Abstract
In a broad range of organisms, Piwi-interacting RNAs (piRNAs) have emerged as core components of a surveillance system that protects the genome by silencing transposable and repetitive elements. A vast proportion of piRNAs is produced from discrete genomic loci, termed piRNA clusters, which are generally embedded in heterochromatic regions. The molecular mechanisms and the factors that govern their expression are largely unknown. Here, we show that Cutoff (Cuff), a Drosophila protein related to the yeast transcription termination factor Rai1, is essential for piRNA production in germline tissues. Cuff accumulates at centromeric/pericentromeric positions in germ-cell nuclei and strongly colocalizes with the major heterochromatic domains. Remarkably, we show that Cuff is enriched at the dual-strand piRNA cluster 1/42AB and is likely to be involved in regulation of transcript levels of similar loci dispersed in the genome. Consistent with this observation, Cuff physically interacts with the Heterochromatin Protein 1 (HP1) variant Rhino (Rhi). Our results unveil a link between Cuff activity, heterochromatin assembly and piRNA cluster expression, which is critical for stem-cell and germ-cell development in Drosophila.
Keywords: cutoff , Drosophila , germline, heterochromatin, piRNA
Introduction
A significant fraction of eukaryotic genomes is made up of repetitive sequences, such as transposable elements (TEs) and tandem repeats, whose deregulation has been linked to DNA damage and sterility (Bucheton, 1990; Belgnaoui et al, 2006; Gasior et al, 2006; Vagin et al, 2006; Brennecke et al, 2007; Klattenhoff et al, 2007; Chambeyron et al, 2008). Germline tissues, which are responsible for transferring the genetic information to the progeny, appear to be the most sensitive targets of TE deregulation (Vagin et al, 2006; Chen et al, 2007; Klattenhoff et al, 2007; Klenov et al, 2007; Pane et al, 2007). A specialized RNAi pathway, centered on small non-coding RNAs known as Piwi-interacting RNAs (piRNAs), guarantees the repression of transposable and repetitive elements and ensures the maintenance of genomic stability during germ-cell division and differentiation (Aravin et al, 2003; Vagin et al, 2006; Brennecke et al, 2007, 2008; Siomi et al, 2010). piRNAs are 23–30-nt-long non-coding RNAs and mainly correspond to sequences in transposable and repetitive elements dispersed in the genome (Aravin et al, 2003). In Drosophila, the production of these molecules relies on the activity of the Argonaute family members Piwi, Aubergine (Aub) and Argonaute-3 (Ago3) (Saito et al, 2006; Brennecke et al, 2007; Gunawardane et al, 2007; Yin and Lin, 2007). Deep-sequencing analyses of the small RNAs associated with these proteins revealed that Piwi and Aub complexes are enriched in antisense piRNAs, while Ago3 is mostly bound by sense piRNAs. Furthermore, a subset of piRNAs displays a 10-nt overlap at their 5′end (Brennecke et al, 2007; Gunawardane et al, 2007). These observations were captured in a model, whereby antisense piRNAs bound to Aub/Piwi can pair with transposon transcripts and catalyse the production of sense piRNA molecules through cleavage of the transposon transcripts. The latter are then loaded into an Ago3 complex, which in turn triggers the production of antisense piRNAs by cleaving cluster-derived antisense transcripts. This cycle is the core of the so-called ‘Ping-pong’ model, a feed-forward loop that amplifies the piRNA population and is thought to reinforce the repression of active transposons dispersed in the genome (Brennecke et al, 2007; Gunawardane et al, 2007). piRNA processing requires additional activities, including the eIF-4A-like translation factor Vasa (Vas), the Heterochromatin Protein 1 (HP1) variant Rhino (Rhi), the helicases Spindle-E and Armitage, the putative nucleases Maelstrom, Zucchini and Squash and the Tudor-domain proteins Tejas and Krimper (Khurana and Theurkauf, 2010; Saito and Siomi, 2010; Senti and Brennecke, 2010). The precise biochemical function of many of these proteins is, however, still largely unclear.
In Drosophila, primary sources of piRNAs are discrete genomic regions termed piRNA clusters (Brennecke et al, 2007). These genomic loci are composed of repetitive sequences, TEs and inactive transposon remnants. The transcripts produced from these loci are processed by cytoplasmic activities to produce the mature piRNAs. The major sources of piRNAs in Drosophila are the cluster 1/42AB located on chromosome 2R in pericentromeric position and clusters 2 and flamenco/COM (flam) located on chromosome X. Cluster 1/42AB and the majority of the piRNA clusters in Drosophila are transcribed from both DNA strands (dual-strand clusters). Conversely, a few loci, including cluster 2 and flam, produce piRNAs only from one genomic DNA strand (uni-strand clusters) (Brennecke et al, 2007; Malone et al, 2009). To date, flam represents the best-characterized piRNA cluster. The flam cluster harbours fragments of the Zam, gypsy and idefix retro-transposons and is required for the silencing of these retroviral elements in the follicle cells of the Drosophila ovaries (Pelisson et al, 1994, 2007; Prud’homme et al, 1995; Sarot et al, 2004; Brennecke et al, 2007; Mevel-Ninio et al, 2007; Desset et al, 2008). Interestingly, the vast majority of the transposon remnants that populate the flam locus are oriented towards the centromere of the X chromosome (Brennecke et al, 2007). Genetic and molecular analyses strongly suggest that the transcription of this cluster occurs from an external promoter on the opposite strand and is likely to produce a long transcript encompassing the entire locus (Brennecke et al, 2007; Mevel-Ninio et al, 2007). The structure of the cluster and the strand bias of the transcriptional events thus generate mostly antisense piRNAs, which do not engage a ping-pong amplification loop (Brennecke et al, 2007; Li et al, 2009; Malone et al, 2009). piRNA clusters in zebrafish, mouse and human also display a clear strand bias similar to flam, whereby piRNAs map only to one DNA strand (Aravin et al, 2006; Girard et al, 2006; Houwing et al, 2007). The mechanism that regulates the expression of the piRNA clusters and the proteins involved in this nuclear process remain largely elusive. It has recently been shown that a germline-specific HP1 variant encoded by the rhino (rhi) gene is essential for the regulation of the dual-strand clusters (Volpe et al, 2001; Klattenhoff et al, 2009). Accordingly, mutations in rhi abolish the expression of these loci, which results in a general depletion of the corresponding piRNA population (Klattenhoff et al, 2009). The activity of Rhi seems to be limited to the dual-strand clusters, since mutations in this protein do not affect the production of piRNAs from the uni-strand clusters. Surprisingly, the expression of the piRNA loci appears to also rely on the activity of the Piwi proteins Aub and Piwi. In mutants of these factors, heterochromatic states spread over the clusters causing their transcriptional downregulation. These findings, therefore, implicate the production of piRNAs in the maintenance of active transcription from the piRNA clusters (Moshkovich and Lei, 2010).
We have previously reported that the protein product of the cutoff (cuff) gene is essential for transposon silencing and germline development (Chen et al, 2007). Mutations in cuff impair the establishment of dorsal–ventral polarity during oogenesis and cause a significant loss of germline cells, possibly due to defects in stem-cell maintenance and/or division. Cuff shows similarity to the yeast transcription termination factor Rai1 (Xue et al, 2000; Kim et al, 2004). In yeast, Rai1 is found in a complex with the 5′–3′ exoribonuclease Rat1/Xrn2 and promotes the termination of transcriptional events initiated by the PolI and PolII RNA polymerases (Kim et al, 2004; El Hage et al, 2008). Different from Rai1, however, Cuff does not display a Dom3Z domain, which is critical for Rai1 activity (Xiang et al, 2009). Given the observation that an additional Rai1 homologue exists in Drosophila which contains the Rai1 catalytic domain, we hypothesized that Cuff might be a germline-specific Rai1 variant, which exerts a novel function in germline cells (Chen et al, 2007).
Here, we show that Cuff localizes to discrete foci in germline nuclei and accumulates in proximity of and within heterochromatic domains. We demonstrate that Cuff is essential for the expression of the piRNA clusters embedded in heterochromatic regions, including the major dual-strand cluster 1/42AB, but not for the flam locus, which is primarily expressed in the soma. In agreement with these observations, we find that Cuff colocalizes and physically interacts with Rhi. We propose that these proteins assemble into a complex, which regulates the production of piRNAs from the dual-strand clusters at a transcriptional level. Mutations in Cuff cause a severe disruption of the nuage, which in turn might contribute to the depletion of piRNAs not only from the dual-strand, but also from some uni-strand clusters, including the major cluster 2. Our results uncover a role for Cuff in piRNA cluster regulation, nuage assembly and piRNA production and highlight a critical function for the piRNA pathway in stem- and germ-cell development during Drosophila oogenesis.
Results
A Zam-based reporter construct is derepressed in piRNA pathway mutants
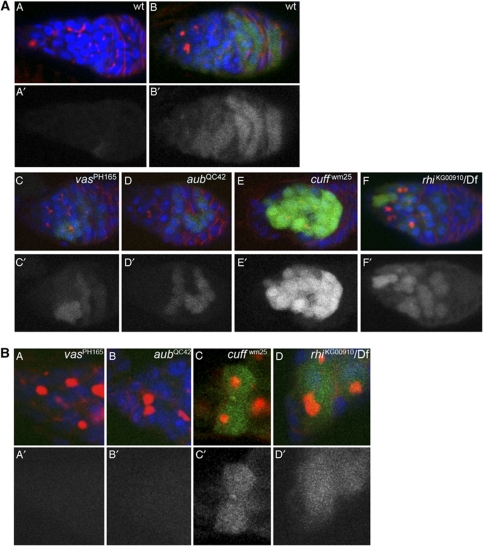
Oogenesis in Drosophila begins in the germarium, which is located at the anterior tip of the ovariole. The germarium harbours two to three stem cells at the very anterior, which divide to give rise to a daughter stem cell and a cystoblast. Each cystoblast begins to differentiate and undergoes four rounds of synchronous mitotic divisions followed by incomplete cytokinesis. The result of this process is the production of a 16-cell cyst in which all the cells are interconnected by ring canals. The 16-cell cyst is then surrounded by follicle cells to give rise to the fully formed egg chamber (Spradling, 1993; St Johnston and Ahringer, 2010). In order to gain insight into the temporal requirement of different piRNA pathway activities, we employed pGFP-ZenvAS reporter lines, where a GFP-encoding cDNA was cloned upstream of a sequence derived from the env gene of the Zam retro-transposon (Desset et al, 2008). The expression of the sensor construct in ovarian tissues was obtained with the Act5C-Gal4 driver. The presence of a sequence from the Zam retro-transposon in this reporter renders the expression of the GFP sensitive to the activity of the piRNA machinery. Accordingly, the pGFP-ZenvAS reporter is silenced in the germline of wild-type females, while a similar sensor construct lacking the zam sequence is constitutively expressed in wild-type germaria (Figure 1A, A′, B and B′; Desset et al, 2008). In contrast, mutations in vas, aub, cuff and rhi cause a severe deregulation of the reporter in germline tissues (Figure 1). In mutants of aub, which encodes a key component of the piRNA biogenesis (Brennecke et al, 2007; Gunawardane et al, 2007), and in vas, which encodes an eIF4A-like translation factor required for piRNA biogenesis (Styhler et al, 1998; Malone et al, 2009), the reporter is highly deregulated in the 8-cell and 16-cell cysts in region 2B of the germarium (Figure 1B, B′, C and C′). In contrast, in the cuff and rhi mutants, high levels of GFP can be seen already at earlier stages of germline development (Figure 1E, E′, F, and F′). The sensor construct is still strongly deregulated in stage 3–6 egg chambers in the cuff and rhi mutants, while only a faint GFP signal is produced in vas and aub after the cyst buds off from the germarium (data not shown). The sensor construct is not deregulated in flies heterozygous for aub, cuff and rhi, while it is partially derepressed in the vas heterozygous backgrounds (Supplementary Figure S1A). A progressive loss of germ cells over time causes cuff mutant females to display empty ovarioles 10–12 days after eclosion (Chen et al, 2007). This observation led us to propose that Cuff function is required for stem-cell maintenance and/or division (Chen et al, 2007). In order to test whether Cuff is active in this cell type, we monitored the expression levels of the pGFP-ZenvAS construct in cuff mutant stem cells. Since mutations in distinct components of the piRNA pathway appear to differentially impact the reporter construct, we also analysed the expression levels of the sensor in stem cells of vas, aub and rhi mutants. This study revealed very limited or no deregulation in vas (Figure 1B, A and A′) and aub stem cells (Figure 1B, B and B′). In the aub mutants, however, we could sporadically find stem cells displaying a clear expression of the construct (Supplementary Figure S1B). By contrast, mutations in Cuff and Rhi strongly deregulate the reporter in stem-cell position (Figure 1B, C, C′, D and D′). Our data suggest that Cuff and Rhi activities are essential for transposon silencing during germline development, while Vas and Aub appear to exert a less prominent role in the silencing mechanism at these early stages.
Figure 1.
(A) The pGFP-ZenvAS reporter is deregulated in the germaria of cuff, aub, rhi and vas mutants. (A, A′) The pGFP-ZenvAS reporter is silenced in wild-type (wt) ovaries. (B, B′) A GFP sensor construct lacking the Zenv sequence is constitutively deregulated in wild-type ovaries. (C, C′) Expression of the pGFP-ZenvAS sensor transgene is visibly increased in stage 4-cell and 8-cell cysts of vasPH165 homozygous mutant germaria. (D, D′) Mutations in aub deregulate the pGFP-ZenvAS reporter in the late mitotic cyst. (E, E′) Expression of the reporter is dramatically upregulated in the cuff mutant germaria. (F, F′) Mutations in rhi cause a significant derepression of the sensor construct. The branching spectrosome/fusome, which interconnects the dividing cells in the mitotic cyst, is visualized with anti-α-Spectrin antibody (red). (A′–F′) GFP single channels. (B) The expression of the pGFP-ZenvAS sensor is differentially affected in stem cells of vas, aub, cuff and rhi mutants. The sensor construct is silenced in vas (A, A′) and aub (B, B′) mutant stem cells. Mutations in cuff (C, C′) and rhi (D, D′) cause a significant derepression of the reporter in stem-cell position. The spectrosome/fusome is visualized with anti-α-Spectrin antibody (red).
Cuff localizes to discrete foci in germ-cell nuclei and is enriched in proximity and within heterochromatic domains
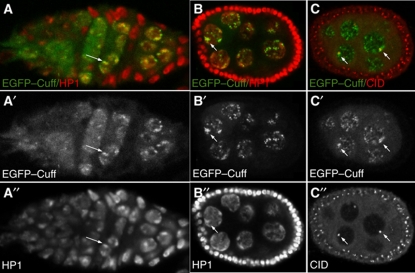
We have previously reported that the Cuff protein displays cytoplasmic/nuage localization when a tripleHA–Cuff fusion protein is expressed in the wild-type germline (Chen et al, 2007). In order to gain further insight into the subcellular localization of the Cuff protein, we produced transgenic lines expressing EGFP-tagged Cuff chimeric proteins, where the EGFP sequence is fused either at the N-terminus of the protein or at the C-terminus. The expression of the EGFP–Cuff in the germline of wild-type individuals was initiated by the Nos-Gal4-VP16 driver and revealed a very intriguing distribution of this protein in the nurse cell nuclei of the developing egg chambers (Figure 2). Different from our observation with the tripleHA–Cuff lines, we noticed that EGFP–Cuff forms distinct nuclear foci, which are readily detectable in the nurse cells (Figure 2A and B). A very similar localization pattern was observed in transgenic lines expressing a Cuff–EGFP fusion protein bearing the EGFP at the C-terminus of Cuff (data not shown). To resolve the apparent discrepancy between the transgenic lines and unambiguously determine the localization of Cuff, we adopted different conditions for our antibody stainings whereby the initial fixation of the ovaries was followed by a mild Proteinase K treatment (Clouse et al, 2008). This approach allowed more access of the reagents to the nuclei and confirmed the punctate Cuff distribution pattern both with anti-Cuff specific antibodies and the previously described tripleHA–Cuff transgenic lines (Figure 2C and data not shown). Cuff-positive foci appear to be often clustered in regions next to the nuclear envelope, where the block of the heterochromatic sequences is generally located. A hallmark of heterochromatic regions is the presence of the Heterochromatin Protein 1A (HP1), which is encoded by the Su(var)205 gene (Vermaak and Malik, 2009). HP1 levels are low early in oogenesis, when the cystoblast undergoes the first mitotic divisions and increase in the late mitotic cyst where HP1 labels the prominent heterochromatic blocks (Yoon et al, 2008). To determine the distribution of Cuff with respect to heterochromatic domains, we labelled EGFP–Cuff expressing ovaries for HP1 (Figure 2). This analysis revealed that the Cuff protein is particularly enriched within and next to heterochromatic regions (Figure 2A–A″ and B–B″). Beginning in the late mitotic cyst and in all the following stages of egg chamber development, Cuff-positive foci extensively overlap with HP1. In fully formed egg chambers (stages 6–10), Cuff appears to be >2-fold enriched in the major heterochromatic blocks of nurse cell nuclei with respect to the rest of the nucleoplasm (Supplementary Figure S2A). Cuff is also found in isolated foci dispersed in the nuclei, where HP1 is not clearly detectable. To gain further insight into Cuff nuclear localization, we analysed the position of the Cuff-positive foci with respect to the centromeres, which are largely composed of constitutive heterochromatic sequences. Drosophila centromeres are marked by the presence of the Histone H3 variant Centromere Identifier (CID) (Blower and Karpen, 2001). Interestingly, immunostainings with anti-CID antibodies in EGFP–Cuff lines unveiled a significant accumulation of Cuff at centromeric/pericentromeric positions (Figure 2C–C″). Approximately, 84% of randomly chosen centromeres (n=50, stage 6–8 egg chambers) that we analysed were tightly associated with distinct Cuff foci, while the remaining centromeres appeared to lie in regions where Cuff is apparently more dispersed or absent. These results were confirmed by co-immunostaining experiments performed with anti-Cuff and anti-CID-specific antibodies in wild-type ovaries (Supplementary Figure S2B). Our data, therefore, point to a role for Cuff in the regulation of genomic regions located in proximity of the centromeres and in general, within heterochromatic domains.
Figure 2.
Cuff localizes to centromeric/pericentromeric and heterochromatic regions. (A–A″) Wild-type germarium expressing the EGFP–Cuff chimeric protein (A, green and A′, single channel) labelled with anti-HP1 antibody (A, red and A″, single channel). EGFP–Cuff localizes to nuclear speckles and appears enriched in heterochromatic regions marked with HP1 (arrows). (B–B″) EGFP–Cuff (B, green and B′, single channel) extensively overlaps with HP1 (B, red and B″, single channel) in nurse cell nuclei of stage 6 egg chambers (arrows). (C–C″) EGFP–Cuff (C, green and C′, single channel) ovaries labelled with anti-CID antibody (C, red and C″, single channel). Cuff is enriched in proximity of the centromeres (arrows).
piRNA cluster expression is affected by mutations in cuff
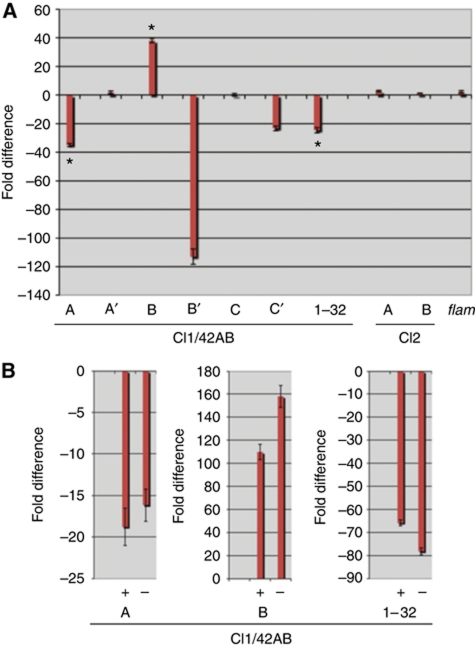
A significant fraction of the ovarian piRNA population is produced from piRNA clusters located in heterochromatic regions (Brennecke et al, 2007). For instance, the dual-strand cluster 1/42AB at the pericentromere of Chromosome 2R accounts for >30% of the uniquely mapping piRNAs (Brennecke et al, 2007). The localization pattern of Cuff prompted us to investigate whether this protein is involved in regulation of the ovarian piRNA master loci. In order to monitor the expression levels for these clusters in the cuff mutant, we performed quantitative reverse transcription (qRT–PCR) assays with a set of specific oligonucleotides (Figure 3A). In addition to the set of primers described by Klattenhoff and colleagues, we employed additional primer pairs along cluster 1/42AB. The full set of primers used in this study allowed the analyses of regions A, A′, B, B′, C, C′ and 1–32 within the cluster 1/42AB. These regions are positioned in a linear fashion within the cluster, with A being closer to the centromere and 1–32 more distal (Klattenhoff et al, 2009). Mutations in cuff appear to differentially affect the expression levels of different regions in cluster 1/42AB. When we compared cuff homozygous mutant ovaries versus heterozygous combination, we could detect a significant reduction in the transcripts produced from region A (∼40-fold), B′ (∼115-fold), C′ (∼20-fold) and 1–32 (∼20-fold) of cluster 1/42AB (Figure 3A). In contrast, region B displays an ∼40-fold upregulation of the transcripts levels (Figure 3A). Finally, transcripts from regions A′ and C appear to be unaffected in the cuff mutant ovaries. Uni-strand clusters 2 and flam are apparently not altered by mutations in cuff. Our data suggest that the regulation of cluster 1/42AB, and possibly of other dual-strand piRNA clusters, differs from the well-characterized uni-strand flam locus. Genetic and molecular studies showed that the somatic flam cluster is likely to be transcribed from an external promoter to produce a transcript encompassing the entire locus (Brennecke et al, 2007; Mevel-Ninio et al, 2007). Different from flam, our data suggest that cluster 1/42AB does not follow this rule and is unlikely to produce a single transcript spanning the entire cluster. Dual-strand clusters are transcribed from both DNA strands in wild-type ovaries (Brennecke et al, 2007). As shown above, in the cuff mutant the expression levels of some regions within the 1/42AB locus are clearly altered. We, therefore, asked whether Cuff controls the expression of one or both DNA strands within the dual-strand clusters. To answer this question, we performed strand-specific qRT–PCR on regions A and B and on the cluster 1–32, which displayed a significant change in the cuff mutant background (Figure 3B). This analysis revealed that in the absence of a functional Cuff protein transcripts from both DNA strands of region A and 1–32 are downregulated, while transcripts produced from both strands of region B appear to be upregulated.
Figure 3.
Cuff regulates piRNA cluster expression. (A) qRT–PCR analysis of the transcripts produced from cluster 1/42AB (regions A, A′, B, B′, C, C′ and 1–32) cluster 2 (regions A and B) and flam. Mutations in cuff cause a downregulation of the regions A, B′, C′ and 1–32 of cluster 1/42AB, while region B is upregulated. Regions A′ and C are not affected by mutations in cuff. Cluster 2 and flam appear unaffected in the cuff mutant. Asterisks mark the region of Cl1/42AB, which were subjected to strand-specific qRT–PCR (B). The fold difference between cuff mutant and wild type is reported on the Y axis. (B) Strand-specific qRT–PCR analysis of cluster 1/42AB regions A, B and 1–32. Mutations in cuff affect the expression levels of both plus and minus genomic DNA strands. The fold difference between cuff mutant and wild type is reported on the Y axis.
Mutations in the cuff gene cause a dramatic depletion of the piRNA population
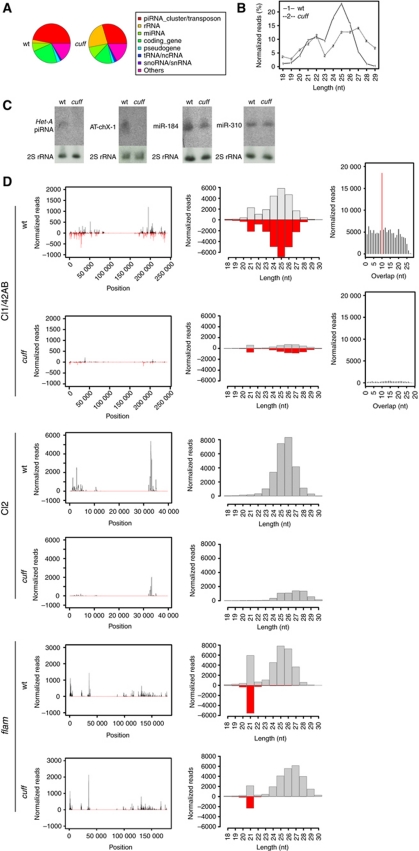
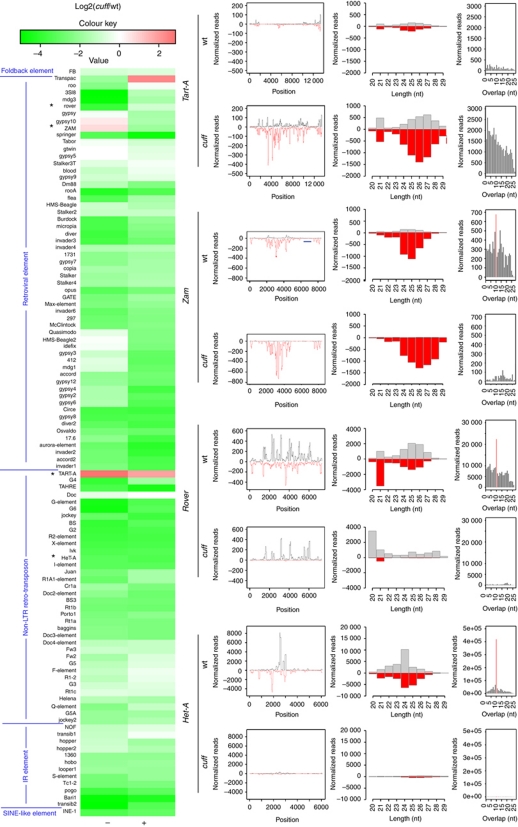
Our analyses suggest that Cuff is involved in the regulation of the dual-strand piRNA cluster 1/42AB, while it is seems to be dispensable for the expression of the uni-strand clusters 2 and flam. The abnormal transcript levels of the cluster 1/42AB in the cuff mutant are expected to impact the piRNA population and alter the production of the corresponding piRNA complement. To test this hypothesis, we generated small RNA libraries from cuff mutant ovaries and compared them with libraries derived from control ovaries. The analysis of the piRNA population revealed that 74.08% of the total piRNAs identified in the wild-type libraries are lost in the cuff mutant (Figure 4A and B). The piRNA population is composed of two classes of molecules, piRNAs that are ‘sense’ with respect to the canonical transposon sequences and piRNAs that are antisense to these elements. Cuff mutations result in a significant reduction of both sense and antisense piRNA populations, whereby 70.92% sense and 79.84% antisense piRNAs are depleted in the absence of the Cuff protein. In order to validate our libraries, we performed northern blot analyses on two piRNAs, which are particularly abundant in the wild-type ovaries, but are depleted in the cuff mutant. Both the Het-A piRNA and the previously described AT-chX-1 piRNA can be detected in the control lane, while they are absent in the cuff lane (Figure 4C). AT-chX-1 was previously shown to target the vas gene in the Drosophila male gonad and to be expressed in an Ago3-dependent fashion in ovaries (Nishida et al, 2007; Li et al, 2009; Nagao et al, 2010). In addition, we tested whether mutations in cuff impact the production of miRNAs in the Drosophila ovaries. To this aim, we performed northern blot analyses on miR-310 and miR-184, which are very abundant in the female germline (Saito et al, 2006; Iovino et al, 2009). Northern blotting analysis with anti-miR-310- and anti-miR-184-specific probes reveals that none of these miRNAs is affected in the cuff mutant ovaries (Figure 4C). These results validate our small RNA libraries. The analysis of piRNAs that map to unique positions in the genome provides a useful approach to define the piRNA clusters and monitor the production of piRNAs from these loci (Brennecke et al, 2007; Li et al, 2009; Malone et al, 2009). piRNA density along the piRNA clusters dispersed in the genome unveiled a substantial (>80%) depletion of both sense and antisense piRNAs corresponding to the major dual-strand clusters 1/42AB and 1–32 (Figure 4D; Supplementary Figures S3 and S5). Of the 27 dual-strand clusters, 24 fail to produce the proper complement of piRNA molecules in the Cuff mutant (Supplementary Figures S3 and S5). piRNAs originating from the dual-strand clusters can be both sense and antisense with respect to transposon sequences (Brennecke et al, 2007). A subset of these molecules engages in a ping-pong mechanism, which is thought to amplify the piRNA population and enhance the silencing of the TEs (Brennecke et al, 2007). Intriguingly, mutations in cuff significantly affect this population and cause a disruption of the ping-pong signal, suggesting that Cuff is involved in the production of piRNAs, which fuel the amplification mechanism (Figure 4D; Supplementary Figure S4). A few loci in the genome, including the uni-strand clusters 2 and flam, are instead populated by polarized transposons and transposon remnants (Prud’homme et al, 1995; Desset et al, 2003; Brennecke et al, 2007; Mevel-Ninio et al, 2007; Malone et al, 2009). Transcription of these loci occurs only from one genomic DNA strand and gives rise mostly to antisense piRNAs (Brennecke et al, 2007). These molecules are not involved in the amplification cycle and the mechanisms underlying their biogenesis are largely unclear. Consistent with a germline-specific function of Cuff, piRNAs produced from the flam locus, which is only expressed in the somatic follicle cells, are not significantly affected by cuff mutations (Figure 4D); their population in the cuff mutant is only 6% lower than in wild type. Surprisingly, piRNAs derived from the cluster 2 are 22.69% of wild type in the absence of the Cuff protein (Figure 4D). This observation suggests that Cuff is required for the production of piRNAs from this locus, even though mutations in cuff do not seem to alter its expression levels.
Figure 4.
Analysis of the piRNA levels in the cuff mutant ovaries. (A) Pie chart displaying the total number of reads of small RNAs in cuff and control (wt) ovaries. (B) Length distribution of the uniquely mapped sequence reads in the cuff and control libraries. Mutations in cuff cause an approximate 74% reduction of the piRNA population. A second peak at 21/22 nt corresponds to a heterogeneous population formed by piRNAs and endo-siRNAs. This class of small RNAs is not dramatically affected in cuff ovaries. (C) Northern blot analyses on Het-A-specific and AT-chX-1 piRNAs in wild-type and cuff mutant ovaries. Both piRNAs are detected in the wild-type lane, while they are absent in the cuff lane. Northern blot analyses on the abundant miR-310 and miR-184 reveal that mutations in cuff do not affect the production/stability of these miRNAs. (D) Analysis of uniquely mapping piRNAs produced by cluster 1/42AB, cluster 2 and flam in wt (left) and cuff mutant (right). For cluster 1/42AB, piRNA densities along the cluster, length distribution and ping-pong signal are displayed. For cluster 2 and flam, piRNA densities along the cluster and length distribution are displayed.
We next sought to determine the impact of mutations in cuff on the global piRNA population. To this aim, we analysed the piRNA density along the canonical transposon families taking into account all the normalized reads mapping to each transposon sequence (Figure 5; Supplementary Figure S6). This analysis revealed that both sense and antisense piRNAs corresponding to 46 transposon families are lost or strongly reduced in the cuff mutant. For these families, the sense and antisense piRNA populations are on average 8.7 and 8.8% (respectively) of those in the wild type. Three such families include the HeT-A, jockey and roo elements (Figure 5; Supplementary Figure S6). piRNAs corresponding to these families display a strong 10 nt overlap and engage in the ping-pong amplification loop. Mutations in cuff cause a strong depletion of piRNAs and disrupt the ping-pong mechanism (Figure 5; Supplementary Figure S6). For 22 transposon families, including gypsy, idefix and Zam, sense piRNAs are downregulated, while the levels of antisense piRNAs appear unaffected or increased (Figure 5; Supplementary Figure S6). On average, the sense piRNA populations for these transposon families are 9.6% of those in the wild type. The gypsy, idefix and Zam elements are active in the somatic follicle cells of the ovary, where they are controlled by the flam locus (Pelisson et al, 1994, 2007; Prud’homme et al, 1995; Desset et al, 2003, 2008; Sarot et al, 2004; Mevel-Ninio et al, 2007). However, Cuff does not appear to have a function in the somatic tissues, thus suggesting that the differences in the piRNA levels are caused by a depletion of a germline piRNA set in the cuff mutant. Surprisingly, the zam sequence employed in the pGFP-ZenvAS reporter (Desset et al, 2008) appears to be devoid of piRNAs, thus suggesting that the regulation of the sensor construct might not directly rely on these molecules (see Discussion). Mutations in cuff significantly reduce the antisense piRNAs corresponding to 20 transposon families, including the rover and Nof transposons, while the decline of the sense piRNAs is less dramatic (Figure 5; Supplementary Figure S5). On average, the antisense piRNA populations for these elements are 11.7% of those in the wild type, with the sense populations varying from 21 to 100% of those in wild type. Finally, only for the TART family of telomere-specific retro-transposons, including the TART-A, TART-B and TART-C variants, did we observe an increase in the corresponding sense and antisense piRNAs (Figure 5; Supplementary Figure S5). While part of these molecules might represent degradation products of the TART transcripts, our data suggest that different transposon classes are differentially affected by mutations in the Cuff protein.
Figure 5.
Mutations in cuff strongly reduce the piRNA levels. Log2 ratio of the normalized number of piRNA reads between the cuff mutant and the wild type for the canonical transposon sequences (left). For each transposon, sense (+) and antisense (−) piRNAs are reported as reduced (green), unaffected (white) or increased (red); for the vast majority of the transposons, the corresponding piRNA set is reduced (colour key and histogram). The HeT-A, TART, rover and Zam transposable elements, which are analysed in more details (right), are highlighted (*). For each of these transposons, piRNA densities along the canonical sequence, length distribution and ping-pong signal are analysed in wt and cuff mutant ovaries. The blue line indicates the position of the Zam fragment adopted in the pGFP-ZenvAS reporter line (Desset et al, 2008).
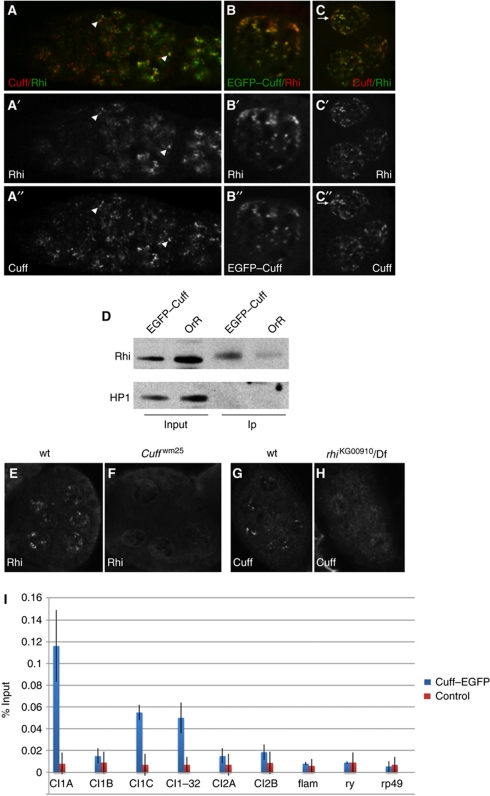
Cuff colocalizes and physically interacts with the HP1 variant Rhino
The role of Cuff in the expression of piRNA clusters and its localization within nurse cell nuclei prompted us to investigate whether this protein is found in a complex with Rhi (Volpe et al, 2001; Klattenhoff et al, 2009). This HP1 variant was recently shown to bind specific regions in the cluster 1/42AB and promote the transcription of dual-strand piRNA clusters (Klattenhoff et al, 2009). Since Rhi displays a similar nuclear localization pattern as Cuff, we first sought to determine whether these proteins colocalize in the Drosophila germline. To this aim, we performed Rhi and Cuff co-immunostainings in wild-type ovaries and found that the two proteins strongly colocalize already in early stages of oogenesis (Figure 6A–A″). Nuclear foci of Cuff/Rhino colocalization are clearly detectable in the late mitotic cysts, while they are less apparent during the first mitotic divisions of the cystoblasts. Cuff and Rhi strongly colocalize throughout oogenesis and in later egg chambers they are readily detectable in discrete nurse cell nuclear foci (Figure 6B–B″ and C–C″). In addition to the extensive colocalization, Cuff-specific staining can also be observed in regions where Rhi is reduced or absent (Figure 6C–C″, arrows), thus suggesting that Cuff may have additional targets during oogenesis. The significant overlap between Cuff and Rhi localization patterns prompted us to investigate whether these proteins are part of the same complex and physically interact. Co-immunoprecipitation assays on EGFP–Cuff expressing ovaries followed by western blotting with anti-Rhi antibodies confirmed this hypothesis (Figure 6D). A signal at ∼47 kDa can be observed in the input lanes of both EGFP–Cuff and control (OrR ovaries) samples (Figure 6D, input lanes), which roughly corresponds to the molecular weight of Rhi (46.3 kDa). A prominent band migrating approximately at the same molecular weight can be detected in EGFP–Cuff immunoprecipitates, which most likely corresponds to the Rhi protein (Figure 6, IP lanes). Conversely, only a faint signal can be detected in the control lane (OrR ovaries), which most likely represents an unspecific signal (Figure 6D, IP lanes). A reciprocal IP using Rhino–GFP and HA–Cuff lines confirmed the interaction (data not shown). In this study, we demonstrate that Cuff is enriched in heterochromatic domains (Figure 2). Therefore, we tested whether the interaction between Cuff and Rhi is specific or whether Cuff is generally associated with heterochromatin proteins. To this aim, we probed the input and IP samples described above with antibodies specific for the HP1 protein (reviewed in Vermaak and Malik, 2009). We could detect a signal at ∼25 kDa in the EGFP–Cuff and OrR input lanes (Figure 6D, input lanes), which roughly corresponds to the molecular weight of HP1 (predicted 23.2 kDa). However, no signal can be detected in the EGFP–Cuff and in the OrR IP lanes (Figure 6D, IP lanes). Cuff and Rhi, therefore, interact specifically and appear to be components of a nuclear complex, which exerts a prominent role in the expression of the piRNA clusters. It is conceivable that the activity of one protein is necessary to recruit the other to specific sites in the genome. Alternatively, both proteins are required to assemble and/or stabilize a piRNA cluster regulatory complex. In order to test these hypotheses, we performed immunostaining experiments on wild-type and cuff mutant ovaries with anti-Rhi-specific antibodies. As previously reported (Klattenhoff et al, 2009), Rhi accumulates in distinct nuclear foci in wild-type egg chambers (Figure 6E). In contrast, Rhi is no longer found in nuclear speckles in cuff mutant egg chambers, where it appears dispersed in the nucleoplasm (Figure 6F). In the reciprocal assay, we asked whether mutations in Rhi abolish the accumulation pattern of Cuff (Figure 6G). Immunostaining experiments on rhi ovaries with anti-Cuff-specific antibodies revealed that Cuff fails to properly localize in the rhi mutant and, similar to Rhi in the cuff ovaries, is dispersed in the nurse cell nucleoplasm (Figure 6H). We conclude that both Cuff and Rhi contribute to the assembly and/or stability of a nuclear complex involved in the regulation of the piRNA clusters.
Figure 6.
Cuff and Rhi extensively colocalize in germline nuclear foci and physically interact. (A–A″) Immunostaining assay with anti-Cuff (A, red and A″, single channel) and anti-Rhi (A, green and A′, single channel) antibodies in wild-type germaria. (B–B″) Nurse cell nuclei of stage 10 egg chambers expressing EGFP–Cuff (B, green and B″, single channel) stained with anti-Rhi antibody (B, red and B′, single channel). (C–C″) Nurse cell nuclei of stage 10 egg chambers immunostained with anti-Cuff (C, red and C″, single channel) and ant-Rhi (C, green and C′, single channel) antibodies. Arrows indicate Cuff-positive foci, which do not colocalize with Rhi. DNA was labelled with Hoechst (A–C, blue). (D) Cuff and Rhi physically interact. IP on control lines (OrR) and EGFP–Cuff ovarian extracts was performed with anti-GFP antibodies and followed by western blotting with anti-Rhi antibodies. A 37-kDa band corresponding to the molecular weight of Rhi can be detected in the input extracts. Rhi is efficiently immunoprecipitated from EGFP–Cuff extracts, while a faint background signal is present in the OrR lane. (E–H) Immunostaining assays on stage 6–10 wt and cuff egg chambers. Rhi localizes to nuclear speckles in wt nurse cell nuclei (E), while it appears dispersed in the nucleoplasm of cuff egg chambers (F). In the reciprocal assay, Cuff localizes to nurse cell nuclear foci in wt stage 6–10 egg chambers (G), while it is dispersed in the nucleoplasm of rhi mutant ovaries. (I) Chromatin immunoprecipitation assay on ovarian extracts obtained from EGFP–Cuff expressing flies. Cuff is enriched in regions 1A, 1C and 1–32, but not in region 1B within cluster 1/42AB. Cuff does not display a significant enrichment at regions 2A and 2B within cluster 2 and in the flam locus. Primers for the rosy (ry) and rpr49 (rp49) genes were used as negative control. Blue bars represent the ChIP assay with anti-GFP antibody on EGFP–Cuff expressing ovaries, while red bars display the result of a control experiment carried on with anti-EGFR antibody. Asterisks mark the reported binding sites for Rhi. Fold enrichment for each analysed region is reported on the Y axis as a percentage of the input chromatin (%Input).
Cuff is enriched at the dual-strand piRNA cluster 1/42AB
The significant similarities between Cuff and Rhi accumulation pattern and function in the piRNA cluster regulation prompted us to investigate whether Cuff is able to bind specific regions within the major piRNA clusters. To this aim, we performed Chromatin immunoprecipitation (ChIP) assays on ovarian extracts isolated from EGFP–Cuff expressing lines (Figure 6I). Control experiments were performed with antibodies specific for proteins not involved in the piRNA pathway. We analysed four regions in cluster 1/42AB, namely 1A, 1B, 1C and 1–32, regions 2A and 2B within cluster 2 and one region in the flam locus (Klattenhoff et al, 2009). In addition, we analysed Cuff enrichment at the euchromatic genes rosy (ry) and rpr49 (rp49). For each assay, the enrichment was calculated as a fraction of the input chromatin. Cuff appears to be enriched in regions 1A, 1C and 1–32 of cluster 1/42AB, but not in region 1B, compared with the ry and rp49 genes. We failed to detect an enrichment of DNA sequences derived from cluster 2, including regions 2A and 2B, and flam in the EGFP–Cuff ChIP assays with respect to the euchromatic genes. Since the Cuff protein does not display any conserved DNA binding domains, it is likely that the interaction between Cuff and DNA sequences in the cluster 1/42AB is mediated by Rhi.
piRNA pathway components are mislocalized in the cuff mutant ovaries
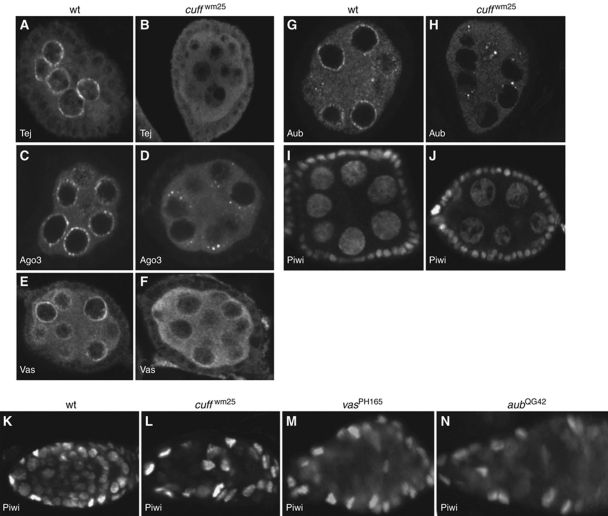
piRNA biogenesis seems to occur mainly in a perinuclear organelle known as ‘nuage’ (Findley et al, 2003; Snee and Macdonald, 2004; Klattenhoff et al, 2007; Lim and Kai, 2007; Pane et al, 2007). This conserved organelle surrounds the nurse cell nuclei in wild-type individuals and hosts a number of activities, including the Tudor-domain protein Tejas (Tej), the translation factor Vas and the Argonaute proteins Aub and Ago3 (Figure 7A, C, E and G), which control the different steps of piRNA production (Hay et al, 1988; Liang et al, 1994; Li et al, 2009; Malone et al, 2009; Patil and Kai, 2010). Immunostaining experiments with anti-Tej, anti-Vas, anti-Aub and anti-Ago3 antibodies revealed that all these proteins lose their perinuclear localization and appear dispersed in the nurse cell cytoplasm in the cuff mutant egg chambers (Figure 7B, D, F and H). These findings indicate that Cuff activity is necessary to preserve the integrity of the nuage. While cytoplasmic Cuff may exert a more direct role in nuage assembly/maintenance (Chen et al, 2007), it is also possible that cluster-derived transcripts, whose production depends on nuclear Cuff, are critical to preserve nuage integrity. In contrast, we found that mutations in vas, aub and ago3 do not appear to affect the nuclear accumulation of Cuff (data not shown).
Figure 7.
Mutations in Cuff affect the subcellular localization of piRNA pathway components. In wild type, Tej, Ago3, Vas and Aub accumulate in the perinuclear nuage (A, C, E and G, respectively). In the cuff mutant, Tej, Ago3, Vas and Aub are dispersed in the cytoplasm, indicating a disruption of the nuage (B, D, F and H). Piwi is normally enriched in the nuclei of nurse cells in wild-type egg chambers (I) and it is not severely affected by mutations in cuff (J). Piwi is prominently nuclear in somatic and germ cells of wild-type germaria (K). In contrast, Piwi is clearly dispersed/downregulated in the germarial germ cells of cuff, vas and aub mutants, while it retains a nuclear accumulation in the somatic cells (L–N, respectively).
Different from the Piwi-clade Argonaute proteins Aub and Ago3, which display a germline-specific function, Piwi is expressed in both germ cells and somatic follicle cells throughout oogenesis (Cox et al, 2000; Saito et al, 2006; Brennecke et al, 2007; Nishida et al, 2007). In these tissues, Piwi primarily localizes to the nucleus (Cox et al, 2000). It has been shown that Piwi nuclear accumulation in the germline relies on the activity of Vas, Aub and other piRNA pathway factors (Klattenhoff et al, 2009; Malone et al, 2009). We, therefore, asked whether cuff is required for the expression/localization of Piwi. Immunostaining experiments revealed that Piwi is apparently unaffected in the fully formed cuff egg chambers (stages 3–10), where it mostly retains the nuclear accumulation pattern (Figure 7I and J). However, Piwi is strongly reduced or dispersed in the cuff mutant germaria (Figure 7K and L). Similarly, we could detect a significant loss of nuclear Piwi in the germaria of the vas (Figure 7M) and aub (Figure 7N) mutants. It is possible that Piwi is particularly sensitive to the loss of piRNAs at this early stage and requires a full complement of piRNAs to be targeted or anchored in the nucleus.
Discussion
In Drosophila, piRNAs are mostly produced from discrete genomic loci, termed piRNA clusters, which are composed of repetitive sequences and inactive transposon remnants (Brennecke et al, 2007). The major piRNA clusters, including the dual-strand cluster 1/42AB and the uni-strand cluster 2 and flam, are located at the border between euchromatic and heterochromatic regions or within the major heterochromatic domains (Brennecke et al, 2007). Heterochromatic states have been generally associated with transcriptionally repressed genomic loci (Vermaak and Malik, 2009). The expression of the piRNA clusters, therefore, needs to be tightly controlled to ensure piRNA production, while the heterochromatin is being assembled. Here, we report the functional characterization of Cuff, a Drosophila protein with similarity to the yeast transcription termination factor Rai1 (Xue et al, 2000; Kim et al, 2004; Chen et al, 2007), and we show that this protein controls piRNA cluster expression and piRNA production during Drosophila germline development. We show that Cuff is critical for the correct levels of transcripts produced from the major dual-strand cluster 1/42AB in the pericentromere of chromosome II. piRNAs originating from this locus and from virtually all the dual-strand clusters dispersed in the genome, are substantially reduced or depleted in the cuff mutant. Consistent with a germline-specific function of Cuff, mutations in this protein do not impact the flam locus, which is known to be active only in the somatic follicle cells (Pelisson et al, 1994; Prud’homme et al, 1995; Desset et al, 2003; Brennecke et al, 2007; Mevel-Ninio et al, 2007; Malone et al, 2009). Our data suggest that Cuff regulates the piRNA master loci presumably acting in a complex with the HP1 variant Rhi. Both Cuff and Rhi are enriched at the cluster 1/42AB, while they do not interact with sequences in the cluster 2 and flam. These observations strongly suggest that a Cuff/Rhi complex regulates the transcription of the dual-strand piRNA clusters. The activity of this complex appears to be crucial for the expression of the dual-strand clusters and might be critical to overcome the complexity of these genomic loci, where bidirectional transcription of the different elements can potentially hinder the production of the proper piRNA population. Different from Rhi, cuff also affects the production of piRNAs from some uni-strand clusters, including the major cluster 2. Our ChIP assays, however, failed to reveal a significant enrichment of Cuff at this locus. It is, therefore, likely that the loss of piRNAs from cluster 2 is not directly caused by mutations in the Cuff protein, but it might rather ensue from the disruption of the nuage observed in the cuff ovaries and the mislocalization of factors required for piRNA production. Nevertheless, the comparison of cuff and rhi piRNA libraries revealed some interesting differences between the two mutants (Supplementary Figure S4). For instance, mutations in cuff appear to have a more prominent impact on the sense piRNA population compared with the rhi mutant, thus suggesting that these proteins might display partially different functions in piRNA cluster expression and piRNA production (Supplementary Figure S4). Intriguingly, the expression levels of the piRNA clusters in the cuff mutant do not precisely mirror the profiles of the corresponding piRNA population. One would predict that the depletion of a specific piRNA set would ensue from a general downregulation of the corresponding cluster-derived transcripts. While some regions within cluster 1/42AB are clearly downregulated in the cuff mutant, other regions are not affected and only one, among those we analysed, appears to be upregulated. These data point to a role for Cuff in the transcriptional control of dual-strand cluster expression, whereby this protein may be required to activate/permit the transcription of these loci. In the absence of Cuff, cluster-derived piRNA precursor transcripts do not accumulate, thus leading to a general collapse of the piRNA population. It is noteworthy that some regions in the cluster 1/42AB are not affected in the cuff mutant. This observation suggests that the dual-strand clusters might not produce single transcripts spanning the entire locus, as it is the case for the flam locus (Brennecke et al, 2007), but they might be rather transcribed by multiple internal and external promoters.
It has been recently reported that mutations in the Argonaute proteins Piwi and Aub reduce the expression levels of genomic regions located within or next to the piRNA clusters (Moshkovich and Lei, 2010). In these mutants, HP1 spreading triggers a significant repression of the piRNA loci, which led to the proposal that in Drosophila, piRNAs ensure the transcription of the clusters by counteracting the spreading of heterochromatic states (Moshkovich and Lei, 2010). Remarkably, the recruitment of Cuff and Rhi to nuclear foci early in oogenesis seems to parallel the assembly of the major heterochromatic blocks. The levels of Cuff, Rhi and HP1 are low in the stem cells and in the early mitotic cyst, while these proteins appear to accumulate in the late mitotic cyst, where they extensively colocalize in specific regions of the nuclei. It is, therefore, tempting to speculate that Cuff and Rhino might protect the clusters from the repressive effects associated with the assembly of the major heterochromatic blocks, which takes place early in oogenesis while the germ cells undergo the mitotic divisions (Yoon et al, 2008).
Mutations in cuff negate the production of both sense and antisense piRNAs corresponding to 46 transposon families. For a significant number of transposons only piRNAs matching one DNA strand seem to be affected in cuff, while piRNAs corresponding to a few transposon families are apparently unaltered. It is noteworthy that the HeT-A and TART retro-transposons, which are involved in the maintenance of telomere integrity, display contrasting piRNA profiles despite the fact that the transcript levels of both these elements are upregulated in cuff ovaries (Chen et al, 2007). Virtually, all the piRNAs corresponding to HeT-A sequences are depleted in the cuff mutant, which might account for the severe deregulation of this retro-transposon in the absence of Cuff. Conversely, we could identify only a limited number of piRNAs matching TART sequences in wild-type ovaries, while a surprising variety of reads corresponding to this element are produced in cuff mutant ovaries. Similar to the piRNA clusters, also the different classes of transposons appear to undergo differential regulation, and it will be a challenge for the future to uncover the underlying rules.
The introduction of a sensor transgene based on the Zam retro-transposon in the germline of cuff, rhi, vas and aub flies allowed us to gain further insight into the temporal requirement of these proteins during Drosophila oogenesis. Zam is an LTR retro-transposon, which is specifically repressed in the ovarian follicle cells by the piRNAs produced from the flam locus (Pelisson et al, 1994; Desset et al, 2003, 2008). While the sensor transgene is consistently repressed in wild-type germ cells, it is significantly deregulated in cuff, rhi, vas and aub germline. The silencing of the reporter appears to be very sensitive to mutations in cuff and rhi, which produce a strong GFP signal in stem and germ cells as well as in fully formed egg chambers. Mutations in vas and aub instead seem to have a milder impact on the sensor construct, whereby a detectable GFP signal is mostly observed in the dividing or late mitotic cysts. The mechanism by which the sensor construct is silenced in wild-type ovaries is unlikely to mirror the piRNA-dependent repression of the Zam retro-transposon in somatic cells, since mutations in cuff do not impact the production of piRNAs from the flam locus. Similarly, flam piRNA levels are unaltered in rhi, aub and vas mutant backgrounds (Klattenhoff et al, 2009; Malone et al, 2009). These observations reveal that Zam sequences can be targeted for silencing also in germline tissues in a flam-independent fashion. Interestingly, we did not detect piRNAs corresponding to the env fragment neither in wild-type nor in the cuff mutant libraries. While we cannot exclude that the expression of the sensor transgene induced by the Act5C-Gal4 driver in the wild-type germline might trigger the production of piRNAs directed against the reporter, it is possible that the silencing of the pGFP-ZenvAS transgene might not proceed through a piRNA-based mechanism. Since nuclear Piwi is dramatically lost in the germaria of all these mutants, it is also unlikely that the strong deregulation of the reporter observed in cuff and rhi mutant stem and germ cells is primarily caused by Piwi mislocalization. Instead, these results suggest that Cuff and Rhi might act upstream of Piwi to regulate the expression of the piRNA clusters and ensure piRNA production early in oogenesis. The mature piRNAs are then loaded into Piwi and serve to drive the nuclear accumulation of this protein. It is conceivable that the activity of Cuff and Rhi protects the piRNA clusters from the repressing effects associated with heterochromatin formation. In later stages, Vas, Aub and most likely other piRNA pathway components might further contribute to the maintenance of active transcription at these loci.
Mutations in cuff cause a progressive loss of germ cells, including stem cells, over time (Chen et al, 2007). In addition, previous studies have implicated germline Piwi in the control of germ-cell division rate, while somatic Piwi is required for stem-cell self-renewal (Cox et al, 2000). Our current data strongly suggest that stem- and germ-cell loss in the cuff mutant is caused by a failure to express the piRNA master loci. In the absence of primary piRNAs, the entire piRNA machinery is dispersed and Piwi fails to accumulate in the stem- and germ-cell nuclei, thus further contributing to the cuff phenotype.
piRNA-based mechanisms appear to be conserved across the phyla and homologues of the Piwi-clade Argonaute proteins are present in organisms as distant as flies and mouse (Khurana and Theurkauf, 2010; Saito and Siomi, 2010; Senti and Brennecke, 2010). Similarly, members of the Rai1 superfamily can be found both in unicellular organisms and in higher Eukaryotes. Therefore, it is conceivable that specific Rai1 and HP1 variants might exist in other organisms and, similar to Drosophila, their interaction might ensure the maintenance of genome integrity during stem- and germ-cell division.
Materials and methods
Fly stocks
cuffWM25, cuffQQ37, cuffRI67, cuffWL25 and cuffRN48 aubQC42 were isolated in female sterile screens on the second chromosome (Schupbach and Wieschaus, 1989, 1991). vasPH165 is a null allele (Styhler et al, 1998). rhi02086 and rhiKG00910 (Volpe et al, 2001) (Bloomington Stock Center) alleles are caused by P-element insertions. In this study, rhi alleles were crossed to a deficiency spanning the rhi locus, Df(2R)Exel17149 (Bloomington Stock Center). OreR, cn bw or cuff heterozygous flies were used as wild-type control. Marker mutations and balancers are described in flybase (http://flybase.org).
pUAS-EGFP_Cuff and pUAS-EGFP lines are described in Supplementary Materials and methods. Reporter lines pUAS-GFP-ZenvAS were kindly provided by Chantal Vaury (Desset et al, 2008).
Immunohistology, immunoprecipitation and western analyses
Antibody stainings were performed as previously described (Neuman-Silberberg and Schupbach, 1996). Mouse anti-α-Spectrin antibody 3A9 at 1:100 (Developmental Studies Hybridoma Bank), mouse anti-HP1 antibody was used at 1:200 (Developmental Studies Hybridoma Bank), rabbit anti-H3K9triMeth was used at 1:200 (Upstate), guinea pig anti-Rhi (a gift from William Theurkauf) was used at 1:1000, goat anti-Vasa antibody (Santa Cruz Biotechnology) was used at 1:1000, chicken anti-CID antibody (a gift from G Karpen) at 1:1000, rabbit anti-Tejas (a gift from T Kai) at 1:1000, rabbit anti-Ago3, rabbit anti-Piwi and rabbit anti-Aub (gifts from Julius Brennecke) at 1:1000.
IP and western analyses were performed as previously described (Chen et al, 2007; Pane et al, 2007). EGFP–Cuff was pulled down with anti-GFP antibodies (Novus Biologicals). Western blotting was carried on with guinea pig anti-Rhino antibody and mouse anti-HP1 diluted 1:1000 in TBST.
qRT–PCR and northern blot
For RNA analysis, total RNA was isolated using Trizol (Invitrogen). Total RNA was treated with Turbo DNA-free kit to remove contaminant DNA (Ambion). Reverse Transcription (RT) was performed with the SuperScript™ III Reverse Transcriptase (Invitrogen).
Quantitative RT–PCR was performed with the ABI Prism® 7900 system (AME Bioscience). Each reaction consisted of 50 ng first-strand cDNA template. For each mutant genotype, three separate RNA samples were prepared. Statistical analysis was performed with Microsoft Office Excel software. For strand-specific qRT–PCR, RT of the template RNA was carried on with primers specific for cluster 1/42AB. Primer sets for the piRNA cluster analyses are described in Klattenhoff et al (2009). Control primers for the Rpr49 RNAs are described in Chen et al (2007). Additional primer pairs specific to cluster 1/42AB are listed below:
Region A′
- Fw
5′-AAGACCCAATTTTTGCGTCGC-3′
- Rev
5′-CAAGGATAGGGATTTGGTCC-3′
Region B′
- Fw
5′-CTATTATTGGCACTGCTATCC-3′
- Rev
5′-GGACCAATTAGCGCGAAGAC-3′
Region C′
- Fw
5′-CTATCGTATAGATAGTGATATTC-3′
- Rev
5′-AACCAAGTTCATCTTGTATAGC-3′.
Northern blot analysis for the detection of small RNAs was performed as previously described (Lagos-Quintana et al, 2002; Pane et al, 2007). DNA probes for the detection of 2S rRNA, AT-chX-1 and miR-310 were previously described (Saito et al, 2006; Nishida et al, 2007; Pane et al, 2007). DNA probes for miR-184 and Het-A piRNAs are listed below:
- miR-184 probe
5′-GCCCTTATCAGTTCTCCGTCCA-3′
- Het-A probe
5′-TCCCGTGTCCTGTTTTTCCTTTCA-3′.
Chromatin immunoprecipitation
ChIP assays on EGFP–Cuff expressing flies were performed according to Blythe et al (2009). Flies were fed on yeast for 2/3 days and 60 ovaries for each assay were isolated in ice-cold PBS. Ovaries were fixed in 1.8% paraformaldehyde for 10′ on ice and rinsed 3 × in PBS. Subsequently, nuclei were extracted in hypotonic buffer and fixed again as described above. Paraformaldehyde fixation was blocked with 125 mM Glycine. After three washes in hypotonic buffer, the nuclei were resuspended in lysis buffer and subjected to three rounds of sonication on a Digital Sonifier (Branson) for 12″ at 20% power input with 2′ on ice between each cycle. After this treatment, DNA fragments displayed an average 200 bp length on a 2% agarose gel. Sheared chromatin was cleared by centrifugation at 13 000 r.p.m. and the supernatant was incubated with rabbit anti-GFP antibodies (Novus Biologicals) overnight at 4°C. Control experiments were performed with rabbit anti-EGFR antibodies (Santa Cruz) under the same conditions. Immunocomplexes were subsequently isolated by incubation with Protein G magnetic beads (Invitrogen) for 4 h at 4°C and washed as described in Blythe et al (2009). Biological duplicates were generated for each experimental sample. DNA samples were subjected to qRT–PCR analyses as described above. Primer pairs adopted in this assay are described in Klattenhoff et al (2009).
Small RNA library production and deep sequencing
Small RNA libraries from the cn bw control line, and from cuffwm25 homozygous ovaries were produced with the Small RNA Library Prep Kit (Illumina). Briefly, 20 μg total RNA extracted from the different genotypes was separated on a 15% denaturing polyacrylamide/urea gel electrophoresis (Invitrogen) for 1 h at 200 V. A gel slice corresponding to 18–30-nt-long RNAs was isolated and small RNAs were eluted and processed with the Small RNA Library Prep Kit as per the manufacturer's instructions. Small RNA libraries were subsequently sequenced on the Solexa Genome Analyzer II platform. The bioinformatic analysis of the piRNA libraries is described in details in Supplementary Materials and methods.
Supplementary Material
Acknowledgments
We would like to thank Paul Lasko, Paul Macdonald and William Theurkauf for providing fly stocks; Julius Brennecke, Gary Karpen and William Theurkauf and Toshie Kai for sharing antibodies. We thank Lance Parsons, Donna Storton and Jessica Buckles for help with small RNA library sequencing and analysis; Shelby Blythe and Germano Cecere for help with chromatin immunoprecipitation assays; Joe Goodhouse for help with confocal microscopy; Gail Barcelo for assistance with molecular techniques. We are grateful to Girish Deshpande, Stefano Di Talia and Elena Domanitskaya for critical reading of the manuscript and members of the Schupbach and Wieschaus laboratories for helpful discussions and suggestions. This work was supported by the Howard Hughes Medical Institute and US Public Health Service Grant RO1 GM 077620 (AP and TS) and NIH grants P50 GM071508 and GM076275 (PJ and MS).
Author contributions: AP and TS conceived the experiments, AP and DYZ carried out the experiments, JP and MS carried out the computational analysis and AP, TS, MS and JP wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Tuschl T (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T (2003) The small RNA profile during Drosophila melanogaster development. Dev Cell 5: 337–350 [DOI] [PubMed] [Google Scholar]
- Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A (2006) Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Karpen GH (2001) The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol 3: 730–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kesler DS, Klein PS (2009) Chromatin Immunoprecipitation in early Xenopus laevis embryos. Dev Dyn 6: 1422–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating Loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucheton A (1990) I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet 6: 16–21 [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Popkova A, Payen-Groschene G, Brun C, Laouini D, Pelisson A, Bucheton A (2008) piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci USA 105: 14964–14969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Pane A, Schupbach T (2007) Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol 17: 637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse KN, Ferguson SB, Schupbach T (2008) Squid, Cup, and PABP55B function together to regulate gurken translation in Drosophila. Dev Biol 313: 713–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H (2000) Piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127: 503–514 [DOI] [PubMed] [Google Scholar]
- Desset S, Buchon N, Meignin C, Coiffet M, Vaury C (2008) In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS One 3: e1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desset S, Meignin C, Dastugue B, Vaury C (2003) COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164: 501–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, Tollervey D (2008) Efficient termination of transcription by RNA polymerase I requires the 5′ exonuclease Rat1 in yeast. Genes Dev 22: 1069–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H (2003) Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130: 859–871 [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL (2006) The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol 357: 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC (2007) A Slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1589 [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN (1988) A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55: 577–587 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF (2007) A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Iovino N, Pane A, Gaul U (2009) miR-184 has multiple roles in Drosophila female germline development. Dev Cell 17: 123–133 [DOI] [PubMed] [Google Scholar]
- Khurana JS, Theurkauf W (2010) piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol 191: 905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S (2004) The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE (2007) Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell 12: 45–55 [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, Seitz H, Zamore PD, Weng Z, Theurkauf WE (2009) The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA (2007) Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res 35: 5430–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12: 735–739 [DOI] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD (2009) Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P (1994) Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120: 1201–1211 [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T (2007) Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci USA 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ (2009) Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevel-Ninio M, Pelisson A, Kinder J, Campos AR, Bucheton A (2007) The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175: 1615–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshkovich N, Lei EP (2010) HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet 6: e1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A, Mituyama T, Huang H, Chen D, Siomi MC, Siomi H (2010) Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 16: 2503–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T (1996) The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech Dev 59: 105–113 [DOI] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC (2007) Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schupbach T (2007) zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil VS, Kai T (2010) Repression of retroelements in Drosophila Germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 20: 724–730 [DOI] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A (2007) A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol 81: 1951–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Song SU, Prud’homme N, Smith PA, Bucheton A, Corces VG (1994) Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J 13: 4401–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme N, Gans M, Masson M, Terzian C, Bucheton A (1995) Flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139: 697–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC (2006) Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Siomi MC (2010) Small RNA-mediated quiescence of transposable elements in animals. Dev Cell 19: 687–697 [DOI] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A (2004) Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E (1989) Female sterile mutations on the second chromosome of Drosophila melanogaster. I. Maternal effect mutations. Genetics 121: 101–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E (1991) Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129: 1119–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J (2010) The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Miyoshi T, Siomi H (2010) piRNA-mediated silencing in Drosophila germlines. Semin Cell Dev Biol 21: 754–759 [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM (2004) Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci 117: 2109–2120 [DOI] [PubMed] [Google Scholar]
- Spradling AC (1993) Germline cysts: communes that work. Cell 72: 649–651 [DOI] [PubMed] [Google Scholar]
- St Johnston D, Ahringer J (2010) Cell polarity in eggs and epithelia: parallels and diversity. Cell 141: 757–774 [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P (1998) vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125: 1569–1578 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD (2006) A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vermaak D, Malik HS (2009) Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu Rev Genet 43: 467–492 [DOI] [PubMed] [Google Scholar]
- Volpe AM, Horowitz H, Grafer CM, Jackson SM, Berg CA (2001) Drosophila rhino encodes a female-specific chromo-domain protein that affects chromosome structure and egg polarity. Genetics 159: 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L (2009) Structure and function of the 5′ → 3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458: 784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Bai X, Lee I, Kallstrom G, Ho J, Brown J, Stevens A, Johnson AW (2000) Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol Cell Biol 20: 4006–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Lin H (2007) An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature 450: 304–308 [DOI] [PubMed] [Google Scholar]
- Yoon J, Lee KS, Park JS, Yu K, Paik SG, Kang YK (2008) dSETDB1 and SU(VAR)3-9 sequentially function during germline-stem cell differentiation in Drosophila melanogaster. PLoS One 3: e2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.