Abstract
H3K4 methylation is associated with active transcription and in combination with H3K27me3 thought to keep genes regulating development in a poised state. The contribution of enzymes regulating trimethylation of lysine 4 at histone 3 (H3K4me3) levels to embryonic stem cell (ESC) self-renewal and differentiation is just starting to emerge. Here, we show that the H3K4me2/3 histone demethylase Jarid1b (Kdm5b/Plu1) is dispensable for ESC self-renewal, but essential for ESC differentiation along the neural lineage. By genome-wide location analysis, we demonstrate that Jarid1b localizes predominantly to transcription start sites of genes encoding developmental regulators, of which more than half are also bound by Polycomb group proteins. Virtually all Jarid1b target genes are associated with H3K4me3 and depletion of Jarid1b in ESCs leads to a global increase of H3K4me3 levels. During neural differentiation, Jarid1b-depleted ESCs fail to efficiently silence lineage-inappropriate genes, specifically stem and germ cell genes. Our results delineate an essential role for Jarid1b-mediated transcriptional control during ESC differentiation.
Keywords: ES cell, histone demethylase, Jarid1b, knockout mouse, neural differentiation
Introduction
Embryonic stem cells (ESCs) have to constantly balance their potential to self-renew and differentiate (Niwa, 2007). Self-renewal and differentiation depend on the gene expression pattern in the cells, which is tightly connected with the chromatin state. Chromatin structure is influenced by post-translational histone modifications, of which a wide variety has been associated with either active transcription or gene silencing (Zhou et al, 2011).
Trimethylation of lysine 4 at histone 3 (H3K4me3) occurs at transcription start sites and high levels have been linked to active transcription (Schübeler et al, 2004; Pokholok et al, 2005; Barski et al, 2007). In concordance, a subunit of the general transcription factor TFIID can bind to this mark, which in turn may facilitate formation of the pre-initiation complex (Vermeulen et al, 2007). Additionally, chromatin-remodelling complexes can recognize H3K4me3 and open the chromatin structure to facilitate transcription (Wysocka et al, 2006; Clapier and Cairns, 2009). Yet, the association of H3K4me3 with promoters is not a definite indication for transcriptional activity (Guenther et al, 2007). In ESCs, the majority of promoters are H3K4me3 positive (Bernstein et al, 2006; Mikkelsen et al, 2007; Meissner et al, 2008); and in addition, many are marked by ‘repressive’ H3K27me3, a chromatin state which is referred to as bivalent (Azuara et al, 2006; Bernstein et al, 2006; Pan et al, 2007). In ESCs, many genes encoding developmental regulators are in this category and it has been hypothesized that bivalency is important to keep the option for all lineage choices (Azuara et al, 2006; Mikkelsen et al, 2007; Stock et al, 2007).
The enzyme responsible for H3K27me3 is the histone lysine methyltransferase (KMT) EZH2. EZH2 together with SUZ12 and EED forms the core of the Polycomb repressive complex 2 (PRC2), which is essential for early embryonic development (Faust et al, 1995; O’Carroll et al, 2001; Pasini et al, 2004). PRC2 binds to 2000–5000 genes in ESCs (Boyer et al, 2006; Pasini et al, 2007). Knockout ESCs of the distinct subunits of PRC2 all display defects in differentiation, including the neural lineage (Pasini et al, 2007; Shen et al, 2008). During differentiation the set of Polycomb group (PcG) target genes changes, indicating that PcG proteins have specific roles at given developmental states (Mohn et al, 2008).
A whole family of histone methyltransferases can catalyse H3K4 methylation (Hublitz et al, 2009; Dambacher et al, 2010), and like the PcG genes, several of these are important for early development and homeotic gene regulation (Yu et al, 1995; Yagi et al, 1998; Glaser et al, 2006). In addition, H3K4 histone methyltransferases function in haematopoiesis (Ernst et al, 2004) and neurogenesis (Lim et al, 2009). Thus, a strict regulation of H3K4 and H3K27 methylation appears to be required for normal differentiation.
Demethylation of H3K4me3 is catalysed by the Jarid1/Kdm5 family (Pedersen and Helin, 2010). There are four Jarid1 proteins in mammals, while most other species have only one homologue. The homologues in Drosophila (lid) and C. elegans (rbr-2) are important for normal development (Christensen et al, 2007; Li et al, 2008; Di Stefano et al, 2011). Whereas Jarid1a knockout mice are viable (Klose et al, 2007) displaying rather mild phenotypes in haematopoiesis and behaviour, it was recently reported that disruption of the Jarid1b locus results in early embryonic lethality (Catchpole et al, 2011). Moreover, high ectopic expression of Jarid1b in mouse ESCs impairs general cell specification and interferes with neural differentiation (Dey et al, 2008).
The role of H3K4me3 and bivalency remains important questions in stem cell biology, particularly with respect to the switch from self-renewal to differentiation. Recently, Jiang et al (2011) showed that decreased levels of H3K4me3 are tolerable in ESCs, but interfere with differentiation. Globally, H3K4me3 decreases during differentiation (Ang et al, 2011) along with the resolution of a proportion of bivalent genes to H3K27me3 only and demethylation of a subset of H3K4me3-positive promoters (Bernstein et al, 2006). Here, we address the role of the H3K4me2/3 demethylase Jarid1b in ESC self-renewal and differentiation.
Results
Jarid1b is dispensable for ESC self-renewal, but required for neural differentiation of ESCs
Jarid1b is predominantly expressed during embryonic development, in ESCs, and in adult testis, brain, eye, spleen and thymus (Supplementary Figure S1A and B; Madsen et al, 2002; Frankenberg et al, 2007; Dey et al, 2008). Since ESCs can be maintained in vitro as pluripotent, self-renewing cells or differentiated into many different cell types of all three germ layers (Young, 2011), they provide a good model system to study lineage choices.
To understand the function of Jarid1b for stem cell self-renewal and differentiation, we knocked down Jarid1b expression by shRNA in ESCs. The knockdown did not compromise ESC morphology, expression of the pluripotency markers Oct4 and Nanog or ESC proliferation (Figure 1B; Supplementary Figure S1C–E).
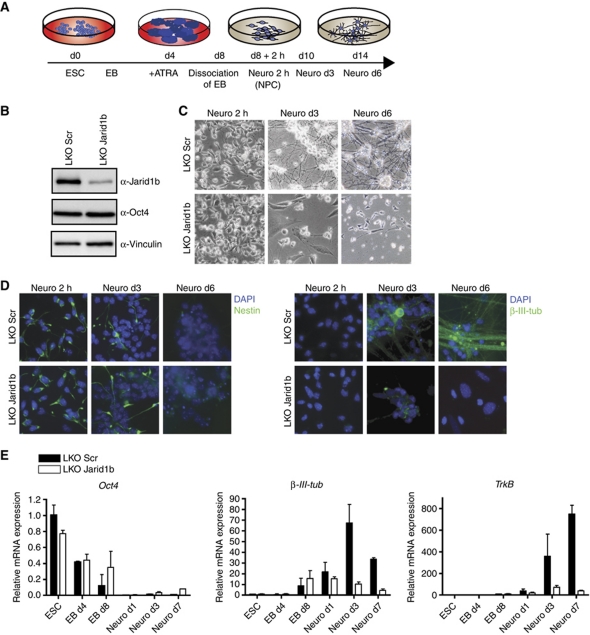
Figure 1.
Neural differentiation of Jarid1b knockdown ESCs is impaired. (A) Schematic representation of experimental outline. ESC, embryonic stem cell; EB, embryoid body; ATRA, all-trans retinoic acid; NPC, neural precursor cell. (B) Immunoblots of stable lentiviral knockdown (LKO) Scramble (Scr) and Jarid1b ESC lines probed for Jarid1b, the stem cell marker Oct4 and Vinculin (loading control). (C) Light microscope images of Scr and Jarid1b knockdown ESCs differentiated into neural progenitor cells (NPCs, Neuro 2 h) and neurons (Neuro d3 and d6). (D) Immunofluorescence staining of differentiated Scr and Jarid1b knockdown ESCs for the neural stem cell marker Nestin and the neuronal marker β-III-Tubulin. DNA is stained with DAPI. (E) Expression of the stem cell marker Oct4 and the neuronal marker genes β-III-Tubulin and TrkB during neural differentiation determined by quantitative RT–PCR analysis (normalized to Rplp0 levels).
While this manuscript was in preparation, another study showed that Jarid1b is required for ESC self-renewal (Xie et al, 2011), and an attempt to establish Jarid1b mutant ESCs from Jarid1b mutant blastocysts was unsuccessful (Catchpole et al, 2011). To unambiguously determine the role of Jarid1b in ESCs, we established conditional targeted Jarid1b ESCs, in which Jarid1b exon 6 is flanked by loxP sites (Supplementary Figure S2). Cre-mediated deletion creates a frameshift and subsequent termination mutation. In conditional Jarid1bF/F; Rosa26::CreERT2 ESC lines Jarid1b could be efficiently deleted by administration of 4-hydroxy-tamoxifen (4OHT; Supplementary Figures S2C and S3C). At global levels, Jarid1b was undetectable 72 h after addition of 4OHT. Expression of other Jarid1-family members remained unchanged after Jarid1b depletion (Supplementary Figure S3B). Importantly, like in Jarid1b knockdown cells, ESC morphology and self-renewal, reflected by Oct4 and Nanog protein expression, and growth rate were unaffected by Jarid1b deletion (Supplementary Figure S3A, C and D). Therefore, we conclude that Jarid1b is not required for ESC self-renewal.
As a general approach to address differentiation potential, we assayed embryoid body (EB) formation in stable Jarid1b knockdown and control cells. Morphologically, there was no difference and both cell lines formed EBs of similar size. However, while control cells expressed genes from all three germ layers after 9 days of differentiation, particularly ectodermal gene expression was not properly induced in Jarid1b knockdown cells (Supplementary Figure S4). In addition, we noticed that expression of the pluripotency markers Oct4 and Nanog was not efficiently silenced in Jarid1b knockdown cells upon differentiation.
To further understand the role of Jarid1b in ectodermal differentiation, we used a protocol which allows the generation of highly homogeneous neural progenitor cells (NPCs) and subsequently terminally differentiated neurons from ESCs (Bibel et al, 2007). Briefly, ESCs are differentiated to EBs, dissociated and plated for neuronal differentiation (schematized in Figure 1A). Jarid1b knockdown EBs had a normal morphology, and NPCs could attach. However, cells with reduced levels of Jarid1b failed to differentiate further towards neurons and did not build a neuronal network. Instead, most Jarid1b knockdown cells detached and died (Figure 1C). Importantly, the same differentiation defect was observed using Jarid1b knockout ESCs (Supplementary Figure S3E). The distinct stages of neural differentiation can be followed by expression of corresponding markers genes: Oct4 (Pou5f1) for ESCs, Nestin for NPCs and β-III-Tubulin and TrkB for neurons. Oct4 was downregulated during differentiation in both Jarid1b knockdown and control cells, even though the downregulation was less efficient in the knockdown. After plating, virtually all cells stained positive for Nestin, identifying them as NPCs (Figure 1D). However, since the plating step selects for neural cells, we analysed cells from dissociated EBs (day 8) before plating for Nestin expression by flow cytometry and found that the number of Nestin-positive cells was slightly but significantly reduced after Jarid1b knockdown (Supplementary Figure S5). During further differentiation, expression of the neuronal markers β-III-Tubulin and TrkB was upregulated in control cells as seen by RT–qPCR and immunofluorescence (Figure 1D and E), while this induction was not observed in the few remaining Jarid1b knockdown cells.
Cell-cycle length and proliferation have been shown to influence cell fate in neural cells (reviewed in Salomoni and Calegari, 2010). Yet, there was no difference in proliferation of Jarid1b knockdown NPCs, as measured by BrdU incorporation (Supplementary Figure S6A and C). Neither did we find a difference in the proportion of cells in different cell-cycle phases (Supplementary Figure S6B and C), pointing to a different mechanism.
Taken together, we conclude that Jarid1b is dispensable for ESC self-renewal, but important for differentiation of ESCs towards the neural lineage. Specifically, Jarid1b seems required for efficient generation of NPCs from ESCs, and the obtained Nestin-expressing cells do not progress further in the absence of Jarid1b.
Differentiation of brain-derived conditional knockout NSCs
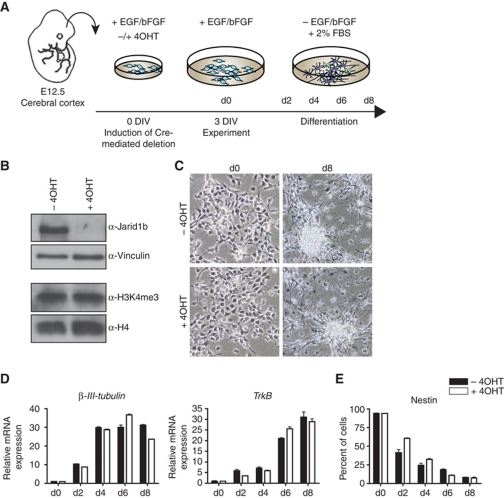
To further dissect at which point during differentiation Jarid1b is required, we derived Jarid1bF/F; Rosa26::CreERT2 neural stem cells (NSCs) from the cerebral cortex of E12.5 mouse embryos. Brain-derived NSCs can be differentiated in vitro into neurons (preferentially during early passages) and astrocytes (later passages) (Hirabayashi and Gotoh, 2010) as illustrated in Figure 2A. In conditional knockout NSCs, Jarid1b was efficiently depleted within 72 h after addition of 4OHT (Figure 2B). Upon withdrawal of the growth factors EGF/bFGF and addition of serum, both control and Jarid1b-deficient early passage NSCs induced expression of the neuronal genes β-III-Tubulin and TrkB and efficiently differentiated into neurons (Figure 2C and D). However, we noticed that downregulation of the NSC marker Nestin was delayed in Jarid1b knockout cells (Figure 2E), which is in analogy to delayed downregulation of stem cell genes during ESC differentiation. This suggests that the crucial effect of Jarid1b is established before the NSC stage and that Jarid1b is dispensable for neuronal differentiation of established NSCs.
Figure 2.
Jarid1b is not required for differentiation of neural stem cells derived from brain. (A) Schematic representation of experimental outline. DIV: days in vitro. (B) Immunoblots of conditional Jarid1b neural stem cells (NSCs) 72 h after induction of Cre-mediated deletion by 4OHT. (C) Phase-contrast images of Jarid1bF/F (−4OHT) and Jarid1b−/− (+4OHT) NSCs at 0 and 8 days of differentiation. (D) Expression of neuronal marker genes during differentiation of Jarid1bF/F (−4OHT) and Jarid1b−/− (+4OHT) NSCs determined by quantitative RT–PCR analysis (normalized to β-actin levels). (E) Immunofluorescence analysis of the neural stem cell marker Nestin performed on an automated screening platform.
Jarid1b occupies developmental regulators in ESCs
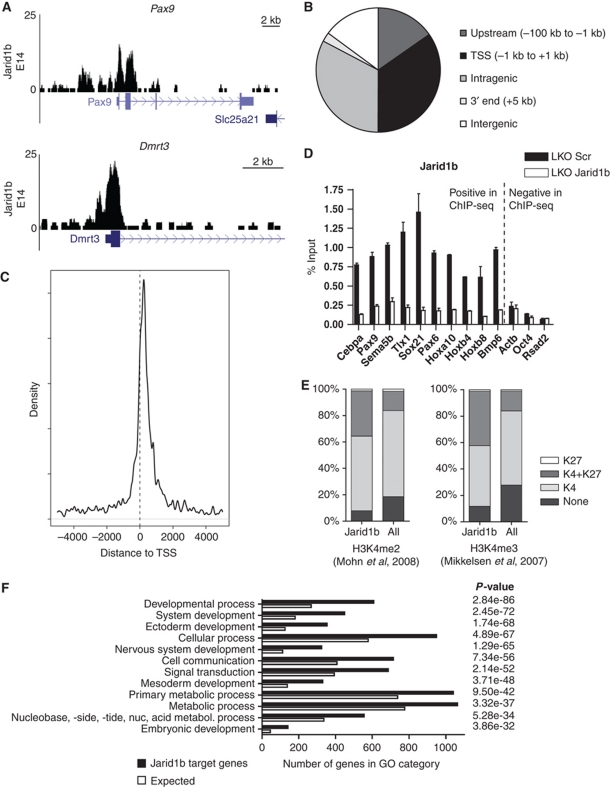
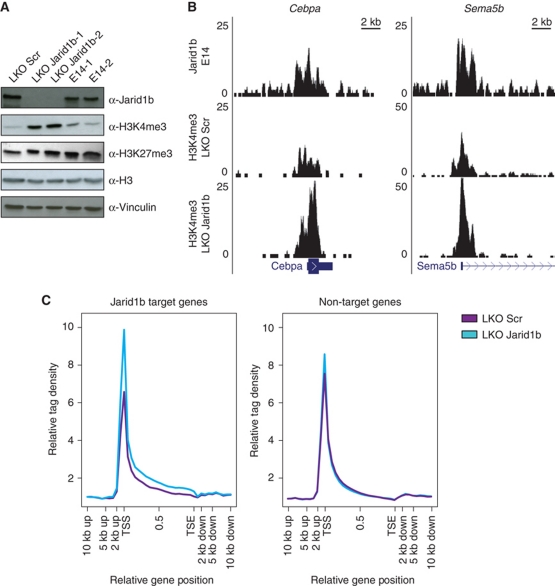
To understand the mechanism by which Jarid1b regulates ESC differentiation, we identified the global binding pattern of Jarid1b by chromatin immunoprecipitation (ChIP) followed by sequencing in ESCs. We generated a rabbit polyclonal antibody against a peptide corresponding to amino acids 1395–1418 of Jarid1b, a region that has very little homology to the other Jarid1-family members. The affinity-purified antibody was specific for Jarid1b as tested in western blot and ChIP in knockdown cells (Figures 1B and 3D). Deep sequencing of DNA from ESCs immunoprecipitated by the Jarid1b antibody resulted in 6.8 million reads. MACS (Zhang et al, 2008) predicted 5232 peaks with a false discovery rate (FDR) below 20% which overlapped 2347 unique genes. Examples of binding profiles are shown in Figure 3A. The majority of peaks were found around transcription start sites (34.8%) and within genes (32.7%) (Figure 3B and C). Few peaks (14.9%) were located in regions that were >100 kb away from any annotated gene. ChIP-qPCR validation in Jarid1b knockdown cells showed that the signal at target genes is strongly reduced (Figure 3D), demonstrating that the enrichment was specific for Jarid1b.
Figure 3.
Jarid1b binds to transcriptional start sites of genes regulating development. (A) Genome-wide binding profiles of Jarid1b in E14 mouse ESCs were determined by ChIP sequencing. Two examples of binding profiles of genes bound by Jarid1b in ESCs are shown. Y-axis denotes number of sequence tag reads. (B) Distribution of Jarid1b peaks upstream of the transcriptional start site (TSS), at the TSS, in intragenic regions, at 3′ ends of genes and in intergenic regions. (C) Mean distribution of tags relative to the TSS (distance in bp). (D) ChIP-qPCR for Jarid1b of genes identified as Jarid1b positive or Jarid1b negative by ChIP-seq in LKO Scr and Jarid1b ESCs. (E) Percentage of H3K27me3, H3K4me2/3 (left/right panel) + H3K27me3, H3K4me2/3 (left/right panel) and unmodified promoters within Jarid1b target genes compared with all genes analysed (Mikkelsen et al, 2007; Mohn et al, 2008). (F) GO term analysis of genes bound by Jarid1b in ESCs. The 12 top scoring categories are presented.
Recently, Jarid1b was reported to bind throughout intragenic regions and suggested to suppress aberrant intragenic transcription (Xie et al, 2011). Although we also detect Jarid1b binding in intragenic regions, this binding reflects individual peaks rather than spreading throughout the gene body as described by Xie et al. Specifically, 22.4% of the intragenic Jarid1b peaks overlap with CpG islands and another 19.4% corresponds to GC-rich regions not annotated as CpG islands (Supplementary Figure S7A and B). This is reminiscent of previous observations for PcG binding sites in ESCs which are highly enriched in CpG islands (Ku et al, 2008; Mohn et al, 2008). Moreover, a strong correlation between PRC2 binding intensity and GC richness has been reported (Mendenhall et al, 2010). Indeed, 94.5% of the intragenic Jarid1b peaks that overlap Suz12 binding sites (Pasini et al, 2010) are GC rich. In addition, a small fraction of intragenic peaks (3.5%) corresponds to alternative transcription start sites, while no specific sequence features were annotated for the remaining peaks (Supplementary Figure S7C and D). To analyse this further, we generated heat maps comparing genes with Jarid1b peaks at the TSS versus in intragenic regions with histone methylation patterns (Supplementary Figure S8). Both groups of Jarid1b peaks overlap with H3K4me3 signals and H3K27me3 marks a large fraction of Jarid1b peaks. In contrast, H3K36me3 is depleted at Jarid1b peaks at the TSS, consistent with previous genome-wide studies, and there is no specific enrichment of H3K36me3 at intragenic Jarid1b peaks.
To address the opposing findings between our study and that published by Xie et al, we tested the specificities of the antibodies used in both studies in Jarid1b knockout ESCs. As shown in Supplementary Figure S9, although all three antibodies confirmed loss of Jarid1b in the knockout ESCs, they showed different levels of specificity. Further, the three antibodies all led to specific enrichment of the regions reported in this manuscript. Importantly, however, only one of the antibodies used by Xie et al (2011) could enrich for the intragenic regions reported in their study, and this enrichment was not reduced in Jarid1b knockout cells (Supplementary Figure S9C). Based on these results, we suggest that the reported association of Jarid1b with intragenic regions (Xie et al, 2011) does not appear to be specific for Jarid1b.
Since H3K4me2/3 is the substrate for Jarid1b, we compared Jarid1b target genes with previously mapped H3K4me2/3-associated genes in ESCs (Mikkelsen et al, 2007; Mohn et al, 2008). To our surprise, only a very small fraction of Jarid1b target genes does not carry H3K4me2/3 (Figure 3E). In ESCs, most promoters are marked by H3K4me2/3 (Mikkelsen et al, 2007; Mohn et al, 2008); however, the proportion of H3K4me2/3-positive transcription start sites is even further enriched within the Jarid1b target gene set compared with all genes, indicating that enzyme and substrate are present at the same promoters. Moreover, the proportion of bivalent genes is notably enriched among Jarid1b target genes. In agreement with this, gene ontology analysis of Jarid1b targets identified developmental regulators including genes involved in neurogenesis and ectoderm development as significantly overrepresented (Figure 3F).
PcG proteins and Jarid1b target a similar set of genes
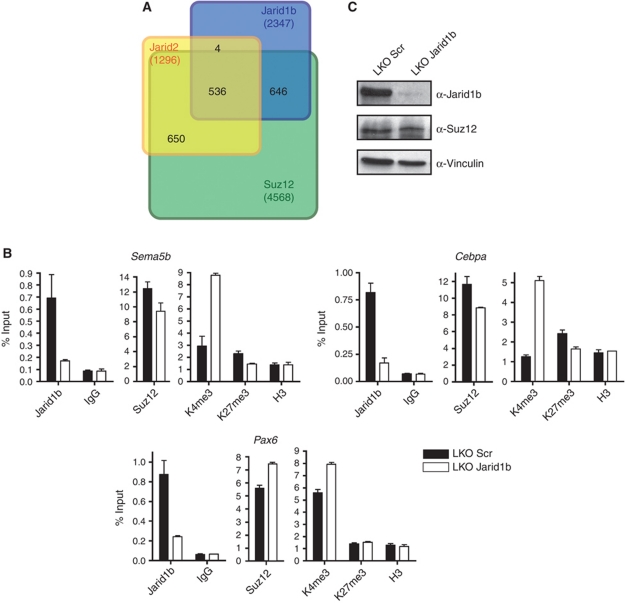
The finding that Jarid1b binds to bivalent regions suggests that there is an overlap between Jarid1b and PcG target genes. Indeed, we found that approximately half of the Jarid1b target genes are bound by Suz12 and in addition many are also bound by Jarid2 (Figure 4A; Pasini et al, 2010). Moreover, comparison with three recently defined ESC regulatory modules (Core, Polycomb and Myc) (Kim et al, 2010) revealed that 13.8% of Jarid1b target genes are bound by five different PcG proteins and are positive for H3K27me3, while there was no significant overlap with the Core (1.5%) or Myc (1.9%) module (Supplementary Figure S10A).
Figure 4.
Polycomb proteins and Jarid1b occupy a similar set of genes in ESCs. (A) Venn diagram of the overlap of Jarid1b, Suz12 and Jarid2 (Pasini et al, 2010) target genes identified by ChIP-seq in ESCs. (B) ChIP-qPCR for Jarid1b, Suz12, H3K4me3, H3K27me3 and H3 in LKO Scr and Jarid1b ESCs. (C) Immunoblots of LKO Scr and Jarid1b ESC lines probed for Jarid1b and the polycomb protein Suz12. Vinculin was used as loading control.
The substantial overlap between target genes raised the question if Jarid1b and PcG proteins mutually influence each other's binding. Knockdown of Jarid1b led to a small but consistent reduction of Suz12 binding at some (e.g., Cebpa, Sema5b) but not all genes (e.g., Pax6) (Figure 4B). Global protein levels of Suz12 remained unchanged (Figure 4C). The reduction of Suz12 also led to a small decrease in H3K27me3 (Figure 4B). Conversely, ChIP for Jarid1b in Eed knockout ESCs showed that enrichment for Jarid1b is lower in these cells than in wild-type ESCs. This effect was even more pronounced in Ring1b knockout ESCs (Supplementary Figure S10B). Moreover, promoter H3K4me3 levels were increased in Ring1b knockout cells and at a subset of tested promoters in Eed knockout cells, probably due to reduced Jarid1b binding. Nevertheless, we could not detect a direct physical interaction between Jarid1b and PRC2 components (Supplementary Figure S10C).
Taken together, we have shown that Jarid1b binds at transcription start sites of developmental regulators, many of which are also bound by PcG proteins. Although Jarid1b and PcG proteins affect the binding of each other to a subset of target genes, neither Jarid1b nor PcG proteins are essential for each other's recruitment.
Jarid1b regulates H3K4me3 levels in ESCs
In agreement with the described catalytic activity for Jarid1b, reduced levels of Jarid1b in ESCs lead to an increase of H3K4me3 at single promoters (Figure 4B) as well as globally (Figure 5A; Supplementary Figure S3C). To extend this observation to the whole genome, we performed ChIP sequencing for H3K4me3 in Jarid1b knockdown and control ESCs. Examples of two Jarid1b target genes are presented in Figure 5B. Jarid1b and H3K4me3 were found to colocalize at the same genomic regions around transcription start sites. The sum of H3K4me3 enrichment over gene bodies shows that there is both an increased signal at the transcription start site and within gene bodies at target genes in Jarid1b knockdown ESCs (Figure 5C), which is in line with Jarid1b peaks at transcription start sites and in intragenic regions. In total, the median number of reads for H3K4me3 increased 2.2-fold around transcription start sites at Jarid1b targets upon knockdown, while H3K4me3 levels at non-target genes were only mildly increased (1.4-fold; Supplementary Figure S11A).
Figure 5.
H3K4me3 levels are increased upon depletion of Jarid1b. (A) Immunoblots of LKO Scr, Jarid1b (J1b-1 and J1b-2) and control ESC lines showing increased global H3K4me3 levels in Jarid1b knockdown ESCs. H3 and Vinculin were used as loading controls. (B) Genome-wide profiles of H3K4me3 were determined by ChIP sequencing in LKO Scr and Jarid1b ESCs and compared with Jarid1b binding profiles. Two examples of Jarid1b target genes are presented. Y-axis denotes number of sequence tag reads. (C) Mean distribution of tags across gene bodies for H3K4me3 ChIP-seq in control and Jarid1b knockdown ESCs at Jarid1b target genes (left panel) and non-target genes (right panel).
Previous studies have shown that high H3K4me3 levels correlate with transcriptional activity (Lorincz and Schübeler, 2007). We wanted to know, how H3K4me3 levels at Jarid1b target promoters compare with genes categorized depending on transcriptional activity (Rahl et al, 2010). On average, H3K4me3 levels at Jarid1b target genes are lower than at highly active genes, and comparable to levels at non-productive promoters (Supplementary Figure S11B). Nonetheless, Jarid1b targets included all three categories (active, non-productive and inactive genes), with a modest enrichment of non-productive genes (Supplementary Figure S11C). The genome-wide analysis supports the idea that Jarid1b keeps H3K4me3 at low or intermediate levels, but does not eliminate the mark in ESCs.
Stem and germ cell-specific genes are incompletely silenced in Jarid1b knockdown NPCs
To understand how Jarid1b contributes to differentiation, we analysed gene expression in ESCs and NPCs with stable Jarid1b knockdown. We did not observe dramatic changes of expression, and in Jarid1b knockdown ESCs only 22 genes were changed >3-fold compared with control ESCs. Confirming our previous results, knockdown of Jarid1b did not affect expression of stem cell genes in ESCs. Despite the substantial global upregulation of H3K4me3 in ESCs, the majority of differentially expressed genes in ESCs were slightly downregulated (Supplementary Figure S12). Furthermore, only 3.4% of genes bound by Jarid1b were differentially expressed, suggesting that Jarid1b does not function as a strong transcriptional regulator in ESCs undergoing self-renewal and that increased levels of H3K4me3 are tolerated in growing ESCs. This is consistent with the finding that Jarid1b is not required for ESC maintenance.
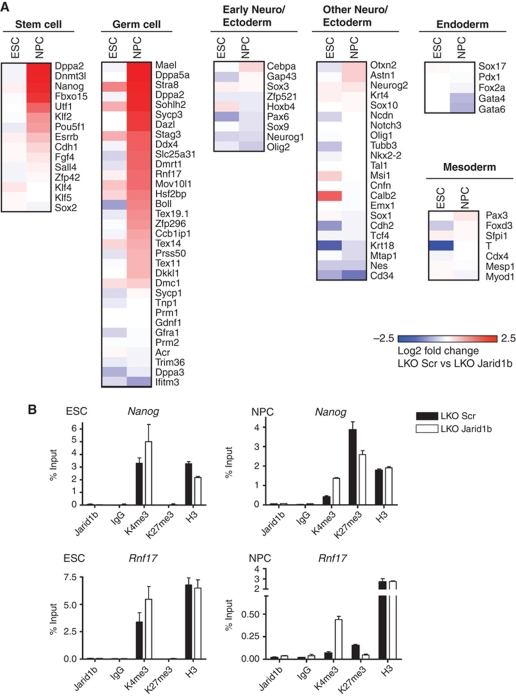
In contrast to ESCs, most of the differentially expressed genes in NPCs were upregulated (Supplementary Figure S12). By categorizing these genes based on reported functions, we found that reproduction-related genes (Weber et al, 2007; Han et al, 2010; Matson et al, 2010; Sabour et al, 2011) were strikingly overrepresented within the group of genes upregulated in Jarid1b knockdown NPCs compared with control NPCs (Figure 6A). In addition, as already noticed during EB differentiation (Supplementary Figure S4), we found that many pluripotency markers (Han et al, 2010; Kamiya et al, 2011) were not completely silenced in NPCs, including Nanog and Fbxo15 (Figure 6A; Supplementary Figure S13A). Moreover, we also tested the expression of markers from the three germ layers (Geijsen et al, 2004; West et al, 2006; Mikkelsen et al, 2007; Han et al, 2010; Kamiya et al, 2011) and defined a subset of early neural marker genes based on RNAPII recruitment from ESCs to NPCs in the same differentiation system used here (Mohn et al, 2008). Early neural genes are similarly expressed in Jarid1b knockdown and control NPCs, which is in agreement with our previous observation that the early neural marker Nestin is induced in Jarid1b knockdown NPCs (Figure 1D). In addition, other ectoderm, endoderm and mesoderm markers did not show differential expression in NPCs with Jarid1b knockdown compared with control (Figure 6A).
Figure 6.
Incomplete downregulation of stem and germ cell genes upon differentiation of Jarid1b knockdown ESCs. (A) Microarray heat map depicting expression of representative self-renewal, germ cell and differentiation markers in ESCs and NPCs. Colouring illustrates log 2 fold changes between LKO Scr and Jarid1b ESCs (first column) and between LKO Scr and Jarid1b NPCs (second column) with blue and red colours representing downregulation and upregulation in knockdown, respectively. (B) ChIP-qPCR for Jarid1b, H3K4me3, H3K27me3 and H3 in LKO Scr and Jarid1b ESCs (left) and NPCs (right) for the stem cell gene Nanog and the germ cell gene Rnf17.
These results indicate that impaired differentiation is probably not due to failed activation of lineage-specific genes or erroneous induction of alternative lineage genes, but rather caused by failure to silence previously expressed genes, including both pluripotency and germ cell-related genes. The majority of these genes normally become silenced during differentiation in all somatic cell types (Meissner et al, 2008; Mohn et al, 2008). In most cases, downregulation is accompanied by loss of H3K4me2/3 and either gain of DNA methylation or de novo H3K27 methylation (Meissner et al, 2008; Mohn et al, 2008). To understand the mechanism underlying incomplete silencing of stem and germ cell genes in Jarid1b knockdown cells, we analysed the chromatin state of Nanog and Rnf17 during neural differentiation (Figure 6B). In agreement with genome-wide chromatin profiles, H3K4me3 marks both genes in ESCs. In control cells, this modification is lost during neural differentiation, while in Jarid1b knockdown NPCs both promoters retain a considerable amount of H3K4me3. This is in agreement with incomplete silencing of these genes. Rnf17 was reported to gain DNA methylation during differentiation and to remain low for H3K27me3, while Nanog acquires H3K27me3 (Meissner et al, 2008; Mohn et al, 2008). Our ChIP data for H3K27me3 confirm these results, and in addition show that H3K27me3 is lower in Jarid1b knockdown NPCs. However, Jarid1b appears to be indirectly required to silence germ cell genes during neural differentiation, because we did not detect Jarid1b binding at the stem and germ cell genes tested, neither in ESCs nor in NPCs (Figure 6B and data not shown).
Discussion
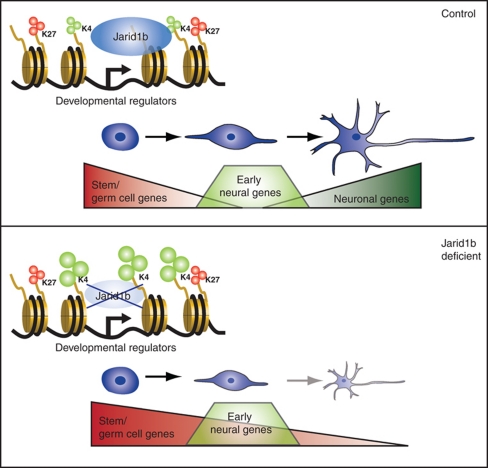
Here, we have shown that Jarid1b is dispensable for ESC self-renewal, that it binds to transcription start sites of genes encoding developmental regulators, and that its inactivation leads to increased H3K4me3 levels at target promoters as well as to a global increase of H3K4me3 levels (Figure 7). Moreover, we have shown that Jarid1b is required for differentiation of ESCs along the neural lineage, which correlates with the lack of silencing of stem and germ cell genes.
Figure 7.
Model for Jarid1b function in ESCs and during neural differentiation. In mouse ESCs, Jarid1b binds to transcription start sites of genes encoding developmental regulators, of which many are marked by bivalent chromatin modifications (upper panel). Jarid1b-deficient ESCs can be maintained despite increased H3K4me3 at Jarid1b target promoters, but fail to differentiate along the neural lineage, which correlates with an inability to silence stem and germ cell-specific genes (lower panel).
Jarid1b is dispensable for ESC self-renewal
Based on siRNA/shRNA knockdown, recent publications propose that Jarid1b is a pluripotency factor required for general ESC self-renewal (Xie et al, 2011) and ESC proliferation (Dey et al, 2008). Moreover, Jarid1b mutant ESCs could not be established (Catchpole et al, 2011). In contrast, we found that stable knockdown of Jarid1b in E14 ESCs using two different shRNA constructs does not compromise ESC proliferation or self-renewal. To exclude phenotypes due to possible shRNA off-target effects or lack of efficient downregulation of Jarid1b, we derived conditionally targeted Jarid1b ESCs. Importantly, complete loss of Jarid1b protein in these ESCs was confirmed using three different Jarid1b antibodies targeting both N- and C-terminal regions. In addition, transcription across the Jarid1b locus in knockout ESCs was very low (Supplementary Figure S2D), suggesting that no truncated or alternatively spliced Jarid1b protein can be produced. Jarid1b knockout ESCs show normal morphology, proliferation and expression of pluripotency markers both upon acute deletion and after several weeks in culture, and we therefore conclude that Jarid1b is dispensable for ESCs self-renewal.
Jarid1b target genes encode developmental regulators
By ChIP-seq, we found that Jarid1b binds predominantly around transcription start sites of genes encoding developmental regulators. These genes have previously been described to be marked by bivalent chromatin modifications in ESCs (Mikkelsen et al, 2007), and consistent with this observation, many of the Jarid1b target genes are also bound by PcG proteins and Jarid2. Jarid2 has been found to form a stable complex with PRC2 and to be essential for the binding of PcG proteins to their target genes (Peng et al, 2009; Shen et al, 2009; Pasini et al, 2010). In contrast, knockdown of Jarid1b only led to a small reduction of Suz12 at a subset of target promoters, indicating that Jarid1b does not recruit PcG proteins. Vice versa, Jarid1b binding is reduced, but not lost, in Eed and Ring1b knockout ESCs. Moreover, we have not detected a direct interaction between Jarid1b and PRC2, suggesting that alterations in H3K4 and H3K27 methylation might, directly or through other proteins binding to these modifications, cause the interdependent modulation of Jarid1b and PcG binding.
Our results are in contrast to a recent study that reported Jarid1b binding within intragenic regions in ESCs and that proposed a role for Jarid1b in the repression of cryptic intragenic transcription (Xie et al, 2011). However, we have not been able to confirm the data published by Xie et al, and in fact our results suggest that the enrichment of the regions published by these authors might not be specific for Jarid1b. Previously, JARID1A has also been proposed to downregulate intragenic H3K4me3 at two genes in HeLa cells, though no cryptic intragenic transcription was detected upon JARID1A deletion (Hayakawa et al, 2007). In contrast to these two reports (Hayakawa et al, 2007; Xie et al, 2011), Jarid1b was found to bind around transcription start sites of cancer-related genes in MCF7 cells (Yamane et al, 2007) and cell-cycle genes in ESCs (Dey et al, 2008). Likewise, Jarid1a was found to localize to promoter regions in ESCs (Pasini et al, 2008), MEFs (Klose et al, 2007) and U937 cells (Lopez-Bigas et al, 2008), and similarly Jarid1d occupied regions close to the transcription start site (Lee et al, 2007), supporting the idea that Jarid1-family members regulate gene expression through binding to transcription start sites.
Jarid1b binds to H3K4me3-positive promoters
Interestingly, the majority of Jarid1b target promoters are positive for H3K4me3, which might appear counterintuitive as Jarid1b catalyses H3K4me3 demethylation. However, the average H3K4me3 levels of Jarid1b target genes are relatively low, comparable to the levels of non-productive promoters (Rahl et al, 2010), suggesting that Jarid1b functions to keep H3K4me3 low rather than eliminating the mark. That is consistent with the observation that Jarid1b binds many bivalent promoters. Likewise, JARID1A has been shown to colocalize with its substrate (Lopez-Bigas et al, 2008), although bivalent domains were underrepresented within JARID1A target genes in U937 cells, probably highlighting a difference to ESCs where bivalent domains are far more abundant (Mikkelsen et al, 2007; Mohn et al, 2008).
Both Jarid1a and the Drosophila H3K4me3 demethylase Lid harbour PHD fingers that have the potential to bind to methylated H3K4 (Wang et al, 2009; Li et al, 2010). Since two crucial residues are conserved between Jarid1a, Lid and Jarid1b (Wang et al, 2009), it is likely that Jarid1b also exhibits H3K4me3 affinity via one of its PHD domains, possibly contributing to its recruitment to H3K4me3-positive promoters. Supporting this idea, Jarid1b binding is reduced in ESCs with knockdown of Dpy-30 (Supplementary Figure S14), a core subunit of the SET1/MLL histone methyltransferase complexes, which regulates H3K4me3 levels in ESCs without affecting ESC self-renewal (Jiang et al, 2011). However, since Jarid1b is not completely lost upon Dpy-30 knockdown and moreover, does not bind to all H3K4 methylated regions, other mechanisms are likely to contribute. Jarid1b contains an Arid domain that is potentially able to directly bind DNA. Interestingly, both the Jarid1a and the Jarid1b ARID domain preferentially bind to GC-rich sequence motifs (Scibetta et al, 2007; Tu et al, 2008), and deletion of the Jarid1b ARID domain affects its histone demethylase activity (Yamane et al, 2007). Nonetheless, additional factors are likely to contribute specificity to cell type-dependent Jarid1b recruitment such as sequence-specific transcription factors, proteins binding to histone tails and/or specific chromatin modifications and possibly long non-coding RNAs.
Loss of Jarid1b leads to increased H3K4me3
Consistent with the previously reported catalytic activity for Jarid1b (Christensen et al, 2007; Yamane et al, 2007), H3K4me3 levels were increased at Jarid1b target genes upon Jarid1b depletion. In addition, global H3K4me3 was increased in Jarid1b knockdown and knockout ESCs. Depletion of Jarid1b in NSCs (this study) or MCF7 cells (Yamane et al, 2007) or depletion of Jarid1a in MEFs (Klose et al, 2007) did not result in a global elevation of H3K4me3 levels, suggesting that the global increase is specific to ESCs. In this respect, it is interesting to note that many promoters lose H3K4 during ESC differentiation (Mikkelsen et al, 2007; Meissner et al, 2008; Mohn et al, 2008) and global H3K4me3 levels decline from ESCs to differentiated cells (Ang et al, 2011), suggesting that this decrease might be instrumental for normal differentiation. It is worth noting that 23.5% of the genes bound by Jarid1b in ESCs lose H3K4me3 during neural differentiation to the NPC stage (Meissner et al, 2008).
Although tolerated, increased H3K4me3 levels in ESCs are accompanied by a number of changes in gene expression, which however do not appear sufficient to alter cell fate. Similarly, alterations in expression of differentiation and signalling genes were observed in ESCs lacking repressive H3K27me3, which however, in analogy to Jarid1b mutant ESCs, did not compromise ESC maintenance (Pasini et al, 2007; Shen et al, 2008). Moreover, ESCs lacking the PRC2 interacting protein Jarid2 (Shen et al, 2009; Pasini et al, 2010) or the PRC2 component Eed (Chamberlain et al, 2008) can be maintained in culture.
In addition, recent studies have addressed the effect of reduced H3K4me3 in ESCs. Both overexpression of Jarid1b (Dey et al, 2008) and depletion of RbBP5 or Dpy-30 (Jiang et al, 2011), core subunits of SET1/MLL H3K4 methyltransferase complexes, induced a number of changes in gene expression and reduced global H3K4me3 levels without affecting self-renewal of ESCs. In contrast, ESC self-renewal defects were observed upon depletion of Wdr5 (Ang et al, 2011), another core subunit of SET1/MLL complexes, which could in part be mediated through the loss of H3K4me3 but might also be attributed to its additional integration into histone acetylase complexes (see comment to Ang et al, 2011, published online).
Therefore, we conclude that perturbations in H3K4 and H3K27 methylation do not compromise the self-renewing capacity of ESCs and propose that methylation of H3K4 and H3K27 function to fine tune transcription rather than inducing cell fate conversions in ESCs.
Jarid1b is required for neural differentiation of ESCs
The effects of the lack of Jarid1b transcriptional control become more evident when ESCs are induced to differentiate. General differentiation of Jarid1b-depleted ESCs assayed by EB formation revealed deficiencies in the silencing of stem cell genes as well as the induction of marker genes representing all three germ layers, with strongest effects observed for the ectodermal lineage. Accordingly, differentiation of Jarid1b-depleted ESCs using a neural differentiation protocol was impaired and these cells did not progress beyond the NPC stage.
Using microarray analysis, we identified differentially expressed genes in NPCs most of which were upregulated, consistent with the reported role of Jarid1 proteins as transcriptional repressors (Tan et al, 2003; Klose et al, 2007). Specifically, we found that stem and germ cell-specific genes, which normally are expressed in ESCs but silenced during neural differentiation (Meissner et al, 2008; Mohn et al, 2008), are not properly switched off in Jarid1b-depleted NPCs. Thus, although NPCs expressing early neural markers could be obtained from Jarid1b mutant ESCs, the inefficient silencing of stem and germ cell genes might prevent the NPCs from further commitment towards mature neuronal cells.
Of the genes upregulated in Jarid1b knockdown NPCs, approximately half have been reported to lose H3K4me2/3 during differentiation in genome-wide chromatin-association studies (Meissner et al, 2008; Mohn et al, 2008). Loss of H3K4 methylation is a strong predictor of inverse changes in DNA methylation (Meissner et al, 2008), and indeed, a substantial proportion of stem and germ cell genes acquire DNA methylation from ESCs to neural cells (Meissner et al, 2008; Mohn et al, 2008). As we could not detect Jarid1b binding at stem and germ cell genes, Jarid1b seems to indirectly affect the demethylation of H3K4, which in turn may prevent CpG de novo methylation to occur (Ooi et al, 2007; Weber et al, 2007; Meissner et al, 2008). In addition, a different subset of stem and germ cell genes is silenced by de novo H3K27me3 (Mohn et al, 2008). A recent study suggests that adjustment of high H3K4me3 to moderate or low levels may modulate PcG-mediated H3K27me3 (Schmitges et al, 2011).
It is conceivable that, during neural differentiation, Jarid1b contributes to active demethylation of genes already bound in ESCs, possible requiring additional cues, or by being recruited to new target genes. Moreover, it has been proposed that poised chromatin states in ESCs are central for the developmental potential of these cells (Pietersen and van Lohuizen, 2008). Thus, inappropriate chromatin priming in Jarid1b knockdown ESCs may interfere with the execution of differentiation programs. Due to complex interrelationships of diverse histone modifications, multiple layers of factors binding to these modifications and feedback from transcriptional activity itself, it is very difficult to dissect the contribution of each individual component in a dynamic differentiation system.
To further refine the requirement of Jarid1b for neural differentiation, we extended our study to Jarid1b knockout NSCs isolated from embryonic brain. We found that Jarid1b is not required for the generation of neurons from these NSCs. Importantly, stem and germ cell-specific genes remained undetectably low upon deletion of Jarid1b in established NSCs, suggesting that once stable silencing mechanisms like DNA methylation or H3K27me3 are in place, Jarid1b is dispensable for maintenance of the silent state. Interestingly, however, we observed a delay in the downregulation of the NSC marker Nestin during differentiation of Jarid1b knockout NSCs. This finding suggests that Jarid1b functions are cell type-specific, in analogy to cell type-specific target genes described for PcG proteins (Mohn et al, 2008). Nonetheless, the observed effect in NSCs is very mild, highlighting the proposed role of Jarid1b as a fine tuner of gene expression.
Jarid1b and development
Recently, it was reported that Jarid1b knockout mice in which exon 1 was replaced by the Neomycin gene are embryonic lethal between E4.5 and E7.5 (Catchpole et al, 2011) a quite severe early embryonic phenotype, also in comparison with described PcG mutants (Faust et al, 1995; O’Carroll et al, 2001; Pasini et al, 2004). In contrast, Jarid1b mutant mice established in our laboratory are viable, although at a lower Mendelian frequency, and fertile (Albert et al, manuscript in preparation). Thus, lack of Jarid1b seems compatible with normal gross development, possibly due to redundant functions and/or compensation by other Jarid1-family members. A detailed analysis of Jarid1b function during brain development remains to be reported, though it can be anticipated from our NSC differentiation experiments that Jarid1b might support efficient differentiation rather than being a determining factor for neuronal differentiation in vivo. It will be interesting to dissect Jarid1b functions in other cell types with high expression, both in normal development and in the context of cancer, where a controversial role for Jarid1b either as oncogene (Lu et al, 1999; Barrett et al, 2002; Yamane et al, 2007; Hayami et al, 2010; Roesch et al, 2010) or as tumour suppressor (Roesch et al, 2006, 2008) has been proposed.
Materials and methods
ESC culture and differentiation
E14TG2a.4 feeder-independent ESCs were cultured on 0.1% gelatin-coated tissue culture plates in standard ESC medium (Pasini et al, 2007). All knockdown experiments were performed using stable shRNAs. Viral transductions were performed using pLKO vectors (NM_152895.1-4880s1c1 (J1b-1), NM_152895.1-4225s1c1 (J1b-2), SHC201 (Scr) (all from Sigma), and NM_024428.2-302s1c1 (Dpy-30) (Open Biosystems)). Unless otherwise indicated differently, LKO-J1b-2 was used. ESCs were transduced with lentiviral particles for 16 h, and selected with 2 μg/μl Puromycin (Invitrogen) 48 h after transduction.
EBs were generated using the ‘hanging drop’ method. Drops of 1000 cells in 20 μl ESC medium with 10% FBS without LIF were placed in the lid of tissue culture dishes. Aggregates were allowed to form for 48 h, harvested and plated into bacterial dishes in the same medium. In all, 0.5 μM ATRA was added to half of the plates. Homogeneous neural cells were generated as previously described (Bibel et al, 2007). Ring1b and Eed knockout cells were previously described (Leeb and Wutz, 2007; Shibata et al, 2008).
Derivation of Jarid1b knockout ESCs
Jarid1b targeted C57BL/6N ESCs were obtained from the European Conditional Mouse Mutagenesis Program (EUCOMM). A lacZ-Neo-reporter cassette flanked by FRT sites was inserted between exons 5 and 6, and Jarid1b exon 6 was flanked by loxP sites. The deletion of exon 6 creates a frameshift and subsequent termination mutation. Jarid1b targeted ESCs were analysed by Southern blotting to confirm correct position of the targeting cassette. Analysis of metaphase spreads confirmed correct karyotype. Two targeted ESC lines, F07 and F08, were selected for injection into BALB/c blastocysts. Both lines resulted in chimeric mice.
Chimeric Jarid1b mice were crossed to Flp-recombinase expressing mice to obtain germline transmission and at the same time generate conditional Jarid1b mice lacking the reporter cassette. Conditional Jarid1b mice were further crossed with Rosa26::CreERT2 mice (obtained from the Jackson Laboratory). Jarid1bF/F; Rosa26::CreERT2 ESCs were derived on feeder cells from blastocysts obtained from superovulated Jarid1bF/+ females mated with Jarid1bF/+; CreERT2/CreERT2 males. The sex and karyotype were determined as described (Pasini et al, 2007). Deletion of Jarid1b was induced by addition of 1 nM 4OHT for at least 48 h.
For genotyping of Jarid1b mice and cells, 5′-CCCTGGGATTGCAGTTAAAG-3′ forward, 5′-TGGCTTCCACAATCTTCAATG-3′ reverse and 5′-TGGCTTCCACAATCTTCAATG-3′ reverse primers were used to discriminate wild-type (527 bp), floxed (609 bp) and deletion (685 bp) alleles. Mice were maintained on a C57BL/6 background. All mouse work was approved by the Danish Animal Ethical Committee (‘Dyreforsøgstilsynet’).
Derivation of Jarid1b knockout NSCs
NSCs were isolated from the frontal cortex of E12.5 Jarid1bF/F; Rosa26::CreERT2 embryos. Briefly, pieces of cerebral cortex were treated with trypsin-EDTA at 37°C for 20 min and mechanically dissociated by pipetting after addition of soybean trypsin inhibitor (Invitrogen). NSCs were plated on poly-D-lysine (Chemicon) and laminin (Sigma)-coated culture plates at a density of 36 000 cells/mm2 in a 1:1 mixture of DMEM/F12 and Neurobasal medium supplemented with 10 ng/ml EGF (BioSource), 20 ng/ml bFGF (BioSource), N2, B27, Penicillin, Streptomycin, Glutamax, Non-Essential Amino Acids, Sodium-Pyruvate, β-mercaptoethanol, Heparin (Sigma), BSA (Sigma) and Hepes (all from Gibco, except where otherwise indicated). Deletion of Jarid1b was induced by addition of 1 nM 4OHT for 48 h. NSCs were induced to differentiate by removal of EGF/bFGF and addition of 2% FBS (Hyclone).
Antibodies
Jarid1b Polyclonal antibody (DAIN) was generated by immunizing rabbits with a peptide corresponding to amino acids 1395–1418 of mouse Jarid1b (NP_690855.2). The antibodies were affinity purified using Sepharose (GE Healthcare) coupled to the immunogen. Antibody specificity was confirmed by immunoblotting and IP.
Additional antibodies used in this study include anti-H3K4me3 (Cell Signaling, C42D8), anti-H3K27me3 (Cell Signaling, D18C8), anti-H3 (Abcam, 1791), anti-Suz12 (Cell Signaling, D39F6), anti-Ring1b (Richly et al, 2010), anti-Vinculin (Sigma, V9131), anti-Nanog (Abcam, 80892), anti-Oct4 (Abcam, 19857), mouse anti-Nestin (BD Biosciences, 611659), goat anti-Nestin (Millipore, MAB353), anti-β-III-tubulin (Sigma, T5076) and anti-BrdU (Becton Dickinson, 347580), and anti-Jarid1b (Abcam, ab50958, Santa Cruz, sc-67035).
Gene expression analysis
Total RNA was isolated using the RNAeasy Minikit (Qiagen) according to the manufacturer's instructions. cDNA was synthesized using the TaqMan Reverse Transcription kit (Applied Biosystems). qPCR was performed using SYBR Green 2 × PCR Master mix (Applied Biosystems) on an ABI Prism 7300 Real-Time PCR system (Applied Biosystems) or on a LightCycler 480 System (Roche Applied Science), using the LightCycler 480 SYBR Green I Master mix (Roche Applied Science) according to the manufacturers’ instructions. Error bars represent standard deviation of three PCR amplifications for each sample. Similar results were obtained in at least three independent experiments. Primer sequences are provided in Supplementary Table I.
For microarray analysis, RNA was extracted with the RNeasy Plus kit (Qiagen). RNA was hybridized on mouse Gene 1.0 ST arrays by the RH Microarray Center at Rigshospitalet, Copenhagen, following Affymetrix procedures and analysis. Data processing and statistics were done in R using the ‘affy’ (RMA) and ‘limma’ packages. The microarray data have been submitted to the Gene Expression Omnibus (GEO) database (GSE31968). Heat maps were produced using matrix2png (Pavlidis and Noble, 2003).
ChIP assays and ChIP-seq
ChIP was performed as described (Pasini et al, 2010). For each IP, 0.5–1 mg of chromatin was used except for histones and histone modifications where 100 μg were used. Primer sequences are provided in Supplementary Table I.
For ChIP-seq analysis, 1 mg total protein was used. Subsequently, the DNA was adaptor-ligated, amplified using a kit from Illumina (IP-102-1001) and analysed by Solexa/Illumina high throughput sequencing. The sequence reads were aligned to the mouse genome (assembly mm9). Peak detection was performed using the MACS algorithm (Zhang et al, 2008) at an FDR cutoff value of <0.2. IgG was used for normalization. Chromosomal positions were annotated to the RefSeq database (mm9) using the UCSC refFlat table (Rhead et al, 2010). Genes not uniquely mapped to the genome were excluded. Gene overlap calculations were performed with the Galaxy browser (Giardine et al, 2005). Gene ontology analysis was performed using PANTHER (Thomas et al, 2003). GC-rich regions were defined as >100 bp long with a GC percentage of >50% and an observed/expected ratio of >0.6 (Ku et al, 2008). For gene body tag density, the total length of the mapped reads was extended in 3′-direction to a total length of 250 bases, which was our estimated mean fragment length after sonication. Gene bodies (defined by the longest Refseq transcript) were divided into 20 windows of equal size per gene as wells 10 windows upstream of the TSS (−10 kb) and 10 windows downstream of the TSE (+10 kb), for which average tag numbers were calculated. Plots showing tag density across gene bodies and peak distance to TSS were generated in R. Heat maps were generated using seqMINER (Ye et al, 2011). The ChIP-seq data have been submitted to the GEO database (GSE31968).
Immunofluorescence and imaging
Immunofluorescence staining was performed using standard procedures. Images were acquired on an Axioplan2 microscope (Carl Zeiss, Inc) equipped with a CoolSNAP cf2 (Photometrics) camera using MetaMorph software (MDS Analytical Technologies).
For unbiased analysis, immunofluorescence of NSC differentiation was performed in 96-well plates. Cells were imaged on an IN Cell Analyzer 1000 (GE Healthcare) utilizing a × 20 objective and 10 images per well, which accounted for 1000–3000 cells. Data were analysed using IN Cell Analyzer Workstation 3.5 software (GE Healthcare). Error bars represent standard deviation of three individual wells for each sample.
Light microscope images were acquired on an Olympus IX71S8F microscope using a LOPlanFl objective.
Flow cytometry
Cells were fixed in 70% ethanol and stained with primary antibody for 1 h, followed by 1 h incubation with Alexa Fluor 488 or 647 anti-rabbit or anti-mouse IgG (Invitrogen). Cells were pulsed with 33 μM bromodeoxyuridine (BrdU) for 30 min. DNA was counterstained by 0.1 mg/ml propidium iodide supplemented with RNase for 1 h at 37°C. Analysis was performed on a FACSCalibur using CellQuest software (BD). Quantification and analysis of cell-cycle profiles were obtained using FlowJo (Tree Star, Inc).
Supplementary Material
Acknowledgments
We thank C Steinhauer for assistance with automated image analysis and F Jia for help with mouse genotyping. We thank Anja Groth and members of the Helin laboratory for critical and fruitful discussions. MA is supported by an EMBO long-term postdoctoral fellowship. LM is supported by a CRG (Novartis) postdoctoral fellowship. The Wilhelm Johannsen Centre is supported by the Danish National Research Foundation, the Lundbeck Foundation and the Danish Ministry of Science, Technology and Innovation. Work in the Helin laboratory is supported by the Danish National Research Foundation, the Danish Cancer Society, the Novo Nordisk Foundation, the Danish Medical Research Council, the Lundbeck Foundation and the Excellence Programme of the University of Copenhagen.
Author contributions: SUS performed experiments in Figures 1, 3, 4, 5, 6 and Supplementary Figures S1C, D, S3A, C–E, S4, S5, S8, S9C, S10–14. MA generated Figures 2, 3B, E, F and Supplementary Figures S2, S3B, S7, S10A and conditional Jarid1b mice and participated in analysis of ChIP sequencing and microarray experiments. MM performed experiments in Supplementary Figures S1E, S3D, S6 and S14A. LM performed experiments in Supplementary Figure S9B. JVJ provided bioinformatics support. MB and NT performed the Solexa DNA sequencing. IA developed and characterized the DAIN antibody, Jarid1b targeted ESCs and other reagents used in this study and performed experiments in Supplementary Figures S1A, B and S2B. SUS, MA and KH wrote the manuscript. MA and KH supervised the project.
Footnotes
KH is a cofounder of EpiTherapeutics and has shares and warrants in the company.
References
- Ang Y-S, Tsai S-Y, Lee D-F, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR (2011) Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Barrett A, Madsen B, Copier J, Lu PJ, Cooper L, Scibetta AG, Burchell J, Taylor-Papadimitriou J (2002) PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen? Int J Cancer 101: 581–588 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (2007) High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Bibel M, Richter J, Lacroix E, Barde Y-A (2007) Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc 2: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG, Burchell JM, Taylor-Papadimitriou J (2011) PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol 38: 1267–1277 [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T (2008) Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells 26: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PAC, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K (2007) RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128: 1063–1076 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Dambacher S, Hahn M, Schotta G (2010) Epigenetic regulation of development by histone lysine methylation. Heredity 105: 24–37 [DOI] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C (2008) The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol 28: 5312–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Walker JA, Burgio G, Corona DFV, Mulligan P, Näär AM, Dyson NJ (2011) Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev 25: 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ (2004) Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell 6: 437–443 [DOI] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T (1995) The eed mutation disrupts anterior mesoderm production in mice. Development 121: 273–285 [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Smith L, Greenfield A, Zernicka-Goetz M (2007) Novel gene expression patterns along the proximo-distal axis of the mouse embryo before gastrulation. BMC Dev Biol 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geijsen N, Horoschak M, Kim K, Gribnau J, Eggan K, Daley GQ (2004) Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 427: 148–154 [DOI] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15: 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang S-L, Stewart AF (2006) Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133: 1423–1432 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DW, Tapia N, Joo JY, Greber B, Araúzo-Bravo MJ, Bernemann C, Ko K, Wu G, Stehling M, Do JT, Schöler HR (2010) Epiblast stem cell subpopulations represent mouse embryos of distinct pregastrulation stages. Cell 143: 617–627 [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Ohtani Y, Hayakawa N, Shinmyozu K, Saito M, Ishikawa F, Nakayama J-I (2007) RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells 12: 811–826 [DOI] [PubMed] [Google Scholar]
- Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue H, Ponder BAJ, Nakamura Y, Hamamoto R (2010) Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Mol Cancer 9: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Gotoh Y (2010) Epigenetic control of neural precursor cell fate during development. Nat Rev Neurosci 11: 377–388 [DOI] [PubMed] [Google Scholar]
- Hublitz P, Albert M, Peters AHFM (2009) Mechanisms of transcriptional repression by histone lysine methylation. Int J Dev Biol 53: 335–354 [DOI] [PubMed] [Google Scholar]
- Jiang H, Shukla A, Wang X, Chen W-Y, Bernstein BE, Roeder RG (2011) Role for Dpy-30 in ES cell-fate specification by regulation of H3K4 methylation within bivalent domains. Cell 144: 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya D, Banno S, Sasai N, Ohgushi M, Inomata H, Watanabe K, Kawada M, Yakura R, Kiyonari H, Nakao K, Jakt LM, Nishikawa S-I, Sasai Y (2011) Intrinsic transition of embryonic stem-cell differentiation into neural progenitors. Nature 470: 503–509 [DOI] [PubMed] [Google Scholar]
- Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y, Kim CG, Cantor AB, Orkin SH (2010) A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG (2007) The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128: 889–900 [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, Adli M, Kasif S, Ptaszek LM, Cowan CA, Lander ES, Koseki H, Bernstein BE (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Norman J, Shilatifard A, Shiekhattar R (2007) Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128: 877–887 [DOI] [PubMed] [Google Scholar]
- Leeb M, Wutz A (2007) Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol 178: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ (2008) Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell 135: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Greer C, Eisenman RN, Secombe J (2010) Essential functions of the histone demethylase lid. PLoS Genet 6: e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim DA, Huang Y-C, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A (2009) Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature 458: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV (2008) Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell 31: 520–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz MC, Schübeler D (2007) RNA polymerase II: just stopping by. Cell 130: 16–18 [DOI] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, Taylor-Papadimitriou J (1999) A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem 274: 15633–15645 [DOI] [PubMed] [Google Scholar]
- Madsen B, Spencer-Dene B, Poulsom R, Hall D, Lu PJ, Scott K, Shaw AT, Burchell JM, Freemont P, Taylor-Papadimitriou J (2002) Characterisation and developmental expression of mouse Plu-1, a homologue of a human nuclear protein (PLU-1) which is specifically up-regulated in breast cancer. Mech Dev 119(Suppl 1): S239–S246 [DOI] [PubMed] [Google Scholar]
- Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D (2010) The mammalian double sex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell 19: 612–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE (2010) GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C et al. (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D (2008) Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Niwa H (2007) How is pluripotency determined and maintained? Development 134: 635–646 [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21: 4330–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SKT, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin S-P, Allis CD, Cheng X, Bestor TH (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell 1: 299–312 [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K (2007) The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol 27: 3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K (2004) Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PAC, Walfridsson J, Olsson L, Bukowski J-P, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K (2010) JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature 464: 306–310 [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PAC, Helin K (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev 22: 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Noble WS (2003) Matrix2png: a utility for visualizing matrix data. Bioinformatics 19: 295–296 [DOI] [PubMed] [Google Scholar]
- Pedersen MT, Helin K (2010) Histone demethylases in development and disease. Trends Cell Biol 20: 662–671 [DOI] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J (2009) Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 139: 1290–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M (2008) Stem cell regulation by polycomb repressors: postponing commitment. Curr Opin Cell Biol 20: 201–207 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA (2010) c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, Pohl A, Pheasant M, Meyer LR, Learned K, Hsu F, Hillman-Jackson J, Harte RA, Giardine B, Dreszer TR, Clawson H et al. (2010) The UCSC Genome Browser database: update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L (2010) Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 468: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Roesch A, Becker B, Schneider-Brachert W, Hagen I, Landthaler M, Vogt T (2006) Re-expression of the retinoblastoma-binding protein 2-homolog 1 reveals tumor-suppressive functions in highly metastatic melanoma cells. J Invest Dermatol 126: 1850–1859 [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M (2010) A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141: 583–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T (2008) RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer 122: 1047–1057 [DOI] [PubMed] [Google Scholar]
- Sabour D, Araúzo-Bravo MJ, Hübner K, Ko K, Greber B, Gentile L, Stehling M, Schöler HR (2011) Identification of genes specific to mouse primordial germ cells through dynamic global gene expression. Hum Mol Genet 20: 115–125 [DOI] [PubMed] [Google Scholar]
- Salomoni P, Calegari F (2010) Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol 20: 233–243 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stützer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, Bunker RD, Wirth U, Bouwmeester T, Bauer A, Ly-Hartig N, Zhao K, Chan H, Gu J, Gut H, Fischle W et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, Bell SP, Groudine M (2004) The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scibetta AG, Santangelo S, Coleman J, Hall D, Chaplin T, Copier J, Catchpole S, Burchell J, Taylor-Papadimitriou J (2007) Functional analysis of the transcription repressor PLU-1/JARID1B. Mol Cell Biol 27: 7220–7235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan G-C, Lee Y, Orkin SH (2009) Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell 139: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu Y-J, Fujiwara Y, Kim J, Mao X, Yuan G-C, Orkin SH (2008) EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32: 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Yokota T, Wutz A (2008) Synergy of Eed and Tsix in the repression of Xist gene and X-chromosome inactivation. EMBO J 27: 1816–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A (2007) Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435 [DOI] [PubMed] [Google Scholar]
- Tan K, Shaw AL, Madsen B, Jensen K, Taylor-Papadimitriou J, Freemont PS (2003) Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J Biol Chem 278: 20507–20513 [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu S, Teng Y-C, Yuan C, Wu Y-T, Chan M-Y, Cheng A-N, Lin P-H, Juan L-J, Tsai M-D (2008) The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat Struct Mol Biol 15: 419–421 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WWMP, van Schaik FMA, Varier RA, Baltissen MPA, Stunnenberg HG, Mann M, Timmers HTM (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo J-L, Patel DJ, Allis CD (2009) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459: 847–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D (2007) Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466 [DOI] [PubMed] [Google Scholar]
- West JA, Park I-H, Daley GQ, Geijsen N (2006) In vitro generation of germ cells from murine embryonic stem cells. Nat Protoc 1: 2026–2036 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD (2006) A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442: 86–90 [DOI] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S (2011) KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 30: 1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Deguchi K, Aono A, Tani Y, Kishimoto T, Komori T (1998) Growth disturbance in fetal liver hematopoiesis of Mll-mutant mice. Blood 92: 108–117 [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y (2007) PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell 25: 801–812 [DOI] [PubMed] [Google Scholar]
- Ye T, Krebs AR, Choukrallah M-A, Keime C, Plewniak F, Davidson I, Tora L (2011) seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res 39: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA (2011) Control of the embryonic stem cell state. Cell 144: 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ (1995) Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378: 505–508 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.