Abstract
Autophagy is an evolutionarily conserved mechanism of cellular self-digestion in which proteins and organelles are degraded through delivery to lysosomes. Defects in this process are implicated in numerous human diseases including cancer. To further elucidate regulatory mechanisms of autophagy, we performed a functional screen in search of microRNAs (miRNAs), which regulate the autophagic flux in breast cancer cells. In this study, we identified the tumour suppressive miRNA, miR-101, as a potent inhibitor of basal, etoposide- and rapamycin-induced autophagy. Through transcriptome profiling, we identified three novel miR-101 targets, STMN1, RAB5A and ATG4D. siRNA-mediated depletion of these genes phenocopied the effect of miR-101 overexpression, demonstrating their importance in autophagy regulation. Importantly, overexpression of STMN1 could partially rescue cells from miR-101-mediated inhibition of autophagy, indicating a functional importance for this target. Finally, we show that miR-101-mediated inhibition of autophagy can sensitize breast cancer cells to 4-hydroxytamoxifen (4-OHT)-mediated cell death. Collectively, these data establish a novel link between two highly important and rapidly growing research fields and present a new role for miR-101 as a key regulator of autophagy.
Keywords: autophagy, breast cancer, microRNA
Introduction
Autophagy is a cellular self-catabolic degradation process in which cytoplasmic constituents are sequestered into double membrane vesicles called autophagosomes, and subsequently degraded via the lysosomal pathway (He and Klionsky, 2009). In addition to its important housekeeping and homeostatic functions at basal levels, autophagy promotes the survival of starved or stressed cells through the recycling of nutrients and through removal of damaged proteins and organelles (Mathew et al, 2007; Mizushima et al, 2008). Autophagy is essential for normal development (Yue et al, 2003; Kuma et al, 2004) and defects in this process are linked with numerous human diseases including neurodegenerative disorders and cancer (Levine and Kroemer, 2008; Chen and Debnath, 2010). In a tumour microenvironment, autophagy can promote cancer cell survival in response to metabolic stress (Degenhardt et al, 2006; Karantza-Wadsworth et al, 2007). However, progressive autophagy can also induce cell death and human cancers often display inactivating mutations in autophagy promoting genes (Corcelle et al, 2009; Chen and Karantza, 2011). Thus, in relation to tumourigenesis, the role of autophagy is complex and likely depends on the genetic composition of the cell as well as the environmental cues the cell is exposed to.
miRNAs are small non-coding RNAs which post-transcriptionally regulate gene expression, predominantly through imperfect base pairing with the 3′-untranslated region (3′UTR) of target mRNAs (Valencia-Sanchez et al, 2006). miRNA-mediated repression of gene expression occurs through complex mechanisms which are not fully understood, including translational inhibition and mRNA degradation (Filipowicz et al, 2008). Substantial evidence implicates a functional role for miRNAs in different cancers and in support of this ∼50% of miRNAs are located at fragile sites of the genome, which are commonly amplified or deleted in human cancers (Calin et al, 2004). Accordingly, miRNA expression is often deregulated in cancer and miRNAs can act as oncogenes or tumour suppressors. In addition, miRNA expression profiling can be used to predict the clinical outcome of cancer patients (Lu et al, 2005; Volinia et al, 2006; Jiang et al, 2008).
Despite recent advances in understanding the molecular mechanisms, which regulate autophagy, several gaps remain (Behrends et al, 2010; Lipinski et al, 2010; Szyniarowski et al, 2011). Considering the widespread implications of both miRNAs and autophagy in cancer-related processes and given the lack of current evidence linking these two rapidly growing fields of research, we were prompted to search for miRNAs which regulate autophagy. Through a luciferase-based functional screening approach, we identified miR-101 as a potent inhibitor of autophagic flux in MCF-7 breast cancer cells. miR-101 is lost in several cancer types including liver, prostate and breast, and emerging evidence suggests a tumour suppressive role for this miRNA (Varambally et al, 2008; Su et al, 2009; Chiang et al, 2010; Buechner et al, 2011). Accordingly, miR-101 induces apoptosis and suppresses tumourigenicity in vitro and in vivo, and was recently reported to inhibit migration and invasion of gastric cancer cells (Varambally et al, 2008; Su et al, 2009; Wang et al, 2010). Established targets for miR-101 include enhancer of zeste homologue 2 (EZH2), cyclooxygenase-2 (COX-2), amyloid precursor protein (APP) and myeloid cell leukaemia sequence-1 (MCL-1) (Varambally et al, 2008; Strillacci et al, 2009; Su et al, 2009; Vilardo et al, 2010). We report three novel miR-101 targets identified by microarray expression analysis, STMN1, RAB5A and ATG4D, which we show to be important autophagic regulators. Importantly, ectopic expression of a miR-101-resistant form of STMN1 alleviates miR-101-mediated repression of autophagy, confirming the functional importance of this target. Finally, in line with recent evidence suggesting that autophagic inhibition sensitizes breast cancer cells to cell death induced by various chemotherapeutic agents (Abedin et al, 2007; de Medina et al, 2009) we show that miR-101, likely through its inhibition of autophagy, can sensitize to 4-OHT treatment by enhancing 4-OHT-mediated cell death.
Results
A functional screen identifies miRNAs affecting autophagy
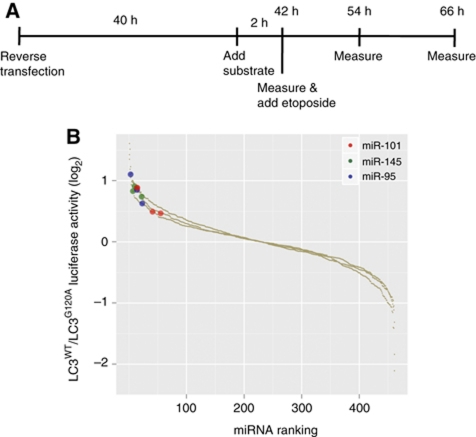
To identify miRNAs regulating autophagy, we employed a functional screening approach using a cell-based assay system in which the autophagosomal marker, MAP1-LC3 (LC3), is fused to a renilla luciferase (RLuc) reporter molecule forming the RLuc–LC3 fusion protein (Farkas et al, 2009). As LC3 itself is specifically degraded by autophagy, the level of autophagy in an MCF-7 reporter cell line stably expressing wild-type RLuc–LC3 (RLuc–LC3WT) can be measured in real time using an in vivo RLuc substrate. As a reference control, MCF-7 cells expressing a mutant fusion protein, RLuc–LC3G120A, which is unable to undergo autophagosomal localization and is thereby not specifically degraded by autophagy, are assayed in parallel. The autophagic flux can hence be evaluated as the change in the relative levels of these two fusion proteins (hereafter denoted as LC3WT/LC3G120A; Farkas et al, 2009).
The reporter cell system was transfected in 96-well format with a library of ∼470 miRNA precursor molecules covering the most abundant human miRNAs following the scheme outlined in Figure 1A. We measured the intrinsic effect of overexpressing the miRNAs on the basal autophagic flux at 42 h post-transfection after which etoposide was added. The autophagy-inducing effect of etoposide is well documented (Shimizu et al, 2004; Katayama et al, 2007; Farkas et al, 2009), and including etoposide treatment in the screen enabled greater sensitivity for the detection of miRNAs blocking autophagy. The RLuc activity was measured again at 12 and 24 h following etoposide addition. Aside from miRNAs, a number of control siRNAs were included in the screen as shown in Supplementary Figure S1. Knockdown of the essential autophagy component Beclin-1 (Supplementary Figure S1A) effectively inhibited autophagy as evident from measurements of the autophagic flux (Supplementary Figure S1B). Transfection efficiency throughout the screen was monitored using a siRNA against RLuc (Supplementary Figure S1C). Furthermore, scrambled control siRNAs scored similarly to the average of the entire miRNA library, ensuring that this negative control was suitable (Supplementary Figure S1D). To monitor and ensure reproducibility, the screening procedure was repeated three times.
Figure 1.
Screening approach for identification of miRNAs regulating autophagic flux in MCF-7 cells. (A) Outline of the timeline used for the screening assay. (B) Combined results from three independent screens, 66 h after transfection. MCF-7 RLuc–LC3WT and RLuc–LC3G120A were reverse transfected with the Ambion Pre-miR Library in three independent screens (see Materials and methods for details). The miRNAs were ranked according to log2 LC3WT/LC3G120A luciferase activity values (y axis). Highlighted in red, green and blue are three miRNAs, which consistently inhibited autophagic flux in the three independent screens (miR-145 P=0.0033, miR-95 P=0.0033 and miR-101 P=0.0305).
Reasoning that autophagy could be induced as a stress response following overexpression of non-physiological levels of miRNAs or from miRNAs expressed outside their normal physiological context, we chose to focus on miRNAs inhibiting autophagy. Figure 1B shows the combined results of all three screens in which the miRNAs have been ranked according to fold change values (LC3WT/LC3G120A). Statistical analysis using a non-parametric rank product method based on ranks of fold changes (Breitling et al, 2004) revealed miR-95, miR-145 and miR-101 as the three most consistent, high-ranking miRNAs, which significantly inhibited autophagic flux in all three screens.
miR-101 is regulated during autophagy
Among the miRNAs identified to repress autophagy, miR-101 and miR-145 were immediately interesting due to well-established links to cancer (Varambally et al, 2008; Su et al, 2009; Kent et al, 2010). Since we have previously seen that miR-145 levels are undetectable in MCF-7 cells (Gregersen et al, 2010) we focused our attention on miR-101. To explore possible links between autophagy and miR-101 expression, we measured the level of endogenous miR-101 under basal growth conditions and following induction of autophagy. Detection of miR-101 in MCF-7 cells by quantitative PCR (qPCR) analysis revealed that endogenous miR-101 expression is increased by various triggers of autophagy including starvation, rapamycin and etoposide treatment (Supplementary Figure S2A and B; top). The mammalian target of rapamycin complex 1 (mTORC1) is a key negative regulator of autophagy signalling and its activation status reflects the level of autophagy in cells (Jung et al, 2010). Phospho-S6-kinase (p-S6K), a direct target of mTORC1, was used to indicate the extent of mTORC1 inactivation caused by these treatments (Supplementary Figure S2A and B; bottom). Considering the differential effects of etoposide and rapamycin, mTORC1-independent signalling could also influence the upregulation of this miRNA. These data indicate a potential physiological role for endogenous miR-101 in autophagy regulation in MCF-7 cells, prompting us to further analyse its function.
Overexpression of miR-101 represses autophagy
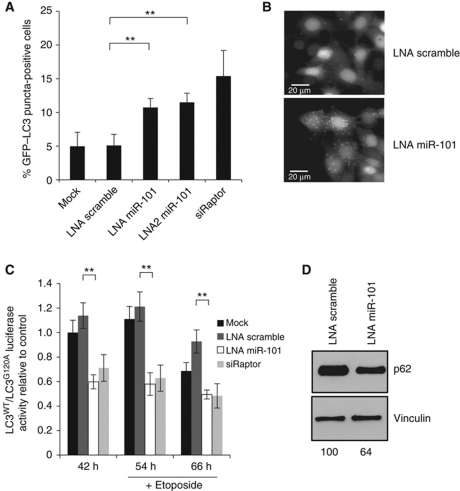
To validate the repressive effect of miR-101 overexpression on autophagy, three independent methods were employed. First, MCF-7 cells stably expressing an eGFP–LC3 fusion protein were used to quantify accumulation of autophagosomes by eGFP–LC3 translocation. As can be seen from Figure 2A, the percentage of eGFP–LC3 puncta-positive cells was significantly reduced in cells overexpressing miR-101, relative to cells transfected with the controls miR-203 and a siRNA against RLuc. miR-203 was chosen as a suitable miRNA-negative control since it had no effect on autophagy, behaving similarly to control siRNAs. Importantly, the effect of miR-101 was comparable to that of cells treated with a siRNA against Beclin-1. Representative images are shown in Figure 2B. Second, the effect of miR-101 on autophagic flux was confirmed using the RLuc assay from the screen. Also in this assay, miR-101 significantly inhibited the autophagic flux (both basal and etoposide induced) to an extent similar to Beclin-1 knockdown and significantly more than the controls (Figure 2C). Finally, we assayed the ability of miR-101 to affect the level of p62. The poly-ubiquitin binding protein p62 binds directly to LC3 and acts as a selective autophagy receptor and molecular carrier of cargo to be degraded by autophagy (Bjorkoy et al, 2005; Pankiv et al, 2007). As p62 itself localizes to autophagosomes and is degraded during autophagy, the level of p62 reflects the autophagic turnover. As evident from Figure 2D, overexpression of miR-101 in MCF-7 cells results in the accumulation of p62, reflecting an inhibition of autophagy. Hence, in accordance with the original screen, all three assays employed validate that overexpression of miR-101 inhibits autophagy in MCF-7 cells. Importantly, this finding extends to other cell lines including T47D, HEK and HeLa cells, as assessed by p62 expression levels, indicating that miR-101 regulation of autophagy is a general phenomenon (Supplementary Figure S3).
Figure 2.
miR-101 overexpression inhibits autophagy. (A) miR-101 overexpression inhibits eGFP–LC3 translocation. MCF-7 eGFP–LC3 cells were transfected with indicated miRNAs or siRNAs and fixed 72 h post-transfection. Percentage of eGFP–LC3 puncta-positive cells was quantified by automated image acquisition and analysis using a threshold of >5 dots/cell. Data are shown as the mean±s.d. of five replicates and is representative of three independent experiments. *P<0.05, **P<0.005. (B) Representative images from the quantification shown in (A). Scale bars represent 20 μM. (C) miR-101 overexpression inhibits autophagic flux. MCF-7 RLuc–LC3WT and RLuc–LC3G120A cells were reverse transfected with indicated miRNAs or siRNAs and 42 h later 50 μM etoposide was added. Luciferase activity was measured at 42, 54 and 66 h after transfection. Data are shown as the mean±s.d. of three replicates and are representative of three independent experiments. *P<0.05, **P<0.005. (D) miR-101 overexpression leads to accumulation of p62 protein. Western blot analysis 72 h after transfection with miR-101 or scramble control. p62 bands were quantified relative to the vinculin loading control using ImageJ software and the relative quantifications are shown. The data are representative of three independent experiments.
Inhibition of endogenous miR-101 induces autophagy
As the screen was performed using miRNA overexpression, an important validation step was to document the physiological relevance of the findings. Towards this, we inhibited the endogenous miR-101 in MCF-7 cells and repeated the three validation assays outlined above. In the eGFP–LC3 translocation assay, two different designs of LNA-derived miR-101 inhibitors significantly increased the number of puncta-positive cells relative to a scrambled LNA or mock-transfected cells (Figure 3A and B). In these experiments, a siRNA against Raptor, an essential component of mTORC1 and a key negative regulator of autophagy signalling (Hara et al, 2002; Jung et al, 2010) served as a positive control. The efficiency of the Raptor knockdown was verified by qPCR (Supplementary Figure S5A). Likewise in the RLuc assay, inhibition of endogenous miR-101 resulted in a clear increase in the autophagic flux similar to the siRNA against Raptor (Figure 3C). Lastly, also the level of the p62 protein was diminished in cultures treated with an LNA targeting miR-101 (Figure 3D). Since depletion of endogenous miR-101 did not affect the growth of the cells (Supplementary Figure S4), the autophagy phenotype is not likely a secondary effect of altered cellular growth. Hence, we demonstrate that endogenous miR-101 affects the process of autophagy, thereby qualifying the physiological relevance of the original findings from the screen.
Figure 3.
Knockdown of endogenous miR-101 induces autophagy. (A) miR-101 inhibition induces eGFP–LC3 translocation. MCF-7 eGFP–LC3 cells were transfected with indicated LNAs or siRNAs and fixed 72 h post-transfection. Percentage of eGFP–LC3 puncta-positive cells was quantified using a threshold of >5 dots per cell. Data are shown as the mean±s.d. of five replicates and are representative of three independent experiments. **P<0.005. (B) Representative images from the quantification shown in (A). Scale bars represent 20 μM. (C) miR-101 inhibition induces autophagic flux. MCF-7 RLuc–LC3WT and RLuc–LC3G120A cells were reverse transfected with indicated LNAs or siRNAs and 42 h later 50 μM etoposide was added. Luciferase activity was measured at 42, 54 and 66 h after transfection. Data are shown as the mean±s.d. of three replicates and are representative of three independent experiments. **P<0.005. (D) miR-101 inhibition leads to decreased p62 expression. Western blot analysis 72 h after transfection with LNA miR-101 or LNA scramble. p62 bands were quantified relative to the vinculin loading control using ImageJ software and the relative quantifications are shown. The data are representative of three independent experiments.
Experimental identification of miR-101 targets
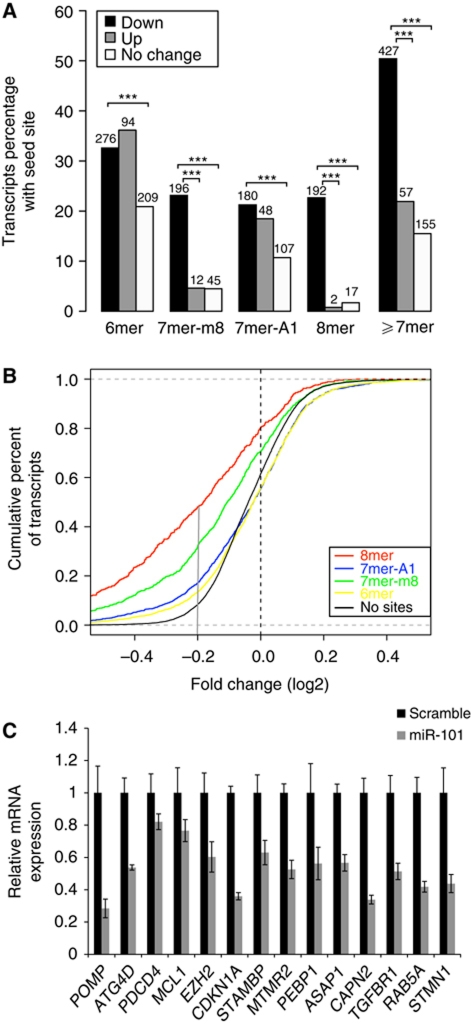
Having established a role for miR-101 in autophagy, we next wanted to clarify the underlying mechanism by identifying the direct downstream targets, which are repressed. We have previously used miRNA inhibition or overexpression coupled to transcriptome profiling to identify biological responses of—and direct targets for—several miRNAs (Frankel et al, 2008; Christoffersen et al, 2010; Gregersen et al, 2010). To identify targets of miR-101, MCF-7 cells were transfected with a miR-101 precursor or scramble control. Transfections were performed in triplicate and total RNA was harvested 24 h post-transfection and analysed using Affymetrix HG-U133 2.0 arrays. Following filtering, normalization and statistical analysis, we found 1107 differentially expressed transcripts of which 847 were downregulated and 260 were upregulated upon miR-101 overexpression (false discovery rate (FDR) ⩽0.1). Comparison of downregulated, upregulated and no change transcript sets revealed a highly significant enrichment for miR-101 seed site occurrence in the 3′UTRs of the downregulated transcripts for four different seed types (Figure 4A). Among the different seed types, the strongest enrichment was observed for the 8mer seed sites. To assess the efficacy of different seed types on mRNA repression, we compared the cumulative distribution functions (CDFs) of fold change for transcripts containing miR-101 seed sites or no seed sites (Figure 4B). The downregulation levels of transcripts with all seed match types were significantly higher than those without seed sites (P-values <0.0005). We validated the microarray study by qPCR analysis of 14 transcripts from the downregulation set, all of which were found to be downregulated in an independent experiment (Figure 4C). Supplementary Table I summarizes the complete list of 1107 differentially expressed transcripts.
Figure 4.
Identification of miR-101 targets by microarray analysis after miR-101 overexpression. (A) Seed site enrichment. The proportion and actual number of transcripts in the up, down and no change sets with different seeds types are shown. The seed types are mutually exclusive (see Materials and methods). Enrichment significance is labelled as ***P<0.0005 for down set versus up set and down set versus no change set. ‘⩾7mer’ includes 8mer, 7mer-m8 and 7mer-A1 sites. P-values for ⩾7mer seed site enrichment were 1.4e−16 (down versus up) and 1.9e−59 (down versus no change). (B) Different seed match types affect the level of transcript downregulation. Cumulative distribution functions for fold changes (log2FC) of transcripts containing different seed types or no seed sites for miR-101. X axis is log2FC from high downregulation to no change (log2FC=0) and from no change to high upregulation and y axis is the fraction of transcripts smaller than or equal to a certain fold change. For example, the solid grey vertical line drawn at log2FC=−0.2 approximately corresponds to 45% of transcripts containing 8mer seed type that were downregulated at least log2FC −0.2, compared with only 10% of transcripts containing 6mer seed type. (C) qPCR validation of 14 downregulated transcripts from an independent transfection experiment. For each transcript, the values are normalized relative to the scramble control sample and to the housekeeping gene. The error bars represent ±s.d. of three replicates.
Knockdown of STMN1, RAB5A and ATG4D inhibits basal and rapamycin-induced autophagy
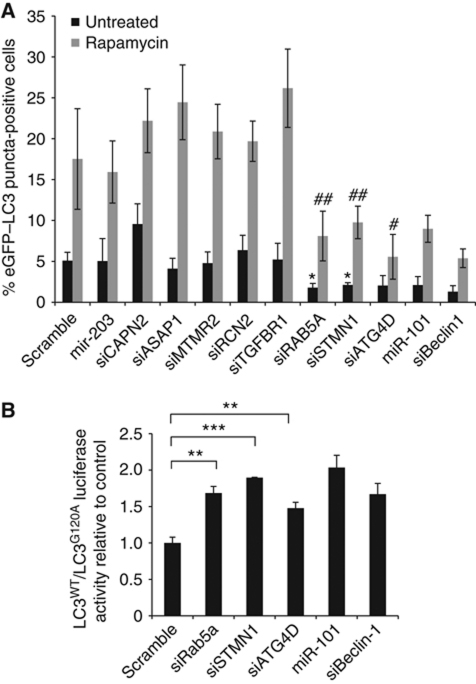
In order to narrow down our search for biologically relevant miR-101 targets, we chose eight genes identified by the array analysis with potentially interesting links to autophagy and knocked them down individually by siRNA. We reasoned that knockdown of functionally important targets downstream of miR-101, should, at least in part, phenocopy the effect of overexpressing miR-101. We tested the effect of these siRNAs on eGFP–LC3 translocation and identified three genes, STMN1, RAB5A and ATG4D, which when knocked down inhibited both basal and rapamycin-induced autophagy (Figure 5A). The knockdown efficiencies of the individual siRNAs were measured by qPCR (Supplementary Figure S5B).
Figure 5.
siRNAs against STMN1, RAB5A and ATG4D inhibit basal and rapamycin-induced autophagy. (A) siRNA knockdown of a panel of genes selected from the array analysis led to identification of STMN1, RAB5A and ATG4D as potentially interesting miR-101 target candidates. MCF-7 eGFP–LC3 cells were transfected with indicated miRNAs or siRNAs and fixed 72 h post-transfection. Two hours prior to fixation, cells were treated with 200 nM rapamycin or left untreated. Percentage of eGFP–LC3 puncta-positive cells was quantified as described previously. Data are shown as the mean±s.d. of five replicates and are representative of two or three independent experiments. *P<0.05 relative to miR-203 untreated, #P<0.05 relative to miR-203 rapamycin, ##P<0.005 relative to miR-203 rapamycin. (B) Knockdown of STMN1, RAB5A and ATG4D decreases autophagic flux. MCF-7 RLuc–LC3WT and RLuc–LC3G120A cells were reverse transfected with indicated miRNAs or siRNAs and 42 h later luciferase activity was measured. Data are shown as the mean±s.d. of three replicates and are representative of at least three independent experiments. **P<0.005, ***P<0.0005.
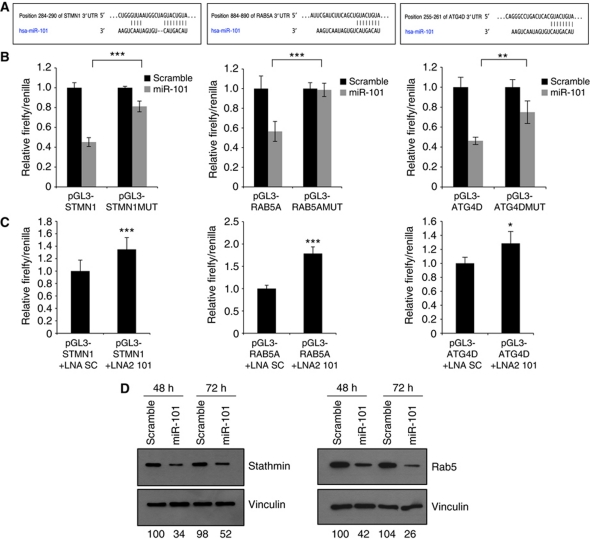
STMN1 encodes Stathmin/Oncoprotein18, a cytosolic phosphoprotein that regulates microtubule dynamics and is found overexpressed in many cancers (Marklund et al, 1996; Rana et al, 2008). In an independent siRNA-based screen, we had previously identified Stathmin as a regulator of autophagy (Maria Høyer-Hansen and Marja Jäättelä, unpublished data). RAB5A, a small GTPase that regulates early endocytosis, was previously suggested to act at an early stage of autophagosome formation (Ravikumar et al, 2008). Finally, ATG4D is a member of the ATG4 family of cysteine proteases, which regulate autophagosome biogenesis through the processing of LC3, allowing its subsequent conjugation to phosphatidylethanolamine (PE) on autophagosomal membranes (Marino et al, 2003; Scherz-Shouval et al, 2007). While overexpression of miR-101 resulted in ∼50% reduction in transcript abundance for STMN1, RAB5A and ATG4D (Figure 4C), specific siRNAs reduced the levels to varying degrees (∼45% for RAB5A, ∼25% for ATG4D and <5% for STMN1; Supplementary Figure S5B).
In addition to regulating steady-state autophagy, knockdown of these three genes partly abrogated the autophagic flux, as shown by a significant increase in the LC3WT/LC3G120A ratio (Figure 5B). In both assays, the extent of autophagic inhibition by these siRNAs was similar to Beclin-1 knockdown or miR-101 overexpression (Figure 5A and B). Since the downregulation of STMN1, RAB5A and ATG4D also decreased eGFP–LC3 translocation in the face of Rapamycin treatment (Figure 5A, grey bars), and since Rapamycin induces autophagy through inhibition of mTORC1, these results indicate a function for these proteins and for miR-101 downstream of mTORC1. Further supporting this data, eGFP–LC3 translocation induced by siRNA-mediated depletion of Raptor is potently abrogated by miR-101 overexpression, additionally suggesting a function for this miRNA downstream of mTORC1 (Supplementary Figure S6).
miR-101 directly targets STMN1, RAB5A and ATG4D
To establish a direct molecular link, we next examined the ability of miR-101 to regulate RAB5A, STMN1 and ATG4D. As evident from Supplementary Table I, STMN1 and ATG4D hold a single 8mer seed match to miR-101 within the 3′UTR while RAB5A contains both an 8mer and a 7mer site. The predicted binding patterns of miR-101 to the 8mer motifs are depicted in Figure 6A. We cloned 300–500 base-pair 3′UTR fragments from STMN1, RAB5A and ATG4D downstream a luciferase reporter and tested the ability of miR-101 to regulate the reporters. As shown in Figure 6B, the 3′UTRs of all three putative target genes responded markedly to miR-101 overexpression relative to a scrambled control. Introducing three point mutations into the predicted miR-101 binding motifs either abolished or significantly reduced this effect, indicating that these interactions are direct. Importantly, the luciferase vectors are also susceptible to repression by the endogenous miR-101 as LNA-mediated inhibition of miR-101 specifically relieves the repression of all three reporters (Figure 6C). Finally, we examined the effect of miR-101 on the endogenous Stathmin and RAB5A proteins (specific antibodies for ATG4D were not available). As evident from Figure 6D, overexpression of miR-101 results in a potent downregulation of both Stathmin and RAB5A protein levels.
Figure 6.
miR-101 directly targets STMN1, RAB5A and ATG4D. (A) Predicted binding between miR-101 and the 8mer seed matches in STMN1, RAB5A and ATG4D 3′UTRs. (B, C) miR-101 regulates STMN1, RAB5A and ATG4D 3′UTR reporters. Luciferase reporter assays 24 h after transfection with indicated pGL3 firefly plasmids and a renilla transfection control plasmid, co-transfected with miR-101, LNA miR-101 or relevant scramble controls. Data shown are the mean±s.d. of four replicates and are representative of two or three independent experiments. *P<0.05, **P<0.005, ***P<0.0005. (D) miR-101 decreases Stathmin and RAB5A protein levels. Western blot analysis 48 or 72 h after transfection with miR-101 or scramble control. The Stathmin and RAB5 bands were quantified relative to the vinculin loading control using ImageJ software and the relative quantifications are shown. The data are representative of three independent experiments.
Stathmin is a functional miR-101 target
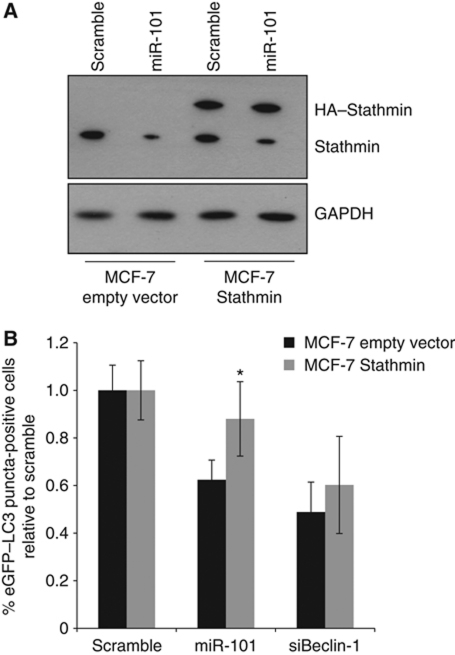
Since both RAB5A and ATG4D have previously established roles in autophagy (Marino et al, 2003; Ravikumar et al, 2008), we focused our attention on the importance of Stathmin as a miR-101 target. To further confirm functional roles for miR-101 and Stathmin in autophagy, transmission electron microscopy was used for detection and quantification of autophagosomes in cellular cross-sections. Figure 7A–C shows representative images of rapamycin-treated MCF-7 cells, either transfected with a scramble control, upon miR-101 overexpression or STMN1 knockdown (see also Supplementary Figure S7). Quantification of autophagosomes per cellular cross-section revealed a significant reduction upon miR-101 overexpression or STMN1 knockdown, relative to the scramble control (Figure 7D), confirming our above findings. To further address the functional importance of STMN1 as a miR-101 target, we developed a stable MCF-7 eGFP–LC3 cell line expressing an HA-tagged Stathmin (MCF-7-Stathmin). HA–Stathmin lacks a 3′UTR and is insensitive to miR-101-mediated downregulation (Figure 8A). Importantly, MCF-7-Stathmin cells were less sensitive to miR-101-mediated inhibition of autophagy, relative to the empty vector control cell line (P=0.006; Figure 8B), strongly suggesting the functional importance of Stathmin as a miR-101 target.
Figure 7.
Transmission electron microscopy confirms repression of autophagy by miR-101 overexpression or Stathmin knockdown. (A–C) Representative images of MCF-7 cells transfected for 72 h as indicated, and treated for 2 h with 200 nM rapamycin prior to fixation. Top panel: overview images (× 3400 magnification). Scale bars represent 5 μM. Bottom panel: close-up images (× 24 500 magnification) of cytoplasmic regions containing autophagosomes (denoted by white arrowheads). Scale bars represent 1 μM. (D) The data were quantified by counting the number of autophagosomes per cross-sectioned cell (scramble n=30, miR-101 n=25, siStathmin n=25). ***P<0.0005.
Figure 8.
Stathmin is an important functional target of miR-101 in autophagy. (A) MCF-7-Stathmin cells stably express a 3′UTR-less HA–Stathmin, which is resistant to miR-101-mediated regulation. Cells were analysed by western blotting 72 h after transfection for levels of HA–Stathmin (upper band) or endogenous Stathmin (lower band). GAPDH was used as a loading control. (B) MCF-7-Stathmin cells are less sensitive to miR-101-mediated inhibition eGFP–LC3 translocation. MCF-7-Stathmin or MCF-7 empty vector control cells were transfected as indicated, fixed 72 h post-transfection and the percentage of eGFP–LC3 puncta-positive cells was quantified as described previously. The data are shown as the mean±s.e.m. of four pooled experiments, each with five replicates and are plotted relative to the Scramble control. *P<0.05 for miR-101 MCF-7-Stathmin versus miR-101 MCF-7 empty vector.
miR-101 synergizes with 4-OHT in inducing MCF-7 cell death
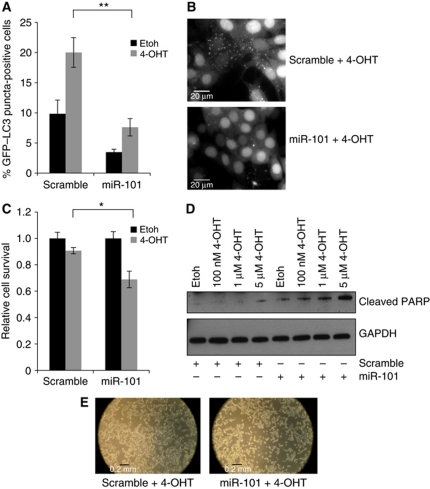
It is well established by several studies that various chemotherapeutics induce autophagy in different cell types including breast cancer cells (Abedin et al, 2007; Kaushal et al, 2008; de Medina et al, 2009). This increased drug-induced autophagy has pro-survival functions in cancer cells, underlining a potential role for autophagy inhibitors in combination with conventional chemotherapeutics in cancer treatment (Abedin et al, 2007; Amaravadi et al, 2007; de Medina et al, 2009). 4-OHT is the active metabolite of tamoxifen, an antagonist of the oestrogen receptor (ER) that has been used as first-line endocrine therapy for ER-positive breast cancer patients for many years (Stuart-Harris and Davis, 2010). Since MCF-7 is an ER-positive breast cancer cell line, we turned to 4-OHT treatment to further establish a functional importance of miR-101 in autophagy-related cellular phenotypes. We demonstrate in Figure 9A and B that autophagy is efficiently induced 48 h after addition of 1 μM 4-OHT. As previously demonstrated (Figure 2A), miR-101 reduces the basal level of eGFP–LC3 translocation (Figure 9A; Etoh), but importantly, miR-101 is also capable of reducing eGFP–LC3 translocation in the face of 4-OHT treatment (Figure 9A and B; 4-OHT). We next examined the ability of miR-101 to enhance 4-OHT-mediated cell death. The combinatorial treatment with 4-OHT and miR-101 significantly reduced cellular survival (Figure 9C) and sensitized cells to 4-OHT-induced apoptosis as evidenced from increased PARP cleavage (85 kDa fragment) and cellular morphology visualized by light microscopy (Figure 9D and E). A similar synergistic effect of miR-101 and 4-OHT on cell survival was observed in another ER-positive breast cancer cell line, T47D (Supplementary Figure S8).
Figure 9.
miR-101-mediated inhibition of autophagy increases 4-OHT-induced cell death. (A) miR-101 decreases 4-OHT-mediated eGFP–LC3 translocation. MCF-7 eGFP–LC3 cells were transfected with miR-101 or a scramble control and the following day cells were treated with 1 μM 4-OHT or EtOH vehicle control. After 3 days of treatment, the percentage of eGFP–LC3 puncta-positive cells was quantified using a threshold of >5 dots per cell. Data shown is the mean±s.d. of five replicates and are representative of three independent experiments. **P<0.005. (B) Representative images from the quantification in (A). Scale bars represent 20 μM. (C) Combined miR-101 and 4-OHT reduces cell viability. Cells were transfected and treated as described in (A) and cellular viability was determined using the MTT assay. The data are normalized to the EtOH control and show the mean ±s.d. of three replicates, representative of three independent experiments. *P<0.05. (D) Combined miR-101 and 4-OHT increases PARP cleavage. Cells were transfected and treated as described in (A) (with indicated 4-OHT concentrations) and analysed by western blotting 72 h after transfection to assess the amount of 85 kDa PARP cleavage fragment. The data are representative of two independent experiments. (E) Phase contrast microscopy images from cells treated as described in (A).
Discussion
miR-101 regulation of autophagy in breast cancer cells
Biochemical studies in yeast and mammalian cells have defined several core protein complexes involved in autophagy during the past years. However, despite recent advances, several gaps remain in our understanding of autophagy regulation. In this study, we performed a functional screen using a recently described assay measuring autophagic flux (Farkas et al, 2009) to identify miRNAs, which could modulate the level of autophagy in MCF-7 cells. We identified miR-101 as a potent regulator of autophagy and validated this observation with several independent assays measuring both steady-state autophagy and autophagic flux. Interestingly, we observed that endogenous miR-101 expression levels are increased in response to various autophagy-inducing stimuli, perhaps serving as an adaptive feedback response to keep the level of autophagy in check.
miRNAs mainly function by translational repression and/or mRNA degradation; however, recent findings have suggested that destabilization of target mRNAs is the predominant reason for miRNA-mediated reduced protein output, while translational repression alone is a less important component than originally suspected (Baek et al, 2008; Guo et al, 2010). Indeed, the repression of many miRNA targets is frequently associated with mRNA destabilization (Bagga et al, 2005; Giraldez et al, 2006; Wu et al, 2006) and we have previously applied microarray profiling as a means of identifying novel miRNA targets (Frankel et al, 2008; Christoffersen et al, 2010; Gregersen et al, 2010). Bioinformatic analysis of our microarray data revealed a significant enrichment for potential miR-101 binding sites among downregulated transcripts after miR-101 overexpression, demonstrating the validity of this experimental approach. Additionally, we found several known miR-101 targets among these transcripts including EZH2, MCL-1, FOS, APP and ATXN1 (Supplementary Table I; Lee et al, 2008; Varambally et al, 2008; Li et al, 2009; Su et al, 2009; Vilardo et al, 2010). Importantly, we identified three novel miR-101 targets, STMN1, RAB5A and ATG4D and showed that siRNA-mediated knockdown of these genes phenocopied the effect of miR-101, strongly suggesting that miR-101 may exert its anti-autophagic function via these targets. In accordance with this, RAB5A and ATG4D have previously established roles in early steps of autophagosome formation (Marino et al, 2003; Ravikumar et al, 2008) while Stathmin's function in autophagy is revealed for the first time in this study.
miR-101 and targets—interplay in the regulation of autophagy
Gene ontology (GO) analysis of the downregulated genes with at least one 6mer seed against all genes on the array as a background revealed a significant enrichment in the molecular function category for GTP-binding proteins (P=1.3e−10) and proteins with GTPase activity (P=5.8e−10), including several members of the RAB GTPase family (Supplementary Table II). RAB GTPases are conserved from yeast to humans and are generally localized to the cytosolic face of intracellular membranes where they regulate processes such as vesicle formation, movement and membrane fusion (Stenmark, 2009). A role for RAB GTPases in the regulation of autophagy is beginning to emerge; RAB7 is implicated in the maturation of late autophagic vacuoles (Jager et al, 2004) and RAB9 has recently been assigned a role in Atg5/Atg7-independent autophagy where it can mediate autophagosome formation through the fusion of isolation membranes with vesicles from trans-Golgi and late endosomes (Nishida et al, 2009). RAB5 is a well-studied GTPase and a key regulator of the early endocytic pathway in mammalian cells (Bucci et al, 1992). Of the three isoforms that exist, namely RAB5A, RAB5B and RAB5C, only RAB5A has a miR-101 binding site in its 3′UTR and accordingly it is the only isoform that is regulated on our array. A recent study showed that the number of LC3-positive autophagic vesicles in COS-7 cells could be regulated by RAB5. Additionally, they showed that RAB5 regulates Atg5–Atg12 conjugation and that it interacts with Beclin-1 in the presence of the PI3-kinase Vps34 (Ravikumar et al, 2008). Together with our data, these findings support an important role for RAB5 in autophagy regulation downstream of the mTORC1 kinase, and upstream of LC3 lipidation, likely in early autophagosome formation.
ATG4D belongs to the ATG4 family of endopeptidases, which are crucial regulatory components of the autophagosome biogenesis pathway. These proteases cleave the C-terminus of newly synthesized LC3 proteins to reveal a glycine residue required for covalent attachment to PE and later, these same enzymes delipidate PE from outer membrane bound LC3 to facilitate autophagosome–lysosome fusion and LC3 recycling (Kaminskyy and Zhivotovsky, 2011). Genetic deletion of ATG4B, but not ATG4C resulted in prominent defects in autophagy, suggesting functional variations among the different homologues of this protease family (Marino et al, 2007, 2010). Comparisons of the four mammalian homologues in vitro and in vivo indicated that while ATG4B is the main human homologue with the broadest spectrum of substrates and the highest activity, ATG4D possessed only weak activity towards some specific ATG8 proteins, including LC3B (Li et al, 2011). However, the precise roles of the individual proteases in different biological settings remain to be characterized further. Interestingly, our data indicate that the ATG4D isoform in particular plays a more pronounced role than previously acknowledged, at least in the regulation of autophagy in MCF-7 cells. Similarly to RAB5A, the precise location of ATG4D function fits well with our indications for a role for miR-101 primarily located downstream mTORC1 and upstream of LC3 lipidation.
Stathmin is a cytosolic phosphoprotein that destabilizes microtubules and plays an important role in cell-cycle regulation (Sobel, 1991; Marklund et al, 1996; Cassimeris, 2002). This is the first report of a role for Stathmin in autophagy; however, interestingly Stathmin was recently identified in a siRNA-based screen for novel autophagy-regulating genes (Maria Høyer-Hansen and Marja Jäättelä, unpublished data). Whether Stathmin's role in autophagy can be attributed to its microtubule-regulating activity remains unknown and further investigation will be required to specify the location and precise mechanism for its autophagy-related action. Interestingly, Stathmin overexpression caused a partial rescue of miR-101-mediated autophagic inhibition, substantiating its importance as a miR-101 target (Figure 8B). Since most miRNAs function through a multitude of targets, and since this rescue is partial, we cannot rule out that other targets, including RAB5A, ATG4D and likely also others, also contribute to the observed effect of miR-101 on autophagy.
miR-101 regulation of autophagy and its relation to tumour suppression
Over the past years, profiling of human cancers has revealed many miRNAs that function as oncogenes including miR-21, miR-155 and the miR-17-92 cluster, or as tumour suppressors such as the miR-34, let-7 and miR-15a/16-1 families (Calin et al, 2002; He et al, 2005, 2007; Iorio et al, 2005; Costinean et al, 2006; Iorio and Croce, 2009). Loss of miR-101 expression is a common characteristic of several cancers and increasing evidence suggests a clear tumour suppressive role for this miRNA (Varambally et al, 2008; Li et al, 2009; Su et al, 2009; Chiang et al, 2010). Overexpression of miR-101 dramatically suppressed the ability of hepatoma cells to form colonies in vitro and to develop tumours in a xenograft mouse model (Su et al, 2009). Additionally, ectopic expression of miR-101 reduced tumour size and sensitized human lung carcinoma cells to radiation treatment (Yan et al, 2010). In support of its anti-tumourigenic function, most of the identified miR-101 targets are well-established oncogenes including EZH2, COX-2, FOS and MCL-1 which are frequently overexpressed in the same malignancies where miR-101 is lost (Varambally et al, 2008; Li et al, 2009; Strillacci et al, 2009; Su et al, 2009). The mechanism for miR-101's anti-tumourigenic potential is obviously complex, mediated by a diverse range of targets that are likely to vary depending on the cell type and environmental setting.
Interestingly, two of the targets identified in this study also have well-documented oncogenic potential. Stathmin, also commonly referred to as Oncoprotein 18, is highly expressed in, among others, prostate, liver, breast cancers and osteosarcomas (Rana et al, 2008) and inhibition of Stathmin by siRNA induces tumour suppressor-like activity, as measured by proliferation, viability and clonogenicity assays (Alli et al, 2007b). Stathmin increases during neoplastic transformation of human mammary epithelial cells (Deangelis et al, 2010) and intratumoural injection of Stathmin-targeting shRNAs effectively abrogated growth of tumour grafts derived from primary melanomas and osteosarcomas (Phadke et al, 2011). Supporting our observations regarding combinatorial effects of miR-101 and 4-OHT treatment, loss of Stathmin expression sensitized a variety of tumour cell lines to chemotherapeutics such as paclitaxel, cisplatin and vinblastine (Iancu et al, 2000; Singer et al, 2007; Alli et al, 2007a). Similarly to Stathmin, upregulation of RAB5A has been observed in different cancers. Its expression has been correlated with the degree of malignancy and metastasis in non-small cell lung carcinoma (Yu et al, 1999) and elevated protein levels are seen in hepatocellular carcinoma and autonomous thyroid adenomas (Croizet-Berger et al, 2002; Fukui et al, 2007). RAB5A was also recently shown to be overexpressed in ovarian cancer where it promotes cellular proliferation (Zhao et al, 2010). Therefore, it is likely that these two targets can account, to some extent, for miR-101 tumour suppressive activity in certain cancers.
In accordance with several studies implicating a predominant pro-survival role for autophagy in MCF-7 cells (Abedin et al, 2007; Qadir et al, 2008; de Medina et al, 2009), we observed that miR-101, possibly via its inhibition of autophagy, could sensitize breast cancer cells to 4-OHT-induced cell death. Therefore, we speculate that part of the mechanism behind miR-101's tumour suppressive effects could be mediated through inhibition of autophagy via targets including RAB5A, STMN1 and ATG4D. In an in vivo tumour setting, progressive loss of miR-101 could contribute to elevated levels of autophagy in cancer cells, enabling long-term tumour cell survival by allowing them to cope with metabolic stress and promoting eventual re-growth following treatment. Therefore, re-introduction of miR-101 into cancer cells in which it is lost may present a potentially very useful therapeutic strategy for future treatment.
Materials and methods
Cell culture
MCF-7 eGFP–LC3 (Hoyer-Hansen et al, 2007), MCF-7 RLuc–LC3WT and MCF-7 RLuc–LC3G120A (Farkas et al, 2009) cells were propagated in RPMI 1640 (Invitrogen) supplemented with 6% fetal bovine serum (FBS) (Hyclone), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). MCF-7-Stathmin and MCF-7 empty vector cell lines were generated by viral transduction of MCF-7 eGFP–LC3 cells with either pBabe-HA–Stathmin or pBabe-empty. HEK and HeLa cells were propagated in DMEM (Invitrogen) supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. T47D cells were cultured in RPMI supplemented with 10% FBS, 1 mM Na-pyruvate, 3.125 ml 40% glucose, 100 units/ml penicillin and 100 μg/ml streptomycin. All cells were incubated at 37°C in 5% CO2.
Cell viability assay
MCF-7 eGFP–LC3 or T47D cells were seeded in 24-well plates and transfected the following day with 50 nM miR-101 or scramble control using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection cells were treated with EtOH or 4-OHT in RPMI 1640 without phenol red and 1% serum. Viability was measured 4 days after transfection by incubation for 4 h at 37°C with 0.5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Sigma-Aldrich) and subsequent dissolving of formazan crystals in a 10% formic acid, 90% isopropanol solution. Absorbance was measured at 560 nm with a reference wavelength of 750 nm.
eGFP–LC3 translocation
MCF-7 eGFP–LC3 cells were seeded in 96-well plates and transfected the following day with 30 nM LNA or 50 nM miRNA/siRNA using Oligofectamine (Invitrogen). Three days after transfection, cells were fixed in 4% formaldehyde and nuclei were stained with Hoechst (33 342). When indicated, cells were treated with 200 nM rapamycin for 2 h prior to fixation or with 1 μM 4-OHT for 2 days prior to fixation. Image acquisition was done using an Amersham InCell1000 High throughput microscope equipped with a × 40 Nikon objective. Fifteen pictures were taken from randomly placed positions within each well (counting ∼1000 cells/well) and analysed using the InCell1000 workstation 3.5 software package. Nuclei were segmented based on the Hoechst signal and cells were segmented from the GFP channel. Cells with >5 eGFP–LC3 puncta were considered positive for eGFP–LC3 translocation.
qPCR analysis
Total RNA from transfected cells was isolated using Trizol reagent (Invitrogen). qPCR was performed using the TaqMan reverse transcription kit (Applied Biosystems) and Fast SYBR Green PCR Master Mix (Applied Biosystems). For PCR primer sequences, see Supplementary data. For miRNA qPCR, total RNA was prepared using Trizol reagent and qPCR analysis was performed with TaqMan miRNA assays (Applied Biosystems) using primers for hsa-miR-101 and RNU6B.
Affymetrix microarray processing
MCF-7 cells were seeded in 6-cm plates and independent triplicate transfections were performed the following day with 50 nM miR-101 or scramble control using Lipofectamine 2000. Total RNA was harvested 24 h after transfection using Trizol reagent. Affymetrix microarray analysis (HG-U133 Plus 2.0 Human) was performed at the Microarray Center, Rigshospitalet, Copenhagen University Hospital. The microarray data from this publication have been submitted to the NCBI GEO database and assigned the identifier GSE31397. Affymetrix probeset intensity of miR-101 and scramble transfection experiments were preprocessed using the affy package in BioConductor (Gentleman et al, 2005), including steps of background correction, between-array normalization and transformation by VSN (Variance stabilization and calibration) method (Huber et al, 2002), and a summarization step by RMA (Robust Multichip Average) method. We then applied a non-specific filtering step to exclude those genes showing low overall expression levels as these genes were unlikely to show downregulation or upregulation after miRNA transfection. To do this, we required the interquartile range of probeset expression levels to be greater than the first quartile value of the interquartile range of expression levels for all probesets. The duplicated probesets that mapped to the same genes and the probesets without entrez gene annotation were also removed by this step. The remaining probesets were subsequently mapped to gene symbols using the Affymetrix hgu133plus2.db annotation Package. Differentially expressed genes were identified by a moderated t-test with FDR ⩽0.1 (1107 transcripts and annotation as listed in Supplementary Table I), using Limma (Smyth, 2005) package in Bioconductor. We defined three data sets: downregulated set (847 transcripts) with FDR ⩽0.1 and log2FC <0, upregulated set (260 transcripts) with FDR ⩽0.1 and log2FC>0, and no change set containing randomly sampled 1000 transcripts from the gene set with FDR >0.9.
Reporter assays
MCF-7 eGFP–LC3 cells were seeded in 96-well plates and transfected the following day with 20 nM miRNA precursor, 30 nM LNA, 50–150 ng pGL3 firefly construct and 10–25 ng renilla construct. Twenty-four hours after transfection, cells were harvested and luciferase activity was measured using the Dual-Glo luciferase assay (Promega). For details on vector construction, see Supplementary data.
RLuc-based screening assay
For a thorough explanation of this assay, see Farkas et al (2009). MCF-7 RLuc–LC3WT and MCF-7 RLuc–LC3G120A cells were reverse transfected side by side in 96-well format with 50 nM of the Pre-miR™ miRNA Precursor Library-Human V3 (Ambion). Transfections were automated and performed by the Hamilton Star Robot using 15 000 cells/well, RPMI without phenol red and Lipofectamine 2000. Forty hours post-transfection 50 nM of Enduren substrate (Promega) was added. Forty-two hours after transfection, RLuc activity was measured immediately followed by addition of 50 μM etoposide. RLuc activity was measured again at 54 and 66 h after transfection. Luciferase measurements were done using the Glomax Multi+luminometer (Promega). The screen was repeated three times and fold change (log2 LC3WT/LC3G120A) of luciferase activity was calculated for the three screens for each time point. For comparison across different time points and different screens, we normalized fold changes by subtracting the median value of fold changes of all miRNAs at each time point for each screen. We applied a non-parametric rank product method based on ranks of fold changes (Breitling et al, 2004) to identify miRNAs associated with differentially expressed fusion proteins RLuc–LC3WT versus RLuc–LC3G120A in MCF-7 cells. The statistical significance of the miRNA ranks were evaluated by 1000 permutations of rank list giving P<0.05 at each time point, using RankProd package in Bioconductor (Hong et al, 2006).
Detection of autophagic flux in all follow-up experiments was performed essentially as described in the screening assay procedure above except that transfections were not automated.
Antibodies and western blot analysis
MCF-7, HEK, HeLa or T47D cells were seeded in 6-well plates and transfected the following day. At indicated time points after transfection, cells were lysed in radioimmune precipitation buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris–HCl, pH 8, 2 mM EDTA) containing 1 mM dithiothreitol, 1 mM Pefabloc (Roche Applied Science), 1 × Complete Mini Protease Inhibitor Cocktail (Roche Applied Science), 1 × Phosphatase Inhibitor Cocktail (Roche Applied Science). In all, 10–20 μg protein/lane was separated on a 4–12% NuPAGE Bis-Tris gel (Invitrogen) or an 8% Tris-Glycine gel and transferred onto a nitrocellulose membrane. The primary antibodies used were p62 (PM045, MBL), LC3 (M152-3, MBL), Stathmin (3352, Cell Signaling), PARP (9542, Cell Signaling), RAB5 (610724, BD Transduction Laboratories), Beclin-1 (612112, BD Transduction Laboratories), Vinculin (V9131, Sigma) and GAPDH (sc-25778, Santa Cruz).
miRNA precursors, siRNAs and LNAs
The library used for screening was the Pre-miR™ miRNA Precursor Library-Human V3 (Ambion) and miRNA precursors used for the remaining experiments were miR-101 (PM11414, Ambion) and miR-203 (PM10152, Ambion). The siRNAs used were Renilla (AM4630, Ambion), Scramble (allstars 1027281, Qiagen), Stathmin/Op18 (sc-36127, Santa Cruz). siRNAs against RCN2 and ATG4D were Dharmacon SMARTpool® siRNAs. All other siRNAs were produced by Ribotask, sequences are as follows: Beclin-1: 5′-CAGUUUGGCACAAUCAAUA-3′, RAB5A: 5′-AACCAGGAAUCAGUGUUGUAG-3′, Raptor: 5′-GAUGAGGCUGAUCUUACAG-3′, CAPN2: 5′-CCGAGGAGGUUGAAAGUAA-3′, ASAP1: 5′-GGGCAAUAAGGAAUAUGGCAGUGAA-3′, MTMR2 5′-AACUCUGACUGUCACGAAUUA-3′ and TGFBRI: 5′-GAACAGAAGUUAAGGCCAA-3′. LNAs were designed by Santaris Pharma and produced by Ribotask.
Transmission electron microscopy
MCF-7 eGFP–LC3 cells were transfected as indicated for 72 h and treated with 200 nM rapamycin for the last 2 h prior to fixation. Samples were fixed in 2% v/v glutaraldehyde in 0.05 M sodium phosphate buffer (pH 7.2) for 24 h. Samples were rinsed three times in 0.15 M sodium cacodylate buffer (pH 7.2) and subsequently post-fixed in 1% w/v OsO4 in 0.12 M sodium cacodylate buffer (pH 7.2) for 2 h. The specimens were dehydrated in a graded series of ethanol, transferred to propylene oxide and embedded in Epon according to standard procedures. Sections, ∼80 nm thick, were cut with a Reichert-Jung Ultracut E microtome and collected on copper grids with Formvar supporting membranes. Sections were stained with uranyl acetate and lead citrate. Imaging was done on a Phillips CM 100 BioTWIN transmission electron microscope (magnifications: × 3400 for overviews and × 24 500 for close-ups). iTEM digital imaging software was used for multiple image alignment of × 24 500 images (Supplementary Figure S7) in order to obtain high-resolution images of cellular cross-sections suitable for accurate identification and counting of autophagosomes. For each sample, 25–30 cellular cross-sections were counted.
Seed enrichment analysis
We retrieved the annotated 3′UTR sequences of the transcripts on the microarrays from the UCSC table browser and included a unique 3′UTR for each transcript by choosing the one possessing the longest 3′UTR length. We searched seed sites of miR-101 against 3′UTR sequences of the up, down and no change sets. Four types of seed match were used as described in Grimson et al (2007): 6mer (miRNA position 2–7), 7mer-m8 (miRNA position 2–8), 7mer-A1 (6mer plus nucleotide A at position 1) and 8mer (7mer-m8 plus nucleotide A at position 1). The seed sites are mutually exclusive. The significance of the proportional differences of the transcripts having seed sites in these three data sets was tested using two-sided Fisher's exact test. The expression change distributions for different seed match type were plotted using CDFs and the distribution differences were evaluated using one-sided Kolmogorov-Smirnov tests. GO enrichment analysis was performed using topGO library (Alexa et al, 2006).
Supplementary Material
Acknowledgments
We wish to thank Morten Lindow and Sakari Kauppinen for the LNA designs, Bettina Mentz for providing technical assistance, and Giorgio Galli for careful reading of the manuscript. Work in the authors’ laboratory was supported by the Danish National Advanced Technology Foundation, the Novo Nordisk Foundation, the Danish National Research Foundation, the EC FP7 programs (ONCOMIRS, grant agreement number 201102 and APO-SYS. This publication reflects only authors’ views. The commission is not liable for any use that may be made of the information herein.), the Lundbeck Foundation, the Danish Medical Research Council and the Danish Cancer Society.
Author contributions: LBF conceived, designed and performed the experiments, analysed the data and wrote the manuscript. JW, ML, MHH, AK and MJ analysed the data. TF developed the screening assay. AHL analysed the data and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A (2007) Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ 14: 500–510 [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Alli E, Yang JM, Ford JM, Hait WN (2007a) Reversal of stathmin-mediated resistance to paclitaxel and vinblastine in human breast carcinoma cells. Mol Pharmacol 71: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Alli E, Yang JM, Hait WN (2007b) Silencing of stathmin induces tumor-suppressor function in breast cancer cell lines harboring mutant p53. Oncogene 26: 1003–1012 [DOI] [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB (2007) Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest 117: 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE (2005) Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563 [DOI] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW (2010) Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70: 715–728 [DOI] [PubMed] [Google Scholar]
- Buechner J, Tomte E, Haug BH, Henriksen JR, Lokke C, Flaegstad T, Einvik C (2011) Tumour-suppressor microRNAs let-7 and mir-101 target the proto-oncogene MYCN and inhibit cell proliferation in MYCN-amplified neuroblastoma. Br J Cancer 105: 296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99: 15524–15529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101: 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L (2002) The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol 14: 18–24 [DOI] [PubMed] [Google Scholar]
- Chen N, Debnath J (2010) Autophagy and tumorigenesis. FEBS Lett 584: 1427–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Karantza V (2011) Autophagy as a therapeutic target in cancer. Cancer Biol Ther 11: 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CW, Huang Y, Leong KW, Chen LC, Chen HC, Chen SJ, Chou CK (2010) PKCalpha mediated induction of miR-101 in human hepatoma HepG2 cells. J Biomed Sci 17: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH (2010) p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ 17: 236–245 [DOI] [PubMed] [Google Scholar]
- Corcelle EA, Puustinen P, Jaattela M (2009) Apoptosis and autophagy: Targeting autophagy signalling in cancer cells – ‘trick or treats’? FEBS J 276: 6084–6096 [DOI] [PubMed] [Google Scholar]
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM (2006) Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA 103: 7024–7029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croizet-Berger K, Daumerie C, Couvreur M, Courtoy PJ, van den Hove MF (2002) The endocytic catalysts, Rab5a and Rab7, are tandem regulators of thyroid hormone production. Proc Natl Acad Sci USA 99: 8277–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Medina P, Payre B, Boubekeur N, Bertrand-Michel J, Terce F, Silvente-Poirot S, Poirot M (2009) Ligands of the antiestrogen-binding site induce active cell death and autophagy in human breast cancer cells through the modulation of cholesterol metabolism. Cell Death Differ 16: 1372–1384 [DOI] [PubMed] [Google Scholar]
- Deangelis JT, Li Y, Mitchell N, Wilson L, Kim H, Tollefsbol T (2010) 2D difference gel electrophoresis analysis of different time points during the course of neoplastic transformation of human mammary epithelial cells. J Proteome Res 10: 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10: 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas T, Hoyer-Hansen M, Jaattela M (2009) Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy 5: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N (2008) Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH (2008) Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem 283: 1026–1033 [DOI] [PubMed] [Google Scholar]
- Fukui K, Tamura S, Wada A, Kamada Y, Igura T, Kiso S, Hayashi N (2007) Expression of Rab5a in hepatocellular carcinoma: possible involvement in epidermal growth factor signaling. Hepatol Res 37: 957–965 [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S (2005) Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY, USA: Springer Science and Business Media Inc. [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Gregersen LH, Jacobsen AB, Frankel LB, Wen J, Krogh A, Lund AH (2010) MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS One 5: e8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP (2007) MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189 [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ (2009) Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43: 67–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ (2007) A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J (2006) RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22: 2825–2827 [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jaattela M (2007) Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell 25: 193–205 [DOI] [PubMed] [Google Scholar]
- Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M (2002) Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1): S96–104 [DOI] [PubMed] [Google Scholar]
- Iancu C, Mistry SJ, Arkin S, Atweh GF (2000) Taxol and anti-stathmin therapy: a synergistic combination that targets the mitotic spindle. Cancer Res 60: 3537–3541 [PubMed] [Google Scholar]
- Iorio MV, Croce CM (2009) MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol 27: 5848–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070 [DOI] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL (2004) Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 117: 4837–4848 [DOI] [PubMed] [Google Scholar]
- Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD (2008) Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res 14: 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Ro SH, Cao J, Otto NM, Kim DH (2010) mTOR regulation of autophagy. FEBS Lett 584: 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminskyy V, Zhivotovsky B (2011) Proteases in autophagy. Biochim Biophys Acta (advance online publication; doi:10.1016) [DOI] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E (2007) Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev 21: 1621–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama M, Kawaguchi T, Berger MS, Pieper RO (2007) DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ 14: 548–558 [DOI] [PubMed] [Google Scholar]
- Kaushal GP, Kaushal V, Herzog C, Yang C (2008) Autophagy delays apoptosis in renal tubular epithelial cells in cisplatin cytotoxicity. Autophagy 4: 710–712 [DOI] [PubMed] [Google Scholar]
- Kent OA, Chivukula RR, Mullendore M, Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A, Mendell JT (2010) Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev 24: 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N (2004) The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Lee Y, Samaco RC, Gatchel JR, Thaller C, Orr HT, Zoghbi HY (2008) miR-19, miR-101 and miR-130 co-regulate ATXN1 levels to potentially modulate SCA1 pathogenesis. Nat Neurosci 11: 1137–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM (2011) Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem 286: 7327–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Fu H, Wang Y, Tie Y, Xing R, Zhu J, Sun Z, Wei L, Zheng X (2009) MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology 49: 1194–1202 [DOI] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J (2010) Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci USA 107: 14164–14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Marino G, Fernandez AF, Cabrera S, Lundberg YW, Cabanillas R, Rodriguez F, Salvador-Montoliu N, Vega JA, Germana A, Fueyo A, Freije JM, Lopez-Otin C (2010) Autophagy is essential for mouse sense of balance. J Clin Invest 120: 2331–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, Lopez-Otin C (2007) Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem 282: 18573–18583 [DOI] [PubMed] [Google Scholar]
- Marino G, Uria JA, Puente XS, Quesada V, Bordallo J, Lopez-Otin C (2003) Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J Biol Chem 278: 3671–3678 [DOI] [PubMed] [Google Scholar]
- Marklund U, Larsson N, Gradin HM, Brattsand G, Gullberg M (1996) Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J 15: 5290–5298 [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E (2007) Role of autophagy in cancer. Nat Rev Cancer 7: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S (2009) Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461: 654–658 [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282: 24131–24145 [DOI] [PubMed] [Google Scholar]
- Phadke AP, Jay CM, Wang Z, Chen S, Liu S, Haddock C, Kumar P, Pappen BO, Rao DD, Templeton NS, Daniels EQ, Webb C, Monsma D, Scott S, Dylewski D, Frieboes HB, Brunicardi FC, Senzer N, Maples PB, Nemunaitis J et al. (2011) In vivo safety and antitumor efficacy of bifunctional small hairpin RNAs specific for the human stathmin 1 oncoprotein. DNA Cell Biol (advance online publication; doi:10.1089) [DOI] [PubMed] [Google Scholar]
- Qadir MA, Kwok B, Dragowska WH, To KH, Le D, Bally MB, Gorski SM (2008) Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat 112: 389–403 [DOI] [PubMed] [Google Scholar]
- Rana S, Maples PB, Senzer N, Nemunaitis J (2008) Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev Anticancer Ther 8: 1461–1470 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC (2008) Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci 121: 1649–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26: 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y (2004) Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 6: 1221–1228 [DOI] [PubMed] [Google Scholar]
- Singer S, Ehemann V, Brauckhoff A, Keith M, Vreden S, Schirmacher P, Breuhahn K (2007) Protumorigenic overexpression of stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology 46: 759–768 [DOI] [PubMed] [Google Scholar]
- Smyth G (2005) Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor, Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S (eds). New York, NY, USA: Springer Science and Business Inc. [Google Scholar]
- Sobel A (1991) Stathmin: a relay phosphoprotein for multiple signal transduction? Trends Biochem Sci 16: 301–305 [DOI] [PubMed] [Google Scholar]
- Stenmark H (2009) Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 10: 513–525 [DOI] [PubMed] [Google Scholar]
- Strillacci A, Griffoni C, Sansone P, Paterini P, Piazzi G, Lazzarini G, Spisni E, Pantaleo MA, Biasco G, Tomasi V (2009) MiR-101 downregulation is involved in cyclooxygenase-2 overexpression in human colon cancer cells. Exp Cell Res 315: 1439–1447 [DOI] [PubMed] [Google Scholar]
- Stuart-Harris R, Davis A (2010) Optimal adjuvant endocrine therapy for early breast cancer. Womens Health (Lond Engl) 6: 383–398 [DOI] [PubMed] [Google Scholar]
- Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM (2009) MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res 69: 1135–1142 [DOI] [PubMed] [Google Scholar]
- Szyniarowski P, Corcelle-Termeau E, Farkas T, Hoyer-Hansen M, Nylandsted J, Kallunki T, Jaattela M (2011) A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy 7: 892–903 [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R (2006) Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 20: 515–524 [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, Brenner JC, Yu J, Kim JH, Han B, Tan P, Kumar-Sinha C, Lonigro RJ, Palanisamy N, Maher CA, Chinnaiyan AM (2008) Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 322: 1695–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Barbato C, Ciotti M, Cogoni C, Ruberti F (2010) MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J Biol Chem 285: 18344–18351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103: 2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT, Xia YJ, Ye ZY, Tao HQ (2010) MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur J Cancer 46: 2295–2303 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG (2006) MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA 103: 4034–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Ng WL, Zhang X, Wang P, Zhang Z, Mo YY, Mao H, Hao C, Olson JJ, Curran WJ, Wang Y (2010) Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One 5: e11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Hui-chen F, Chen Y, Zou R, Yan S, Chun-xiang L, Wu-ru W, Li P (1999) Differential expression of RAB5A in human lung adenocarcinoma cells with different metastasis potential. Clin Exp Metastasis 17: 213–219 [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N (2003) Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA 100: 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Liu XF, Wu HC, Zou SB, Wang JY, Ni PH, Chen XH, Fan QS (2010) Rab5a overexpression promoting ovarian cancer cell proliferation may be associated with APPL1-related epidermal growth factor signaling pathway. Cancer Sci 101: 1454–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.