Abstract
While the pro-differentiation and tumour suppressive functions of Notch signalling in keratinocytes are well established, the underlying mechanisms remain poorly understood. We report here that interferon regulatory factor 6 (IRF6), an IRF family member with an essential role in epidermal development, is induced in differentiation through a Notch-dependent mechanism and is a primary Notch target in keratinocytes and keratinocyte-derived SCC cells. Increased IRF6 expression contributes to the impact of Notch activation on growth/differentiation-related genes, while it is not required for induction of ‘canonical’ Notch targets like p21WAF1/Cip1, Hes1 and Hey1. Down-modulation of IRF6 counteracts differentiation of primary human keratinocytes in vitro and in vivo, promoting ras-induced tumour formation. The clinical relevance of these findings is illustrated by the strikingly opposite pattern of expression of Notch1 and IRF6 versus epidermal growth factor receptor in a cohort of clinical SCCs, as a function of their grade of differentiation. Thus, IRF6 is a primary Notch target in keratinocytes, which contributes to the role of this pathway in differentiation and tumour suppression.
Keywords: keratinocytes, Notch, p63, squamous cell carcinoma, tumour suppression
Introduction
Notch signalling is an important form of intercellular communication with a key role in cell-fate determination and differentiation (Bray, 2006). In many mammalian systems, this pathway enhances stem cell potential and suppresses differentiation, while in others, notably keratinocytes, it exerts an opposite role suppressing tumour development (Dotto, 2008). The Notch gene family encodes evolutionarily conserved type-1 transmembrane receptors that are activated by ligand binding and proteolytic cleavage, with release of the Notch intracellular domain (Artavanis-Tsakonas et al, 1999). The ‘canonical’ pathway involves translocation of the activated Notch cytoplasmic domain to the nucleus, where it associates with the DNA-binding protein CSL (CBF-1 in human and RBP-Jκ in mouse) and an ancillary protein, Mastermind-like 1 (MamL1) or related family members (Nam et al, 2006; Wilson and Kovall, 2006), forming a complex that is required for CSL-dependent transcription. The best-characterized targets of Notch/CSL/MamL-mediated activation are members of the HES and HERP families of bHLH transcriptional repressors (Iso et al, 2003). However, a number of other direct targets of Notch/CSL transcription have been identified, which can be induced by Notch activation in a cell-type-specific manner.
Keratinocytes express mainly Notch1 and Notch2 receptors. Deletion of the Notch1 gene is by itself sufficient to alter keratinocyte growth/differentiation (Rangarajan et al, 2001) and enhance susceptibility to skin cancer formation (Nicolas et al, 2003). While loss of the Notch2 gene by itself does not result in any detectable skin phenotype, together with loss of Notch1 it elicits a dramatic hair follicle/skin phenotype (Pan et al, 2004) that recapitulates to a large extent that caused by deletion of the CSL/RBP-Jκ gene (Yamamoto et al, 2003; Blanpain et al, 2006). Combined Notch1 and Notch2 deletions result in alterations of both intracellular and paracrine control mechanisms, with a defective skin barrier function and increased inflammatory reaction (Demehri et al, 2009; Dumortier et al, 2010). In embryonic skin, the differentiation-promoting function of Notch signalling has been implicated downstream of the establishment of asymmetric cell division (Williams et al, 2011) and of cilia formation (Ezratty et al, 2011). In the mature epidermis, expression of Notch ligands of the Delta-like family in putative keratinocyte stem cell populations can promote neighbouring Notch1-expressing cells to enter a ‘transit amplifying’ phenotype and commit to differentiation (Lowell et al, 2000; Estrach et al, 2008). On the other hand, concomitantly increased expression of Notch receptors and ligands of the Jagged family in the suprabasal epidermal layers can be part of a positive-reinforcement paracrine mechanism for synchronization of differentiation and skin homeostasis (Rangarajan et al, 2001; Nickoloff et al, 2002; Ambler and Watt, 2010).
The molecular mechanisms downstream of Notch activation that elicit differentiation remain elusive. Previous work showed that activation of Notch signalling in mouse keratinocytes is associated with suppression of specific Wnt family members, through CSL/RBP-Jκ-dependent up-regulation of p21WAF1/Cip1 (Devgan et al, 2005). However, in human keratinocytes (HKCs), Notch1 activation leads to a lesser increase of p21WAF1/Cip1 expression, and causes more long-term suppression of growth and induction of differentiation that can be explained, in part, by decreased expression of p63 (Nguyen et al, 2006), a p53 family member with a master regulatory function in epidermal development, morphogenesis and/or stem cell maintenance (Koster and Roop, 2004; McKeon, 2004). Increased Notch signalling in keratinocytes down-modulates p63 gene expression through an indirect mechanism independent of ‘canonical’ targets like Hes/Hey family members and p21Waf1/Cip1. Suppression of p63 depends instead on the down-modulation by Notch of interferon responsive factors, specifically interferon regulatory factor (IRF)3 and IRF7, through an as yet uncharacterized mechanism (Nguyen et al, 2006).
Nine members of the interferon regulatory family of transcription factors (IRFs) have been identified (see for review Taniguchi et al, 2001). All IRFs share similar helix-loop-helix domains, in their N-terminal regions, that recognize common DNA-binding elements called IRF elements (IRF-E) and interferon-stimulated response elements (ISRE), present in a wide variety of genes with different functions. The C-terminal region of IRFs is less conserved and mediates their different interactions with other family members, unrelated transcription factors, co-activators and co-repressors (Taniguchi et al, 2001).
IRF function is best understood in the context of innate immunity and interferon signalling. However, a number of these transcription factors, including IRF1, 2, 3 and 7, have also been implicated in control of cell proliferation and tumour development, in a context- and cell-type-specific fashion (Tamura et al, 2008). Another family member, IRF6, appears without function in innate immunity and interferon signalling, but is essential for normal epidermal development and differentiation (Ingraham et al, 2006; Richardson et al, 2006). Mutations of this gene have been found in two human syndromes: Van der Woude and popliteal pterygium syndrome, which are characterized by cleft palate and lip pits, skin folds, syndactyly and oral adhesions (Kondo et al, 2002). In mice, homozygous loss-of-function mutations of IRF6 result in severe defects in limb and skin development with compromised differentiation of keratinocytes in the interfollicular epidermis (Ingraham et al, 2006; Richardson et al, 2006). A link between p63 and IRF6 has been established in epidermal development, with p63 binding to an IRF6 enhancer and positively controlling its expression, while IRF6 negatively regulates p63 levels (Moretti et al, 2010). A basic function of IRF6 in suppressing growth and promoting differentiation of keratinocytes has also been indicated by in vitro and in vivo studies (Moretti et al, 2010; Thomason et al, 2010), and it has been extended to mammary carcinoma cells, in connection with Maspin, a protease implicated in cancer progression (Bailey et al, 2005, 2008).
In the present communication, we show that IRF6 is a primary Notch target in keratinocytes, which is involved in its more indirect ‘non-canonical’ effects on differentiation, including induction of terminal differentiation markers and suppression of pro-proliferative genes like p63 and integrins. These findings are of likely clinical significance, as suppression of IRF6 expression promotes oncogenic behaviour of ras-expressing human primary keratinocytes, and expression of this gene in cutaneous SCCs parallels that of Notch1 as a function of differentiation.
Results
The IRF6 gene is under positive Notch control in keratinocytes
Notch signalling promotes commitment of keratinocytes towards differentiation through a mechanism that depends, in part, on down-modulation of IRF3 and IRF7 expression (Nguyen et al, 2006). The essential and specific role of IRF6 in epidermal development (Ingraham et al, 2006; Richardson et al, 2006) suggested that this gene may also be involved in this context.
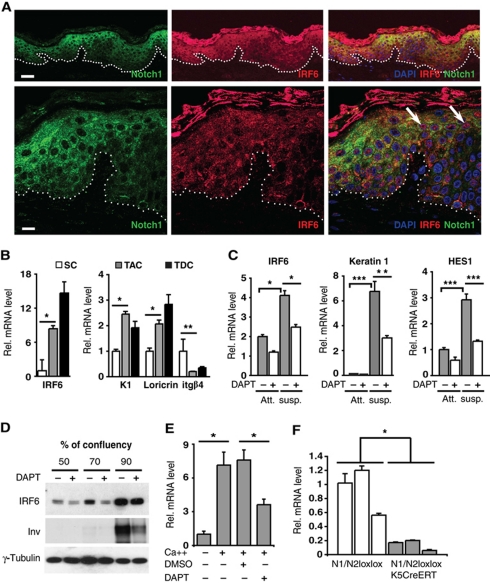
Double immunofluorescence analysis of human skin showed coincidentally increased expression of the Notch1 and IRF6 proteins in the upper layers of the epidermis (Figure 1A, upper panels). Consistent with the previous reports (Rangarajan et al, 2001; Ingraham et al, 2006; Richardson et al, 2006; Bailey et al, 2008), the two proteins were found to localize to the cytoplasm, as both are subject to rapid degradation upon nuclear entry (Bailey and Hendrix, 2008; Kopan and Ilagan, 2009). However, in some cells of the suprabasal layers, nuclear localization of IRF6 could also be detected (Figure 1A, lower panels; Supplementary Figure S1).
Figure 1.
IRF6 is induced during keratinocyte differentiation through a Notch-dependent mechanism. (A) Confocal double-immunofluorescence analysis of Notch1 and IRF6 expression in human skin. (Upper panels) Representative low-magnification images showing concomitantly increased expression of the proteins in the suprabasal epidermal layers. (Lower panels) High-magnification images showing prevalent cytoplasmic localization of the two proteins, with nuclear localization of IRF6 being also detectable in some cells of the outer layers (indicated with arrows). The prevalent cytoplasmic staining of the two proteins is consistent with previous publications (Rangarajan et al, 2001; Ingraham et al, 2006; Richardson et al, 2006). Images are representative of several independent fields. The IRF6 results were confirmed by additional immunofluorescence/confocal imaging analysis of human skin (Supplementary Figure S1). Bars=30 and 9.1 μm, upper and lower panels, respectively. (B) Stem cell (SC), transit amplifying cell (TAC) and terminally differentiated cell (TDC) populations were isolated from human epidermis as described in Materials and methods and expression of the indicated genes was determined by real-time RT–PCR, using 36β4 for normalization. *P<0.002, **P<0.05. Similar results were observed in two other independent experiments using cells from different donors. (C) HKCs, treated for a total of 3 days with DAPT (10 μM) or DMSO vehicle control were either kept under growing conditions (att.) or induced to differentiate by suspension culture for the last 12 h of the experiment (susp.). Expression of the indicated genes was determined by real-time RT–PCR, with 36β4 for normalization. *P<0.002, **P<0.01, ***P<0.0001. Induction of IRF6 expression under these conditions was observed five times, utilizing three independent strains of HKCs, at either RNA or protein level, and the counteracting effects of DAPT were also confirmed. (D) HKCs treated with DAPT (10 μM) or DMSO vehicle control were grown to the indicated densities followed by immunoblot analysis of IRF6 and involucrin expression using γ-tubulin for normalization. Similar results were observed four times, utilizing three independent strains of HKCs. (E) MKCs were induced to differentiate by increased extracellular calcium (2 mM) plus/minus treatment with DAPT (20 μM), followed by real-time RT–PCR analysis of IRF6 expression, using 36β4 mRNA levels for normalization. *P<0.001. Up-regulation of IRF6 levels was similarly observed three times, with separate primary mouse keratinocyte preparation, at either RNA or protein level and counteracting effects of Notch signalling inhibition repeated twice. (F) The epidermis of three mice with keratinocyte-specific deletion of the Notch1 and Notch2 genes (Notch1loxP−loxP/Notch2loxP−loxP × K5 CreERT) versus three littermate controls (Notch1loxP−loxP/Notch2loxP−loxP) was analysed for levels of IRF6 mRNA expression by real-time RT–PCR with GAPDH for normalization. To delete the Notch1 and Notch2 genes, mice were given five OH-TAM injections starting at days 6 of age, with skin samples being taken 4 weeks after first injection. As previously reported, this protocol resulted in >70% deletion of the Notch1 and Notch2 genes (Dumortier et al, 2010). *P<0.05.
As an alternative biochemical method to assess levels of IRF6 expression, keratinocytes from freshly dissociated human epidermis were separated on the basis of their rate of attachment to the substrate, which can enrich for undifferentiated keratinocytes with high self-renewal potential (quickly adhering) versus cells at an intermediate or late stage of differentiation (adhering after longer time or failing to adhere) (Jones and Watt, 1993; Dazard et al, 2000). Immediate RNA preparation without culturing and real-time RT–PCR analysis showed markedly higher expression of IRF6 in the differentiating versus proliferative populations, which paralleled the up-regulation of differentiation marker expression (keratin 1 and loricrin) and the down-regulation of the basal layer integrin β4 (Figure 1B).
Consistent with previous work (Moretti et al, 2010), expression of IRF6, as well as the differentiation marker keratin 1 and the Notch target gene HES1, was also up-regulated in cultured primary HKCs upon induction of differentiation by suspension or high density conditions. Induction of these genes was suppressed by treatment with DAPT, a γ-secretase inhibitor that is widely used to suppress endogenous Notch activation (Rizzo et al, 2008) (Figure 1C and D). Even in mouse primary keratinocytes (MKCs) induced to differentiate by increased extracellular calcium, enhanced IRF6 expression was counteracted by DAPT treatment (Figure 1E). A genetic link between Notch and IRF6 expression was further indicated by analysis of mice with keratinocyte-specific deletion of the Notch1 and Notch2 genes (Dumortier et al, 2010). A drastic reduction of IRF6 expression was found in the epidermis of these mice relative to wild-type littermate controls (Figure 1F).
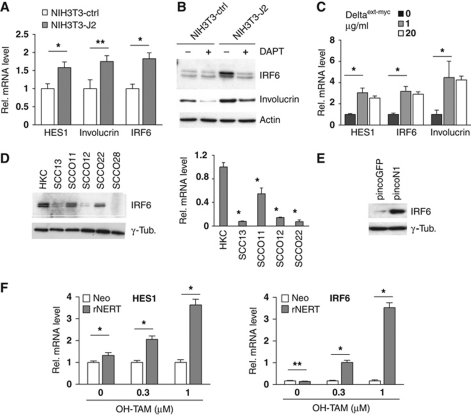
To assess whether activation of endogenous Notch signalling is sufficient to induce IRF6 expression, HKCs were co-cultured with fibroblasts expressing the Jagged2 ligand versus controls. Notch activation, as assessed by induction of the ‘canonical’ target gene HES1 as well as the differentiation marker involucrin, was paralleled by increased expression of IRF6 with this up-regulation being counteracted by DAPT treatment (Figure 2A and B). Similar induction of gene expression was found with HKCs cultured in presence of the immobilized Notch ligand Deltaext-myc (Ohishi et al, 2000) (Figure 2C).
Figure 2.
IRF6 expression is under positive Notch control in keratinocytes. (A) HKCs were co-cultured with either NIH3T3 cells overexpressing full-length Jagged2 (NIH3T3-J2) or control NIH3T3 cells carrying the empty expression vector (NIH3T3-ctrl) as described in the Materials and methods. HKCs were collected 48 h later for expression analysis of the indicated genes by real-time RT–PCR. *P<0.0001, **P<0.0005. (B) HKCs co-cultured as in the previous panel plus/minus treatment with DAPT (20 μM) for the last 24 h were analysed by immunoblotting with antibodies against the indicated proteins. Induction of IRF6 expression similar to the one presented here and in the previous panel was observed a total of four times, utilizing two independent strains of HKCs. (C) HKCs were plated on dishes coated with increasing concentrations of purified Delta1 ligand (Deltaext-myc) followed, 72 h later, by real-time RT–PCR analysis of the indicated genes. Similar results were obtained in a second independent experiment with the Delta1 ligand as well as in an other experiment with a different strain of HKCs plated on dishes coated with the Notch ligand Jagged1. *P⩽0.0001. (D) Early passage HKCs under low-confluency conditions were analysed in parallel with the indicated keratinocyte-derived SCC cell lines for levels of IRF6 expression by immunoblotting (left panel). The same set of cells was analysed for levels of IRF6 mRNA by real-time RT–PCR using 36β4 mRNA levels for normalization (right panel). *P<0.0001. Similar down-modulation of IRF6 expression was observed in two independent sets of freshly excised skin SCC versus normal epidermis as shown in Figure 9A. (E) SCC13 cells were infected with a recombinant retrovirus expressing constitutively active Notch1 together with GFP (pincoN1), or with a virus expressing GFP (pincoGFP) alone followed, 72 h later, by immunoblot analysis of IRF6 expression. Similar results were observed three other times, including an experiment with adenoviral-mediated activated Notch1 expression. (F) SCC13 cells were stably infected with a retroviral vector expressing a flag-tagged activated Notch1 protein fused to the human oestrogen receptor (rNERT), or empty vector control (Neo). Cells were subsequently treated with OH-TAM at the indicated concentrations, collected 30 h later and analysed for HES1 and IRF6 expression by real-time RT–PCR. *P<0.0001, **not significant. A similar induction of IRF6 expression in rNERT cells upon OH-TAM treatment was observed in at least three other independent experiments.
In a number of keratinocyte-derived SCC cell lines, in which Notch signalling is down-modulated (Lefort et al, 2007; Kolev et al, 2008; Mandinova et al, 2009), IRF6 expression was significantly decreased (Figure 2D). Constitutive expression of activated Notch1 in these cells by retroviral infection caused strong IRF6 induction (Figure 2E). In these same cells engineered to express a constitutively active form of Notch1 fused to the oestrogen receptor (rNERT) (Schroeder and Just, 2000), treatment with 4-hydroxytamoxifen (OH-TAM) also resulted in a substantial induction of IRF6 expression, in a dose-dependent manner that paralleled that of HES1 expression (Figure 2F).
IRF6 is a primary target of Notch/CSL-dependent transcription
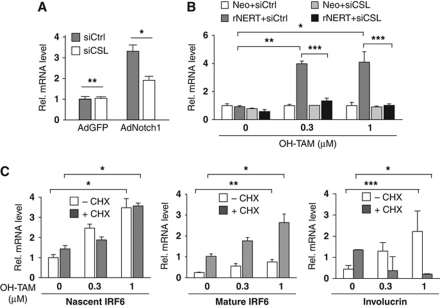
The induction of IRF6 by activated Notch1 in HKCs as well as in SCC cells was counteracted by siRNA-mediated knockdown of CSL, implicating the ‘canonical’ pathway of Notch-dependent transcription (Figure 3A and B). In SCC cells expressing the rNERT protein, expression of the nascent IRF6 transcript, as detected by primers corresponding to the first exon–intron junction, was similarly induced by OH-TAM treatment in the presence or absence of the cycloheximide protein synthesis inhibitor, indicating that the IRF6 gene is a primary target of Notch/CSL activation (Figure 3C, left panel). Interestingly, the levels of mature IRF6 mRNA were up-regulated as a consequence of protein synthesis inhibition, pointing to an additional post-transcription mode of IRF6 regulation (Figure 3C, middle panel). In contrast to IRF6, expression of the involucrin differentiation marker was not induced in cells concomitantly treated with OH-TAM and cycloheximide, indicating that up-regulated IRF6 expression by increased Notch activity is not coincidental but precedes differentiation (Figure 3C, right panel).
Figure 3.
IRF6 is a primary target of Notch/CSL-dependent transcription. (A) HKCs were transfected with siRNAs against CSL in parallel with scrambled siRNA control followed, 48 h later, by infection with recombinant adenoviruses expressing activated Notch1 (AdN1) or GFP control (AdGFP). IRF6 expression was assessed by real-time RT–PCR 24 h later. *P<0.02, **not significant. Similar up-regulation of IRF6 was obtained in three independent experiments, including one with retroviral-mediated activated Notch1 expression. Similar counteracting effects of CSL knockdown were also observed in a second independent experiment with MKCs. (B) SCC13 cells expressing the rNERT protein and control cells (Neo) were transfected with anti-CSL or scrambled siRNAs for 48 h. Cells were subsequently treated with OH-TAM at the indicated concentrations for an additional 24 h followed by analysis of IRF6 expression by real-time RT–PCR. *P<0.0001, **P<0.0005, ***P=0.001. Similar results were observed in a second independent experiment based on SCC13 cells with adenoviral-mediated activated Notch1 expression. (C) SCC13 cells expressing the rNERT protein and control cells (Neo) were treated with cycloheximide (+CHX; 10 μM) or DMSO (−CHX) followed, 2 h later, by OH-TAM treatment at the indicated concentrations for 24 h. Levels of nascent IRF6, mature IRF6 and involucrin transcripts were assessed by real-time RT–PCR using primers, respectively, for the first exon–intron junction, a downstream coding exon of IRF6 and for the involucrin gene. *P<0.005, **P<0.001, ***P<0.0001. Similar results were obtained in a similar experiment with rNERT-expressing HKCs or control cells, plus/minus OH-TAM and cycloheximide treatment.
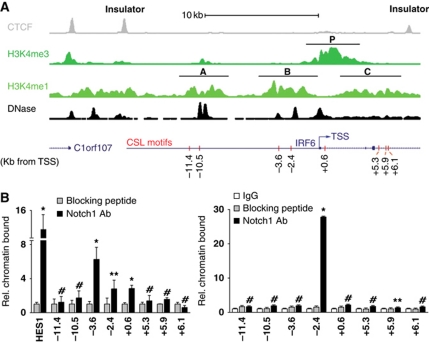
Sequence analysis of the proximal region of the human IRF6 gene promoter revealed the presence of a ‘canonical’ CSL-binding site located at −2.4 kb from the transcription start site (TSS). A luciferase reporter construct encompassing this region displayed constitutive high promoter activity that could not be further modulated by Notch activation or differentiation. Chromatin configuration and regulatory elements located at large distance from the TSS play a critical role in transcription of genes. For insights into regulation of the IRF6 locus, we analysed publicly available data of chromatin immunoprecipitation—high-throughput sequencing (ChIP-seq) and genome-wide DNase I hypersensitivity mapping of human primary keratinocytes (produced by the ENCODE group at the Broad Institute and Massachusetts General Hospital and University of Washington, respectively). Because insulator elements restrict enhancer function and segregate genomic regulatory units (Cuddapah et al, 2009), we focused on an ∼25 kb region of the IRF6 locus containing the TSS and delimited by ChIP-seq peaks for the insulator protein CTCF (Figure 4A, top line, insulator).
Figure 4.
Endogenous Notch1 binds to the IRF6 locus within specific regions of chromatin organization. (A) Diagrammatic illustration of the ChIP-seq results obtained with human primary keratinocytes with antibodies against the CTCF insulator and the indicated methylated forms of Histone H3, in parallel with the mapping of DNase I hypersensitivity sites. Note the correspondence between the DNase I hypersensitivity peaks and ‘dips’ (nucleosome-depleted regions) within the H3K4me1 peaks. Positions of the promoter (P) and putative enhancers (A–C) regions discussed in the text are indicated, alongside the location of the IRF6 TSS and the predicted CSL-binding motifs (red bars). (B) Human primary keratinocytes (left panel) or total extracts of human epidermis (right panel) (see Materials and methods) were processed for ChIP assays using an antibody specific for Notch1, utilizing antibodies preincubated with the corresponding blocking peptide and/or non-immune IgGs as control. PCR amplification of the various regions of the human IRF6 promoter encompassed the eight following predicted CSL-binding sites: −11.4 kb: 5′-GGGGTGGGAACAG-3′; −10.5 kb (two overlapping sites): 5′-CATGTGGGAATGTGAGAAAAC-3′; −3.6 kb: 5′-ATGATGGGAGCATTG-3′; −2.4 kb: 5′-GTCATGGGAATTTCA-3′; +0.6 kb: 5′-TTTTGGGAAACTGGAG-3′; +5.3 kb: 5′-GGCCTGGGAATGG-3′; +5.9 kb: 5′-GTTGTGGGAAAGG-3′; +6.1 kb: 5′-GGGTTGGGAAAGG-3′. Un-precipitated chromatin preparations were similarly analysed and used as ‘input DNA’ control. The nucleotide sequence of the PCR primers is given in Materials and methods. The results are representative of two independent experiments. The relative amount of precipitated DNA, expressed in arbitrary units, was calculated after normalization for total input chromatin, according to the following formula (Frank et al, 2001): % total=2ΔCt × 5 where ΔCt=Ct (input)–Ct (immunoprecipitation). Ct, cycle threshold. Statistical significance of the results was determined by unpaired Student's t-test, comparing the ratio Notch1/IgG signal for each binding site relative to the one for the binding site at position −11.4. *P<0.0001, **P<0.05, #not significant. Similar results were observed in a total of four experiments, with two different strains of cultured HKCs and human epidermal extracts from two different donors.
Previous studies have demonstrated that distal regulatory regions (e.g. enhancers) show enrichment for monomethylated histone H3 lysine 4 (H3K4me1), while promoter regions are enriched for trimethylated histone H3 lysine 4 (H3K4me3) (Heintzman et al, 2007). As expected, the IRF6 promoter region showed strong enrichment for H3K4me3 (Figure 4A, second line from the top, P). Two regions upstream of the TSS, the first lying between −11.5 and −8.5 kb from the TSS, and the second from −4.5 to −1 kb, showed strong enrichment for H3K4me1, suggesting that they contain enhancers or distal promoter regulatory elements acting on IRF6; a third, weakly H3K4me1-enriched region was also present within the transcribed region of the IRF6 gene (Figure 4A, third line from the top, regions A, B, C). Analysis of DNase I hypersensitivity demonstrated sites of increased chromatin accessibility within the promoter and upstream A and B regions, providing a further indication of their potential regulatory function (Figure 4A, fourth line from the top).
A motif search identified several consensus CSL-binding sites within the 25-kb region of the IRF6 locus, with four sites mapping within the two upstream H3K4me1-enriched regions, and three of them lying within or adjacent to DNase I hypersensitivity peaks (Figure 4A, fifth line from the top). To assess experimentally to which of these sites Notch1 may bind, we performed ChIP assays with extracts from human primary keratinocytes under high-confluence differentiating conditions. The analysis showed binding of the endogenous Notch1 protein at both of the predicted CSL motifs within region B (−3.6 and −2.4 kb position), with little or no binding to more distal sites (Figure 4B, left panel). Positive binding was also found at a TSS proximal site (+0.6 kb position) with little or no binding further downstream. Importantly, a similar ChIP assay performed with extracts of intact human epidermis showed strong Notch1 binding only at the −2.4 kb site (Figure 4B, right panel), suggesting that this may be the predominant site of Notch1 regulation within the chromatin context of in vivo fully differentiating keratinocytes.
IRF6 is a mediator of the ‘non-canonical’ Notch pro-differentiation function
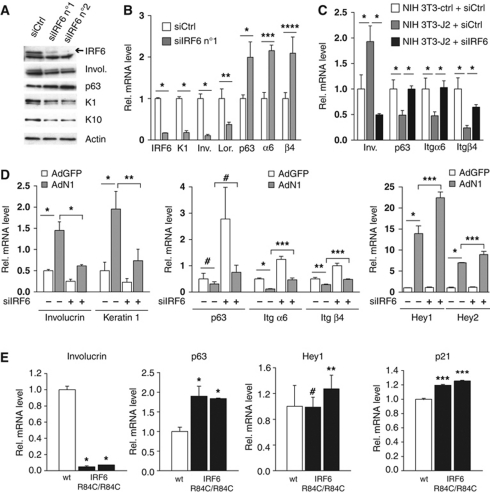
To gain insights into the functional significance of the above findings, we evaluated the consequences of suppressed and increased IRF6 expression. As we previously reported for loss of Notch signalling (Lefort et al, 2007), siRNA-mediated knockdown of IRF6 in HKCs caused down-modulation of terminal differentiation markers (involucrin, keratin 1 and 10, loricrin) and up-regulation of markers of the proliferative compartment (p63, integrin α6/β4) (Figure 5A and B; Supplementary Figure S2A). In HKCs co-cultured with Jagged2-expressing fibroblasts, modulation of these markers by Notch activation was counteracted by IRF6 knockdown (Figure 5C). Similar effects were observed with IRF6 knockdown in MKCs. Even in this case, suppression of IRF6 expression caused down-modulation of terminal differentiation markers and up-regulation of markers of the proliferative compartment, and counteracted their opposite modulation by activated Notch1 expression (Figure 5D, left and middle panels). Interestingly, induction of the ‘canonical’ HEY1 and HEY2 targets by activated Notch1 expression was slightly enhanced, rather than suppressed, by IRF6 knockdown (Figure 5D, right panel), indicating that IRF6 is not required for up-regulation of these genes or, if anything, may negatively control it. Similar modulation of differentiation-related genes with a slight up-regulation of ‘canonical’ Notch targets (HEY1, p21WAF1/Cip1) was also found in vivo, in the epidermis of late gestation IRF6 mutant embryos (Richardson et al, 2006) (Figure 5E).
Figure 5.
IRF6 is a mediator of Notch1 pro-differentiation function in keratinocytes. (A) HKCs were transfected with two different siRNAs against IRF6 (siIRF6 n°1 and n°2) in parallel with scrambled siRNA control (siCtrl) for 72 h followed by immunoblot analysis for the indicated proteins. (B) HKCs were transfected with siRNAs against IRF6 (siIRF6 n°1) in parallel with scrambled siRNA control (siCtrl) for 72 h followed by real-time RT–PCR analysis for the indicated genes. *P⩽0.0001, **P<0.02, ***P<0.001, ****P<0.005. Similar results with a second set of siRNAs (siIRF6 n°2) are shown in Supplementary Figure S2A. Results similar to those shown in this and previous panel were observed at least four times with a total of four different strains of HKCs. (C) HKCs transfected with siRNAs against IRF6 (siIRF6) in parallel with scrambled siRNA control (siCtrl) were co-cultured with NIH3T3 fibroblasts expressing Jagged2 (NIH3T3-J2) or control fibroblasts (NIH3T3-ctrl) for 48 h. HKCs were analysed for expression of the indicated genes by real-time RT–PCR. *P⩽0.0001. Similar results were observed in second independent experiment with a different strain of HKCs plus/minus retroviral-mediated activated Notch1 expression. (D) MKCs were transfected with anti-IRF6 or scrambled siRNAs for 48 h followed by infection with an adenovirus expressing the activated cytoplasmic form of Notch (AdN1) (Rangarajan et al, 2001) or GFP control (AdGFP) for additional 24 h. Expression of the indicated genes was analysed by real-time RT–PCR with β-actin for normalization. *P<0.0001, **P<0.01, ***P<0.001, #not significant. Similar results were observed in three experiments with separate preparations of MKCs, by either RNA or protein analysis. (E) Skin samples from E16.5 mouse embryos wild-type versus homozygous for the IRF6 mutation R84C (IRF6 R84C/R84C) (Richardson et al, 2006) (white and black bars, respectively) were analysed by real-time RT–PCR for the indicated genes. Statistical significance of the results was determined for differences in gene expression values in the mutant versus control mice. *P<0.0001, **P<0.05, ***P<0.005, #not significant. Similar results were observed by analysis of mutant versus wild-type embryos of younger age (E14).
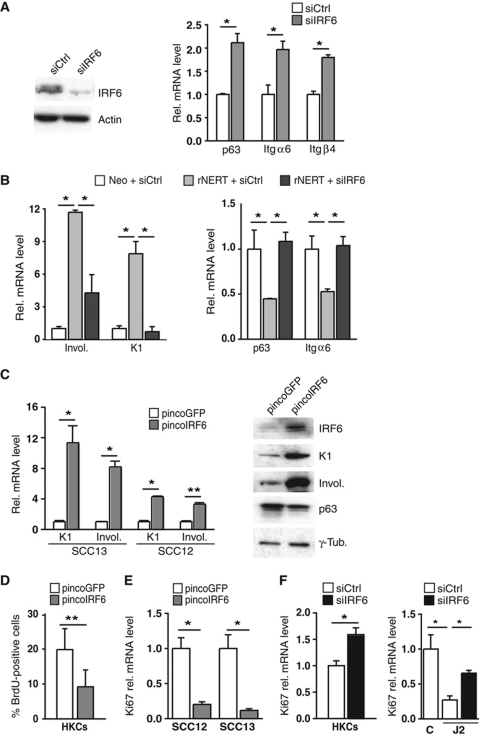
The analysis was extended to SCC cells, which express lower levels of IRF6 than HKCs. Integrin α6/β4 and p63 expression was up-regulated in SCC13 cells upon further IRF6 knockdown (Figure 6A). In SCC13 cells expressing rNERT, induction of differentiation markers and down-modulation of p63 and integrin α6/β4 in response to OH-TAM treatment was counteracted by IRF6 knockdown (Figure 6B; Supplementary Figure S2B). Opposite modulation of these genes was caused by increased IRF6 expression in SCC13 cells, as well as a second SCC cell line (SCC12) (Figure 6C).
Figure 6.
IRF6 is a mediator of Notch signalling in SCC cells. (A) SCC13 cells were transfected with siRNA against IRF6 in parallel with scrambled siRNA control. IRF6 silencing was confirmed by immunoblot (left panel) and the other genes were analysed by real-time RT–PCR (right panel). *P<0.0001. Similar results were observed in two other independent experiments. (B) SCC13 cells expressing the rNERT protein and control cells (Neo) were transfected with siRNAs against IRF6 (siIRF6) in parallel with scrambled siRNA control (siCtrl) for 48 h and treated with 4-hydroxytamoxifen (OH-TAM; 1 μM) for additional 24 h. Expression of the indicated genes was analysed by real-time RT–PCR. *P<0.0001. Similar results were obtained with a second set of siRNA as shown in Supplementary Figure S2B. (C) SCC13 and SCC12 cells were infected with an IRF6-expressing retrovirus (pincoIRF6) or empty vector control (pincoGFP) for 72 h followed by analysis of the indicated genes by either real-time RT–PCR or immunoblotting (left and right panels, respectively). *P⩽0.0001, **P<0.006. Similar results were observed in a total of three experiments with SCC13 cells and twice with SCC12 cells. (D) HKCs were infected with an IRF6-expressing retrovirus (pincoIRF6) or empty vector control (pincoGFP) for 48 h. Cells were analysed by BrdU labelling. **P<0.01. (E) SCC12 and SCC13 cells were infected with an IRF6-expressing retrovirus (pincoIRF6) or empty vector control (pincoGFP) for 48 h and levels of Ki67 expression were analysed by real-time RT–PCR. *P⩽0.0001. (F) HKCs were transfected with siRNAs against IRF6 (siIRF6) in parallel with scrambled siRNA control (siCtrl) for 48 h (left panel) or HKCs plus/minus siIRF6 were co-cultured with Jagged2 (J2) or control (C) expressing NIH3T3 fibroblast for 48 h (right panel). Levels of Ki67 expression were analysed by real-time RT–PCR. *P<0.0001.
In parallel with effects on differentiation, IRF6 overexpression in both HKCs and SCC cells led to a significant decrease in proliferation, as measured by BrdU labelling or Ki67 expression (Figure 6D and E). Conversely, knockdown of IRF6 expression had a significant positive effect on proliferation of HKCs under basal conditions as well as when co-cultured with Jagged2-expressing fibroblasts (Figure 6F).
IRF6 exerts an essential pro-differentiation function in HKCs in vivo, and contributes to tumour suppression
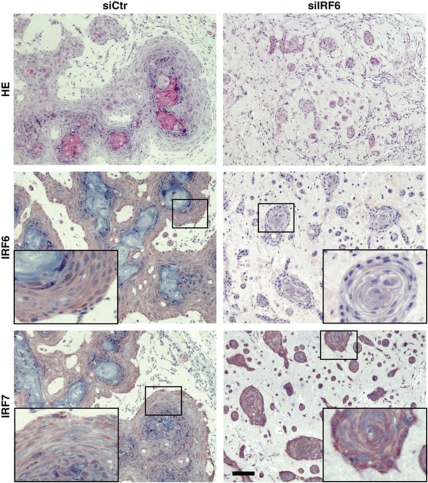
To assess whether IRF6 plays a similar regulatory function in vivo, HKCs were transfected with siRNA against IRF6 in parallel with scrambled siRNA control, followed by intradermal injection into NOD/SCID mice. Under these conditions, control cells formed large epidermal islands with evident granular and squamous differentiation 7 days after injection. By contrast, cells in which IRF6 had been knocked down remained separated in small nests with no signs of terminal differentiation (Figure 7). Immunohistochemical analysis confirmed the IRF6 silencing in the nests formed by HKCs with IRF6 knockdown, which was associated with elevated levels of IRF7 and p63 as well as decreased levels of the differentiation markers keratin 1 and loricrin (Figure 7; Supplementary Figure S3).
Figure 7.
Silencing of IRF6 expression in HKCs alters the differentiation process in vivo. HKCs transfected with siRNA against IRF6 or scrambled siRNA control for 3 days were collected, admixed with Matrigel and injected intradermally into the skin of NOD/SCID mice. Cells plus/minus IRF6 knockdown were injected in parallel in the right and left flank of mice, to minimize individual animal variations. A week later, nodules formed at the sites of injection were excised and tissues were processed for H&E staining and immunohistochemical analysis with antibodies against the indicated proteins. The results are representative of at least 10 nodules formed by each type of cells. Corresponding high-magnification images are shown as inserts. Bar=50 μm.
Experiments were complemented by grafting onto nude mice of human primary keratinocytes plus/minus IRF6 knockdown. Control keratinocytes formed a fully stratified and differentiated epidermis by 11 days of grafting, whereas keratinocytes with down-modulation of IRF6 expression showed no ordered stratification, forming instead nests of cells that remained separated from each other, and with only limited signs of suprabasal differentiation marker expression (Supplementary Figure S4).
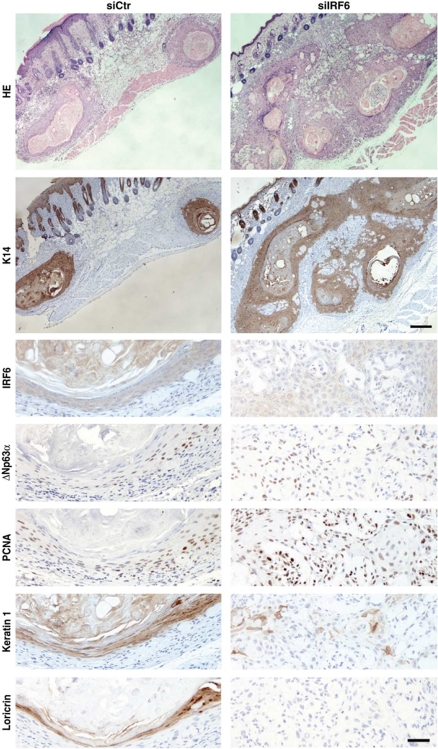
To assess the possible consequences of IRF6 down-modulation on tumour formation, HKCs plus/minus IRF6 knockdown were infected with an oncogenic H-rasV12-transducing retrovirus prior to intradermal injection into mice. As we recently reported (Wu et al, 2010), control ras-expressing HKCs formed under these conditions only differentiated squamous cysts. By contrast, cells with concomitant IRF6 knockdown gave rise to cysts with areas of high cellularity, with defective differentiation marker expression and elevated expression of proliferation markers like p63 and PCNA (Figure 8).
Figure 8.
Inhibition of IRF6 expression in HKCs promotes tumour formation. HKCs transfected with anti-IRF6 or scrambled siRNAs for 24 h were subsequently infected with a H-rasV12-transducing retrovirus (LZRS-rasV12) (Lazarov et al, 2002), admixed with Matrigel and injected intradermally into the skin of NOD/SCID mice. The two types of cells were injected in parallel in the right and left flank of mice, to avoid the risk of individual animal variations. Nodules formed at the sites of injection were excised 8–10 days later and tissues were processed for H&E staining. The results are representative of six nodules formed by each type of cells. Retrieved tissues were analysed by immunohistochemistry with antibodies against the indicated proteins in parallel with HE staining. Upper bar=100 μm, lower bar=25 μm.
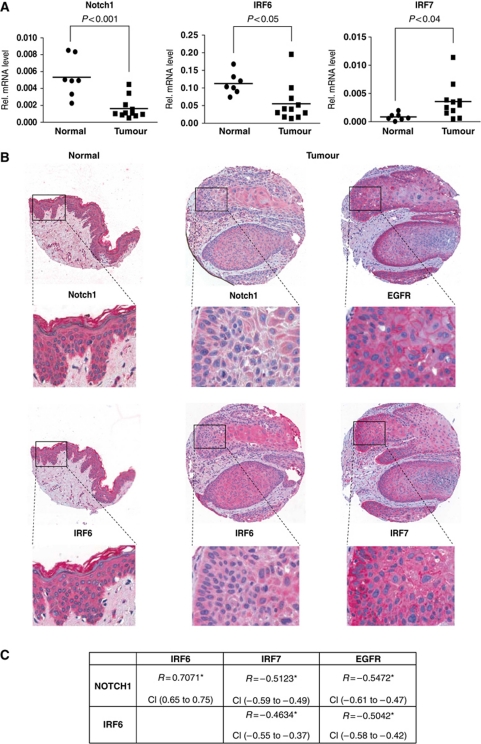
To assess the clinical significance of the above findings, we examined the pattern of expression of the various genes in a set of clinically occurring tumours versus samples of normal epidermis. Real-time RT–PCR analysis showed a significant down-modulation of the IRF6 gene, in parallel with Notch1, in a set of SCCs versus normal epidermis samples, with an opposite up-regulation of IRF7, which we previously showed to be under Notch negative control (Nguyen et al, 2006) (Figure 9A). The results were complemented by tissue array/immunohistochemical analysis of a larger cohort of cutaneous SCCs. In the vast majority of tumours, in the areas of tumours with greater versus lesser differentiation, there was an inverse relation between expression of Notch1 and IRF6 versus IRF7 and epidermal growth factor receptor (EGFR), which plays an opposite role to Notch signalling in keratinocyte proliferation and tumourigenesis (Kolev et al, 2008) (Figure 9B and C).
Figure 9.
Opposite pattern of Notch1 and IRF6 versus EGFR and IRF7 expression in human SCCs. (A) RNA samples from a panel of freshly excised cutaneous SCCs and normal epidermis were analysed by real-time RT–PCR for the indicated genes. Values are expressed in arbitrary units after internal normalization with 36β4 mRNA levels and indicated on a plot (black squares) together with the calculated average (bar). Statistical significance of the results was calculated by t-test. (B) Expression pattern of the indicated proteins was assessed by immunohistochemical analysis of tissue microarrays containing 246 human SCCs as well as 11 samples of normal skin with corresponding antibodies. Shown is a representative staining pattern of normal skin (left column) versus one tumour (right columns), together with higher-magnification images (inserts). Consistent with the immunofluorescence results (see Figure 1A), the pattern of Notch1 and IRF6 staining (as detected by the red chromogenic reaction) is largely cytoplasmic. The nuclear signal (blue) is due to haematoxylin counterstaining. Note the higher levels of Notch1 and IRF6 expression in the normal epidermis versus SCC and, within the SCC, the coincident pattern of staining of the two proteins opposite from that of EGFR and IRF7. (C) Statistical analysis of the tissue array data. For this, tumours were individually analysed and assigned arbitrary units of staining for each of the proteins, followed by calculation of the Spearman's rank correlation coefficient (r). Positive versus negative values correspond to direct versus inverse correlations. Correlation is considered low for r-values between 0.3 and 0.4, moderate between 0.4 and 0.6 and high above 0.6. All correlation coefficient values were highly significant (*P<0.0001). Robustness of the correlation is further supported by the narrow confidence interval (CI) values, that is, the range of r-values with a likelihood of 95%.
Discussion
The pro-differentiation and tumour suppressive functions of Notch signalling in keratinocytes have been well established (Dotto, 2008; Watt et al, 2008). However, the underlying mechanisms remain to be clarified. Notch activation has been proposed to function through both a ‘canonical’ pathway dependent on CSL, and a ‘non-canonical’ pathway independent of this protein (Rangarajan et al, 2001; Blanpain et al, 2006; Nguyen et al, 2006). However, this is likely to be an oversimplification, since expression of putative mediators of the ‘non-canonical’ pathway, like Deltex, can be under CSL control (Bray, 2006). Thus, a more accurate view of Notch signalling is one that involves both CSL-direct and indirect mechanisms. Our previous work established that Notch activation is involved in cell-cycle control of keratinocytes with p21WAF1/Cip1 as a direct Notch/CSL target (Rangarajan et al, 2001). Notch activation induces also differentiation of these cells through a more indirect mechanism, involving down-modulation of integrins of the basal layer, p63 as well as two IRF family members with a widespread role in innate immunity and growth control, IRF3 and IRF7 (Nguyen et al, 2006). We have shown here that IRF6, another IRF family member with a specific role in epidermal development and differentiation (Ingraham et al, 2006; Richardson et al, 2006), is a primary Notch target in keratinocytes, which is involved in the more indirect effects of this pathway on control of differentiation-related genes. This combined Notch-IRF6 regulatory function extends to in vivo keratinocyte differentiation and SCC formation.
Like p63, IRF6 plays a crucial role in epidermal development and a recently described cross-regulation helps to explain the partially overlapping phenotype of human syndromes resulting from mutation of the two genes (Moretti et al, 2010; Thomason et al, 2010). In parallel with suppression of differentiation, loss of IRF6 function results in up-regulation of p63 expression. In turn, IRF6 expression is down-regulated as a consequence of p63 deficiency (Ingraham et al, 2006; Richardson et al, 2006). This occurs not only in the transition between simple and stratified epithelium but also in mature keratinocytes, consistent with previous observations that p63, besides enhancing self-renewal of keratinocytes, promotes entry into differentiation (Nguyen et al, 2006; Truong et al, 2006). However, p63 itself is strongly down-regulated with differentiation and is up-regulated in SCCs (Westfall and Pietenpol, 2004; Dotto, 2009), while IRF6 exhibits the opposite pattern of expression. Therefore, other key determinants of IRF6 expression must exist.
We have shown here that IRF6 gene transcription is induced in differentiating keratinocytes by a Notch-dependent mechanism. IRF6 nascent transcripts are induced by Notch activation even under conditions of protein synthesis inhibition, indicating that this gene is a primary Notch target. The fact that levels of mature IRF6 mRNA are super-induced under these conditions points to the possibility of a further level of regulation, typical of genes regulated at the post-transcriptional mRNA stability level. In contrast to IRF6, differentiation marker expression is not induced by increased Notch activity under conditions of protein synthesis inhibition. This supports the overall conclusion that IRF6 is a primary Notch target gene in keratinocytes, which is in turn involved in control of differentiation. In SCC cells, the decreased IRF6 levels can be explained by compromised Notch signalling, with differentiation being a secondary cause. This however does not rule out the likely possibility of an amplification mechanism, whereby differentiation is also reinforcing IRF6 expression and function. In fact, positive feedback loops of this kind are very often employed in important cell-fate decisions and, of relevance to the present situation, expression of Notch1 receptor as well as Jagged1/2 ligands are both under positive control of Notch pathway activation (Bray, 2006; Yashiro-Ohtani et al, 2009).
We used a combination of multi-modality chromatin analysis to identify regions of likely regulatory function within the IRF6 locus. While endogenous Notch1 protein binds to one such region upstream of the IRF6 promoter (region B in Figure 5A), our data point to a complex mode of regulation that depends on overall chromatin configuration and the concerted action of Notch with other positive and negative regulators of transcription. Analysis of families with non-syndromic cleft lip has identified a single-nucleotide polymorphism in an enhancer region of the IRF6 gene that disrupts binding of the AP-2 transcription factor (Rahimov et al, 2008). Interestingly, this previously established AP-2 binding site maps within one of the putative regulatory regions that we have identified in our analysis, which is not bound by Notch1 (region A in Figure 4). This raises the possibility of a convergent control of IRF6 expression, which would be consistent with a previous report of a synergistic function of AP-2 and Notch in keratinocytes (Wang et al, 2008). However, such a mode of regulation may be limited to development, as knockdown of AP-2 expression in mature keratinocytes had no effect on IRF6 expression.
In both human primary keratinocytes and SCC cells, increased IRF6 expression elicits the same modulation of basal and suprabasal differentiation marker genes as Notch activation, with the effects of the latter being dependent on IRF6 expression. IRF3 and IRF7 are two other IRF family members that can dimerize with each other and form a large transcriptional complex with NF-κB, AP-1 and CREB transcription factors. Besides their role in innate immunity, these IRFs have been implicated in oncogenesis (Tamura et al, 2008). As with NF-κB and Notch, they may play a tumour suppressing or promoting function, depending on cell type and context. Increased IRF3 expression and activity can promote apoptosis or inhibit growth of a number of cancer cell types, while IRF7 is a mediator of the latency and oncogenic function of EBV latent membrane protein 1 (Zhang and Pagano, 2002). Both transcription factors can control inflammatory cytokine production, which can have determining consequences for tumour development in vivo (Tamura et al, 2008). In keratinocytes, the combined knockdown of the two proteins suppresses p63 expression, while persistently elevated IRF7 expression counteracts the down-modulation of p63 by Notch activation (Nguyen et al, 2006). A positive role of IRF7 in keratinocyte-derived tumours is further supported by the strikingly opposite modulation of IRF7 versus Notch1 and IRF6 in a large set of clinically occurring SCCs, and the up-regulation of IRF7 that accompanies the enhancement of keratinocyte tumour formation by IRF6 knockdown.
Recent work has showed that the negative regulation of p63 by Notch is not limited to the keratinocyte system, but applies also to the mammary gland (Mazzone et al, 2010; Yalcin-Ozuysal et al, 2010). This gland is composed of two closely juxtaposed cell types, myoepithelial and luminal, with increased Notch signalling during development being mutually exclusive with p63 and Notch activation suppressing p63 expression, as well as p63 controlled proteins like integrins (Mazzone et al, 2010; Yalcin-Ozuysal et al, 2010). In another context, secondary palate development, a genetic convergence of the IRF6 and Notch signalling, as affected by Jagged ligand mutations, has also been established, even if the molecular basis for this interplay remains to be defined (Richardson et al, 2009).
The mechanism of action of IRF6 remains to be elucidated (Bailey and Hendrix, 2008). The major phenotypic consequences of genetic single point mutations abrogating its DNA-binding activity are consistent with IRF6 functioning as a direct modulator of transcription. However, since a large fraction of this protein is in the cytoplasm, an attractive possibility is that it may also affect transcription indirectly, for instance by preventing other proteins with which it interacts from entering the nucleus. This could be a phosphorylation-regulated phenomenon, as a number of IRFs need to be phosphorylated for nuclear translocation and activation and, in the case of IRF6, phosphorylation also enhances proteosome-dependent degradation (Bailey et al, 2008).
Materials and methods
Cell culture
Conditions for culturing of primary mouse and HKCs and induction of differentiation were as previously reported (Nguyen et al, 2006). Stem cell, transit amplifying cell and terminally differentiated cell populations were isolated as described previously (Dazard et al, 2000). SCCO11, O12, O22 and O28 cells were kindly provided by Dr J Rocco (Massachusetts General Hospital), and SCC13 and SCC12 cells by Dr J Rheinwald (Brigham and Women's Hospital), while other cells were from ATCC. The LZRS-H-rasV12 (Dajee et al, 2003) retrovirus was provided by Dr P Khavari (Stanford University). NIH3T3 fibroblasts expressing full-length Jagged2 or carrying an empty vector control (Luo et al, 1997) and purified IgG-Deltaext-myc (Ohishi et al, 2000) were generous gifts of J Aster (Brigham and Women's Hospital) and I Bernstein (Fred Hutchinson Cancer Research Center, Seattle), respectively. For co-culture experiments, NIH3T3 fibroblasts expressing full-length Jagged2 or carrying an empty vector control were pretreated with mitomycin (5 μg/ml) for 2 h before adding HKCs for co-culture as described (Lowell et al, 2000). For plate-bound Delta experiments, plates were preincubated with 10 μg/ml of rabbit anti-human IgG (Sigma), blocked with BSA 2% and incubated with IgG-Deltaext-myc ligand or the control human IgG at the indicated concentrations for 2 h. Cells were plated on top for 72 h. DAPT was purchased from Calbiochem. For cell proliferation measurement, cells were incubated with BrdU (Amersham) (pulse of 2 h) followed by immunofluorescence analysis with anti-BrdU antibodies (BD Biosciences) according to the manufacturer's instructions.
Plasmids and viruses
The plasmids rNERT-neo and rNeo (Schroeder and Just, 2000) were kindly provided by Dr U Just (Christian-Albrechts-University of Kiel, Germany). The pincoNotch1 plasmid was obtained by inserting the cDNA of activated Notch1 (from digestion of the pcDNA3/hNIC by BamHI/XhoI) into the BamHI/EcoRI sites of the pincoGFP vector (Nocentini et al, 1997). The pincoIRF6 plasmid was obtained by cloning the myc-tagged full-length cDNA of human IRF6 from the pcDNA3.2-myc-IRF6 plasmid (kindly provided by B Schutte) into the vector AttR1–AttR2-pinco (kindly provided by C Missero, Naples, Italy) using the Gateway™ cloning kit (Invitrogen) and the following primers: 5′-CACCATGGAGGAGCAGAAGCTGATCTCAGAGGAGGA-3′ (forward) and 5′-TCACTGGGCCAGAGCCTGT-3′ (reverse). The viruses AdGFP, AdNotch1 were previously described (Capobianco et al, 1997). Conditions for retrovirus and adenovirus production and infection were as previously reported (Rangarajan et al, 2001; Nguyen et al, 2006).
Skin SCC samples
Skin and cutaneous squamous cell carcinoma samples were obtained at the Department of Dermatology of the Zurich University Hospital, Switzerland, from clinical biopsies. Parts not needed for histological diagnosis were further processed with institutional review board approval. The epidermis was mechanically separated from the underlying dermis by a brief heat treatment (Kolev et al, 2008). Tissues were homogenized in TRI Reagent (Sigma) for RNA preparation.
Quantitative real-time RT–PCR
Conditions for RNA preparation and real-time RT–PCR were as previously described (Lefort et al, 2007). The list of gene-specific primers is provided in Supplementary Table S1.
Immunodetection techniques and antibodies
Conditions for immunohistochemistry (Rangarajan et al, 2001), immunoblotting (Lefort et al, 2007) were as described. The following antibodies were used: p21 (sc-6246), Notch1 (sc-6014), actin (sc-1616), p63 (sc-8343), PCNA (sc-56) (Santa Cruz), involucrin (ab68) and IRF3 (ab76409) (Abcam), keratin 1 (PRB149P), loricrin (PRB145P) and keratin 14 (PRB155P) (Covance), IRF6 (custom ordered from Eurogentec using the two previously reported KLH-coupled peptides (EDELEQSQHHVPIQDTFPF and SPEASWPKTEPLEMEV; Bailey et al, 2005)), IRF7 (LS-B2945) (Lifespan Biosciences), EGFR (ab52894: Abcam) and γ-tubulin (T-6557) (Sigma).
siRNA transfection for in vitro and in vivo assays
Primary HKCs or SCC13 cells were transfected with 200 nM of Stealth validated siRNAs (Invitrogen) for human IRF6 n°1 (HSS105511), IRF6 n°2 (HSS105512) and CSL (HSS142635) and control (45-2001) using lipofectamine reagent (Invitrogen). Primary mouse keratinocytes were transfected with 200 nM of validated siRNA (Qiagen) for mouse IRF6 (SI01077762) or control (SI03650325). For in vivo intradermal and grafting assays, siRNA-transfected primary HKCs plus/minus infection with the LZRS-H-rasV12 retrovirus for 16 h were collected, admixed with Matrigel (BD Biosciences) and injected at the epidermal–dermal junction or into back-skin implanted graft chambers (2.5 × 106 cells per injection) into NOD/SCID mice as previously reported (Wu et al, 2010).
Chromatin configuration analysis
ENCODE ChIP-seq and DNase I hypersensitivity tracks, and CSL motif locations were mapped to the human genome (NCBI36/hg18) through the UCSC genome browser. Image post-processing was performed with Adobe Illustrator. ENCODE data and experimental protocols are available online (http://genome.ucsc.edu/ENCODE/).
ChIP assays
Human epidermis was separated from the underlying dermis by a brief heat treatment (Kolev et al, 2008) and was minced finely in ice-cold PBS. Confluent primary HKCs as well as tissue samples were then cross-linked with 37% formaldehyde to a final concentration of 1% for 10 min at RT followed by the addition of glycine (final concentration 125 mM). After cross-linking, tissues were washed twice with 10 ml PBS plus protease inhibitors. Tissue pellets were processed for ChIP assays as previously described (Lefort et al, 2007) using the rabbit anti-Notch1 antibody (Santa Cruz, C-20) in parallel with affinity-purified non-immune IgGs. When specified, anti-Notch1 antibody was preincubated for 2 h at RT in the presence of neutralizing amounts of the corresponding blocking peptide (sc-6014P, Santa Cruz). Primers used for real-time PCR of various regions of the human IRF6 promoter were: 5′-TGGGTGCCCTGTTTTGATGA-3′ and 5′-ACTTCTAACCCAAGCCTAGC-3′ (−11.4 kb); 5′-TCCAACCTGACAGACTCCTA-3′ and 5′-CCTAGAGAACTGAGACAGGA-3′ (−10.5 kb); 5′-TCAATGGAGGGCAAAATGAT-3′ and 5′-ACGCCTCATCTGCTTGATCT-3′ (−3.6 kb); 5′-ACCCTCCCAGCTTGAGTTTT-3′ and 5′-AAACCCCAGTGGCATACAAG-3′ (−2.4 kb); 5′-ACTATCCGGTAGAGCTAAAG-3′ and 5′-CCTCACTCCAGTTTCCCAAA-3′ (+0.6 kb); 5′-GAGAGCCCTCTATACCAATC-3′ and 5′-CTTGCTATGCAGAGAGTTGC-3′ (+5.3 kb); 5′-AGACAGAAGTAGGTGGACAG-3′ and 5′-AGAAAGAAGCTGGTGTGGAG-3′ (+5.9 kb); 5′-ATGCTTTGGGTCCTTGCTGA-3′ and 5′-TGAGTTGGGTGGGAAACATC-3′ (+6.1 kb). Primers used for a region of the human HES1 promoter were: 5′-CCTCCCATTGGCTGAAAGTT-3′ and 5′-CCTGGCGGCCTCTATATATA-3′.
Statistics
All statistical evaluations were carried out using GraphPad Prism 5.0. All analyses are paired, two-tailed Student's t-test unless otherwise specified. All real-time RT–PCR samples were tested in triplicate and error bars represent s.d. P-values of <0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Drs B Schutte, U Just, J Aster, P Khavari, J Rocco, J Rheinwald for gifts of plasmids and cells and I Bernstein for its generous gift of IgG-Deltaext-myc ligand. This work was supported by grants from NIH (Grants AR39190 and AR054856), the European Union (Epistem, Sixth Framework Program, LSHB-CT-2005-019067), the Swiss National Foundation (Grant 3100A0-122281/1) and Oncosuisse (Grant 02361-02-2009) to GPD, and by a grant from the Olga-Mayenfisch-Stiftung to GH and by grants from Wellcome Trust (082868) and MRC (G 0901539) to MJD.
Author contributions: The overall study was conceived and designed by GPD and KL, with important contributions from GR, BCN, PD, ER and GFLH. KL, GR, BCN, ER and OYO performed the experiments and analysed the data. RJHR designed and analysed the data relative to the ChIP sequencing. PD and GFLH designed and analysed the data performed on skin SCC samples. FR, MDP and MJD provided critical mouse material. GPD and KL wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ambler CA, Watt FM (2010) Adult epidermal Notch activity induces dermal accumulation of T cells and neural crest derivatives through upregulation of jagged 1. Development 137: 3569–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 [DOI] [PubMed] [Google Scholar]
- Bailey CM, Abbott DE, Margaryan NV, Khalkhali-Ellis Z, Hendrix MJ (2008) Interferon regulatory factor 6 promotes cell cycle arrest and is regulated by the proteasome in a cell cycle-dependent manner. Mol Cell Biol 28: 2235–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CM, Hendrix MJ (2008) IRF6 in development and disease: a mediator of quiescence and differentiation. Cell Cycle 7: 1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CM, Khalkhali-Ellis Z, Kondo S, Margaryan NV, Seftor RE, Wheaton WW, Amir S, Pins MR, Schutte BC, Hendrix MJ (2005) Mammary serine protease inhibitor (Maspin) binds directly to interferon regulatory factor 6: identification of a novel serpin partnership. J Biol Chem 280: 34210–34217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Pasolli HA, Fuchs E (2006) Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev 20: 3022–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ (2006) Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7: 678–689 [DOI] [PubMed] [Google Scholar]
- Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM (1997) Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol 17: 6265–6273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K (2009) Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res 19: 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, Marinkovich MP, Tao S, Lin Q, Kubo Y, Khavari PA (2003) NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature 421: 639–643 [DOI] [PubMed] [Google Scholar]
- Dazard JE, Piette J, Basset-Seguin N, Blanchard JM, Gandarillas A (2000) Switch from p53 to MDM2 as differentiating human keratinocytes lose their proliferative potential and increase in cellular size. Oncogene 19: 3693–3705 [DOI] [PubMed] [Google Scholar]
- Demehri S, Morimoto M, Holtzman MJ, Kopan R (2009) Skin-derived TSLP triggers progression from epidermal-barrier defects to asthma. PLoS Biol 7: e1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan V, Mammucari C, Millar SE, Brisken C, Dotto GP (2005) p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev 19: 1485–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP (2008) Notch tumor suppressor function. Oncogene 27: 5115–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP (2009) Crosstalk of Notch with p53 and p63 in cancer growth control. Nat Rev Cancer 9: 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A, Durham AD, Di Piazza M, Vauclair S, Koch U, Ferrand G, Ferrero I, Demehri S, Song LL, Farr AG, Leonard WJ, Kopan R, Miele L, Hohl D, Finke D, Radtke F (2010) Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of notch signaling in the murine skin. PLoS One 5: e9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrach S, Cordes R, Hozumi K, Gossler A, Watt FM (2008) Role of the Notch ligand Delta1 in embryonic and adult mouse epidermis. J Invest Dermatol 128: 825–832 [DOI] [PubMed] [Google Scholar]
- Ezratty EJ, Stokes N, Chai S, Shah AS, Williams SE, Fuchs E (2011) A Role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell 145: 1129–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B (2001) Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15: 2069–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, Murray JC, Schutte BC (2006) Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet 38: 1335–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 194: 237–255 [DOI] [PubMed] [Google Scholar]
- Jones PH, Watt FM (1993) Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73: 713–724 [DOI] [PubMed] [Google Scholar]
- Kolev V, Mandinova A, Guinea-Viniegra J, Hu B, Lefort K, Lambertini C, Neel V, Dummer R, Wagner EF, Dotto GP (2008) EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol 10: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, McDonald-McGinn DM, Zackai EH, Lammer EJ, Aylsworth AS, Ardinger HH, Lidral AC, Pober BR, Moreno L, Arcos-Burgos M, Valencia C et al. (2002) Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet 32: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Roop DR (2004) The role of p63 in development and differentiation of the epidermis. J Dermatol Sci 34: 3–9 [DOI] [PubMed] [Google Scholar]
- Lazarov M, Kubo Y, Cai T, Dajee M, Tarutani M, Lin Q, Fang M, Tao S, Green CL, Khavari PA (2002) CDK4 coexpression with Ras generates malignant human epidermal tumorigenesis. Nat Med 8: 1105–1114 [DOI] [PubMed] [Google Scholar]
- Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, Neel V, Garlick J, Chiorino G, Dotto GP (2007) Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev 21: 562–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell S, Jones P, Le Roux I, Dunne J, Watt FM (2000) Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol 10: 491–500 [DOI] [PubMed] [Google Scholar]
- Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J (1997) Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol 17: 6057–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandinova A, Kolev V, Neel V, Hu B, Stonely W, Lieb J, Wu X, Colli C, Han R, Pazin M, Ostano P, Dummer R, Brissette JL, Dotto GP (2009) A positive FGFR3/FOXN1 feedback loop underlies benign skin keratosis versus squamous cell carcinoma formation in humans. J Clin Invest 119: 3127–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone M, Selfors LM, Albeck J, Overholtzer M, Sale S, Carroll DL, Pandya D, Lu Y, Mills GB, Aster JC, Artavanis-Tsakonas S, Brugge JS (2010) Dose-dependent induction of distinct phenotypic responses to Notch pathway activation in mammary epithelial cells. Proc Natl Acad Sci USA 107: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon F (2004) p63 and the epithelial stem cell: more than status quo? Genes Dev 18: 465–469 [DOI] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, Botti E, Giunta A, Spallone G, Garaffo G, Vernersson-Lindahl E, Merlo G, Mills AA, Ballaro C, Alema S, Chimenti S, Guerrini L, Costanzo A (2010) A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest 120: 1570–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC (2006) Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124: 973–983 [DOI] [PubMed] [Google Scholar]
- Nguyen BC, Lefort K, Mandinova A, Antonini D, Devgan V, Della Gatta G, Koster MI, Zhang Z, Wang J, di Vignano AT, Kitajewski J, Chiorino G, Roop DR, Missero C, Dotto GP (2006) Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev 20: 1028–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff BJ, Qin JZ, Chaturvedi V, Denning MF, Bonish B, Miele L (2002) Jagged-1 mediated activation of notch signaling induces complete maturation of human keratinocytes through NF-kappaB and PPARgamma. Cell Death Differ 9: 842–855 [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, Van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F (2003) Notch1 functions as a tumor suppressor in mouse skin. Nat Genet 33: 416–421 [DOI] [PubMed] [Google Scholar]
- Nocentini G, Giunchi L, Ronchetti S, Krausz LT, Bartoli A, Moraca R, Migliorati G, Riccardi C (1997) A new member of the tumor necrosis factor/nerve growth factor receptor family inhibits T cell receptor-induced apoptosis. Proc Natl Acad Sci USA 94: 6216–6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K, Varnum-Finney B, Flowers D, Anasetti C, Myerson D, Bernstein ID (2000) Monocytes express high amounts of Notch and undergo cytokine specific apoptosis following interaction with the Notch ligand, Delta-1. Blood 95: 2847–2854 [PubMed] [Google Scholar]
- Pan Y, Lin MH, Tian X, Cheng HT, Gridley T, Shen J, Kopan R (2004) gamma-secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell 7: 731–743 [DOI] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, Melbye M, Jugessur A, Lie RT, Wilcox AJ, Fitzpatrick DR, Green ED, Mossey PA, Little J, Steegers-Theunissen RP, Pennacchio LA et al. (2008) Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet 40: 1341–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, Miele L, Aguet M, Radtke F, Dotto GP (2001) Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J 20: 3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Jiang R, Dixon MJ (2009) Integration of IRF6 and Jagged2 signalling is essential for controlling palatal adhesion and fusion competence. Hum Mol Genet 18: 2632–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson RJ, Dixon J, Malhotra S, Hardman MJ, Knowles L, Boot-Handford RP, Shore P, Whitmarsh A, Dixon MJ (2006) Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet 38: 1329–1334 [DOI] [PubMed] [Google Scholar]
- Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L (2008) Rational targeting of Notch signaling in cancer. Oncogene 27: 5124–5131 [DOI] [PubMed] [Google Scholar]
- Schroeder T, Just U (2000) Notch signalling via RBP-J promotes myeloid differentiation. EMBO J 19: 2558–2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T (2008) The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26: 535–584 [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N (2001) IRF family of transcription factors as regulators of host defense. Annu Rev Immunol 19: 623–655 [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J (2010) Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Invest 120: 1561–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA (2006) p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev 20: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Pasolli HA, Williams T, Fuchs E (2008) AP-2 factors act in concert with Notch to orchestrate terminal differentiation in skin epidermis. J Cell Biol 183: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt FM, Estrach S, Ambler CA (2008) Epidermal Notch signalling: differentiation, cancer and adhesion. Curr Opin Cell Biol 20: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall MD, Pietenpol JA (2004) p63: Molecular complexity in development and cancer. Carcinogenesis 25: 857–864 [DOI] [PubMed] [Google Scholar]
- Williams SE, Beronja S, Pasolli HA, Fuchs E (2011) Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature 470: 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA (2006) Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124: 985–996 [DOI] [PubMed] [Google Scholar]
- Wu X, Nguyen BC, Dziunycz P, Chang S, Brooks Y, Lefort K, Hofbauer GF, Dotto GP (2010) Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature 465: 368–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin-Ozuysal O, Fiche M, Guitierrez M, Wagner KU, Raffoul W, Brisken C (2010) Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ 17: 1600–1612 [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Tanigaki K, Han H, Hiai H, Honjo T (2003) Notch/RBP-J signaling regulates epidermis/hair fate determination of hair follicular stem cells. Curr Biol 13: 333–338 [DOI] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Intlekofer AM, Blacklow SC, Zhuang Y, Pear WS (2009) Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes Dev 23: 1665–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Pagano JS (2002) Structure and function of IRF-7. J Interferon Cytokine Res 22: 95–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.