Abstract
Clustered regularly interspaced short palindromic repeats (CRISPRs) and Cas proteins represent an adaptive microbial immunity system against viruses and plasmids. Cas3 proteins have been proposed to play a key role in the CRISPR mechanism through the direct cleavage of invasive DNA. Here, we show that the Cas3 HD domain protein MJ0384 from Methanocaldococcus jannaschii cleaves endonucleolytically and exonucleolytically (3′–5′) single-stranded DNAs and RNAs, as well as 3′-flaps, splayed arms, and R-loops. The degradation of branched DNA substrates by MJ0384 is stimulated by the Cas3 helicase MJ0383 and ATP. The crystal structure of MJ0384 revealed the active site with two bound metal cations and together with site-directed mutagenesis suggested a catalytic mechanism. Our studies suggest that the Cas3 HD nucleases working together with the Cas3 helicases can completely degrade invasive DNAs through the combination of endo- and exonuclease activities.
Keywords: Cas3, CRISPR, HD domain, Methanocaldococcus jannaschii , nuclease
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPRs) and their associated proteins (Cas) represent a highly adaptive and heritable defence system against exogenous nucleic acids (Makarova et al, 2006; Barrangou et al, 2007). Cas proteins incorporate sequences derived from the invader DNA into the host chromosome, which are then transcribed and processed into short CRISPR RNAs (crRNAs), which direct the cleavage of the foreign nucleic acids by other Cas proteins (van der Oost et al, 2009; Deveau et al, 2010; Horvath and Barrangou, 2010; Karginov and Hannon, 2010; Marraffini and Sontheimer, 2010). CRISPR loci (or clusters) comprise up to 100 identical direct repeats (25–50 nt) separated by variable sequences (spacers) of similar size, some of which are homologous to sequences from viral or plasmid genomes. A role of CRISPR spacers in defence against the phage or plasmid invasion has been demonstrated in the natural systems of Streptococcus thermophilus, Staphylococcus epidermidis, and Sulfolobus solfataricus, as well as in an engineered strain of Escherichia coli (Barrangou et al, 2007; Brouns et al, 2008; Marraffini and Sontheimer, 2008; Manica et al, 2011). In general, there are three main steps in the CRISPR-based immunity: adaptation (or immunization), expression/processing, and interference (van der Oost et al, 2009; Deveau et al, 2010; Horvath and Barrangou, 2010; Karginov and Hannon, 2010). The adaptation stage involves the recognition and insertion of small pieces of foreign nucleic acid as novel spacers into a CRISPR locus. During the expression and processing step, the long primary CRISPR transcripts are processed into short crRNAs, which direct the recognition and cleavage of the invader DNA or RNA by the associated Cas proteins in the interference step.
Comprehensive bioinformatics analyses of the CRISPR-harbouring genomes has identified at least 65 different Cas proteins, most of which are predicted to have nuclease or nucleic acid binding activity and can be organized into up to 45 protein families (Haft et al, 2005; Makarova et al, 2006, 2011). Six Cas protein families (Cas1–6) represent the core group of the CRISPR-associated proteins, whose genes are found close to the CRISPR clusters in most CRISPR-containing genomes. Based on the phylogeny and composition of the Cas operons, three major types (I, II, and III) of the CRISPR/Cas systems have been proposed and eight subtypes have been named after the particular genome where it is the only CRISPR system (E. coli, Ypest, Nmeni, Dvulg, Tneap, Hmari, Apern, and Mtube subtypes) (Haft et al, 2005; Makarova et al, 2006, 2011). The major CRISPR system I includes the Cas1, Cas2, and Cas3 proteins, as well as the Cascade complex of 4–5 Cas proteins involved in the processing of the crRNAs and recognition of DNA targets (Brouns et al, 2008; Haurwitz et al, 2010; Lintner et al, 2011; Makarova et al, 2011). CRISPR types II and III have no Cas3 proteins, but include the large multidomain protein Cas9 (type II) or Cas6+RAMP (Cmr or Csm) module (type III) and can target DNA (types II and III-A) or RNA (type III-B) (Marraffini and Sontheimer, 2008; Hale et al, 2009; Haurwitz et al, 2010).
Cas3 proteins are widespread and are found in most CRISPR subtypes (except Mtube and Nmeni). Based on amino-acid sequence, Cas3 proteins are predicted to contain the DExD/H helicase and HD domains, which can be covalently fused (Cas3) or encoded as individual proteins (Cas3′ or Cas3″, respectively) (Haft et al, 2005; Makarova et al, 2006, 2011; van der Oost et al, 2009; Deveau et al, 2010). The HD domain proteins represent a large group of enzymes (over 22 000 sequences in databases, IPR003607), which catalyse phosphomonoesterase or phosphodiesterase reactions on a broad range of substrates including nucleotides and nucleic acids and function primarily in nucleic acid metabolism and signal transduction (Aravind and Koonin, 1998). The biochemically and structurally characterized microbial HD domain proteins include the RelA/SpoT protein from Streptococcus equisimilis, the Thermus thermophilus dNTPase, and the E. coli 5′-deoxyribonucleotidase YfbR (Hogg et al, 2004; Kondo et al, 2007; Zimmerman et al, 2008). Helicases are abundant proteins with ATPase activity, which unwind DNA or RNA and contain seven conserved ‘helicase’ motifs distributed across an ∼400 residue core domain (Gorbalenya and Koonin, 1993).
In the CRISPR-based immunity, the Cas3 proteins have been proposed to function together with the Cascade–crRNA complex (Brouns et al, 2008; Jore et al, 2011; Lintner et al, 2011; Makarova et al, 2011; Sinkunas et al, 2011). The E. coli Cascade–crRNA complex specifically recognizes and binds to the target DNA or RNA, but does not cleave them, suggesting that another nuclease (possibly Cas3) is recruited to degrade the target nucleic acids (Jore et al, 2011). In addition, a recent work has revealed that the Pseudomonas aeruginosa Cas3 protein functions downstream of CRISPR RNA processing and that both the Cas3 HD and helicase domains are required for the CRISPR function in the suppression of biofilm formation by phage-infected cells (Cady and O’Toole, 2011). The purified Cas3 HD domain protein SSO2001 from S. solfataricus cleaved both double-stranded (ds) DNAs and dsRNAs and showed much lower activity against single-stranded (ss) DNAs and ssRNAs (Han and Krauss, 2009). In contrast, the biochemical characterization of the S. thermophilus DGCC7710 Cas3 protein revealed the presence of a metal-dependent ssDNase activity in the HD domain, whereas the associated helicase domain harboured the ATP-dependent helicase and ssDNA-stimulated ATPase activities (Sinkunas et al, 2011). Recently, the purified E. coli Cas3 protein has been shown to catalyse annealing of RNA with DNA in the absence of ATP forming R-loops, whereas in the presence of ATP it exhibited helicase activity and unwound the model R-loop substrate (Howard et al, 2011). Crystal structure of the HD domain of the Cas3 protein TTHB187 from T. thermophilus has revealed one-metal ion in the active site, and the cleavage of ssDNA by this HD domain was not activated by Mg2+, but by Mn2+ or Ni2+ (Mulepati and Bailey, 2011).
Although the Cas3 proteins play a key role in the CRISPR interference, the molecular mechanisms of their activity remain not fully characterized. Here, we show that the Cas3 HD domain protein MJ0384 from the methanogenic archaeon Methanocaldococcus jannaschii cleaves endonucleolytically and exonucleolytically (3′–5′) the ssDNAs and ssRNAs, as well as the R-loop, 3′-flap, and splayed arm (SA) structures, which represent the potential intermediates of DNA degradation. The crystal structure of MJ0384 revealed an active site with two bound metal cations and together with site-directed mutagenesis suggest a catalytic mechanism for Cas3 nucleases.
Results
The M. jannaschii CRISPR system and the Cas3 HD domain protein MJ0384
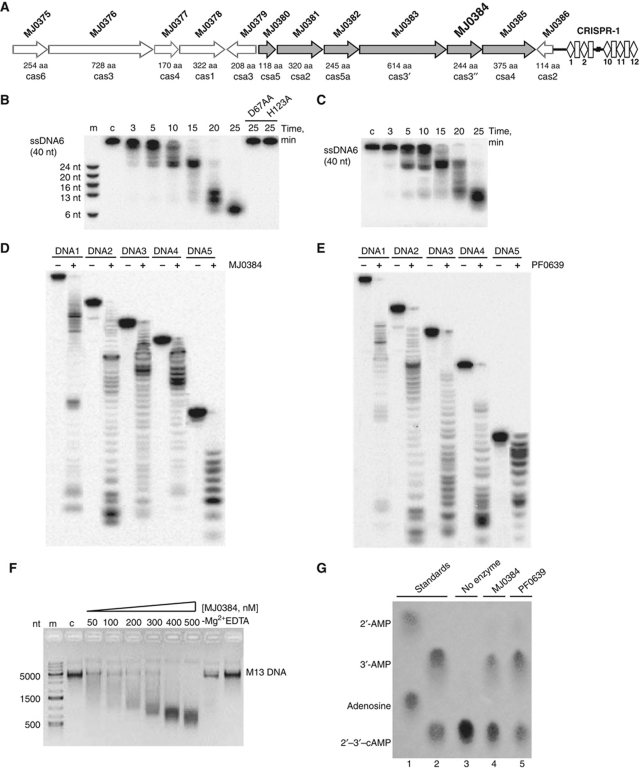
M. jannaschii has one of the most complex CRISPR systems known, which comprises 20 CRISPR clusters with 178 spacers total and at least 20 cas genes associated with the CRISPR clusters 1 and 2 (Bult et al, 1996; Deveau et al, 2010). The M. jannaschii genome encodes nine core Cas proteins: Cas1 (MJ0378), Cas2 (MJ0386), Cas3 (MJ0376, MJ0383, MJ0384), Cas4 (MJ0377), Cas5 (MJ0382), and Cas6 (MJ0375, MJ1234). The other 11 Cas proteins represent the CRISPR subtype groups Apern (MJ0379, MJ0380, MJ0381, and MJ0385) and Mtube (MJ1667, MJ1668, MJ1669, MJ1670, and MJ1672), as well as two other CRISPR-associated proteins (MJ1666 and MJ1674). The gene encoding the Cas3 HD domain protein MJ0384 (Cas3″, 244 aa) is associated with the CRISPR cluster 1 and is located in a potential six-gene operon with five other cas genes, which are likely to be co-transcribed (Figure 1A). Four cas genes located upstream of the MJ0384 gene encode the core Cas protein MJ0383 (a Cas3 helicase or Cas3′, 614 aa), MJ0382 (Cas5, 245 aa), MJ0381 (Csa2, 320 aa), and MJ0380 (Csa5, 118 aa), whereas the MJ0385 (Csa4, 375 aa) gene is located downstream of MJ0384 (Figure 1A). MJ0376 is a predicted Cas3 helicase (728 aa) with the degenerate N-terminal HD domain, so MJ0384 appears to be the only active Cas3 HD nuclease in M. jannaschii. The sequence of MJ0384 shows 52.2% identity to the Cas3 HD domain protein PF0639 from Pyrococcus furiosus and lower similarity (17–35% identity) to the homologous proteins from S. solfataricus (Supplementary Figure S1). Both MJ0384 and PF0639 sequences contain the typical HD domain signature motif H-HD-D (His20, His66, Asp67, and Asp219 in MJ0384) (Supplementary Figure S1).
Figure 1.
M. jannaschii CRISPR cluster 1 and nuclease activity of MJ0384 and PF0639. (A) Schematic representation of the M. jannaschii cas genes associated with the CRISPR cluster 1. (B, C) Time course of the linear ssDNA hydrolysis. The 5′-[32P]-labelled ssDNA (40 nt) was incubated without protein (lane c) or with wild-type and inactive MJ0384 mutant proteins (B; 200 nM) or PF0639 (C; 300 nM) for the indicated times. (D, E) Cleavage of several ssDNAs with various sequences and lengths. The 5′-[32P]-labelled ssDNAs (0.1 μM; 17–92 nt) were incubated without (−) or with MJ0384 (200 nM, 10 min) or PF0639 (300 nM, 20 min). (F) Cleavage of the circular M13mp18 ssDNA by MJ0384. The M13 ssDNA (5 nM) was incubated for 30 min without enzyme (lane c) or with various amounts of MJ0384 (50–500 nM) and the reaction products were analysed by agarose gel electrophoresis and SYBR Green staining. The last two lanes show additional controls without Mg2+ addition (−Mg2+) or in the presence of 10 mM EDTA. Lane m, DNA markers. (G) Hydrolysis of 2′,3′-cAMP by MJ0384 and PF0639: cellulose TLC analysis of the reaction products. Lanes 1 and 2, standards (1: adenosine and 2′-AMP; 2: 2′,3′-cAMP and 3′-AMP). Lanes 3–5, 2.5 mM 2′,3′-cAMP was incubated for 20 min at 60 °C in the absence (3) or in the presence of MJ0384 (1 μg) or PF0639 (1 μg). After the incubation, the samples (6 μl) were separated on the cellulose TLC plate and visualized under UV light.
Cas3 HD domain proteins MJ0384 and PF0639 cleave ssDNAs and ssRNAs
Since the CRISPR systems have been proposed to target both DNAs and RNAs (Brouns et al, 2008; Marraffini and Sontheimer, 2008; Hale et al, 2009; Horvath and Barrangou, 2010), we screened purified MJ0384 and PF0639 for nuclease activity against ssDNAs and ssRNAs of various lengths and sequences and found nuclease activity against both types of substrates (Figure 1B–E; Supplementary Figure S2A–E). DNase activity was approximately two times higher than RNA cleavage. In ssDNA cleavage, both proteins showed a strict dependence on Mg2+, but their activity was not supported by other divalent metal cations (Mn2+ or Ca2+) and was not stimulated by the addition of KCl or NaCl (Supplementary Figure S2F–I). With ssRNA as a substrate, both MJ0384 and PF0639 showed significant cleavage without the addition of monovalent or divalent metal cations, which was completely inhibited by EDTA (Supplementary Figure S2H and I). Both MJ0384 and PF0639 displayed no apparent sequence or substrate length selectivity in the cleavage of various ssDNAs and ssRNAs and showed the exonuclease-like cleavage pattern with both substrates (Figure 1; Supplementary Figures S2 and S3A and B). Product profiles of both MJ0384 and PF0639 (Figure 1B–E) reveal temporal accumulation of the intermediate cleavage products, which shows no dependence on substrate sequence and is likely caused by substrate secondary structures and/or a protein ‘footprint’ on the substrate molecule. The ability of MJ0384 to cleave ssDNA endonucleolytically was demonstrated using the circular ssDNA of the M13mp18 and φX174 phages. MJ0384 effectively cleaved both the M13mp18 (Figure 1F) and φX174 (not shown) ssDNA in a reaction, which was dependent on Mg2+ and inhibited by EDTA. The cleavage of the circular M13mp18 ssDNA by MJ0384 produced a broad range of products, suggesting that the initial endonucleolytic cleavage was followed by the exonuclease digestion of the linear ssDNA (Figure 1F). Thus, the Cas3 HD domain proteins MJ0384 and PF0639 exhibit both endonuclease and exonuclease activities against ssDNAs and ssRNAs.
The position of the phosphodiester bond cleavage (on 3′-side or on 5′-side) by MJ0384 was analysed using the T4 polynucleotide kinase (PNK) catalysed reactions of ssDNA phosphorylation at the 5′-hydroxyl termini (forward reaction) and the phosphate exchange between the oligonucleotide 5′-phosphate and ATP (Supplementary Figure S3C). After cleavage of the unlabelled ssDNA substrate by MJ0384, the reaction products were incubated with PNK and [γ-32P]ATP and analysed by denaturing PAGE (polyacrylamide gel electrophoresis) and autoradiography. High product labelling in the PNK phosphorylation reaction and low labelling in the PNK exchange reaction suggest that the MJ0384 products have no phosphate on their 5′-ends and, therefore, MJ0384 appears to produce the nucleotide products containing 3′-phosphates and 5′-hydroxyls (Supplementary Figure S3C). This type of phosphodiester bond cleavage can be considered a more efficient way of invasive nucleic acid degradation because these cleavage products cannot be directly re-ligated by polynucleotide ligases and need to be first repaired (3′-phosphate removed and 5′-hydroxyl phosphorylated).
Several RNases cleaving substrates on the 5′-side (RNase A, RNase T1, Ire1p, RNase Bi) have been shown to release the products containing a 2′,3′-cyclic phosphodiester bond, which is subsequently cleaved by the same enzyme or by an auxiliary protein to produce a 3′-phosphate end (Thompson et al, 1994; Okorokov et al, 1997; Gonzalez et al, 1999). In addition, the 2′,3′-cyclic phosphodiesterase activity has been demonstrated in several HD domain proteins (Yakunin et al, 2004; Blondal et al, 2005). We found that both MJ0384 and PF0639 exhibit significant phosphodiesterase activity against 2′,3′-cAMP and 2′,3′-cGMP (Figure 1G; Supplementary Figure S3D). This activity was stimulated by Mn2+ and produced 3′-NMP (3′-AMP) as a final product (Figure 1G). The role of Cas3 HD nuclease 2′,3′-cyclic phosphodiesterase activity in the CRISPR mechanism is presently unclear, but it is worth mentioning that the mature E. coli crRNA contains a 2′,3′-cyclic phosphate terminus (Jore et al, 2011), which might represent a potential substrate for Cas3 HD nucleases.
Cleavage of branched DNA and RNA substrates by MJ0384
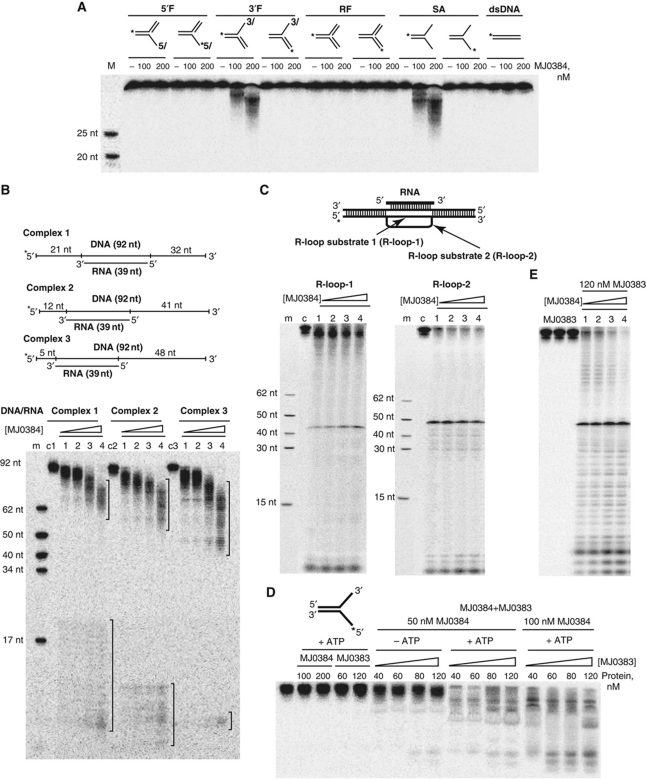
Purified MJ0384 was also tested for nuclease activity against linear dsDNA or dsRNA, as well as against several branched DNA and RNA substrates including 5′-flap (5′F), 3′-flap (3′F), SA, and replication fork (RF), which represent the potential intermediates of the degradation of dsDNA. These experiments revealed the cleavage of substrates containing free single-stranded 3′-ends (3′F and SA) and no activity against 5′F, RF, dsDNA, or dsRNA (Figure 2A; Supplementary Figure S4). The presence of double-stranded regions in the long ssDNA substrates prevented the cleavage of these sequences by MJ0384 (Figure 2B). In this experiment, three different DNA strands (92 nt long, 32P-labelled on the 5′-end) were annealed to a shorter unlabelled RNA (39 nt) complementary to three different positions on the DNA strand (Figure 2B). The cleavage of these substrates by MJ0384 produced two groups of products: the long products are produced by the exonucleolytic cleavage of DNA from the 3′-end of the DNA strand, whereas the short products are the result of both the endo- and exonucleolytic cleavage of the ssDNA close to its 5′-end (Figure 2B). The size of the product group was proportional to the length of ssDNA strand accessible for the cleavage by MJ0384, whereas the central part of the labelled DNA strand was protected from the cleavage by the annealed RNA fragment (Figure 2B).
Figure 2.
Nuclease activity of MJ0384 against complex DNA substrates. (A) Cleavage of complex DNA substrates: 5′-flap (5′F), 3′-flap (3′F), RF, SA, and dsDNA. The substrates were 5′-[32P]-labelled on the indicated strand (*) and incubated (15 min at 42 °C) in the absence (−) or in the presence of MJ0384. M, markers (5′-32P-labelled oligonucleotides). (B) Cleavage of DNA/RNA complexes. The complexes were incubated for 20 min at 42 °C in the absence (c1, c2, c3) or in the presence of MJ0384 (1) 50 nM; (2) 100 nM; (3) 150 nM; (4) 200 nM. The brackets on the top of the gel indicate the products of the exonucleolytic (3′–5′) substrate cleavage by MJ0384, whereas those at the gel bottom designate the endo- and exonucleolytic products. (C) Cleavage of R-loop substrates. The substrates were incubated for 60 min at 42 °C in the absence (c) or in the presence of MJ0384 (1, 50 nM; 2, 100 nM; 3, 200 nM; 4, 300 nM). (D, E) Effect of the Cas3 helicase protein MJ0383 and ATP on the cleavage of the SA (D) and R-loop-2 (E) substrates by MJ0384. The 5′-[32P]-labelled (*) SA substrate (20 nM) was incubated for 20 min at 42 °C in the presence of 5 mM MgCl2 and 1 mM ATP with MJ0384 alone or MJ0383 alone, or with a mixture of MJ0384 and MJ0383 in the presence of 5 mM MgCl2 without or with the addition of ATP (1 mM). The 5′-[32P]-labelled (*) R-loop-2 substrate (20 nM) was incubated for 35 min at 42 °C in the presence of 5 mM MgCl2 and 1 mM ATP with MJ0383 alone, or with the mixture of MJ0383 (120 nM) and MJ0384 (1) 50 nM; (2) 100 nM; (3) 200 nM; (4) 300 nM.
Thus, in contrast to the Cas3 HD nuclease SSO2001 (Han and Krauss, 2009), both MJ0384 and PF0639 show nuclease activity against ssDNAs and ssRNAs, but not dsDNAs or dsRNAs. The nuclease activity of MJ0384 and PF0639 appears to be similar to that of the Cas3 proteins from S. thermophilus (Sinkunas et al, 2011) and T. thermophilus (Mulepati and Bailey, 2011), although the ability of the latter enzymes to cleave RNA has not yet been demonstrated. In addition, the 3′ → 5′ direction of the MJ0384 exonuclease activity (Figure 2A) correlates with the 3′ → 5′ polarity and 3′-overhang requirement of the S. thermophilus Cas3 helicase activity (Sinkunas et al, 2011). It is expected that the cooperating Cas3 HD nuclease and helicase domains or proteins are likely to exhibit the same polarity in their activities.
MJ0384 cleaves R-loops
Recent work on the E. coli Cascade–crRNA complex has demonstrated that it specifically recognizes complementary sequences in target dsDNAs and displaces the noncomplementary DNA strand, producing an R-loop that cannot be cleaved by the Cascade complex and has been proposed to be targeted by Cas3 proteins (Jore et al, 2011). R-loops are bubble-like structures that form when one DNA strand of a double helix is displaced by the annealing of a complementary RNA strand (Thomas et al, 1976). R-loop structures are associated with various biological processes including replication of plasmid, viral, and bacterial genomic DNA, transcription, and recombination (Kogoma, 1997; Gnatt et al, 2001). R-loops can be resolved by the activity of several enzymes including: the RecG helicase, DNA polymerase I, topoisomerase I, or cleaved by topoisomerase III (Fukuoh et al, 1997; Wilson-Sali and Hsieh, 2002; Li and Manley, 2005). Recently, the helicase domain of the E. coli Cas3 has been reported to catalyse R-loop formation in the absence of ATP and R-loop unwinding in the presence of ATP (Howard et al, 2011).
To determine if MJ0384 can cleave R-loops, we prepared two R-loop-like substrates by hybridization of an unlabelled 92 nt ssDNA with an unlabelled short ssRNA (39 nt) following by hybridization with a long 32P-labelled DNA (Figure 2C). In the R-loop-1 substrate, the second (labelled) DNA strand is completely complementary to the first DNA strand, whereas in the R-loop-2 substrate, the loop area ssDNA is not complementary to the first DNA strand and it has a 10 nt insert in the loop area (Figure 2C). This is supposed to increase the R-loop ssDNA area as might be expected in the Cascade–crRNA/target DNA complex. Purified MJ0384 exhibited significant nuclease activity against both R-loop substrates, with higher activity towards the R-loop-2 (Figure 2C). With both substrates, the major cleavage product is located in the centre of the gel and corresponds to the product of endonucleolytic cleavage by MJ0384 in the ssDNA region of the R-loop (Figure 2C). This cleavage is expected to produce a 3′-flap-like structure, which is also a substrate for MJ0384 and can be cleaved exonucleolytically (3′–5′) (Figure 2A). This activity is represented by multiple minor bands located below the major cleavage product of R-loop-2 (Figure 2C). The low molecular weight bands at the bottom of the gel appear to be products of the endonucleolytic cleavage close to the labelled 5′-ends, which likely become single stranded for a short time during the reaction. Thus, the Cas3 HD nuclease MJ0384 can cleave R-loops in vitro, suggesting that it can potentially contribute to the cleavage of the target nucleic acids bound to the Cascade–crRNA complex.
Cas3 helicase MJ0383 stimulates the cleavage of SA and R-loops by MJ0384
In most CRISPR-containing genomes, the genes encoding the Cas3 HD domain and helicase proteins are co-localized, and in many cases even fused, suggesting a functional interaction between these proteins in vivo. Likewise, the MJ0384 nuclease gene is located next to the MJ0383 helicase gene in the M. jannaschii genome (Figure 1A). We hypothesize that the helicase activity of MJ0383 can unwind the double-stranded substrates producing single-stranded sequences, which can be degraded by MJ0384. Therefore, we determined the effect of purified Cas3 helicase protein MJ0383 on the cleavage of the SA and R-loop substrates by MJ0384. As shown in Figure 2D, the 5′-[32P]-labelled DNA strand of the SA substrate was not cleaved by MJ0384 or MJ0383 alone and showed low cleavage when these two proteins were added without ATP. However, SA substrate cleavage by the mixture of MJ0384 and MJ0383 was greatly increased in the presence of ATP (Figure 2D), implying that the ATP-dependent helicase activity of MJ0383 unwinds the SA substrate, making the 3′-end of the labelled DNA strand available for cleavage. In the same way, the addition of the MJ0383 helicase and ATP significantly increased the cleavage of the R-loop-2 substrate by MJ0384 (Figure 2E). The cleavage pattern of this substrate in the presence of MJ0383 and ATP indicates that the entire sequence of the labelled DNA strand becomes accessible for cleavage by MJ0384. Thus, the helicase activity of MJ0383 expands the range of substrates cleavable by the Cas3 HD nuclease MJ0384 allowing it to degrade double-stranded sequences as well.
Crystal structure of MJ0384 and active site
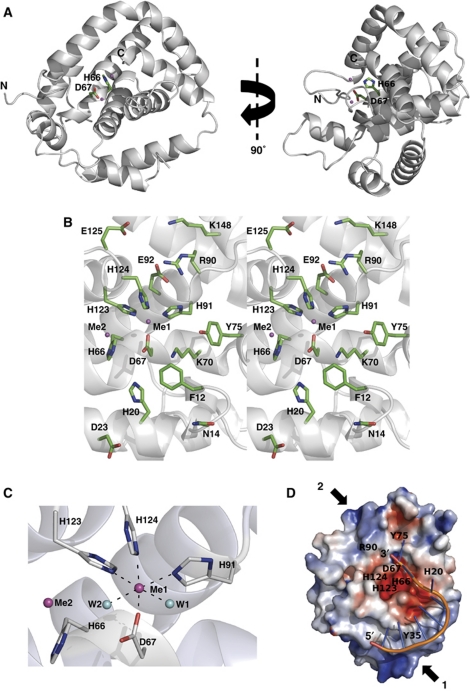
MJ0384 was crystallized and its crystal structure was solved at 2.3 Å resolution (PDB code 3S4L; Supplementary Table SI). The structure revealed a globular monomeric protein composed of eight α-helices and two short β-strands connected by extended loops (Figure 3A). Most available structures of the HD domains show all-α proteins with the exception of the Agrobacterium tumefaciens Atu1052 (2GZ4), which like MJ0384 contains two short β-strands. Gel-filtration experiments using a Superdex S200 column demonstrated that both MJ0384 and PF0639 (predicted monomer molecular mass 28.4 and 27.1 kDa, respectively) exist mainly as monomers in solution (∼80%; 25.5–27.1 kDa) with a small amount of the dimeric form (10–20%; 42–57.2 kDa) (data not shown). The monomeric state of MJ0384 and PF0639 is consistent with their nuclease activity against ssDNA and ssRNA.
Figure 3.
Crystal structure of MJ0384. (A) Overall structure of the MJ0384 monomer: two views related by a 90° rotation around y axis. The position of the potential active site is indicated by the side chains of the HD motif residues His66 and Asp67 (shown as sticks) and bound metal ions (shown as magenta-coloured balls). The C-terminal tail (residues 217–244) is absent due to a limited proteolysis step performed prior to MJ0384 crystallization. (B) Close-up stereo view of the MJ0384 active site. The protein side chains are shown as green sticks along a MJ0384 ribbon (grey). Two bound metal ions are shown as the magenta-coloured balls. (C) Close-up view of the MJ0384 active site showing the position of the two-metal cations (Me1 and Me2, magenta-coloured balls) and the coordinating residues (shown as sticks). Two water molecules (W1 and W2) are shown as blue balls. (D) Superposition of the ssDNA fragment (8 nt) from the E. coli topoisomerase III DNA-binding site (1I7D) onto the potential DNA-binding site of MJ0384 (basic patch-1). The surface charge distribution near the MJ0384 active site is shown with the basic patches coloured in blue and the negatively charged areas coloured in red. The positions of several active site residues are indicated with the labels, the potential DNA-binding sites are indicated with the arrows (basic patch-1 and basic patch-2), and DNA is shown as sticks. The DNA docking is performed using the HADDOCK server (de Vries et al, 2010). This model represents the potential binding of ssDNA for exonucleolytic cleavage by MJ0384.
A Dali search for structurally similar proteins using the MJ0384 coordinates identified the HD domain of the T. thermophilus Cas3 protein TTHB187 (17% sequence identity to MJ0384) at the top match (PDB codes 3SK9 and 3SKD, Z-score 9.1 and 9.3, r.m.s.d. 3.2 and 3.3). Compared with MJ0384, the TTHB187 HD domain structure contains one additional α-helix and two β-strands at the C-termini, but it is unclear if they belong to the HD domain as the SMART database predicts that the TTHB187 HD domain comprises the residues 17–219. Overall, both MJ0384 and TTHB187 show no high structural similarity to other HD domain proteins, suggesting that the Cas3 HD nucleases represent a distinct group of the HD domain proteins. The Dali search also identified the E. coli exopolyphosphatase PPX domain III (PDB codes 2FLO and 1U6Z) and the human phosphodiesterase PDE2A catalytic domain (3IBJ) as structurally similar to MJ0384 (Z-score 6.4 and 5.4, r.m.s.d. 3.8 and 3.1 Å, respectively). The all-α domain III of the E. coli PPX exhibits low structural similarity to MJ0384 (no sequence homology), but it has no signature HD motif and is proposed to contribute to polyphosphate binding (Alvarado et al, 2006; Rangarajan et al, 2006). The catalytic domain of the human PDE2A contains the conserved HD motif, which is located in the small active site and is involved in the coordination of an Mg2+ ion (Pandit et al, 2009).
In the MJ0384 structure, the side chains of the signature HD motif residues His66 and Asp67 (3.7 Å apart) are located on the long α2 helix, which forms the bottom of the large, open active site (Figure 3B). This is similar to the TTHB0187 HD domain (3SKD), but differs from the location of the HD motifs in the structures of other HD domain proteins, which are usually positioned on the loops connecting two α-helices (3IBJ, 2PAQ, 2PQ7). The side chain of another conserved His (His20) of the MJ0384 HD motif is positioned near the His66–Asp67 couple (4.4 Å from Asp67) on the long α1 helix, which creates the wall of the active site. The fourth HD motif residue of MJ0384 (Asp219) is located on the C-terminal tail, which was cleaved during the pre-crystallization treatment with thermolysin (aa 217–244). It can be expected that in MJ0384, the flexible C-terminal strand functions as a clamp closing the bound substrate in the active site and positioning the Asp219 side chain close to other three HD motif residues. In the TTHB187 HD domain structure (3SKD), the homologous Asp205 is located at the end of the last α-helix, but this protein shares low sequence similarity to MJ0384 (17% identity). In addition, the catalytic cavity of MJ0384 accommodates several conserved (Tyr35, Lys70, Tyr75) and semi-conserved (Arg90, His91, Glu92, His123, His124, Lys148) charged and polar residues, which can potentially contribute to substrate binding and/or hydrolysis (Figure 3B).
The structure of MJ0384 revealed the presence of two-metal ions (Me1 and Me2, 5.7 Å apart), which were interpreted as Ca2+ because 5 mM CaCl2 was used in the pre-crystallization treatment with thermolysin. The Me1 ion is close to the HD motif Asp67 (2.26 Å) and likely corresponds to a catalytic metal ion, whereas the Me2 cation is loosely associated with the protein (Figure 3C; Supplementary Figure S5A). The Me1 ion is hexacoordinated by the side chains of the conserved Asp67 (part of the HD motif; 2.26 Å), three semi-conserved His91 (2.31 Å), His123 (2.37 Å), and His124 (2.32 Å), and two water molecules (2.32 and 2.82 Å) (Figure 3C). The structure of the TTHB187 HD domain (3SKD) revealed the presence of one-metal ion (Ni2+) tetracoordinated by the four residues of the HD motif (site A) (Mulepati and Bailey, 2011). The MJ0384 Me1 site is similar to site B in the unpublished structure of the unknown HD domain protein Mes0020 (2PQ7) (Mulepati and Bailey, 2011), whereas the MJ0384 Me2 site can be potentially transformed to the TTHB187 site A in the presence of the Asp219 (which is missing in the MJ0384 structure). In the active site of the human PDE2A, the HD motif residues (His660, His696, Asp697, and Asp808) coordinate two different metal cations (Zn2+ and Mg2+) (Iffland et al, 2005), whereas one-metal ion (Co2+) was coordinated by the HD motif residues in the E. coli HD domain nucleotidase YfbR (Zimmerman et al, 2008). When compared with the available structures of HD domain proteins, the structures of MJ0384 and the TTHB187 HD domain reveal a distinct mode of the metal ion coordination and location of the HD motif residues. In addition, the active sites of the HD nucleases seem to be larger and more open compared with the active sites of nucleotide hydrolyzing HD domain enzymes.
Site-directed mutagenesis of MJ0384 and potential catalytic mechanism
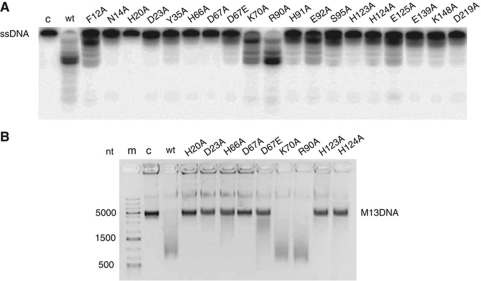
To identify the MJ0384 residues important for nuclease activity, we performed site-directed mutagenesis of the 18 conserved and semi-conserved residues located in the active site. As expected, the alanine replacement of the four conserved residues of the HD domain motif (H20A, H66A, D67A, and D219A) produced proteins essentially inactive in the exonuclease cleavage of linear ssDNA (40 nt), whereas the conserved replacement of Asp67 to Glu resulted in a protein with low residual activity (Figure 4A). Similarly, alanine replacement of the HD motif residues of the Cas3 proteins from S. thermophilus and T. thermophilus also eliminated their nuclease activities (Mulepati and Bailey, 2011; Sinkunas et al, 2011). In addition, the alanine replacement of the metal ion coordinating His residues (H91A, H123A, and H124A) also had a strong negative effect on the cleavage of linear ssDNA by MJ0384, suggesting that Me1 plays an important role in the MJ0384 catalytic mechanism (Figure 4A). As well, low nuclease activity against ssDNA was observed in the N14A, D23A, Y35A, E139A, and K148A proteins, whereas F12A, K70A, R90A, E92A, S95A, and E125A showed reduced or wild-type activity (Figure 4A). Similar results were obtained for the MJ0384 mutant proteins in the endonuclease assay with the circular M13mp18 ssDNA (Figure 4B) and in ssRNA (39 nt) cleavage (Supplementary Figure S3H), suggesting that all activities are associated with the same active site.
Figure 4.
Site-directed mutagenesis of MJ0384. (A, B) Cleavage of the linear ssDNA (A; exonuclease activity) and circular M13 ssDNA (B; endonuclease activity) by purified wild-type and mutant MJ0384 proteins. The 5′-[32P]-labelled ssDNA (40 nt) or M13 ssDNA were incubated at 45 °C without enzyme (c) or with purified proteins for 10 or 30 min, respectively.
Surface charge distribution analysis of the MJ0384 structure using the adaptive Poisson–Boltzmann solver (Dolinsky et al, 2007) revealed the presence of two prominent patches of positively charged residues located near the MJ0384 active site and representing the potential binding sites for nucleic acid substrates (Figure 3D; Supplementary Figure S5B). The basic patch-1 is located at the bottom of the MJ0384 molecule and is mainly associated with the long α1 helix (Lys26, Arg30, Arg34, Lys37, Arg41), whereas the patch-2 is positioned to the top of the molecule and contains the basic residues from the α4 (Arg90) and α6 (Lys148, Lys150) helices (Figure 3D). The two basic patches are separated by the negatively charged zone of the MJ0384 active site, so the binding of ssDNA or ssRNA to the basic patches would place the cleavable phosphodiester bond adjacent to the catalytic residues and bound metal cation. Docking experiments using the HADDOCK web server (de Vries et al, 2010) revealed that the short ssDNA fragment (8 nt long) from the E. coli topoisomerase III structure (PDB 1I7D) fits well into the MJ0384 basic patch-1 with most of the substrate bound to the protein through the phosphodiester backbone (Figure 3D). In this model, the substrate 3′-terminal nucleotide is positioned close to the catalytic Asp67 and might represent the DNA binding for the exonucleolytic cleavage (Figure 3D). In an endonuclease reaction, the substrates (ssDNAs or ssRNAs) can be expected to bind to both basic patches 1 and 2 with the phosphodiester backbone extended over the MJ0384 active site.
The general catalytic mechanism of the phosphodiester bond cleavage includes the activation of the catalytic water molecule by a general base in the nuclease active centre, which usually contains one, two, or three metal cofactors (Nishino and Morikawa, 2002; Dupureur, 2008). Although di-metal centres are well characterized in nucleases and proposed to function in the TTHB187 HD domain (Mulepati and Bailey, 2011), there is also significant experimental evidence for a one-metal model as a general mechanism of the metal-dependent hydrolysis of a phosphodiester bond (Dupureur, 2008). Our results support a one-metal-based mechanism for the nucleic acid cleavage by MJ0384 with one divalent metal cation (Mg2+ or Mn2+) coordinated by two residues of the HD motif (His66 and Asp67), three semi-conserved His side chains (His91, His123, and His124), and one water molecule. Although the MJ0384 structure shows that the His66 side chain is not involved directly in the Me1 coordination, the H66A mutant protein exhibited a negligible nuclease activity, suggesting a catalytic role for this residue (Figures 3B and C and 4). In the inactive conformation of the HD domain of the Streptococcus disgalactiae RelA, the HD motif Asp78 is also not involved in the metal coordination, but it moves close to the metal ion upon substrate binding (Hogg et al, 2004). We propose that in the MJ0384–substrate complex, the His66 side chain joins the coordination sphere of the Me1 ion (probably displacing the water-2 molecule). The binding of the nucleic acid substrate to MJ0384 might be facilitated by its C-terminal tail (Met217–Ile244, not present in the MJ0384 apo-structure) containing the HD motif residue Asp219, which is important for the nuclease activity of MJ0384 (Figure 4A). We propose that the C-terminal tail of MJ0384 clamps down the substrate in the active site with the metal ion and catalytic water molecule positioned close to the cleavable phosphodiester bond. As in other nucleases (Nishino and Morikawa, 2002), the divalent metal ion in the MJ0384 active site potentially contributes to the activation of the water nucleophile (by reducing pKa and inducing deprotonation), phosphate coordination, and transition state stabilization. The activated hydroxyl performs a nucleophilic attack on the phosphorus atom of the scissile phosphodiester bond, cleaving this bond with the formation of the 3′-phosphate and 5′-hydroxyl ends.
Discussion
Most of the characterized HD domain proteins exhibit phosphomonoesterase or phosphodiesterase activity against various nucleotide substrates (Yakunin et al, 2004; Blondal et al, 2005; Ryan et al, 2006). Our studies, together with three previous works (Han and Krauss, 2009; Mulepati and Bailey, 2011; Sinkunas et al, 2011) have revealed that the Cas3 HD domain proteins represent a novel subfamily and the first large group of these enzymes with nuclease activity. The structure of MJ0384 shows a distinct mode of catalytic metal coordination and a wide open active site, potentially accessible to large polynucleotide substrates (Figure 3). Accordingly, the five biochemically characterized Cas3 HD domain proteins: SSO2001; S. thermophilus Cas3; TTHB187; MJ0384; and PF0639 show nuclease activity against ssDNAs, ssRNAs, and dsDNAs (Han and Krauss, 2009; Mulepati and Bailey, 2011; Sinkunas et al, 2011) (Figure 1). Because these enzymes appear to cleave substrates nonspecifically in vitro, their in vivo activities would be expected to be tightly regulated (e.g. by other proteins).
Cas3 proteins have been proposed to play a central role in the interference step of the CRISPR mechanism and function as its effector (killer) enzyme through the direct cleavage of invasive nucleic acids (Jore et al, 2011; Sinkunas et al, 2011). Our results show that the Cas3 HD nucleases MJ0384 and PF0639 can cleave both ssDNAs and ssRNAs in vitro, and therefore can potentially target both invader DNA and mRNA. However, these proteins exhibit no apparent sequence selectivity in nucleic acid cleavage, suggesting that the interaction with other Cas proteins is required for specific target recognition and degradation. The E. coli and S. solfataricus Cascade–crRNA complexes can specifically bind to target DNA and have been proposed to recruit a Cas3 protein for target DNA degradation (Brouns et al, 2008; Jore et al, 2011; Lintner et al, 2011). The S. solfataricus Cascade complex is composed of four Cas proteins encoded by the CRISPR cluster 4 operon: SSO1443 (Csa5); SSO1442 (Csa2); SSO1441 (Cas5a); and SSO1437 (Cas6) (Lintner et al, 2011). The S. solfataricus CRISPR cluster 4 is similar to the CRISPR cluster 1 of M. jannaschii (and P. furiosus), which includes the genes encoding the homologous proteins Csa5 (MJ0380), Csa2 (MJ0381), Cas5a (MJ0382), and Cas6 (MJ0375), as well as the Cas3 proteins MJ0384 (a Cas3″ HD nuclease) and MJ0383 (a Cas3′ helicase) (Figure 1A). We hypothesize that like in S. solfataricus, one of the CRISPR pathways of M. jannaschii (and P. furiosus) comprises a Cascade complex (MJ0380, MJ0381, MJ0382, and MJ0375), which functions together with the Cas3 proteins MJ0384 and MJ0383 directing them to target nucleic acids.
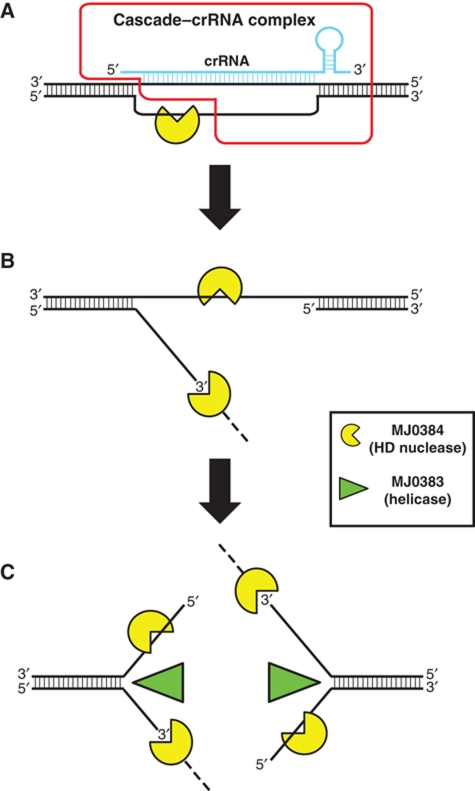
In E. coli, the Cascade–crRNA complex has been shown to bind to target dsDNA and generate an R-loop with significant area of the ssDNA exposed and susceptible to cleavage by the endonuclease P1 (Jore et al, 2011). Interestingly, the E. coli Cas3 protein was found to be able to generate R-loops in vitro in the absence of ATP and unwind them in the presence of ATP (Howard et al, 2011). We suggest that the M. jannaschii Cascade–crRNA complex also forms an R-loop, whose exposed ssDNA can be cleaved by MJ0384, first endonucleolytically producing the 3′-flap structure, which is then cleaved exonucleolytically (in the 3′–5′ direction) (Figure 5A and B). Since the Cas3 helicase MJ0383 promotes the cleavage of double-stranded sequences in the R-loop and SA substrates by MJ0384 in vitro (Figure 2D and E), we propose that target DNA degradation by a Cas3 HD nuclease is not limited by the ssDNA area produced by a Cascade–crRNA complex. After degradation of the ssDNA in the R-loop by MJ0384, the Cascade–crRNA complex is likely to be released from the target DNA, and MJ0384 can cleave endonucleolytically the ssDNA area on the exposed second DNA strand, producing a double-strand break and two SA-like structures (Figure 5C). The Cas3 helicase MJ0383 can unwind target dsDNA fragments producing ssDNAs, which will be cleaved by MJ0384: exonucleolytically (on the 3′-end strand) and endonucleolytically (on the 5′-end strand) (Figure 5C). Potentially, the Cas3 HD nucleases, working together with the Cas3 helicase and other Cas proteins (Cascade), can completely degrade the invader dsDNA. The proposed function of the Cas3 helicase and nuclease activities in the CRISPR mechanism bears a resemblance to the classical RecB helicase-nuclease protein from E. coli, a central enzyme of DNA recombination, which also contributes to the degradation of foreign dsDNAs through the combination of 3′–5′ exonuclease and helicase activities (Dillingham and Kowalczykowski, 2008). In addition, the E. coli Cascade–crRNA complex can bind to the target ssDNA or ssRNA (Jore et al, 2011), which might also recruit a Cas3 nuclease for target degradation. Many CRISPR-containing genomes encode several Cas3 HD domain proteins with low sequence similarity to each other, suggesting that these nucleases might have different substrate specificities or preferences. Future work will reveal the molecular details of substrate cleavage by Cas3 nucleases and their interactions with other Cas proteins in the CRISPR mechanism.
Figure 5.
Proposed role of MJ0384 in the CRISPR interference mechanism. (A) The M. jannaschii Cascade–crRNA complex binds to the invader dsDNA and forms the R-loop with the exposed ssDNA area, which is cleaved by MJ0384, first endonucleolytically and then exonucleolytically (3′–5′). The Cascade–crRNA image (outlined in red) is adapted from Jore et al (2011). (B) After the release of the Cascade–crRNA complex, MJ0384 cleaves endonucleolytically the exposed ssDNA area on the second target DNA strand producing a double-strand break. (C) The Cas3 helicase MJ0383 unwinds the dsDNA fragments, which are continuously degraded by MJ0384. Working together, MJ0384 and MJ0383 completely degrade the invader DNA.
Materials and methods
Protein expression and purification
Gene cloning and protein purification of the 6His-tagged MJ0384, MJ0383, and PF0639 were carried out as described previously (Beloglazova et al, 2008) (Supplementary Figure 3E–G). Site-directed mutagenesis of MJ384 was performed using a protocol based on the QuikChange site-directed mutagenesis kit (Stratagene).
Preparation of DNA and RNA substrates
The ssDNA or RNA oligonucleotide substrates (17–92 nt; Supplementary Table SI) were purchased from IDT (USA). The oligonucleotides were 5′-end labelled using [γ-32P]ATP (6000 Ci/mmol; Perkin-Elmer) and T4 PNK (Fermentas) and purified using denaturing PAGE (15% polyacrylamide/8 M urea). The labelled oligonucleotides were eluted from the gel, precipitated with 2% LiClO4 in acetone, washed with acetone, dried, and dissolved in DEPC-treated Milli-Q water. The synthetic dsDNAs or RNAs (Figure 2A; Supplementary Figure S4) were prepared by annealing the oligonucleotides DNA6+DNA10 or RNA4+RNA10; the 5′-flap (5′F) DNA or RNA substrates with oligos DNA6+DNA7+DNA8 or RNA4+RNA7+RNA8, respectively; the 3′-flaps (3′F) with oligos DNA6+DNA7+DNA8 or RNA4+RNA7+RNA9; the RF-like substrates with oligos DNA6+DNA7+DNA8+DNA9 or RNA4+RNA7+RNA8+RNA9; the SA substrates with oligos DNA6+DNA7 or RNA4+RNA7; the DNA/RNA complexes 1, 2, and 3 with oligos DNA1+RNA3, or DNA11+RNA3, or DNA12+RNA3, respectively; and the R-loop substrates 1 and 2 with oligos DNA1+RNA3+DNA13 or DNA1+RNA3+DNA14 (Supplementary Table SII).
Enzymatic assays
The reaction mixture for ssDNase assays contained (in a final volume 10 μl) 50 mM Tris–HCl (pH 8.5), 20 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol (DTT), 0.1 μM 5′-[32P]-labelled ssDNA, and 200 nM of MJ0384 or PF0639. The solutions were incubated at 45 °C for the indicated time and quenched by the addition of equal volume of the formamide loading buffer (Beloglazova et al, 2008). The reaction products were separated using the electrophoresis in 15% polyacryalamide (PAA)/8 M urea gels and visualized by autoradiography as previously described (Beloglazova et al, 2008). The reaction mixture for ssRNase assays contained (10 μl) 50 mM Tris–HCl (pH 7.5), 50 mM KCl, 5 mM MnCl2, 1 mM DTT, 0.1 μM 5′-[32P]-labelled ssRNA, and MJ0384 or PF0639 (200 nM). The solutions were incubated at 45 °C for 20 min and quenched by the addition of equal volume of the formamide loading buffer. The reaction products were resolved in 15% PAA/8 M urea gels using the TBE running buffer and visualized by autoradiography (Beloglazova et al, 2008). Endonuclease assays were performed at 42 °C for 30 min using the circular ssDNA of the M13mp18 or φX174 phages (New England BioLabs) as substrates and the reaction mixtures containing 50 mM Tris–HCl (pH 8.5), 20 mM KCl, 10 mM MgCl2, 1 mM DTT, 5 nM DNA (M13mp18 or φX174), and 50–500 nM MJ0384. The reactions were incubated with Proteinase K (Fermentas; 37 °C, 10 min) and stopped by the addition of the agarose gel loading buffer (10% glycerol, 0.025% bromophenol blue, 10 mM EDTA pH 8.0, 0.5% SDS). The reaction products were separated by electrophoresis in 0.9% (w/v) agarose gels and visualized by SYBR Green staining.
For the analysis of the ssDNA product ends, the unlabelled DNA products were precipitated with 2% LiClO4, washed by acetone, dried, dissolved in Milli-Q water, and phosphorylated using [γ-32P]ATP, T4 PNK, and reaction conditions for the forward (labelling) or phosphate exchange reactions according to the manufacturer's protocol (Fermentas). After the incubation with PNK, the phosphorylated products were analysed using 15% PAA/8 M urea gels and radiography. Cleavage of the branched DNA substrates was analysed in a reaction mixture containing 50 mM Tris–HCl (pH 8.5), 20 mM KCl, 10 mM MgCl2, 1 mM DTT, 20 nM 5′-[32P]-labelled substrate, and MJ0384 (100 or 200 nM). Reaction mixtures for branched RNA cleavage contained 50 mM Tris–HCl (pH 7.5), 50 mM KCl, 5 mM MnCl2, 1 mM DTT, 20 nM 5′-[32P]-labelled substrate, and MJ0384 (100 or 200 nM). The reaction products were resolved using denaturing 15% PAA/8 M urea gels and the TBE running buffer and visualized by autoradiography (Beloglazova et al, 2008).
The effect of the MJ0383 helicase protein on the nuclease activity of MJ0383 was analysed using the SA and R-loop-2 DNA substrates prepared by the annealing of the unlabelled oligo DNA6 and the 5′-[32P]-labelled oligo DNA10 or DNA1 and RNA3 and 5′-[32P]-labelled DNA14 for R-loop-2 (Supplementary Table SII). The SA substrate contains the unlabelled 3′-arm and [32P]-labelled 5′-arm (Figure 2E). The cleavage reactions were performed at 42 °C for 15 min in a reaction mixture containing 10 mM Tris–HCl (pH 8.5), 20 mM KCl, 5 mM MgCl2, 1 mM ATP, 20 nM SA substrate, and indicated amounts of MJ0383 and MJ0384. The reactions were terminated by the addition of equal volume of the formamide loading buffer, the products were resolved in 15% PAA/8 M urea gels and visualized by autoradiography. Phosphodiesterase activity against 2′,3′-cyclic nucleoside monophosphates and the reaction products of the phosphodiesterase reaction were analysed as previously described (Yakunin et al, 2004).
Protein crystallization and data collection
Prior to crystallization, the purified MJ0384 (2 mg/ml) was incubated overnight with thermolysin (1 mg/ml, 1/100 v/v, 5 mM CaCl2, 4 °C) and separated from the protease (and untreated MJ0384) using gel-filtration (Superdex 200 16/60; 20 mM HEPES-K, pH 8.0, 200 mM NaCl, 10% glycerol, 1 mM TCEP, 0.1 mM PMSF). It appears that thermolysin treatment of MJ0384 (28.4 kDa) removed its flexible C-terminal tail (Met217–Ile244, 28 aa) producing a truncated variant with an electrophoretic mobility on SDS-gels corresponding to an M.w. of ∼24 kDa and with an intact His-tagged N-termini (based on its ability to bind to an Ni2+-NTA affinity column). MJ0384 was crystallized at room temperature using the sitting-drop vapour diffusion method by mixing 1 μl of the protein solution (15 mg/ml) with 1 μl of the crystallization solution containing 25% PEG 5000 MME, 0.2 M (NH4)2SO4, and 0.1 M succinate–phosphate–glycine buffer (pH 6.0). The crystals were cryoprotected in Paratone-N prior to flash-freezing in liquid nitrogen for data collection at the beamline ID19 (Structural Biology Center, Advanced Photon Source, Argonne National Laboratory) using an ADSC Quantum 315 detector. Data set was collected at the peak wavelength of the selenium absorption edge (λ=0.979 Å) and reduced with XDS.
Structure determination
After data reduction, six heavy atoms were identified based on the anomalous differences using SHELXD (Sheldrick, 2008). Initial phases from SHELXE were optimized by histogram matching and solvent flattening with RESOLVE (Adams et al, 2010) and yielded a high-quality electron density map. The partial model built using Autobuild (Adams et al, 2010) was manually completed using COOT. The last refinement using TLS (Winn et al, 2001, 2003) was also performed with Phenix. The two remaining ‘strong’ unmodelled blobs highlighted by simulated annealing omit maps at 6.0 sigma were subsequently filled in with Ca2+ ions, which are presumed to have come from the pre-crystallization treatment with thermolysin. In all, 97.9% of residues of the final model are in the most-favoured regions and 1.1% are in allowed regions of the Ramachandran plot (Chen et al, 2010). The final model yielded an Rwork and Rfree of 23 and 27%, respectively. Data collection and refinement statistics are summarized in Supplementary Table SI. The atomic coordinates have been deposited at the Protein Data Bank, with accession code 3S4L.
Supplementary Material
Acknowledgments
We thank all members of the Structural Proteomics in Toronto (SPiT) Centre and the personnel of the SBC-CAT beamline at Argonne National Laboratory for help in conducting the experiments. We are grateful to Xiaohui Xu for help in protein crystallization, and Jerzy Osipiuk is thanked for help in diffraction data collection. This work was supported by the Government of Canada through Genome Canada and the Ontario Genomics Institute (2009-OGI-ABC-1405; AY and AS), NSERC Discovery team grant (AY and AS), and by the National Institutes of Health Grant GM074942 (AS).
Author contributions: NB designed the experiments, purified proteins, performed biochemical characterization, analysed the data, and contributed to manuscript preparation. PP purified and crystallized MJ0384, solved the crystal structure, and contributed to manuscript preparation. RF purified proteins and contributed to biochemical characterization and manuscript preparation. GB performed gene cloning and mutagenesis. AS supervised the crystallographic process. AFY coordinated the project, designed the experiments, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66 (Pt 2): 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado J, Ghosh A, Janovitz T, Jauregui A, Hasson MS, Sanders DA (2006) Origin of exopolyphosphatase processivity: fusion of an ASKHA phosphotransferase and a cyclic nucleotide phosphodiesterase homolog. Structure 14: 1263–1272 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (1998) The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci 23: 469–472 [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, Koonin EV, Edwards AM, Savchenko A, Yakunin AF (2008) A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem 283: 20361–20371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T, Hjorleifsdottir S, Aevarsson A, Fridjonsson OH, Skirnisdottir S, Wheat JO, Hermannsdottir AG, Hreggvidsson GO, Smith AV, Kristjansson JK (2005) Characterization of a 5′-polynucleotide kinase/3′-phosphatase from bacteriophage RM378. J Biol Chem 280: 5188–5194 [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, Kerlavage AR, Dougherty BA, Tomb JF, Adams MD, Reich CI, Overbeek R, Kirkness EF, Weinstock KG, Merrick JM, Glodek A et al. (1996) Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273: 1058–1073 [DOI] [PubMed] [Google Scholar]
- Cady KC, O’Toole GA (2011) Non-identity targeting of Yersinia-subtype CRISPR-prophage interaction requires the Csy and Cas3 proteins. J Bacteriol 193: 3433–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66 (Pt 1): 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AM (2010) The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 5: 883–897 [DOI] [PubMed] [Google Scholar]
- Deveau H, Garneau JE, Moineau S (2010) CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol 64: 475–493 [DOI] [PubMed] [Google Scholar]
- Dillingham MS, Kowalczykowski SC (2008) RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev 72: 642–671, Table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA (2007) PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res 35 (Web Server issue): W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupureur CM (2008) Roles of metal ions in nucleases. Curr Opin Chem Biol 12: 250–255 [DOI] [PubMed] [Google Scholar]
- Fukuoh A, Iwasaki H, Ishioka K, Shinagawa H (1997) ATP-dependent resolution of R-loops at the ColE1 replication origin by Escherichia coli RecG protein, a Holliday junction-specific helicase. EMBO J 16: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD (2001) Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science 292: 1876–1882 [DOI] [PubMed] [Google Scholar]
- Gonzalez TN, Sidrauski C, Dorfler S, Walter P (1999) Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J 18: 3119–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV (1993) Helicases: amino acid sequence comparisons and structure-function relationships. Curr Opin Struct Biol 3: 419–429 [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE (2005) A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D, Krauss G (2009) Characterization of the endonuclease SSO2001 from Sulfolobus solfataricus P2. FEBS Lett 583: 771–776 [DOI] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA (2010) Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329: 1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R (2004) Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117: 57–68 [DOI] [PubMed] [Google Scholar]
- Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327: 167–170 [DOI] [PubMed] [Google Scholar]
- Howard JL, Delmas S, Ivancic-Bace I, Bolt EL (2011) Helicase dissociation and annealing of RNA-DNA hybrids by Escherichia coli Cas3 protein. Biochem J 439: 85–95 [DOI] [PubMed] [Google Scholar]
- Iffland A, Kohls D, Low S, Luan J, Zhang Y, Kothe M, Cao Q, Kamath AV, Ding YH, Ellenberger T (2005) Structural determinants for inhibitor specificity and selectivity in PDE2A using the wheat germ in vitro translation system. Biochemistry 44: 8312–8325 [DOI] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders AP, Dickman MJ, Doudna JA, Boekema EJ, Heck AJ, van der Oost J, Brouns SJ (2011) Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18: 529–536 [DOI] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ (2010) The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 37: 7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogoma T (1997) Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61: 212–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Nakagawa N, Ebihara A, Chen L, Liu ZJ, Wang BC, Yokoyama S, Kuramitsu S, Masui R (2007) Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis. Acta Crystallogr D Biol Crystallogr 63 (Pt 2): 230–239 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL (2005) Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 122: 365–378 [DOI] [PubMed] [Google Scholar]
- Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copie V, Young MJ, White MF, Lawrence CM (2011) Structural and functional characterization of an archaeal CASCADE complex for CRISPR-mediated viral defense. J Biol Chem 286: 21643–21656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006) A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manica A, Zebec Z, Teichmann D, Schleper C (2011) In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 80: 481–491 [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322: 1843–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ (2010) CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 11: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulepati S, Bailey S (2011) Structural and biochemical analysis of the nuclease domain of the clustered regularly interspaced short palindromic repeat (CRISPR) associated protein 3(CAS3). J Biol Chem 286: 31896–31903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Morikawa K (2002) Structure and function of nucleases in DNA repair: shape, grip and blade of the DNA scissors. Oncogene 21: 9022–9032 [DOI] [PubMed] [Google Scholar]
- Okorokov AL, Panov KI, Offen WA, Mukhortov VG, Antson AA, Karpeisky M, Wilkinson AJ, Dodson GG (1997) RNA cleavage without hydrolysis. Splitting the catalytic activities of binase with Asn101 and Thr101 mutations. Protein Eng 10: 273–278 [DOI] [PubMed] [Google Scholar]
- Pandit J, Forman MD, Fennell KF, Dillman KS, Menniti FS (2009) Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc Natl Acad Sci USA 106: 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan ES, Nadeau G, Li Y, Wagner J, Hung MN, Schrag JD, Cygler M, Matte A (2006) The structure of the exopolyphosphatase (PPX) from Escherichia coli O157:H7 suggests a binding mode for long polyphosphate chains. J Mol Biol 359: 1249–1260 [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM (2006) Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc Natl Acad Sci USA 103: 6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A 64 (Pt 1): 112–122 [DOI] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V (2011) Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 30: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, White RL, Davis RW (1976) Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci USA 73: 2294–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JE, Venegas FD, Raines RT (1994) Energetics of catalysis by ribonucleases: fate of the 2′,3′-cyclic phosphodiester intermediate. Biochemistry 33: 7408–7414 [DOI] [PubMed] [Google Scholar]
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ (2009) CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 34: 401–407 [DOI] [PubMed] [Google Scholar]
- Wilson-Sali T, Hsieh TS (2002) Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc Natl Acad Sci USA 99: 7974–7979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr 57 (Pt 1): 122–133 [DOI] [PubMed] [Google Scholar]
- Winn MD, Murshudov GN, Papiz MZ (2003) Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol 374: 300–321 [DOI] [PubMed] [Google Scholar]
- Yakunin AF, Proudfoot M, Kuznetsova E, Savchenko A, Brown G, Arrowsmith CH, Edwards AM (2004) The HD domain of the Escherichia coli tRNA nucleotidyltransferase has 2′,3′-cyclic phosphodiesterase, 2′-nucleotidase, and phosphatase activities. J Biol Chem 279: 36819–36827 [DOI] [PubMed] [Google Scholar]
- Zimmerman MD, Proudfoot M, Yakunin A, Minor W (2008) Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J Mol Biol 378: 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.