determined with CTFTILT 1.4 (Mindell and Grigorieff, 2003). Around 10 700 tilt pairs were picked in 320 pixel boxes in JWEB and imported into SPIDER. Effects of the CTF were corrected on the individual particles by inverting the phases (Penczek et al, 1997), taking tilt and astigmatism into account. Images were Fermi filtered and binned to 4.76 Å/pixel and the box size reduced to 112 pixels in SPIDER. The untilted images were subjected to single-particle analysis. For an optimal result, reference-free alignment and MSA with correspondence analysis and hierarchical ascendant classification in the first round was followed by three rounds of multi-reference alignment in IMAGIC. After this, 16 characteristic views were selected as references for alignment and MSA classification of the original particles in SPIDER. The particles were subjected to a final round of alignment to eight references in SPIDER and MSA with hierarchical ascendant classification into 150 classes in IMAGIC. Further image processing was performed with scripts from the SPIDER installation package and SPIDER website (http://www.wadsworth.org/spider_doc/spider/docs/spider.html) that were adapted and combined according to requirements. Class volumes were reconstructed by random conical tilt (Radermacher et al, 1987), applying the tilt parameters of each pair, but using both tilted and untilted views to improve the quality of the volumes and reduce the effect of the missing cone. Class volumes were refined by projection matching and subsequently classes with similar class volumes were merged. To avoid merging of wrong classes, mainly similar classes differing only slightly in their orientations were aligned to each other and merged into a new reconstruction. Eventually, a volume with clear structural features for the matrix arm of complex I with its subdomains, and the matrix domains of complex III, was chosen as the initial model. This initial model was refined by projection matching of the whole data set with ∼21 400 projections and the final reconstruction was calculated with SIRT (Penczek et al, 1992) omitting 30% of the particles with the lowest cross-correlation coefficient. The resolution of the final map was determined by Fourier shell correlation as 19 Å at FSC=0.143 (Rosenthal and Henderson, 2003) (Supplementary Figure S1A). To agree with the handedness of the negative-stain model (Schäfer et al, 2007) and the recently published X-ray structures of the NADH dehydrogenase (Efremov et al, 2010; Hunte et al, 2010), the final volume was mirrored on the y axis.
Map visualization and analysis
3D volumes were visualized in UCSF Chimera (Pettersen et al, 2004) and the X-ray structures of the individual complexes were placed manually according to distinct structural features. Surface potentials were calculated with the APBS (Baker et al, 2001) plugin (MG Lerner and HA Carlson, APBS plugin for PyMOL, University of Michigan, Ann Arbor (2006)) in PyMOL Molecular Graphics System (Version 1.3, Schrödinger, LLC).
Supplementary Material
Acknowledgments
We gratefully acknowledge the advice of Sean Connell and Janet Vonck on image processing. We thank Florent Rouvière and Fabrice Giusti for synthesizing amphipol A8-35 batches FRH4 and FGH20, respectively. This research was partly supported by the Cluster of Excellence ‘Macromolecular Complexes’, Frankfurt. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH Grant P41 RR-01081).
The EM map has been deposited in the EM data bank (http://www.emdatabank.org) under the entry code EMD-1876. The fitted model with bovine complex III and IV and bacterial complex I has been deposited in the Protein Data Bank (http://www.pdb.org) under the accession number 2YBB.
Author contributions: WK initiated the study. TA designed and performed the research. DJM helped with cryo-EM. J-LP provided amphipol A8-35 and contributed to the discussion. TA and WK analysed the data and wrote the paper.
Footnotes
J-LP holds patents on amphipols and their use. All other authors declare that they have no conflict of interest.
References
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA (2008) Respiratory active mitochondrial supercomplexes. Mol Cell 32: 529–539 [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426: 630–635 [DOI] [PubMed] [Google Scholar]
- Castellani M, Covian R, Kleinschroth T, Anderka O, Ludwig B, Trumpower BL (2010) Direct demonstration of half-of-the-sites reactivity in the dimeric cytochrome bc1 complex: enzyme with one inactive monomer is fully active but unable to activate the second ubiquinol oxidation site in response to ligand binding at the ubiquinone reduction site. J Biol Chem 285: 502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B, Williams GR (1955) Respiratory enzymes in oxidative phosphorylation. III. The steady state. J Biol Chem 217: 409–427 [PubMed] [Google Scholar]
- Clason T, Ruiz T, Schägger H, Peng G, Zickermann V, Brandt U, Michel H, Radermacher M (2010) The structure of eukaryotic and prokaryotic complex I. J Struct Biol 169: 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs DH, Watts NR (1985) Generating sucrose gradients in three minutes by tilted tube rotation. Anal Biochem 148: 254–259 [DOI] [PubMed] [Google Scholar]
- Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kühlbrandt W (2011) Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA 108: 14121–14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Grip WJ, Van Oostrum J, Bovee-Geurts PH (1998) Selective detergent-extraction from mixed detergent/lipid/protein micelles, using cyclodextrin inclusion compounds: a novel generic approach for the preparation of proteoliposomes. Biochem J 330 (Part 2): 667–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demel RA, De Kruyff B (1976) The function of sterols in membranes. Biochim Biophys Acta 457: 109–132 [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Eubel H, Keegstra W, Boekema EJ, Braun HP (2005) Structure of a mitochondrial supercomplex formed by respiratory-chain complexes I and III. Proc Natl Acad Sci USA 102: 3225–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremov RG, Baradaran R, Sazanov LA (2010) The architecture of respiratory complex I. Nature 465: 441–445 [DOI] [PubMed] [Google Scholar]
- Flötenmeyer M, Weiss H, Tribet C, Popot JL, Leonard K (2007) The use of amphipatic polymers for cryo-electron microscopy of NADH:ubiquinone oxidoreductase (complex I). J Microsc 227: 229–235 [DOI] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Gohon Y, Dahmane T, Ruigrok RW, Schuck P, Charvolin D, Rappaport F, Timmins P, Engelman DM, Tribet C, Popot JL, Ebel C (2008) Bacteriorhodopsin/amphipol complexes: structural and functional properties. Biophys J 94: 3523–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohon Y, Giusti F, Prata C, Charvolin D, Timmins P, Ebel C, Tribet C, Popot JL (2006) Well-defined nanoparticles formed by hydrophobic assembly of a short and polydisperse random terpolymer, amphipol A8-35. Langmuir 22: 1281–1290 [DOI] [PubMed] [Google Scholar]
- Gohon Y, Pavlov G, Timmins P, Tribet C, Popot JL, Ebel C (2004) Partial specific volume and solvent interactions of amphipol A8-35. Anal Biochem 334: 318–334 [DOI] [PubMed] [Google Scholar]
- Gomez B Jr, Robinson NC (1999) Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry 38: 9031–9038 [DOI] [PubMed] [Google Scholar]
- Grandier-Vazeille X, Guerin M (1996) Separation by blue native and colorless native polyacrylamide gel electrophoresis of the oxidative phosphorylation complexes of yeast mitochondria solubilized by different detergents: specific staining of the different complexes. Anal Biochem 242: 248–254 [DOI] [PubMed] [Google Scholar]
- Grigorieff N, Beckmann E, Zemlin F (1995) Lipid location in deoxycholate-treated purple membrane at 2.6 A. J Mol Biol 254: 404–415 [DOI] [PubMed] [Google Scholar]
- Hackenbrock CR, Chazotte B, Gupte SS (1986) The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J Bioenerg Biomembr 18: 331–368 [DOI] [PubMed] [Google Scholar]
- Hite RK, Li Z, Walz T (2010) Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J 29: 1652–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Cobessi D, Tung EY, Berry EA (2005) Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol 351: 573–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Zickermann V, Brandt U (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329: 448–451 [DOI] [PubMed] [Google Scholar]
- Iwata S, Lee JW, Okada K, Lee JK, Iwata M, Rasmussen B, Link TA, Ramaswamy S, Jap BK (1998) Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281: 64–71 [DOI] [PubMed] [Google Scholar]
- Krause F, Reifschneider NH, Goto S, Dencher NA (2005) Active oligomeric ATP synthases in mammalian mitochondria. Biochem Biophys Res Commun 329: 583–590 [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W (1982) Discrimination of protein and nucleic acids by electron microscopy using contrast variation. Ultramicroscopy 7: 221–232 [DOI] [PubMed] [Google Scholar]
- Kuonen DR, Roberts PJ, Cottingham IR (1986) Purification and analysis of mitochondrial membrane proteins on nondenaturing gradient polyacrylamide gels. Anal Biochem 153: 221–226 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lange C, Nett JH, Trumpower BL, Hunte C (2001) Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J 20: 6591–6600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Maire M, Champeil P, Moller JV (2000) Interaction of membrane proteins and lipids with solubilizing detergents. Biochim Biophys Acta 1508: 86–111 [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol 128: 82–97 [DOI] [PubMed] [Google Scholar]
- McKenzie M, Lazarou M, Thorburn DR, Ryan MT (2006) Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol 361: 462–469 [DOI] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol 142: 334–347 [DOI] [PubMed] [Google Scholar]
- Muench SP, Huss M, Song CF, Phillips C, Wieczorek H, Trinick J, Harrison MA (2009) Cryo-electron microscopy of the vacuolar ATPase motor reveals its mechanical and regulatory complexity. J Mol Biol 386: 989–999 [DOI] [PubMed] [Google Scholar]
- Nield J, Orlova EV, Morris EP, Gowen B, van HM, Barber J (2000) 3D map of the plant photosystem II supercomplex obtained by cryoelectron microscopy and single particle analysis. Nat Struct Biol 7: 44–47 [DOI] [PubMed] [Google Scholar]
- Penczek P, Radermacher M, Frank J (1992) Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy 40: 33–53 [PubMed] [Google Scholar]
- Penczek PA, Zhu J, Schröder R, Frank J (1997) Three dimensional reconstruction with contrast transfer compensation from defocus series. Scanning Microsc 11: 147–154 [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem 278: 52873–52880 [DOI] [PubMed] [Google Scholar]
- Popot JL, Althoff T, Bagnard D, Baneres JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, Cremel G, Dahmane T, de la Maza LM, Ebel C, Gabel F, Giusti F, Gohon Y, Goormaghtigh E, Guittet E, Kleinschmidt JH et al. (2011) Amphipols from A–Z. Annu Rev Biophys 40: 379–408 [DOI] [PubMed] [Google Scholar]
- Radermacher M, Ruiz T, Clason T, Benjamin S, Brandt U, Zickermann V (2006) The three-dimensional structure of complex I from Yarrowia lipolytica: a highly dynamic enzyme. J Struct Biol 154: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J (1987) Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. J Microsc 146: 113–136 [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R (2003) Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333: 721–745 [DOI] [PubMed] [Google Scholar]
- Rossmann MG (2000) Fitting atomic models into electron-microscopy maps. Acta Crystallogr D Biol Crystallogr 56: 1341–1349 [DOI] [PubMed] [Google Scholar]
- Sazanov LA, Hinchliffe P (2006) Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311: 1430–1436 [DOI] [PubMed] [Google Scholar]
- Schäfer E, Dencher NA, Vonck J, Parcej DN (2007) Three-dimensional structure of the respiratory chain supercomplex I1III2IV1 from bovine heart mitochondria. Biochemistry 46: 12579–12585 [DOI] [PubMed] [Google Scholar]
- Schäfer E, Seelert H, Reifschneider NH, Krause F, Dencher NA, Vonck J (2006) Architecture of active mammalian respiratory chain supercomplexes. J Biol Chem 281: 15370–15375 [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak E, Robinson NC (1999) Phospholipase A(2) digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38: 14966–14972 [DOI] [PubMed] [Google Scholar]
- Solmaz SR, Hunte C (2008) Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. J Biol Chem 283: 17542–17549 [DOI] [PubMed] [Google Scholar]
- Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J 27: 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpower BL (1990) The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J Biol Chem 265: 11409–11412 [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science 272: 1136–1144 [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M (1996) A new generation of the IMAGIC image processing system. J Struct Biol 116: 17–24 [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schägger H (2006) Blue native PAGE. Nat Protoc 1: 418–428 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Huang L, Shulmeister VM, Chi YI, Kim KK, Hung LW, Crofts AR, Berry EA, Kim SH (1998) Electron transfer by domain movement in cytochrome bc1. Nature 392: 677–684 [DOI] [PubMed] [Google Scholar]
- Zoonens M, Catoire LJ, Giusti F, Popot JL (2005) NMR study of a membrane protein in detergent-free aqueous solution. Proc Natl Acad Sci USA 102: 8893–8898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.