Abstract
The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Central Repository makes data and biospecimens from NIDDK-funded research available to the broader scientific community. It thereby facilitates: the testing of new hypotheses without new data or biospecimen collection; pooling data across several studies to increase statistical power; and informative genetic analyses using the Repository’s well-curated phenotypic data. This article describes the initial database plan for the Repository and its revision using a simpler model. Among the lessons learned were the trade-offs between the complexity of a database design and the costs in time and money of implementation; the importance of integrating consent documents into the basic design; the crucial need for linkage files that associate biospecimen IDs with the masked subject IDs used in deposited data sets; and the importance of standardized procedures to test the integrity data sets prior to distribution. The Repository is currently tracking 111 ongoing NIDDK-funded studies many of which include genotype data, and it houses over 5 million biospecimens of more than 25 types including serum, plasma, stool, urine, DNA, red blood cells, buffy coat and tissue. Repository resources have supported a range of biochemical, clinical, statistical and genetic research (188 external requests for clinical data and 31 for biospecimens have been approved or are pending). Genetic research has included GWAS, validation studies, development of methods to improve statistical power of GWAS and testing of new statistical methods for genetic research. We anticipate that the future impact of the Repository’s resources on biomedical research will be enhanced by (i) cross-listing of Repository biospecimens in additional searchable databases and biobank catalogs; (ii) ongoing deployment of new applications for querying the contents of the Repository; and (iii) increased harmonization of procedures, data collection strategies, questionnaires etc. across both research studies and within the vocabularies used by different repositories.
Database URL: http://www.niddkrepository.org
Background
In 2003, the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) at the National Institutes of Health (NIH) established data, biosample and genetic repositories to increase the impact of current and previously funded NIDDK studies by making their data and biospecimens available to the broader scientific community (see www.niddkrepository.org). These repositories, collectively known as the ‘NIDDK Central Repository’, enable scientists not involved in the original study to test new hypotheses without new data or biospecimen collection, and the Repository provides the opportunity to pool data across several studies to increase the power of statistical analyses. In addition, most NIDDK-funded studies collect genetic biospecimens and some carry out high-throughput genotyping, making it possible for other scientists to use Repository resources to perform informative genetic analyses using well-curated phenotypic data.
In this article, we describe: the ambitious initial design of the Repository; the subsequent simplification of that design to better accommodate the needs of users and the constraints of available resources; the current status of the Repository; the data and biospecimens offered to researchers; and examples of the use made of Repository resources for biomedical research. We conclude by describing some of the key lessons we learned in the evolution of the Repository and the bioinformatic enhancements we are currently making to the Repository.
An ambitious database proposal in 2002
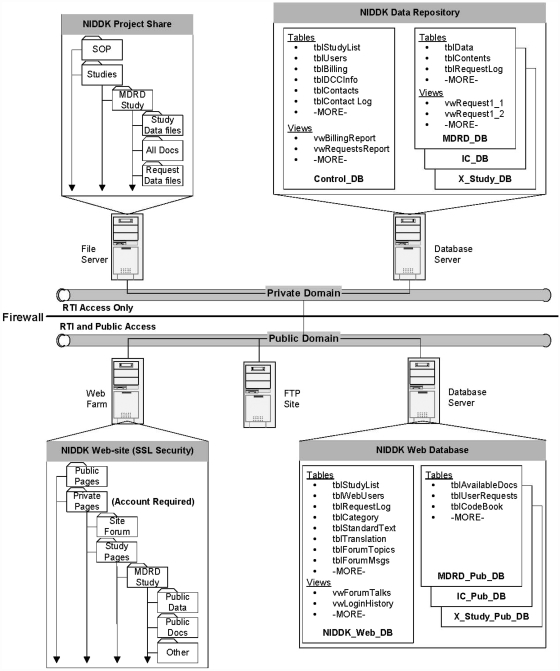
We envisioned that the NIDDK Data Repository would be a large system consisting of primary databases in the private domain (shown in Exhibit 1 as NIDDK Data Repository), and support databases in the public domain (shown in Exhibit 1 as NIDDK Web Databases). Creating databases in both domains was deemed necessary for providing security and accessibility for authorized project and public users.
Exhibit 1.
Initial Plan for NIDDK Data Repository.
The primary databases in the private domain were planned to include a project management (Control) database and individual study databases. The Control database (Control_DB) was intended to have tables and views (stored queries) that would help manage project functions, track and manage study databases and provide information for reports. The study databases (Study_DB) was intended to have tables and views that contain the study data, code books and information that will assist in database management, track researcher requests and provide data in response to researcher requests.
The support databases were intended to include any databases necessary to support the public website. It was anticipated that a primary database (NIDDK_Web_DB) would have the tables and views that support the website's ability to inform researchers of available studies, manage researcher access to the private pages, support a hosted user forum and support researcher requests for data. Additional study databases (Study_Pub_DB) would be created to contain study–specific tables for codebooks, documentation lists, user request logs, etc. These databases would be used to provide study–specific information and to facilitate methods for researcher requests for data based on available fields.
Revision of design
Our initial plan was ambitious, complex and expensive. Upon the award of the contracts to build the Repository and supporting database tools, we conducted a requirements analysis that considered both NIDDK’s and the scientific community’s interests and needs. This analysis concluded that our proposed approach was inappropriate for a number of reasons the most important being development cost and lag time in bringing the Repository online. This formal review of the perspectives of all repository stakeholders (i.e. NIDDK, the research centers contributing the data, the subjects providing the data and the data consumers) identified the following core requirements for developing and maintaining a large repository of the scale we envisioned.
A public website to support communication functions including informing users about: how to identify the contents of the Repository, how to obtain repository products, how to contribute products to the Repository and how to access Repository personnel.
A screening process for data and specimen requesters to control access to Repository resources. Accordingly, if a user was interested in obtaining Repository products, they would be obligated to provide a research plan that identifies how the products are to be used and this plan would be reviewed and approved or disapproved by NIDDK.
A hierarchal view of available data, biospecimens and supporting documentation. This hierarchy begins with an overview of the study that identifies its purpose, outcomes and design features; a detailed description of how the study operated (protocol and MOOP); and the nuts and bolts of how the data were captured (data collection forms).
A mechanism for supplying information on subsets of study variables (and therefore data) since a non-trivial percentage of those variables would be of little general interest to potential users.
Rigorous procedures to insure that data distributed by the Repository were checked for completeness, accuracy and compliance with HIPAA regulations.
A simpler design for the Repository
To fulfill these requirements, we revised our plan for the design and implementation of the Repository to include:
A standard template for the documentation for each study that included: (i) a general description of the study, (ii) manuals of Operations and Protocols (descriptions of the procedures used to collect clinical data and samples), (iii) all Data Capture Forms used in collecting clinical data, (iv) Data Descriptions (including summary statistics on distributions of variables and SAS variable descriptions and (v) Links to the study’s publications.
Placement of data and biospecimens in different physical locations and restricted domains from their documentation. The data (in contrast to data descriptions) were resident in the data archive section of the Repository and were accessible only to Repository staff. These data were only distributed to approved researchers. This meant that there would be reduced security issues involving unapproved access to the data because these data would be behind the Repository firewall. The documentation was also part of the data archive but unlike the archive it was viewable from the public component of the website.
Development of a series of semi-automated applications that permitted users to submit requests for data and samples online. These processes were modified over time to support higher levels of automation.
A data curation process that provided a standard directory layout for organizing the data into data archives and adding documentation to promote usability.
Development of a pre-release data checking procedure that selects published peer-reviewed manuscripts from a study and independently reproduces tables and statistical analyses using the data deposited in the repository. This process helps insure the integrity of the data distributed by the Repository.
Over time—as the number of studies housed at the Repository has grown—we have recognized an additional requirement for efficient ways of searching the Repository contents and retrieving relevant documents. New tools for that purpose are being rolled out during 2011. (In a later section of this article, we describe these tools.)
Major components of the Repository in 2011
At present the NIDDK Central Repository has five major components:
an archive of clinical data and documentation from NIDDK-sponsored studies;
a collection of biospecimens and an associated database that identifies specimens collected from ongoing and completed studies funded by NIDDK and links them to the associated phenotypic data;
a Web portal that makes study-specific information within the Repository easily viewable and that accepts electronic requests for biospecimens and data; and
a collection of genotyping data from genome-wide association studies (GWAS) and sequencing studies housed at the National Center for Biotechnology Information’s (NCBI’s) database of Genotypes and Phenotypes (dbGaP; see http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap).
a library of study- and site-specific consent forms that govern the release and use of study data and specimens.
Status of Repository
Studies
As of 9 March 2011, the Repository was tracking 111 NIDDK-funded studies. From these studies, the Repository offers resources for clinical, biochemical, statistical and genetic research especially in the areas of diabetes, kidney disease, liver disease and inflammatory bowel disease. At present, the Repository offers clinical data from 29 completed NIDDK-funded studies—15 of which currently offer biospecimens and 7 of which have available genotype data. Table 1 provides descriptions of these studies, the specimens available from each study, and the number of subjects enrolled. Since there is substantial variability in the types of clinical data available from each study, it is not feasible to summarize it in this article. Suffice it to say that the collection of clinical data is large, diverse and carefully curated. As an example of the studies included in this collection, we would note the DCCT-EDIC study which is continuing to follow a cohort of Type 1 diabetic patients recruited in 1983. The clinical data include the results of physical examinations with extensive measurements at regular intervals of retinopathy, nephropathy, neuropathy and cardiovascular status along with metabolic and lipid profiles. (Biospecimens available from DCCT-EDIC include DNA, plasma, RNA, serum, urine and peripheral blood mononuclear cells [PBMC].). (Samples may include multiple aliquots of the same unique specimens.)
Table 1.
Studies currently offering clinical data from the NIDDK Central Repository
| Acronym | Study | Conditions | Study type | Biospecimens | Genotype available | Sample sizes |
|---|---|---|---|---|---|---|
| A2ALL | Adult Living Donor Liver Transplantation | Liver disease, End stage liver disease, kidney disease | Retrospective records review | DNA, serum, tissue, whole blood | 819 | |
| AASK | The African American Study of Kidney Disease and Hypertension Study | Kidney disease, Hypertension | Clinical trial | buffy coat, serum, urine | 2802 screened, 1094 randomized | |
| ATN | Acute Renal Failure Trial Network | Kidney disease, End stage kidney disease | Clinical trial | 1124 | ||
| BACH | Boston Area Community Health Study | Urogynecologic symptoms, Incontinence, Interstitial cystitis, Chronic pelvic pain, Prostatitis, Hypogonadism, Sexual dysfunction | Epidemiologic survey | 5506 | ||
| BE-DRI | Behavior Enhances Drug Reduction of Incontinence | Incontinence | Clinical trial | 4043 screened, 307 randomized | ||
| CDS | Comprehensive Dialysis Study | Kidney disease, Dialysis | Prospective cohort study | PBMC, plasma, serum | 1677 | |
| CPCRN | Chronic Prostatitis Collaborative Research Network Cohort Study | Prostatitis, Chronic Pelvic Pain | Prospective cohort study | 488 | ||
| CPCRN RCT #1 | Chronic Prostatitis Collaborative Research Network Clinical Trial | Prostatitis, Chronic pelvic pain | Clinical trial | 272 | ||
| CRISP | Consortium for Radiologic Imaging Studies of PKD | Kidney disease, PKD | Prospective cohort study | DNA, plasma, serum, urine, whole blood | Yes | 241 |
| DPP | Diabetes Prevention Program | Type 2 diabetes, Impaired glucose tolerance | Clinical trial | DNA, Plasma | 3819 | |
| DPT-1 | Diabetes Prevention Trial—Type 1 | Type 1 diabetes, | Clinical trial | DNA, Plasma, Serum | 711 | |
| EDIC | Epidemiology of Diabetes Interventions and Complications | Type 1 diabetes, Coronary heart disease, Kidney disease, Neuropathy, Retinopathy | Longitudinal Follow-up of Participants in DCCT | DNA, Plasma, serum, urine | Yes | 1297 active in 2008a |
| DCCT | The Type 1 Diabetes Control and Complications Trial | Type 1 diabetes, Coronary heart disease, Kidney disease, Neuropathy, Retinopathy | Clinical trial | DNA, Plasma, PBMC, RNA, Serum | 1441 | |
| FIND | Family Investigation of Nephropathy and Diabetes | Type 2 diabetes, Kidney disease | (i) Family-based Linkage analysis, and (ii) genetic case-control study using mapping by admixture linkage disequilibrium (MALD) | DNA, serum, urine, whole blood | Yes | 9031 including family dyads and triads for linkage analysis |
| GoKind | The Genetics of Kidneys in Diabetes | Type 1 diabetes, Kidney disease | Genetic case–control study | DNA, Plasma, serum, urine | Yes | 3079 including both singletons and trios |
| HALT-C | The Hepatitis C Antiviral Long-term Treatment against Cirrhosis | Hepatitis C, Liver disease, Cirrhosis | Clinical trial | tissue | Yes | 1145 |
| HEMO | Hemodialysis Study | Kidney disease, Dialysis | Clinical trial | serum | 1846 | |
| IBD | Inflammatory Bowel Disease Genetics | Inflammatory bowel disease, Crohn’ s disease | Genetic case–control study | DNA, serum, whole blood | Yes | 4761 including cases, controls and triosb |
| ICCTG RCT#1 | Interstitial Cystitis Clinical Trail # 1 | Interstitial cystitis, Chronic pelvic pain | Clinical trial (Pilot Study) | urine | 121 | |
| ICCTG RCT#2 | Interstitial Cystitis Clinical Trail # 2 | Interstitial cystitis, Chronic pelvic pain | Clinical trial | urine | 265 randomized | |
| ICDB | Interstitial Cystitis cohort study | Interstitial cystitis, Chronic pelvic pain | tissue | |||
| LTD | Liver Transplantation Database | Liver disease, End stage liver disease | Prospective cohort study | 916 | ||
| LTD2 | Liver Transplantation Database Follow Up | Liver disease, End stage liver disease | Prospective cohort study | 728 survivors of LTD cohort of 916 | ||
| MDRD | Modification of Diet in Renal Disease | Kidney disease | Two clinical trials | buffy coat, plasma, serum, urine | 585 and 255 | |
| MTOPS | Medical Therapy of Prostatic Symptoms | Benign prostatic hyperplasia, Enlarged prostate | Clinical trial | serum | 3047 | |
| NANS | National Analgesic Nephropathy Study | Kidney disease, End-stage renal disease | Case–control study | c | 240 ESRD cases 206 controls | |
| SISTEr | The Stress Incontinence Surgical Treatment Efficacy Trial | Urinary incontinence, Stress urinary incontinence | Clinical trial | 655 | ||
| T1DGC | Type 1 Diabetes Genetics Consortium | Type 1 diabetes | Genetic case–control study | DNA, plasma, serum | Yes | 14 350, including sib-pairs and trios |
| Virahep-C | Viral Resistance to Antiviral Therapy of Chronic Hepatitis C | Hepatitis C, Liver disease | Clinical trial | DNA, PBMC, plasma, RNA, Serum | 401 |
aSource: www2.bsc.gwu.edu/bsc/oneproj.php?pkey=10 (18 July 2011, date last accessed).
bAs of November 2007; Source: www.niddkrepository.org/niddk/jsp/public/IBD/IBDMetadata.jsp (18 July 2011, date last accessed).
cDigitized CT scans of kidneys are available for 207 EDRD subjects and 26 normal cases.
A complete catalog of all of the clinical data sets available from the Repository can be found at https://www.niddkrepository.org/niddk/jsp/public/dataset.jsp
Biospecimens
The Repository houses biospecimens both from studies for which we have clinical data sets and studies that have not yet deposited clinical data sets. As a result, the number and range of Repository biospecimens is substantially greater than those shown in Table 1. In Table 2, we present a tabulation of the different types of biospecimens available from the Repository and the studies that contributed each type of specimen. It will be seen from Table 2 that the Repository offers more than 20 different types of biospecimens with over 5 million samples in storage. The most common biospecimens are serum, plasma, urine, DNA and buffy coat, plus the more than 470 000 stool samples collected by The Environmental Determinants of Diabetes in the Young (TEDDY) study.
Table 2.
Biospecimens currently banked at the NIDDK Repositorya
| Specimenb | Studiesc | No. of Specimens |
|---|---|---|
| Bile | BARC, PALF | 156 |
| Blood | CLiC, CRISP II, DAC, FHN, FSGS/FONT_FONT, FSGS/FONT_FONT II, FSGS/FONT_FSGS, HALT PKD II, HALT PKD I, HBRN, PALF, RIVUR_CUTIE, RIVUR_RIVUR, SIGT, TrialNet_TN07 Oral Insulin | 24 252 |
| Blood, Peripheral Blood Smear | TEDDY | 6924 |
| Buffy coat | AASK_MAIN, AASK_PILOT, CRIC, FAVORIT, MDRD, SIGT, TEDDY | 80 936 |
| Cell Pack DNA | T1DGC | 17 013 |
| Cells | Virahep-C | 74 |
| DNA | DPP, DPT1_Denver, DCCT-EDIC, FIND, IBD, TrialNet, TrialNet_TN01 NH, TrialNet_TN02 MMF/DZB | 131 535 |
| Extracted mRNA | TEDDY | 528 |
| Fibroblasts, skin | PALF | 165 |
| Frozen plasma | AASK_COHORT, AASK_MAIN | 9226 |
| Hair | CKiD | 399 |
| Nail clipping | CKiD, TEDDY | 5512 |
| Nasal swab | TEDDY | 32 748 |
| Peripheral blood mononuclear cells (PBMC) | CDS, CITC_CIT-02, CITC_CIT-03, CITC_CIT-04, CITC_CIT-0501, CITC_CIT-07, CITC_CIT-99, TEDDY, TrialNet, TrialNet_TN01 NH, TrialNet_TN02 MMF/DZB, TrialNet_TN04 T cell assay validation, TrialNet_TN05 antiCD20, TrialNet_TN07 Oral Insulin, TrialNet_TN08 GAD new onset, TrialNet_TN09 CTLA4-Ig, TrialNet_TN12 Metabolic Control, Virahep-C | 50 577 |
| Plasma | AALF-AALI, AALF-AALF, AASK_COHORT, ASSESS-AKI, BARC, CDS, CITC_CIT-02, CITC_CIT-03, CITC_CIT-04, CITC_CIT-0501, CITC_CIT-06, CITC_CIT-07, CITC_CIT-99, CKiD, CLiC, CRIC, CRISP II, CRISP I, DAC, DILIN_Prospective, DILIN_Retrospective, DPP, DPT1_Seattle, FAVORIT, FBEC, FHN, FSGS/FONT_FONT, FSGS/FONT_FONT II, FSGS/FONT_FSGS, GoKind, GpCRC, HALT PKD II, HALT PKD I, HBRN, HFMC, LABS, MDRD, NASH, NASH_NAFLD_A_DB2, NASH_NAFLD_DB, NASH_NAFLD_P_DB2, NASH_PIVENS, NASH_TONIC, PALF, PEDS-C, RIVUR_RIVUR, SIGT, SyNCH PK, SyNCH, T1DGC, TEDDY, Teen-LABS, TrialNet, TrialNet_TN01 NH, TrialNet_TN02 MMF/DZB, TrialNet_TN05 antiCD20, TrialNet_TN07 Oral Insulin, TrialNet_TN08 GAD new onset, TrialNet_TN09 CTLA4-Ig, TrialNet_TN12 Metabolic Control, TrialNet_TN14 Anti-IL-1 Beta, Virahep-C | 1 455 363 |
| Red Blood Cells | FAVORIT, SIGT, TEDDY | 95 669 |
| RNA | CITC_CIT-02, CITC_CIT-03, CITC_CIT-04, CITC_CIT-0501, CITC_CIT-07, CITC_CIT-99, TrialNet_TN01 NH, TrialNet_TN02 MMF/DZB, Virahep-C | 9536 |
| Saliva | TEDDY | 3994 |
| Serum | A2ALL, AALF-AALI, AALF-AALF, AASK_COHORT, AASK_MAIN, AASK_PILOT, ASSESS-AKI, BARC, CAMUS, CDS, CITC_CIT-02, CITC_CIT-03, CITC_CIT-04, CITC_CIT-0501, CITC_CIT-06, CITC_CIT-07, CITC_CIT-99, CKiD, CLiC, CRIC, CRISP II, CRISP I, DAC, DILIN_Prospective, DPT1_Seattle, DPT1_Florida, DPT1_Boston, FAVORIT, FHN, FSGS/FONT_FONT, FSGS/FONT_FONT II, FSGS/FONT_FSGS, GoKind, GpCRC, HALT PKD II, HALT PKD I, HBRN, HEMO, HFMC, IBD, LABS, MDRD, MTOPS, NASH, NASH_NAFLD_A_DB2, NASH_NAFLD_DB, NASH_NAFLD_P_DB2, NASH_PIVENS, NASH_TONIC, PALF, PEDS-C, RIVUR_CUTIE, RIVUR_RIVUR, SyNCH, T1DGC, TEDDY, Teen-LABS, TrialNet, TrialNet_TN01 NH, TrialNet_TN02 MMF/DZB, TrialNet_TN09 CTLA4-Ig, TrialNet_TN14 Anti-IL-1 Beta, Virahep-C | 2 132 215 |
| Stool | TEDDY | 470 863 |
| Stool (PBS washed) | TEDDY | 1770 |
| Tissue | A2ALL, AALF-AALI, AALF-AALF, BARC, CLiC, DILIN_Prospective, HBRN, HFMC, ICDB, NASH, NASH_NAFLD_A_DB2, NASH_NAFLD_DB, NASH_NAFLD_P_DB2, NASH_PIVENS, PALF | 55 785 |
| Urine | AALF-AALI, AALF-AALF, AASK_COHORT, AASK_MAIN, AASK_PILOT, ASSESS-AKI, BARC, CKiD, CLiC, CRISP II, DILIN_Prospective, DCCT-EDIC, FAVORIT, FSGS/FONT_FONT, FSGS/FONT_FONT II, FSGS/FONT_FSGS, GoKind, ICCRN_ICCRN RCT #2, ICCRN_ICCTG RCT#1, LABS, MDRD, MaGIC, PALF, RICE, RIVUR_CUTIE, RIVUR_RIVUR, SyNCH PK, SyNCH, Teen-LABS, UITN_TOMUS, UITN_ValUE, CRIC, CRISP I, MDRD, HALT PKD I, HALT PKD II, AALF-AALF, PALF, HALT PKD I, HALT PKD II, CRISP II, MDRD, CRIC, CRISP I, HALT PKD I, HALT PKD II | 432 315 |
| Whole blood DNA | T1DGC | 326 |
| Whole genome- amplified DNA | T1DGC | 1436 |
| Total | 5 019 951 |
aBiospecimens from some studies are available for sharing now; others will be available in the future. For availability dates, see www.niddkrepository.org/niddkdocs/resources/Sample_Availability_Dates.pdf.
bTable excludes specimens, if N < 10, and a few specimen types of indeterminate status.
cStudies are: A2ALL, Adult Living Donor Liver Transplantation; AALF, Adult Acute Liver Failure Study Group; AASK, The African American Study of Kidney Disease and Hypertension Study; ASSESS-AKI, ASsessment, Serial Evaluation and Subsequent Sequelae in Acute Kidney Injury; BARC, Biliary Atresia Research Consortium; CAMUS, Complementary and Alternative Medicine for Urological Symptoms; CDS, Comprehensive Dialysis Study; CITC, Clinical Islet Transplantation Consortium, substudies; CKiD, Cohort Study of Kidney Disease; CLiC, Longitudinal Study of Genetic Causes of Intrahepatic Cholestasis; CRIC, Chronic Renal Insufficiency Cohort Study; CRISP, Consortium for Radiologic Imaging Studies of PKD, 1 & 2; DAC, Dialysis Access Consortium; DCCT-EDIC, Diabetes Control and Complications Trial and Epidemiology of Diabetes Interventions and Complications follow-up study; DILIN, DILIN 1: Idiosyncratic Liver Injury Associated with Drugs, Prospective and Retrospective; DPP, Diabetes Prevention Program; DPT-1, Diabetes Prevention Trial–Type 1, site specific; FAVORIT, Folic Acid for Vascular Outcome Reduction in Transplantation Trial;
FBEC, Familial Barrett's Esophagus; FHN, Frequent Hemodialysis Network; FSGS/FONT, Focal Segmental Glomerulosclerosis, substudies; GoKind, The Genetics of Kidneys in Diabetes; GpCRC, Gastroparesis Registry; HALT PKD, The Polycystic Kidney Disease Treatment Network, 1 & 2; HBRN, Hepatitis B Research Network; HEMO, Hemodialysis Study; HFMC, Hemodialysis Fistula Maturation Consortium; IBD, Inflammatory Bowel Disease Genetics; ICCRN RCT 1 & 2, Interstitial Cystitis Clinical Resarch Network, Trials 1 & 2; ICDB, Interstitital Cystitis cohort study; LABS, Longitudinal Assessment of Bariatric Surgery; MaGIC, Maryland Genetics of Intersitial Cystitis Study; MDRD, Modification of Diet in Renal Disease; MTOPS, Medical Therapy of Prostatic Symptoms; NASH, Nonalcoholic Steatohepatitis Clinical Research Network, substudies; PALF, Pediatric Acute Liver Failure; PEDS-C, Pegylated Interferon +/– Ribavirin for Children with HCV; RICE, RAND Interstitial Cystitis Epidemiology Study, substudies; RIVUR, Randomized Intervention for Children with VesicoUreteral Reflux;
SIGT, Screening for Impaired Glucose Tolerance; Synch, Silymarin Trial for Hepatitis C and NASH, substudies; T1DGC, Type 1 Diabetes Genetics Consortium; TEDDY, Consortium for Identification of Environmental Triggers of Type 1 Diabetes; Teen-LABS, Adolescent Bariatrics: Assessing Health Benefits & Risks; TrialNet, TrialNet, substudies; UITN, Urinary Incontinence Treatment Network, substudies; Virahep-C, Viral Resistance to Antiviral Therapy of Chronic Hepatitis C.
Use of Repository
Since 2004, the Repository website (http://www.niddkrepository.org) has provided the public with access to details of all studies included in the NIDDK Central Repository, including study summaries, protocols, manuals of operation, data collection forms and lists of publications, available data sets and biospecimens. In addition, the Website allows investigators to apply electronically for access to data and biospecimens. Although the Repository Website provides an efficient and easily accessible portal for obtaining information on archived studies, Repository staff and statisticians frequently provide scientists with additional information prior to formal requests for data or biospecimens. So, for example, a researcher might send the Repository an e-mail saying: ‘I understand that a subset of patients in the Modification of Diet in Renal Disease (MDRD) study had polycystic kidney disease (PKD). How can I obtain information regarding the number of PKD patients in the MDRD database?’ The Repository has responded to numerous such requests for detailed information. Between 2008 and 2010, an average of 28 such requests were received annually via the ‘Ask the Repository’ link on the Repository’s Website. Additional requests were received via the NIDDK telephone help line, the ‘Contact Us’ page of the Repository Website, and by e-mails sent directly to Repository staff.
As of 9 March 2011, a total of 188 external requests for archived data sets and 31 external requests for biospecimens either have been approved or are pending. The number of requests has increased over time as the Repository has become better known in relevant scientific communities. In the Repository’s first 2 years of operation (2003–04), there were no approved data set or biospecimen requests; by 2010 requests had increased to an annual rate of 31.
As Table 3 shows, there has been substantial variation in the popularity of data sets and biospecimens from different studies. The most frequently requested data sets involved studies of type 1 and type 2 diabetes. DCCT/EDIC ranks first in popularity, with 42 approved or pending requests for data and biospecimens from this landmark study of type 1 diabetes. The Diabetes Prevention Program for type 2 diabetes ranks second, with 21 approved or pending requests for data sets. Data sets and biospecimens from the Type 1 Diabetes Genetics Consortium (T1DGC; 20 requests) and Genetics of Kidneys in Diabetes (GoKinD; 13 requests) rank third and seventh, respectively. In addition, the Diabetes Prevention Trial of Type 1 Diabetes (DPT-1) ranks ninth (10 requests). These diabetes studies accounted for almost one-half (106 of 219) of the approved requests for Repository data sets and biospecimens.
Table 3.
Frequency in rank order of approved and pending requests for data sets and biospecimens in NIDDK Data Repository (as of 9 March 2011)
| Rank | Acronym | Study title | Data requests | Specimen requests | Total |
|---|---|---|---|---|---|
| 1 | DCCT/EDIC | Type 1 Diabetes Control and Complications Trial & Epidemiology of Diabetes Interventions and Complications Follow-up | 36 | 6 | 42 |
| 2 | DPP | Diabetes Prevention Program | 21 | 0 | 21 |
| 3 | T1DGC | Type 1 Diabetes Genetics Consortium | 18 | 2 | 20 |
| 4 | MDRD | Modification of Diet in Renal Disease | 18 | 1 | 19 |
| 5 | VIRAHEP-C | Viral Resistance to Antiviral Therapy of Chronic Hepatitis C | 9 | 5 | 14 |
| 6 | HEMO | Hemodialysis Study | 11 | 3 | 14 |
| 7 | GoKinD | Genetics of Kidneys in Diabetes | 10 | 3 | 13 |
| 8 | CRISP | Consortium for Radiological Imaging Studies of Polycystic Kidney Disease | 10 | 3 | 13 |
| 9 | DPT-1 | Diabetes Prevention Trial of Type 1 Diabetes | 8 | 2 | 10 |
| 10 | AASK | The African American Study of Kidney Disease and Hypertension Study | 9 | 0 | 9 |
| 11 | MTOPS | Medical Therapy of Prostatic Symptoms | 8 | 0 | 8 |
| 12 | ICDB | Interstitial Cystitis Data Base | 6 | 1 | 7 |
| 13 | LTD | Liver Transplantation Database | 6 | 0 | 6 |
| 14 | ATN | The Acute Renal Failure Trial Network | 5 | 0 | 5 |
| 15 | IBDGC | Inflammatory Bowel Disease Genetics Consortium | 4 | 0 | 4 |
| 16 | HALT-C | Hepatitis C Antiviral Long-term Treatment against Cirrhosis | 3 | 0 | 3 |
| 17 | LTD-Follow-up | Liver Transplantation Database Follow-up | 2 | 0 | 2 |
| 18 | NANS | National Analgesic Nephropathy Study | 2 | 0 | 2 |
| 19 | AALF | Adult Acute Liver Failure Triala | 0 | 2 | 2 |
| 20 | CKiD | Prospective Cohort Study of Kidney Disease in Children | 0 | 2 | 2 |
| 21 | BACH | Boston Area Community Health Study | 1 | 0 | 1 |
| 22 | PROBE | Prospective Database of Infants with Cholestasis | 0 | 1 | 1 |
| 23 | SISTEr | The Stress Incontinence Surgical Treatment Efficacy Trial | 1 | 0 | 1 |
| Total approved and pending requests | 188 | 31 | 219 |
Tabulation from NIDDK Central Repository Web site on 9 March 2011. The numbers in Table 1 reflect only approved external or pending requests not including requests for NIDDK ancillary studies or internal requests from members of study consortia.
aThe Web site for this study uses the acronym ‘ALF’. We use AALF and PALF to distinguish the adult and pediatric trials.
Studies of renal disease were the second most requested category of data sets and biospecimens. These included the MDRD study (19 requests); the African American Study of Kidney Disease and Hypertension (AASK; nine requests); the Hemodialysis Study (HEMO; 14 requests); the Consortium for Radiologic Imaging Studies of PKD (CRISP; 13 requests); the Acute Renal Failure Trial Network (ATN; five requests); and the National Analgesic Nephropathy Study (NANS; two requests). Studies of liver disease and transplantation were the next most requested data sets and biospecimens (Table 3).
In addition to the aforementioned requests from external researchers, the Repository also supports ancillary research by investigators participating in the original study group or collaborating with them who wish to use archived biospecimens to address research questions beyond the funded scope of the original study. As of 9 March 2011, 113 requests have been approved or are pending to provide biospecimens for such ancillary studies.
Sharing non-renewable resources
While digital data sets can be copied ad infinitum, some biospecimens stored in the Repository are not renewable. This creates unique challenges. In January 2010, NIDDK issued a program announcement (PAR-10-090) that was ‘intended to facilitate equitable and appropriate distribution of biosamples from the NIDDK Central Repositories.’ Investigators requesting nonrenewable biospecimens are required to consult with the Repository to determine whether a sufficient quantity of the samples is available and whether the proposed use of the biospecimens is consistent with the informed consent used in the research study. Investigators seeking nonrenewable biospecimens from the Repository are then required to submit an application describing ‘the background and rationale for request; a list of specific objectives; detailed information about the proposed studies; detailed information about the amount and type of samples needed and documentation from the Repository confirming that samples are available; plans for sample management; a description of follow-up plans.’ Requestors are also required to ‘explain how the proposed research will take advantage of the large amount of associated phenotypic data.’
Cost
Maintaining repositories of data and biospecimens is not cheap, but their costs pale in comparison to the costs of original data collection. From 2003 to 2013, NIDDK will spend a total of approximately $73 million for the NIDDK Repositories (1). Costs are most expensive for archiving biospecimens ($28 million) and genetic samples ($33 million), while data archiving is less expensive ($12 million). The costs for acquisition of biosamples has ranged from ∼$0.70 to $7 per tube while production of DNA or a cell line and DNA have ranged from ∼$70 to $800. Maintaining these samples in the Repository has cost ∼$0.01 per tube per year for biosamples and $10 to $16 per cell line per year.
The cost of the original data collection is, however, much more expensive. The DCCT-EDIC, for example, has cost more than $200 million since its inception, while the archiving and distribution costs for genetic samples and immortalized cell lines, biospecimens and multiple data sets have been less than $3 million.
Expectations for future use
The NIDDK Central Repository was established to improve the scientific yield of NIDDK-funded research by making valuable data and specimens available to the wider scientific community. At present, the Repository is being used by a widening community of researchers, and it is also providing valuable archival services for the original research teams. We expect that the use of the NIDDK Central Repository should increase not only with growing awareness of its resources by the scientific community but also with the issuance of RFAs for research that can effectively use this resource. So, for example, NIDDK solicited grant applications in 2009 to form a multicenter consortium to ‘discover or validate biomarkers for well-defined human chronic kidney diseases (CKD) (RFA-DK-08-015).’ Discovery and testing of candidate biomarkers requires biological samples (tissues, cells, or body fluids) from subjects whose disease status has been well characterized. As the RFA notes, the NIDDK Central Repository can provide the resources needed for such research.
Examples of Repository’s impact on biomedical research
Repository resources have supported a range of biochemical, clinical, statistical and genetic research. Genetic research has included GWAS, validation studies, studies of Mendelian disease inheritance patterns, studies of genotype–phenotype correlations, development of methods to improve statistical power of GWAS, and testing of new statistical methods for genetic research. This research was spurred by investigators who responded to the 2006 NIDDK request for applications (RFA-DK-06-005) for ‘applications that implement large-scale studies and innovative analytical designs using samples from EDIC or GoKinD (or both) to identify genes and even specific genetic variants that confer susceptibility or resistance to diabetic complications’.
In addition to facilitating new genetic and biochemical research using extant biospecimens, the Repository offers important opportunities for clinical research to scientists who were not members of the original study teams. They can request data sets from the Repository to both explore new and extend prior clinical research. Such ‘secondary analyses’ serve many important scientific purposes (2), including insuring efficient use of clinical data produced by studies that required a large investment of funds and effort, facilitating replication and extension of the analyses of the original investigators, and providing a ready resource for inexpensive testing of hypotheses not incorporated in the original study. The latter benefit can be particularly valuable because it can allow research advances without the immediate need for new data collection. Such uses can also provide pilot results that will motivate new studies—or they may dissuade investigators from pursuing an unpromising line of future research. By lowering the cost of entry into a research area, secondary analyses of archived data can be particularly valuable to junior scientists and others without resources for primary data collection.
NIH mandates data sharing (3). The Repository supports that mandate by providing a vehicle for researchers to access curated and well-maintained archival data sets and biospecimens and by assisting requestors seeking to understand these data and specimens. Below we provide a few examples of biomedical research that has used the Repository’s resources.
Statistical re-analyses
Using EDIC data archived in the Repository and DCCT data made available to the public prior to establishment of the Repository, Kilpatrick and colleagues have published nine articles that replicate and explore possible extensions to work reported by the original DCCT/EDIC investigators (4–12). The conclusions reached by these investigators include that:
Blood glucose variability (within and between days) does not predict the development of retinopathy or nephropathy in type 1 diabetics when mean blood glucose is accounted for (7, 8, 11). Longer-term fluctuations in HbA1c, however, may contribute to these risks (8).
In addition to HbA1C, mean blood glucose and within-day blood glucose variability are associated with risk of hypoglycemia (12).
Mean blood glucose is a better predictor of cardiovascular risk than HbA1c (5).
The relationship between mean plasma glucose levels and HbA1c is not constant. In the DCCT study, subjects in the conventional treatment condition had consistently higher mean plasma glucose levels than intensively treated patients at any given level of HbA1c (9).
Higher levels of insulin resistance (estimated glucose disposal rate; eGDR) at baseline in DCCT was predictive of increased risk of retinopathy, nephropathy and cardiovascular complications (10).
Without passing judgment on the relative merits of arguments about these conclusions, we note that the secondary analyses of Kilpatrick and colleagues provided examples of some of the expected benefits of data sharing. First, as acknowledged by Kilpatrick himself, the availability of archived data—among other factors—means that ‘large grant application success is not always required to perform meaningful research in [clinical biochemistry]’ (13; p. 28). None of the DCCT/EDIC articles published by Kilpatrick and colleagues prior to 2009 reported external funding. Second, these new analyses of archive data provoked productive (if sometimes testy) scientific debate (11, 14–20) as well as re-examination of the original statistical analyses (21).
Biochemical analyses
The NIDDK Central Repository’s biospecimens have been used for a variety of biochemical studies including research in lipidomics, metabolomics and chemoenzymatic analysis. Ding and colleagues (22), for example, used biospecimens from the NIDDK Central Repository to apply an accurate mass and time (AMT) tag approach for a lipidomics analysis on the plasma, erythrocyte and lymphocyte samples obtained in the Screening for Impaired Glucose Tolerance (SIGT) project (www.med.emory.edu/research/GCRC/SIGT). Ding and colleagues’ study concluded that the AMT tag approach was able to create lipid profiles in different sample types and detect ‘qualitative and quantitative differences in lipid abundance.’
Genetic research
Nancy Cox, Andrzej Krolewski and Andrew Paterson were funded under the 2006 RFA and have published a wide range of findings. Using DCCT/EDIC and GoKinD clinical and genetic data, they have conducted a series of GWAS. They have, for example,
used the DCCT/EDIC sample to discover a major locus near SORCS1 that was associated with HbA1c and mean glucose levels in the conventional treatment condition (23);
found that multiple variations in SOD1 are associated with microalbuminuria and serious nephropathy in DCCT/EDIC subjects (type 1 diabetics) (24);
found two new loci in UBASH3A and BACH2 that were associated with type 1 diabetes (25);
found ELMO1 locus that predicted susceptibility to diabetic nephropathy in type 1 diabetes mellitus in the GoKinD study cohort of 820 type 1 diabetes subjects and 885 control subjects and in 1,304 DCCT/EDIC subjects (26, 27);
found two loci associated with diabetic nephropathy in both mice and humans (28); and
contrary to published results for type 2 diabetes, found no association between diabetic nephropathy and ‘D18S880 microsatellite and other polymorphisms of the CNDP2-CNDP1 region’ (29).
Increasing statistical power
The Repository has provided the opportunity for both the combination of samples to increase statistical power and for the development and testing of new statistical methods. Barrett and colleagues (30), for example, combined two previously published genome-wide association analyses of type 1 diabetes involving 1601 cases from the NIDDK GoKinD study; 1704 controls from the National Institute of Mental Health (NIMH) study (31); and 5272 cases and controls from the Wellcome Trust Case Control Consortium (WTCCC) Study (32), along with their own 7982 cases and controls from the NIDDK T1DGC study. Combining these studies provided improved statistical power enabling the authors to identify more than 40 loci associated with type 1 diabetes—with 27 newly identified regions—after excluding previously reported associations.
Lessons learned as Repository evolved
Many lessons have been learned in the 8 years of Repository operation. We offer below four important lessons that may be of benefit to others who undertake similar efforts. These lessons involve: the folly of overly ambitious and complex database designs, the need to regularly remind coordinating centers of the need to scrupulously maintain and archive linkage files, the benefits of planning in advance to link study data to the consent documents that specify how these data may be used, and the value of well-conducted data set integrity checks.
Ambition and complexity
It became clear in the first months of the Repository’s life that our initial plan was overly ambitious, complex and expensive. Maintaining the archival data (the data that is distributed) in a relational database for flexible processing was both expensive and unnecessary. If this level of flexibility were needed, it can be readily and (relatively) inexpensively handled by maintaining a database of metadata that is derived from the archived study data.
Linkage files
Clinical studies typically use one set of subject IDs for internal study purposes, and—as a privacy precaution—create ‘masked’ IDs when depositing data with the Repository. While Data Coordinating Centers (DCCs) maintain ‘linkage files’ identifying which study biospecimen IDs belong to which study subject IDs, the shared data need an additional linkage file that allows these biospecimen IDs to be linked to the ‘masked’ IDs. Early in the operations of the Repository we discovered that some study DCCs did not include such linkage files with the study documentation when they archived data and biospecimens with the Repository. The Repository PI and staff undertook a campaign to remind extant and new biospecimen depositors of the crucial need for accurate and well maintained linkage files to be deposited along with their biospecimens.
Database of consent documents
Study consent documents are generated by methods that make them awkward to automate. Typically, they may vary by study, clinical site, study subpopulation and time interval and different restrictions may apply to different uses of the data or biospecimens (e.g. only for use in diabetes research). These consent documents are nonetheless crucial to Repository operations since they specify the conditions under which data and biospecimens from a study may be released.
Inadequate attention was given in Repository planning to the need for a database of subject consent forms for each study. At the outset of Repository operations, consent forms were on file with the sample collection institutions as well as the NIDDK funding office, but the Repository staff did not have direct access to these consent forms. In order to have accountability for data and sample distribution, the Repository began requesting copies of paper consent forms from NIDDK. However, storage and retrieval of more than 10,000 multi-page paper consent forms was problematic. The Repository ultimately created a standalone database in which to store, upload and retrieve subject consent forms for each study. This consent form database includes specific study and site information for each consent form, disease states and other critical data which are searchable—plus a PDF of the paper consent forms. This database allows secure access to consent forms by Repository staff and the NIDDK funding office, and it helps ensure that only samples and data which were ‘approved for sharing’ and approved for particular ‘types’ of research are shared.
Development of the consent database required a sustained effort during normal Repository operations to separate, scan and assign filenames for each paper consent form by study and collection site, and then to enter into the database the relevant data from each consent form including; approval and expiration dates, disease state(s), exceptions to sharing, plus ‘approved only for specific research’ and ‘not approved for genetic research’ restrictions. This was hardly an optimal solution. If the need for such a consent database had been better anticipated, we would have conducted a comprehensive review of the information and design requirements for a consent database immediately upon award of the Repository contract. A ‘consent forms database’ would then have been developed in conjunction with the data and biospecimen databases. The resultant consent forms database would have been co-located in the main database and accessible alongside of and linked to the sample data instead of adjacent to the sample data.
Data set integrity checks
As a partial check of the integrity of the data sets archived in the NIDDK data Repository, prior to data release, we perform a set of tabulations and statistical analyses to verify that published results from the study can be reproduced using our archived data sets. The intent of these data set integrity checks is to provide confidence that the data sets distributed by the NIDDK Repository is a true copy of the study data. These analyses have helped us avoid serious problems including, for example, distribution of data sets that were missing a sizable number of cases and distribution of data sets that included subsamples of subjects who had refused consent for data distribution beyond the original study team.
Future Repository enhancements
We anticipate that the future impact on disease research of the studies archived in the Repository will be enhanced by: (i) cross-listing of Repository biospecimens in other searchable databases; (ii) roll out of a suite of applications for querying the contents of the Repository; and (iii) over time, an increased harmonization of procedures, data collection strategies, questionnaires etc. across both research studies and within the vocabularies used by different repositories (see e.g. the DataSHaPER tools for harmonization developed by the P3G network; see www.datashaper.org/).
Cross-listing of resources
To make the Repository resources visible to a broad user community, our available biospecimens are listed in the catalogs of other biobanks. Currently, we list approximately 500 000 biospecimens of six sample types for five diseases in the NIH Office of Rare Diseases Research Biospecimens/Biorepositories Rare Disease-HUB (RD-HUB). Biospecimens from four studies of renal diseases (total of 6855 subjects) are listed in the P3G Renal Biobank and biospecimens from one diabetes study (3075 subjects) are listed in the P3G Diabetes Biobank. The biospecimen resources of these partner biobanks are also cross-listed at the Repository Website under ‘Related Websites’ (see www.niddkrepository.org/niddk/jsp/public/websites.jsp). The Repository plans to expand our efforts to cross-list study biospecimens in a wide range of biobanks catalogs.
We are also in the process of registering the Repository as a biobank within the Common Biorepository Model (CBM) network (see: cabig.nci.nih.gov/workspaces/TBPT/CBM/). This will permit the NIDDK Central Repository to be accessible using the NCI Specimen Resource Locator (SRL)—a service that allows researchers to locate human biospecimens (tissue, serum, DNA/RNA, other specimens) for their research.
Query tools
The need for adequate tools to search the expanding contents of the NIDDK Repository was recognized in our initial proposal. The simplification of the Repository’s design at its birth required both a different suite of search tools and more time to understand our users’ needs and to develop the require tools. The slow accretion of studies in the archive’s early years also diminished the urgency of the need for such tools. Below we briefly describe both our initial plan and the search tools that are currently being rolled out.
2002 plan
We initially planned to establish cross–reference relationships between specific fields from multiple study databases, and to create translation tables to standardize similar field values into a single code and description. These translation tables would have been separate from the study tables and might be created using data dictionaries and/or code books. The following tables are an example of the planned translation tables:
tblCategory—general category area of interest
tblStandardText—specific standardized text under a general category
tblTranslation—creates a relationship between tblStandardData and specific fields in study tables
These translation tables would standardize the criteria used in the search requests on similar fields across all study databases. This methodology would also eliminate the need to know the synonyms for similar fields across studies. Where possible, we anticipated that semantically equivalent fields in different databases would be identified in advance of any data requests on study databases. We expected that all finished databases in the Repository would be reviewed to identify fields that match existing relationships. As the translation tables grow, we expected this search and cross–reference capacity of the search interface will increase.
This planned search strategy was abandoned when we choose to simplify the Repository design (see ‘Revision of Design’ section).
Current query tools
To provide a search capability for the current Repository, we are rolling out a suite of applications referred to as the public query tool (PQT). To provide greater flexibility and enhance searching capabilities for the user, we developed a series of publicly accessible query tools whose main intent was to address the question, ‘What’s in the NIDDK Central Data Repository (CDR)?’. The PQT provides public viewers/users of the contents of the CDR with an easy to use interface that supports a wide variety of user interests (e.g. what studies have family history data for Type I diabetes and/or contain a minimum of 150 African-American subjects older than 50 years of age). The PQT includes four distinct search engine tools.
The first tool—the Keyword Metadata Search tool—allows users to select keywords from drop down menus that identify the studies with those specific features. The keywords are obtained from study specific metadata examples of which include diagnosis and type of study. The tool searches the metadata to define studies that link to the keywords. Users who are not familiar can quickly identify studies with a variety of useful properties. No specific knowledge of the studies is required to use this tool which is currently available on the website.
The second tool—the Ontology based keyword search engine—uses study variables that have been identified as scientifically important. To support this and the other tools below, variables of scientific interest have been extracted from the data archive (into a curated database) and can be accessed by the tools. In the case of the Ontology tool, it is designed to search ‘free text’ keywords provided by the user as contrasted with structured text from ‘drop down’ controls used by the basic search tool. The user supplied key words will link to an ontology that has been mapped to the curated database. The keywords will use the mappings to identify studies that exhibit the traits implied by the keywords.
The third tool—codebook variable engine—will allow a user to highlight a study and the important variables that have been included in the curated database. Each variable included in the list can be ‘clicked’ to generate a variable description and an associated set of frequencies.
The fourth tool—the crosstab tool—will allows users to obtain crosstabulations both within and across specified studies. Such crosstabulations will allow users to identify, for example, studies that have 35 or more African American subjects that survived liver transplants for a minimum of 5 years; or to learn before requesting study data whether a given study has at least 50 subjects between the ages of 40 and 60 with fasting glucose or 140 mg/dl or higher.
Our tools are intended to represent three perspectives:
A Study perspective that identifies specific traits that identify the purpose of the study, its principal findings and the main design elements. These elements would include design elements and/or treatment features that might provide insights for the design of new studies, using existing studies as a starting point.
The disease domain perspective identifies data across a variety of clinical sources that present a unified view of individual patients within a specific disease domain that have different protocols. This user is interested in viewing studies administered by different protocols that are about the same disease type. From this perspective data from multiple studies are linked and pooled (if possible) for a reexamination of the underlying and undiscovered properties related to a disease and the treatment of that disease. The data represented by his perspective will identify features of the severity disease. These variables may include serum creatinine levels (for the kidney disease domain), disease confounders (e.g. blood pressure and age, diet and lifestyle) and primary disease-related outcomes from multiple studies within a given disease domain. The potential for linking multiple studies with common data elements within a disease domain is an important feature from the perspective of this user.
A common data element perspective that uses data with broad level attributes for the purpose of comparing all types of clinical studies from a common set of measures (i.e. age, gender, diagnosis does genotype data exist? Is medical history available?). This level will include data elements defined by the NCI Common Biorepository Model (CBM). There are 30 variables in the CBM (cabig.nci.nih.gov/workspaces/TBPT/CBM/). We will include all 30 variables for each NIDDK study in the Repository. All CBM variables will be harmonized to a standard set of ontologies that are included in the Cancer Biomedical Informatics Grid (caBIG) (see cabig.nci.nih.gov/workspaces/VCDE).
Increased harmonization
While good query tools are extremely helpful, there is no substitute for the use of a universal set of standards during the study design phase that incorporates a standard vocabulary and nomenclature into the design process. Potentially useful coding systems include:
Logical Observation Identifiers Name and Code (LOINC) used in diagnostic reports, survey instruments, laboratory tests and clinical measurements (loinc.org/) and
Systemized Nomenclature of Medicine—Clinical Terms (SNOMED) used to assign codes for organisms, anatomic parts, specimens, diagnoses and symptoms (www.nlm.nih.gov/research/umls/Snomed/snomed_main.html).
However, most legacy studies have not incorporated such standards into their design. Considerable efforts are under way to standardize both procedures and terminology in biomedical research, with special attention to studies that will provide data and biospecimens for secondary analysis. The ability to pool data is crucially dependent on the equivalence of research methods used to obtain and store data and biospecimens. The ability to discover common data elements across studies, in turn, depends upon the use of a standard vocabulary or the development of automated thesauruses that permit identification of potentially equivalent measurements or specimens. Standardizing procedures and terminology will provide important benefits, but standardizing variable measurements will be a major endeavor that will require both substantial time and resources to complete. Such harmonization efforts are, however, crucial to increasing the usage and realizing the full scientific value of the NIDDK Central Repository and other data and biobanks. Current efforts by others include the P3G DataSHaPER (33), Phoenix (34, 35) and the CBM (cabig.nci.nih.gov/workspaces/TBPT/CBM/).
Conclusion
The NIDDK Central Repository was established to increase the impact of valuable data and biospecimens by making these materials available to the broader scientific community. The available evidence suggests that the Repository is beginning to fulfill this promise. Development of new bioinformatic tools to query the availability of data or biospecimens within the Repository together with the expanding reputation of the Repository and ongoing harmonization efforts should lead to increased use of this valuable resource.
Funding
National Institute of Diabetes and Digestive and Kidney Diseases; National Institutes of Health, Department of Health and Human Services, under Contracts (HHSN: 267200800015C, 267200800016C and 267200800018C). Funding for open access charge: HHSN267200800016C.
Conflict of interest. None declared.
Acknowledgements
This article draws upon material included in (i) RTI International’s Technical Proposals for the Contracts for Initial Award (RFP NIH-NIDDK-02-04) and Continuation of Funding (RFP NIH-NIDDK-07-07) for the NIDDK Central Repository; (ii) an RTI contract modification proposal submitted in February of 2009; (iii) descriptions of Repository content and procedures posted on current and previous versions of the Repository Website (www.niddkrepository.org/niddk/jsp/public/dataset.jsp), and (iv) a presentation on the Repository by Rasooly, Eggers, et al. (1) at the 2011 meetings of the International Society for Biological and Environmental Repositories. With the exception of this note, we do not identify text that excerpts or draws upon these sources.
References
- 1.Rasooly RS, Eggers PS, Akolkar B, Karp R. The NIDDK Central Repositories: An NIH resource for research on diabetes, endocrine, digestive, liver, kidney, and urogenbital diseases. (Abstract of presentation at 2011 Annual Meeting of International Society for Biological and Environmental Repositories.) Biopreserv Biobanking. 2011;9:1. [Google Scholar]
- 2.Fienberg SE, Martin ME, Straf ML, et al. Sharing Research Data. Washington D.C: National Academy Press; 1985. [Google Scholar]
- 3.National Institutes of Health. Final NIH statement on sharing research data. Notice: NOT-OD-03-032, released date: 26 February 2003. grants.nih.gov/grants/guide/notice-files/NOT-OD-03-032.html (2 March 2010, date last accessed) [Google Scholar]
- 4.Kilpatrick ES, Rigby AS, Atkin SL. The relationship between mean glucose and HbA1c in premenopausal women compared with males in the Diabetes Control and Complications Trial. Diabet. Med. 2008;25:112–113. doi: 10.1111/j.1464-5491.2007.02305.x. [DOI] [PubMed] [Google Scholar]
- 5.Kilpatrick ES, Rigby AS, Atkin SL. Mean blood glucose compared with HbA1c in the prediction of cardiovascular disease in patients with type 1 diabetes. Diabetologia. 2008;51:365–371. doi: 10.1007/s00125-007-0883-x. [DOI] [PubMed] [Google Scholar]
- 6.Kilpatrick ES, Rigby AS, Atkin SL. The diabetes control and complications trial: the gift that keeps giving. Nat. Rev. Endocrinol. 2009;5:537–545. doi: 10.1038/nrendo.2009.179. [DOI] [PubMed] [Google Scholar]
- 7.Kilpatrick ES, Rigby AS, Atkin SL. Effect of glucose variability on the long-term risk of microvascular complications in type 1 diabetes. Diabetes Care. 2009;32:1901–1903. doi: 10.2337/dc09-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilpatrick ES, Rigby AS, Atkin SL. A1C variability and the risk of microvascular complications in type 1 diabetes: data from the Diabetes Control and Complications Trial. Diabetes Care. 2008;31:2198–2202. doi: 10.2337/dc08-0864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilpatrick ES, Rigby AS, Atkin SL. Variability in the relationship between mean plasma glucose and HbA1c: implications for the assessment of glycemic control. Clin. Chem. 2007;53:897–901. doi: 10.1373/clinchem.2006.079756. [DOI] [PubMed] [Google Scholar]
- 10.Kilpatrick ES, Rigby AS, Atkin SL. Insulin resistance, the metabolic syndrome, and complication risk in type 1 diabetes: ‘double diabetes’ in the Diabetes Control and Complications Trial. Diabetes Care. 2007;30:707–712. doi: 10.2337/dc06-1982. [DOI] [PubMed] [Google Scholar]
- 11.Kilpatrick ES, Rigby AS, Atkin SL. The effect of glucose variability on the risk of microvascular complications in type 1 diabetes. Diabetes Care. 2006;29:1486–1490. doi: 10.2337/dc06-0293. [DOI] [PubMed] [Google Scholar]
- 12.Kilpatrick ES, Rigby AS, Goode K, et al. Relating mean blood glucose and glucose variability to the risk of multiple episodes of hypoglycaemia in type 1 diabetes. Diabetologia. 2007;50:2553–2561. doi: 10.1007/s00125-007-0820-z. [DOI] [PubMed] [Google Scholar]
- 13.Kilpatrick ES. The hitchhiker's guide to research in clinical biochemistry. Clin. Biochem. Rev. 2010;31:25–28. [PMC free article] [PubMed] [Google Scholar]
- 14.Bolli GB. Glucose variability and complications. Diabetes Care. 2006;29:1707–1709. doi: 10.2337/dc06-0716. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch IB, Brownlee M. The effect of glucose variability on the risk of microvascular complications in type 1 diabetes. Diabetes Care. 2007;30:186–187. doi: 10.2337/dc06-1610. author reply 188–189. [DOI] [PubMed] [Google Scholar]
- 16.Monnier L, Colette C, Leiter L, et al. The effect of glucose variability on the risk of microvascular complications in type 1 diabetes. Diabetes Care. 2007;30:185–186. doi: 10.2337/dc06-1594. author reply 187–188. [DOI] [PubMed] [Google Scholar]
- 17.Diabetes Research in Children Network Study Group. The effect of glucose variability on the risk of microvascular complications in type 1 diabetes. Diabetes Care. 2007;30 185; author reply 187–189. [Google Scholar]
- 18.Service FJ, O'Brien PC. The effect of glucose variability on the risk of microvascular complications in type 1 diabetes. Diabetes Care. 2007;30 doi: 10.2337/dc06-1782. 186; author reply 187–188. [DOI] [PubMed] [Google Scholar]
- 19.Kilpatrick ES, Rigby AS, Atkin SL. The effects of glucose variability on the risk of microvascular complications in Type 1 diabetes. Diabetes Care. 2007;30:2. doi: 10.2337/dc06-0293. [DOI] [PubMed] [Google Scholar]
- 20.Bolli GB, Gerstein HC, Rosenstock J. The effects of glucose variability on the risk of microvascular complications in Type 1 diabetes. [Letter to Editor] Diabetes Care. 2007;30:1. [Google Scholar]
- 21.Lachin JM, Genuth S, Nathan DM, et al. Effect of glycemic exposure on the risk of microvascular complications in the diabetes control and complications trial–revisited. Diabetes. 2008;57:995–1001. doi: 10.2337/db07-1618. [DOI] [PubMed] [Google Scholar]
- 22.Ding J, Sorensen CM, Jaitly N, et al. Application of the accurate mass and time tag approach in studies of the human blood lipidome. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008;871:243–252. doi: 10.1016/j.jchromb.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paterson AD, Waggott D, Boright AP, et al. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes. 2010;59:539–549. doi: 10.2337/db09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Kateb H, Boright AP, Mirea L, et al. Multiple superoxide dismutase 1/splicing factor serine alanine 15 variants are associated with the development and progression of diabetic nephropathy: the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Genetics study. Diabetes. 2008;57:218–228. doi: 10.2337/db07-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant SF, Qu HQ, Bradfield JP, et al. Follow-up analysis of genome-wide association data identifies novel loci for type 1 diabetes. Diabetes. 2009;58:290–295. doi: 10.2337/db08-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pezzolesi MG, Katavetin P, Kure M, et al. Confirmation of genetic associations at ELMO1 in the GoKinD collection supports its role as a susceptibility gene in diabetic nephropathy. Diabetes. 2009;58:2698–2702. doi: 10.2337/db09-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pezzolesi MG, Poznik GD, Mychaleckyj JC, et al. Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009;58:1403–1410. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsaih SW, Pezzolesi MG, Yuan R, et al. Genetic analysis of albuminuria in aging mice and concordance with loci for human diabetic nephropathy found in a genome-wide association scan. Kidney Int. 2010;77:201–210. doi: 10.1038/ki.2009.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wanic K, Placha G, Dunn J, et al. Exclusion of polymorphisms in carnosinase genes (CNDP1 and CNDP2) as a cause of diabetic nephropathy in type 1 diabetes: results of large case-control and follow-up studies. Diabetes. 2008;57:2547–2551. doi: 10.2337/db07-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller PW, Rogus JJ, Cleary PA, et al. Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J. Am. Soc. Nephrol. 2006;17:1782–1790. doi: 10.1681/ASN.2005080822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fortier I, Burton PR, Robson PJ, et al. Quality, quantity and harmony: the DataSHaPER approach to integrating data across bioclinical studies. Int. J. Epidemiol. 2010;39:1383–1393. doi: 10.1093/ije/dyq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stover PJ, Harlan WR, Hammond JA, et al. PhenX: a toolkit for interdisciplinary genetics research. Curr. Opin. Lipidol. 2010;21:136–140. doi: 10.1097/MOL.0b013e3283377395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamilton CM, Strader LC, Pratt JG, et al. The PhenX toolkit: get the most from your measures. Am. J. Epidemiol. 2011;174:253–260. doi: 10.1093/aje/kwr193. [DOI] [PMC free article] [PubMed] [Google Scholar]