Abstract
TGF-β signalling is regulated by post-translational modifications of Smad proteins to translate quantitative difference in ligand concentration into proportional transcriptional output. Previous studies in cell culture systems suggested that Smad ubiquitination regulatory factors (Smurfs) act in this regulation by targeting Smads for proteasomal degradation, but whether this mechanism operates under physiological conditions is not clear. Here, we generated mice harbouring a target-disrupted Smurf2 allele. Using primary mouse embryonic fibroblasts and dermal fibroblasts, we show that TGF-β-mediated, Smad-dependent transcriptional responses are elevated in the absence of Smurf2. Instead of promoting poly-ubiquitination and degradation, we show that Smurf2 actually induces multiple mono-ubiquitination of Smad3 in vivo. Phosphorylation of T179, immediately upstream of the Smad3 PY motif, enhances Smurf2 and Smad3 interaction and Smad3 ubiquitination. We have mapped Smurf2-induced Smad3 ubiquitination sites to lysine residues at the MH2 domain, and demonstrate that Smad3 ubiquitination inhibits the formation of Smad3 complexes. Thus, our data support a model in which Smurf2 negatively regulates TGF-β signalling by attenuating the activity of Smad3 rather than promoting its degradation.
Keywords: Smad3, Smurf2, TGF-β, ubiquitination
Introduction
TGF-β is the founding member of a large family of cytokines and developmental morphogens that regulate a wide array of cellular processes ranging from specification of developmental fate during embryogenesis to tissue homeostasis in the adult (Derynck and Miyazono, 2008; Wu and Hill, 2009). A detailed depiction of TGF-β signalling mechanisms will have broad significance in understanding human development as well as pathogenesis of major human diseases, including cancer, fibrosis and autoimmunity.
In the current paradigm, TGF-β and its related factors signal through a complex of membrane-bound kinase receptors, which induce phosphorylation of Smad2 and Smad3 in the TGF-β/activin pathway or Smad1, Smad5, and Smad8 in the BMP pathway at two serine residues in the C-terminus (Shi and Massague, 2003; Feng and Derynck, 2005). The C-terminal phosphorylation creates an interacting interface that enables these receptor-activated Smads to form either homomeric complexes or heteromeric complexes with Smad4, which is common to all ligand-specific pathways in the TGF-β superfamily. The activated Smads are then dissociated from the membrane-bound receptors and become accumulated in the nucleus, where they regulate a wide range of transcriptional responses in cooperation with other transcription factors in various cellular contexts. A range of other intracellular signalling pathways, including different MAPKs, RhoA, and PI3K/AKT, can also be activated by the TGF-β receptors in non-canonical, Smad-independent manners (Zhang, 2009). It is established that the duration and strength of Smad-dependent TGF-β signal are determined by the level of R-Smads in the nucleus, which is maintained by a delicate balance of influx and efflux (Schmierer et al, 2008). In both resting and activated states, Smad proteins continuously shuttle between the nucleus and cytoplasm. The nuclear accumulation of Smads induced by ligand/receptor complexes is the consequence of an imbalance in this shuttling movement, which is regulated by phosphorylation at the C-terminus and formation of protein complex (Clarke et al, 2006; Schmierer et al, 2008; Varelas et al, 2008). Accumulating evidence indicates that after prolonged exposure to ligands, TGF-β receptors and Smads become ubiquitinated, and a number of ubiquitin E3 ligases that mediate this post-translational modification have been identified (Lonn et al, 2009). At least in cultured cells, ubiquitination of Smads leads to degradation in proteasomes, which may affect their nuclear levels (Lo and Massague, 1999; Zhu et al, 1999; Lin et al, 2000; Fukuchi et al, 2001; Zhang et al, 2001). However, there is compelling evidence that the bulk of Smad2 and Smad3, which accumulate in the nucleus during TGF-β signalling, are not degraded there, but rather are exported out of the nucleus upon dephosphorylation and dissociation of the Smad complexes (Inman et al, 2002; Lin et al, 2006).
Ubiquitination is a major form of protein post-translational modification that regulates essentially all aspects of cellular functions (Hershko and Ciechanover, 1998). In addition to marking proteins for degradation with poly-ubiquitin chains linked via Lys48, ubiquitination also leads directly to cell signalling when protein substrates are modified by other types of poly-ubiquitin chains via Lys63 or singular ubiquitin moiety on one (mono) or multiple (multi) Lys residues (Haglund and Dikic, 2005). Regardless of the linkage type or length, the transfer of ubiquitin moieties to protein substrates is carried out by sequential actions of the ubiquitin-activating enzyme (E1), ubiquitin conjugase (E2), and ubiquitin ligase (E3). The E3 ubiquitin ligase, which makes a direct physical contact with substrates and plays a crucial role in substrate selection, is of the most importance in this chain of reactions. Hundreds of E3 ubiquitin ligases are estimated to be encoded by a typical mammalian genome and their identities and functions are highly diverse, but mechanistically all E3 ligases can be classified into one of two categories: RING domain-bearing E3s that act as scaffolds to enhance ubiquitin transfer from an E2 to a substrate and HECT-domain-containing E3s that catalyse ubiquitin transfer via a thioester intermediate formed on a conserved cysteine residue in the enzyme.
Smad ubiquitin regulatory factors (Smurfs) are HECT-domain-containing E3 ligases. Two highly related Smurfs exist in vertebrates, Smurf1 and Smurf2 (Zhu et al, 1999; Kavsak et al, 2000; Lin et al, 2000; Zhang et al, 2001), which share 80% amino-acid sequence identity and several distinctive structural features including a phospholipid/calcium-binding C2 domain at the amino terminus, two to three copies of WW repeats that mediate interactions with PY motifs of substrate proteins, and a catalytic HECT-domain at the carboxyl terminus. Both Smurf1 and Smurf2 have the ability to interact directly with Smad1 and Smad5, whereas Smurf2 was also reported to interact with Smad2 and Smad3 (Zhu et al, 1999; Lin et al, 2000; Zhang et al, 2001). In addition, Smurf1 and Smurf2 can target the activated TGF-β type I receptor for degradation through binding to Smad7, an inhibitory Smad, thereby turning off receptor signalling and decreasing the nuclear level of R-Smads (Kavsak et al, 2000; Ebisawa et al, 2001). Besides Smurfs, several other HECT E3 ligases, such as WWP1, Itch, and Nedd4L, are also implicated in Smad ubiquitination (Bai et al, 2004; Komuro et al, 2004; Seo et al, 2004; Kuratomi et al, 2005; Gao et al, 2009).
To address the functions of Smurf2 in a physiological setting, we generated a strain of Smurf2-deficient mice through gene targeting. These mice are viable and grow up healthy to adulthood. We observed on rare occasions a few Smurf2-deficient mice displaying phenotypes related to defects in planar cell polarity, but focused our investigation on defining the role of Smurf2 in TGF-β signalling. Here, we report that Smurf2 negatively regulates TGF-β/Smad signalling by blocking the formation of homotrimeric or heterotrimeric Smad3 complexes via multiple mono-ubiquitination of Smad3 instead of marking it for degradation.
Results
Targeted disruption of the Smurf2 gene
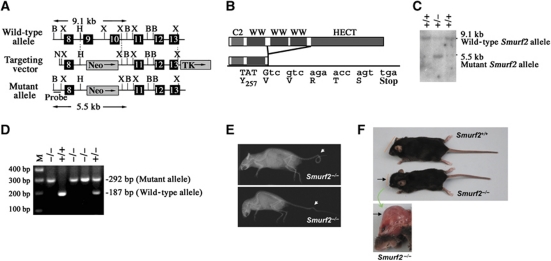
A standard strategy of gene targeting in ES cells through homologous recombination (Deng et al, 1996; Yang et al, 1998) was used to generate a Smurf2 null allele by deleting the exons 9 and 10 that encode the second and third WW domain (Figure 1A). This manipulation forces exon 8 to be spliced directly onto exon 11 and creates a premature stop codon that terminates protein translation prior to the last two highly conserved WW domains and the HECT ubiquitin ligase domain (Figure 1B; Supplementary Figure S1A). Southern blot analysis of genomic DNA with a probe derived from intron 7 confirmed the replacement of exons 9 and 10 by a neomycin resistance sequence. This was evident by the appearance of a 5.5-kb BamHI fragment from the mutant Smurf2 locus in lieu of a 9.1-kb BamHI fragment from the wild-type (WT) locus (Figure 1C). RT–PCR experiments demonstrated a significant reduction of the mutated Smurf2 mRNA expression while no overt compensatory changes in Smurf1 or other HECT-domain E3 ligase messages (Supplementary Figure S1B). We constructed the mutated Smurf2 cDNA expression construct and found that the residual Smurf2 protein fragment neither binds Smad3 nor represses the transcriptional activity of Smad3 (Supplementary Figure S1C and D). These data indicate that we have created a Smurf2 null allele.
Figure 1.
Mice with target-disrupted Smurf2 allele develop normally. (A) Schematic representation of the Smurf2 locus and the targeting construct. N, NotI; X, XhoI; H, HpaI; B, BamHI. (B) Illustration of Smurf2 domain map. (C) Southern blot showing WT and targeted Smurf2 alleles. The 5′ external probe is shown in (A). (D) PCR genotyping of Smurf2 alleles. (E) X-ray images showing vertebra malformation in a few isolated Smurf2−/− mice. Upper panel: a looped tail; bottom panel: a kinky tail. (F) Appearance of a Smurf2−/− mouse with hydrocephalus.
Homozygous Smurf2−/− pups were born at the expected Mendelian ratio (Figure 1D and data not shown), in agreement with a recently reported Smurf2-deficient line generated by deletion of exon 9 alone (Narimatsu et al, 2009). Like the Smurf1-deficient mice, the majority (>99%) of Smurf2−/− mice exhibited no overt developmental defect during embryogenesis and they were phenotypically indistinguishable from their WT or heterozygous littermates at weaning. Occasionally, we observed kinky or looped tail (Figure 1E) and hydrocephalus (Figure 1F) in a few Smurf2−/− mice (<1%), but with reduced severity as compared with those that lack three of the four Smurf alleles (i.e. Smurf1−/−; Smurf2+/− or Smurf1+/−; Smurf2−/−, also in Narimatsu et al, 2009). This phenotype was partially attributed to the impairment of planar cell polarity (Narimatsu et al, 2009). Since none of the Smurf1−/− mice exhibited such gross anatomic defects (Yamashita et al, 2005), our observation suggests that Smurf2 plays a more prominent role in regulating planar cell polarity.
Smurf2−/− cells exhibit enhanced responses to TGF-β
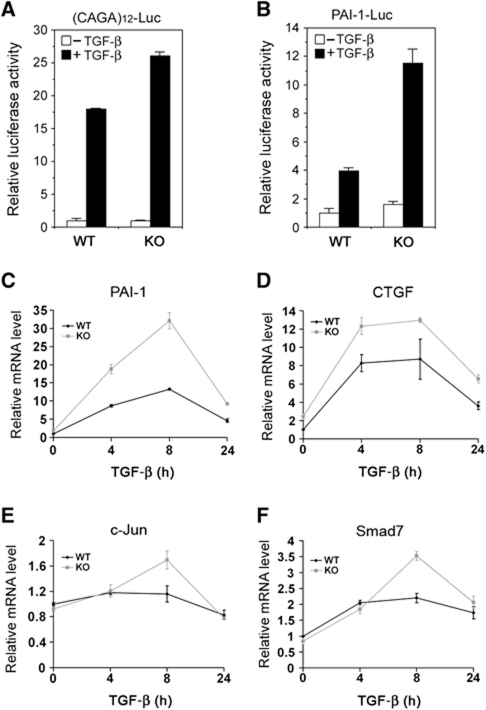
A major objective for this study is to determine if Smurf2 regulates TGF-β signalling under physiological conditions. Towards this end, we examined TGF-β-induced transcriptional responses in Smurf2−/− mouse embryonic fibroblasts (MEFs) by assaying for Smad-dependent luciferase reporters introduced through transfection. We found that TGF-β-induced response from (CAGA)12-Luc (Dennler et al, 1998), an artificial luciferase reporter for measuring Smad-dependent transcription, was significantly stronger in Smurf2−/− MEFs than in WT MEFs (Figure 2A). Likewise, TGF-β-induced response from plasminogen activator inhibitor-1 (PAI-1)-Luc (Keeton et al, 1991), another Smad-responsive reporter containing an ∼800 bp natural promoter sequence of PAI-1, was also much higher in Smurf2−/− MEFs (Figure 2B). In fibroblasts, TGF-β is a well-known and potent inducer of genes such as PAI-1 and connective tissue growth factor (CTGF) that are required for extracellular matrix production (Mori et al, 2004). To further assess Smurf2 control over TGF-β-induced transcriptional responses at the endogenous gene level, we examined PAI-1 and CTGF expression in primary dermal fibroblasts by real-time PCR over a course of 24 h under TGF-β treatment. Both TGF-β-induced PAI-1 and CTGF expression followed similar kinetic time courses in either WT or Smurf2−/− primary dermal fibroblasts, but the amplitude of induction was much higher in Smurf2−/− cells (Figure 2C and D). Two other well-established TGF-β targets, c-Jun and Smad7, also showed a stronger induction in Smurf2−/− primary dermal fibroblasts 8 h after TGF-β treatment while following similar kinetic time courses (Figure 2E and F). These results demonstrate that Smurf2 normally functions to attenuate Smad-dependent transcriptional responses to TGF-β, thus, confirming the status of Smurf2 as a negative regulator of TGF-β/Smad signalling.
Figure 2.
Elevated TGF-β/Smad transcriptional responses in Smurf2−/− cells. (A) (CAGA)12-Luc and (B) PAI-1-Luc reporter assays in WT and Smurf2−/− (KO) MEFs. SB431542 was used to remove all TGF-β activation (–TGF-β). Quantification of endogenous PAI-1 (C) CTGF (D), c-Jun (E), or Smad7 (F) mRNA levels in WT and KO dermal fibroblasts by real-time PCR. The level of HPRT transcript is used as an internal control for standardization and the results are presented as relative expression±s.d.
Smurf2 does not regulate the protein stability of Smad2 or Smad3 in vivo
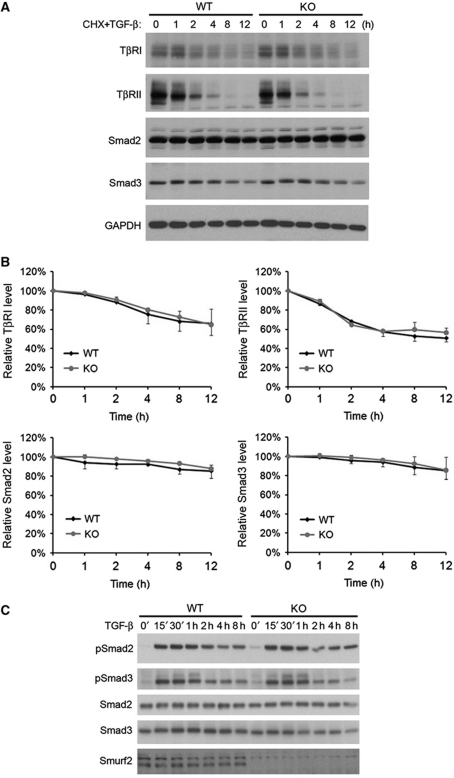
It was reported previously based on biochemical and cell culture studies that Smurf2 regulates TGF-β signalling by targeting Smads or the type I receptor for proteasomal degradation (Zhu et al, 1999; Kavsak et al, 2000; Lin et al, 2000; Zhang et al, 2001). Thus, loss of Smurf2 would be expected to increase the levels of Smads and the type I receptor, as well as the level of phosphorylated Smads. Surprisingly, we failed to observe any difference between the WT and the Smurf2−/− MEFs in the turnover rates of TβRI, TβRII, Smad2, or Smad3 with (Figure 3A and B) or without TGF-β treatment (data not shown). Moreover, the levels of phosphorylated Smad2 and Smad3 were not significantly different between WT and Smurf2−/− MEFs following TGF-β treatment (Figure 3C; Supplementary Figure S2). These results indicate that removing Smurf2 does not alter the protein levels of Smad2, Smad3, or the type I receptor, so the Smurf2-specific effect on TGF-β signalling is likely elicited downstream of Smad2/3 activation.
Figure 3.
Loss of Smurf2 does not affect protein stability of Smad2/3 or TGF-β-induced phosphorylation. (A) Western blot analyses and (B) quantification of TβRI, TβRII, Smad2, and Smad3 turnover in MEFs following cycloheximide (CHX) and TGF-β treatment. Data were collected from two different experiments using primary MEFs prepared from two different litters. (C) Time course of Smad2/3 phosphorylation in response to TGF-β stimulation. Figure source data can be found with the Supplementary Information.
Smurf2 induces multiple mono-ubiquitination of Smad3
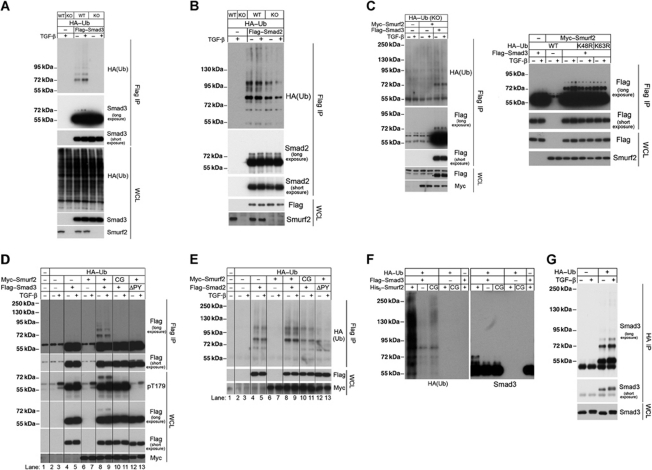
Since Smurf2 does not control Smad2/3 stability in vivo, we reexamined whether it is still required for ubiquitin modification of Smads under physiological conditions. For this purpose, we reintroduced Flag-tagged Smad3 along with HA-tagged ubiquitin in Smurf2−/− MEFs by transfection. After isolating Flag–Smad3 by immunoprecipitation and blotting for HA-tagged ubiquitin, we detected a ladder of discretely modified Smad3 in WT but not Smurf2−/− MEFs (Figure 4A). The lowest molecular weight step of this ladder is a 72-kDa band and the increment is about 17 kDa, which corresponds to the molecular weight of two ubiquitin moieties. This indicates that Smurf2 has a preference for transferring two ubiquitins at a time to Smad3 since the latter has a nominal molecular weight of 55 kDa. TGF-β treatment only increased the level of modified Smad3 slightly. On the Flag–Smad3 western blot, however, we detected only the 72-kDa modified band after prolonged exposure (Figure 4A), implying that the 72-kDa band is the ubiquitin-modified Smad3 rather than a ubiquitinated protein associated with Smad3. A similar pattern of ubiquitinated Smad2 was also detected in WT MEFs, but its level decreased only slightly in the absence of Smurf2 (Figure 4B).
Figure 4.
Smurf2 induces multiple mono-ubiquitination of Smad3. (A) Smurf2 is required for ubiquitination of Smad3. Total Flag–Smad3 was immunoprecipitated from transfected MEFs and resolved by SDS–PAGE. Western blot analyses were carried out to detect HA–ubiquitin (top) and Flag–Smad3 (middle two panels) in the precipitates. The levels of total HA–Ub, Flag–Smad3, and endogenous Smurf2 in the whole-cell lysates were also analysed and are shown in the bottom panels. (B) Loss of Smurf2 has little impact on the ubiquitination of Smad2, which was analysed similarly as in (A). (C) Smurf2 induces mono-ubiquitination of Smad3. Flag–Smad3 was analysed as in (A) except HA–Ub(KO) (left panel), or HA–Ub(K48R) and HA–Ub(K63R) (right panel) were used in the co-transfection. The asterisk indicates the Flag–Smad3. (D) Both the E3 ligase of Smurf2 and the PY motif of Smad3 are required for Smurf2-induced mono-ubiquitination of Smad3. WT Myc–Smurf2 and Flag–Smad3 and their mutants Myc–Smurf2(CG) and Flag–Smad3ΔPY were transfected into the MEFs along with HA–Ub. Following TGF-β treatment for 1 h and immunoprecipitation, these proteins were analysed by IP-western blot. Note that T179 of endogenous Smad3 was phosphorylated in response to TGF-β while T179 of exogenous Flag–Smad3 had a higher basal level of phosphorylation. (E) Smurf2 exhibits only a slight effect on ubiquitination of Flag–Smad2, which was analysed similarly as in (D). (F) Ubiquitination of Flag–Smad3 isolated from transfected HEK293 cells in a reconstituted system containing ATP, purified E1, E2, and recombinant His6-Smurf2 or -Smurf2CG. The reaction was carried out at 37 °C for 1 h. (G) Ubiquitin modification pattern of endogenous Smad3 in the WT MEF. Figure source data can be found with the Supplementary Information.
The concerted attachment of two ubiquitin moieties to Smad3 could be accomplished either through adding two branched ubiquitins on a single lysine residue (di-ubiquitination) or attaching two separate ubiquitins to two lysine residues (dual mono-ubiquitination) to produce the 72-kDa ubiquitin-modified Smad3. To distinguish between these two possibilities, we reintroduced Smurf2 along with Flag–Smad3 and a mutant ubiquitin, HA–Ub(KO), that lacks all lysine residues and is unable to support the growth of any ubiquitin chain into Smurf2−/− MEFs. If Smurf2 catalyses addition of two ubiquitin moieties onto one substrate lysine site, the above experiment would produce a single ubiquitin-modified Smad3 with a normal molecular weight of 63.5 kDa. Following immunoprecipitation of Flag–Smad3 and western blot analysis for HA–ubiquitin and Flag–Smad3, we detected only the 72-kDa ubiquitinated Smad3 in the presence of Myc–Smurf2 (Figure 4C, left panel), thus, indicating mono-ubiquitination of two lysine residues. We further tested two less stringent ubiquitin mutants, HA–Ub(K48R) and HA–Ub(K63R), which do not support the formation of either K48 or K63-linked ubiquitin chain, respectively. This time, we detected the same 72 kDa band as the major ubiquitin-modified Smad3 species in addition to minor presence of one higher ladder of ubiquitin-modified Smad3 (Figure 4C, right panel). This suggests that, when allowed, the attached first ubiquitin can be extended. Taken together, these data show that the predominant form of ubiquitin-modification of Smad3 by Smurf2 is likely the mono-ubiquitination on two lysine residues, but these attached ubiquitins can be further extended to produce oligo-ubiquitin modification. Nevertheless, the length of ubiquitin chains on the bulk of modified Smad3 is not extended sufficiently to mark for proteasomal degradation (Figure 4A and C).
To determine if rendering mono-ubiquitination of Smad3 is a specific property of the Smurf2 E3 ligase activity, we introduced the ligase-deficient Smurf2 mutant (C716G) (Zhang et al, 2001) along with Flag–Smad3 into Smurf2−/− MEFs. We found that ubiquitinated Smad3 was only detected in cells that received the WT but not the C716G mutant Smurf2 (Figure 4D). However, even the WT Smurf2 failed to cause ubiquitination of a mutant Smad3 lacking the PY motif required for Smurf2 binding (Figure 4D). The specificity of Smurf2 for Smad3 ubiquitination was also illustrated in the experiment with Smurf1−/− MEFs. We found that there was no difference in the Smad3 ubiquitination pattern between WT and Smurf1−/− MEFs (Supplementary Figure S3A).
In contrast, Smurf2 only caused slightly more ubiquitination of Smad2 following expression of Myc–Smurf2 and Flag–Smad2 in Smurf2−/− MEFs (Figure 4E). It appears that Smurf2−/− MEFs still retain high level of E3 ligase activities that promote ubiquitination of Smad2, presumably due to actions of other HECT-domain E3 ligase family members, such as Nedd4L, WWP1, and Itch (Bai et al, 2004; Seo et al, 2004; Kuratomi et al, 2005; Gao et al, 2009). Nevertheless, the C716G Smurf2 mutant did not support further ubiquitination of Smad2 beyond the basal level and the ΔPY Smad2 mutant was not ubiquitinated at all (Figure 4E). Furthermore, assaying for ubiquitination of TβRI, Smad4, and Smad7 in Smurf2−/− MEFs produced ubiquitin modification patterns that are identical to those in WT MEFs (Supplementary Figure S4). Interestingly, TβRI exhibited a remarkably strong pattern of poly-ubiquitination even in the absence of Smurf2 (Supplementary Figure S4). Thus, it is unlikely that Smurf2 is a specific ubiquitin ligase for these pathway components either.
To further demonstrate that Smurf2 causes mono-ubiquitination of Smad3, we carried out a reconstituted in-vitro ubiquitination assay with recombinant His6-Smurf2, E1, E2, HA–Ub, and Flag–Smad3 that was purified from transfected HEK293 cells. Following immuno-isolation of Flag–Smad3 and western blot analysis for HA–Ub, we detected a smear of ubiquitinated proteins, which was only seen when recombinant His6-Smurf2 but not the mutant His6-Smurf2(CG) was included in this system (Figure 4F, left panel). However, on the anti-Flag–Smad3 western blot, we detected the discrete 72 kDa band as the only modified Smad3 species (Figure 4F, right panel). This suggests that Smad3 received two ubiquitin moieties whereas the anti-HA–Ub smear was probably due to auto-ubiquitination of Smurf2 that associated with Smad3 or other non-specific proteins.
Finally, we asked to what extent Smad3 mono-ubiquitination occurs at endogenous level by transfecting HA–Ub into WT MEFs to facilitate the detection of ubiquitinated proteins, followed by immunoprecipitation with anti-HA antibody and western blot analysis specifically for Smad3. The results clearly showed mono-ubiquitinated 72 kDa Smad3 in addition to oligo- and poly-ubiquitinated Smad3 (Figure 4G), but the level of the last was significantly lower than the former two. TGF-β treatment increased the levels of both mono-ubiquitinated and oligo-ubiquitinated Smad3 but not high molecular weight poly-ubiquitinated Smad3 (Figure 4G). Taken together, our results firmly establish a multi mono-ubiquitination nature of Smurf2-induced Smad3 ubiquitination, which explains the lack of any discernible change in Smad3 stability in Smurf2−/− MEFs despite a conspicuous sensitization of Smad3 transcriptional responses to TGF-β (Figure 2A and B).
Phosphorylation of T179 of Smad3 is required for Smurf2 binding
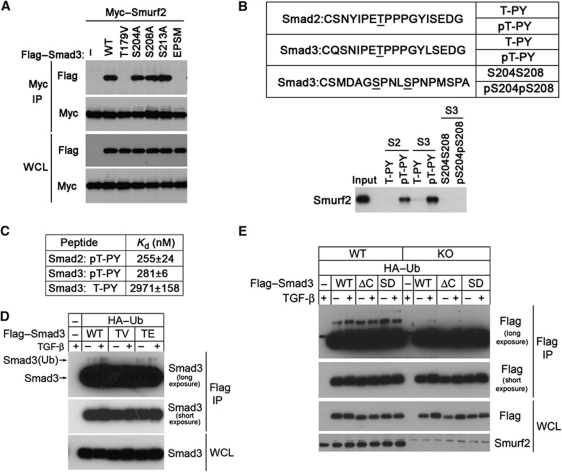
Published data from analyses of Smad1 in the BMP pathway indicate that phosphorylation of several sites in the linker region recruits Smurf1 to activated Smad1 (Sapkota et al, 2007). There are four known linker phosphorylation sites in Smad3, namely T179, S204, S208, and S213, among which T179, S204, and S208 can become phosphorylated in response to TGF-β (Millet et al, 2009; Wang et al, 2009). However, when over-expressed, these linker sites are phosphorylated even in the absence of TGF-β stimulation (Figure 4D; also in Gao et al, 2009). To determine if binding of Smurf2 to Smad3 requires phosphorylation in the linker region, we mutated all four sites and found that this abolished Smurf2 binding (Figure 5A). Further analyses indicated that replacing T179 with valine was sufficient to eliminate Smurf2 binding, whereas other single-site mutations at S204, S208, or S213 were not (Figure 5A). T179 lies immediately upstream to the PY motif and the requirement of T179 phosphorylation for Smurf2 binding is consistent with a recent report that this site is required for binding to the WW domains of Nedd4L, another HECT-domain-containing E3 ligase (Gao et al, 2009). Moreover, we could readily detect T179 phosphorylation in the modified 72 kDa Smad3 (Figure 4D). To determine if phosphorylation at T179 (or T220 in case of Smad2) is sufficient to induce Smurf2 binding, we synthesized several pairs of phosphorylated and unphosphorylated peptides with 18 residues centred around T179 (or T220 of Smad2) or covering S204 and S208 for peptide-binding assays (Figure 5B). After incubation with cell lysates prepared from HEK293 cells transfected with Myc–Smurf2, only phosphorylated peptides pT179-PY of Smad3 and pT220-PY of Smad2 were able to pull down Smurf2 (Figure 5B). To quantify the affinity of Smurf2 to these peptides, we purified a recombinant WW2–WW3 fragment of Smurf2 and performed isothermal titration calorimetry (ITC). The results revealed very high affinities of Smurf2 towards the phosphorylated Smad2 and Smad3 peptides, with dissociation constants of 255 and 281 nM, respectively (Figure 5C). The affinity of Smurf2 to the unphosphorylated Smad3 peptide was about 10 times lower at 2971 nM (Figure 5C). Consistent with the requirement of T179 phosphorylation for binding, the Smad3 (T179V) mutant, also showed decreased ubiquitination compared with the WT Smad3 or phosphorylation mimic Smad3 (T179E) mutant (Figure 5D). In contrast, it appears that Smad3 C-terminal phosphorylation does not affect the Smurf2-induced ubiquitination of Smad3, because deletion of C-terminal SSVS motif (ΔC) or substitution of the last two serines to phosphorylation mimicking aspartic acid residues (SD) had little effect on Smurf2-induced Smad3 ubiquitination in MEFs (Figure 5E).
Figure 5.
Phosphorylation of T179 in the linker region is required for Smad3 to bind Smurf2. (A) IP-western blot analyses of the ability of various Smad3 mutants to bind Smurf2 in transfected HEK293 cells. EPSM, Smad3 mutant with all four phosphorylation sites (T179, S204, S208, and S213) mutated. (B) In-vitro peptide-binding assays showing requirement of phosphorylation for Smurf2 binding. (C) ITC analysis of binding affinity between the WW2–WW3 fragment of Smurf2 and Smad2/3 peptides. (D) IP-western blot analysis for ubiquitination of the T179 mutant of Smad3 in WT MEFs. (E) IP-western blot analysis for ubiquitination of C-terminal phosphorylation site mutants of Smad3 in WT or Smurf2−/− MEFs. Figure source data can be found with the Supplementary Information.
Smurf2 induces ubiquitination of Smad3 in the MH2 domain
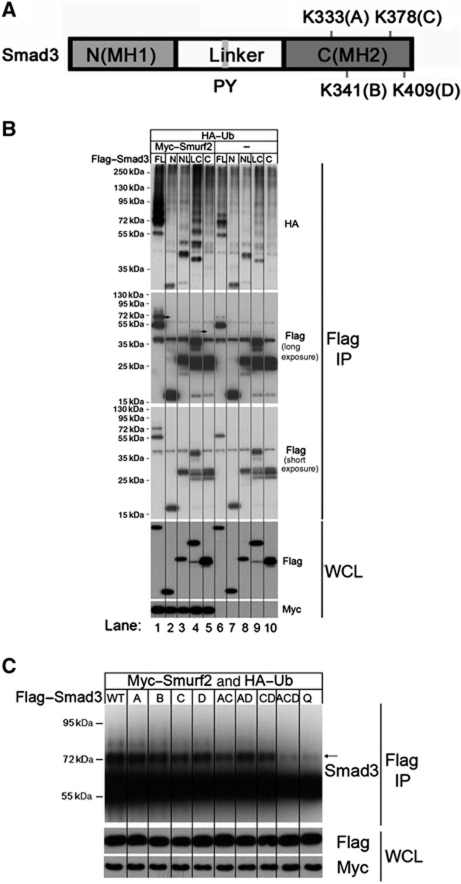
To investigate the structural basis for Smurf2-mediated ubiquitination of Smad3, we set out to identify the lysine residue(s) of Smad3 to which the ubiquitin moiety is attached. To this end, a series of Smad3 deletion mutants were constructed and their ubiquitination patterns were analysed in Smurf2−/− MEFs with or without expression of Myc–Smurf2. In the absence of Smurf2, both Smad3 NL and LC were ubiquinated as detected by HA–(Ub)-blot (Figure 6B, top panel, lanes 8 and 9). Adding back Myc–Smurf2 to Smurf2−/− MEFs significantly enhanced the ubiquitination of Smad3LC (Figure 6B, lane 4 versus 9) and only slightly so that of Smad3NL (lane 3 versus 8). In contrast, Smurf2 had no effect on Smad3 mutants containing only the MH1 (N) or the MH2 (C) domain (Figure 6B, top panel, comparing lanes 2 and 5 to lanes 7 and 10), which is consistent with the role of the linker in mediating the Smad3 and Smurf2 interaction. Of note is the poly-ubiquitin modification of Smad3LC, which likely resulted from aggregation of other ubiquitinated proteins onto Smad3LC in the proteasomes. In the light of this, we carried out western analysis with Flag–Smad3 and only detected mono- or oligo-ubiquitin modification of full-length Smad3 and Smad3LC, which was further enhanced in the presence of Smurf2 (Figure 6B, middle panel, comparing lanes 1 and 4 to lanes 6 and 9, respectively). Since there is no lysine residue in the linker region, the major ubiquitin attachment site(s) probably lies in the MH2 domain.
Figure 6.
Smad3 is ubiquitinated at the C-terminal (MH2) domain. (A) Schematic representation of Smad3 structure. Four potential Ub acceptor lysines are denoted as A, B, C, or D. (B) IP-western blot analyses of various Smad3 fragments in Smurf2−/− MEFs. FL, full-length Smad3; N, MH1 domain; NL, MH1 domain+linker region; LC, linker region+MH2 domain; C, MH2 domain. (C) IP-western blot analyses of the C-terminal Ub acceptor site lysine mutants in Smurf2−/− MEFs. The arrow indicates the dual mono-ubiquitinated Smad3. Figure source data can be found with the Supplementary Information.
Within the MH2 domain of Smad3, there are four lysine residues, K333, K341, K378, and K409 (Figure 6A). Crystal structures show that Smad3 can form a heterotrimeric complex with Smad4, comprising two molecules of phosphorylated Smad3 and one molecule of Smad4, or a homotrimeric complex of three phosphorylated Smad3 (Chacko et al, 2001, 2004). These complexes are stabilized by the positively charged K333 and K378 interfacing with two C-terminal phosphoserine residues of an adjoining subunit. Although K409 is not directly involved in intermolecular hydrogen bonding, it is positioned at the interface where D408 makes bifurcated hydrogen bonding to R268 of the opposing Smad3 and/or R361 of Smad4. Our data from complemented ubiquitination assays in Smurf2−/− MEFs showed that neither single substitutions of any of these four lysine residues nor double replacement of K333 and K409 or K378 and K409 with arginine had any effect on ubiquitination of Smad3 (Figure 6C). Double replacement of K333 and K378, the two lysines that are directly involved in hydrogen bonding detectably reduced the level of ubiquitin modification of Smad3; however, simultaneously replacing three lysines, K333, K378, and K409 (ACD mutant) or all four lysines (Q mutant) abolished Smurf2-induced Smad3 mono-ubiquitination entirely (Figure 6C; Supplementary Figure S6). These data suggest that K333 and K378 are likely the major sites for concerted attachment of two ubiquitins while other lysines can be selected in case one of these two is not available. However, a single lysine (ACD mutant) is not sufficient to support ubiquitin modification, as we noted earlier that Smurf2 has a preference of transferring two ubiquitins to its substrate simultaneously (Figure 6C). This reinforces our conclusion that the major form of Smurf2-mediated ubiquitin modification of Smad3 is dual mono-ubiquitination.
Ubiquitination blocks formation of Smad3 complexes
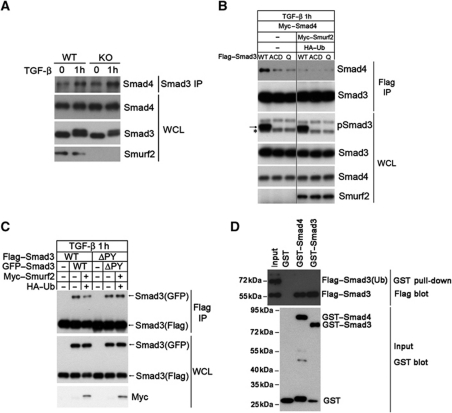
In the light of the critical roles of the MH2 domain lysines at the intermolecular interface of Smad3 complexes, ubiquitin modification at these sites would be expected to block the hydrostatic interaction that supports complex formation. To test this, we isolated endogenous Smad3 by immunoprecipitation from WT or Smurf2−/− MEFs using an antibody specific to Smad3, and examined the content of Smad4 by western blot analysis. The results showed a marked increase in the amount of Smad4 that was co-precipitated with Smad3 from Smurf2−/− MEFs compared with that from WT MEFs in response to TGF-β treatment (Figure 7A). Next, we examined the ability of Smurf2 to influence Smad3 to complex with Smad4 upon TGF-β simulation. In the absence of Myc–Smurf2 and HA–Ub, Flag–Smad3 but not the ACD and Q mutants readily formed complexes with Myc–Smad4 (Figure 7B). In contrast, such complexes could hardly be detected in the presence of Myc–Smurf2 and HA–Ub (Figure 7B).
Figure 7.
Mono-ubiquitination of Smad3 destabilizes Smad complexes. (A) Loss of Smurf2 enhances the formation of the endogenous Smad3/Smad4 heteromeric complex. MEFs were treated±TGF-β for 1 h before subjecting to IP-western blot analysis. (B) Reintroducing Smurf2 into Smurf2−/− MEFs disrupts the exogenous Smad3/Smad4 complex. The asterisk denotes the endogenous phosphorylated Smad3, and the arrow denotes the exogenous phosphorylated Smad3. Note that the triple and quadruple mutants are defective in receptor activation and Smad4 interactions. (C) Reintroducing Smurf2 back to Smurf2−/− MEFs reduces formation of the exogenous Smad3 homomeric complex. Note the changes in the amount of Smad3(GFP) brought down by anti-Flag IP in the absence or presence of Smurf2 and HA–Ub or with the ΔPY mutant. (D) GST pull-down assay showing that mono-ubiquitinated Smad3 does not bind the purified Smad4 or phospho-Smad3. The 1:1 mixture of ubiquitinated and non-ubiquitinated Smad3SD was prepared from transfected HEK293 cells, and incubated with GST, GST–Smad4, or phosphorylated GST–Smad3, respectively, before being processed for GST pull-down and western blot analysis. Figure source data can be found with the Supplementary Information.
To determine if Smurf2-induced ubiquitination affects formation of the Smad3 homotrimeric complex, we made use of Flag–Smad3 and GFP–Smad3 to visualize the intermolecular interaction. Once again, the Flag–Smad3 and GFP–Smad3 complex was readily detected following their expression in Smurf2−/− MEFs, but the amount of such complex was reduced in the presence of added Myc–Smurf2 and HA–ubiquitin (Figure 7C). Moreover, deletion of the PY motif made the mutant Smad3 refractory to the inhibitory effect of Smurf2-mediated ubiquitination (Figure 7C). Thus, formation of the Smad3 homotrimeric complex is also subject to control of ubiquitination induced by Smurf2.
To definitely prove that ubiquitination imposes a steric hindrance to the formation of Smad3 complexes, we took the advantage of the Smad3SD mutant, which has the last two serine residues replaced with aspartic acids to mimic C-terminal phosphorylation (Feng et al, 1998). This mutant behaves as the activated Smad3 when expressed in HEK293 cells and it gives rise to both an unmodified 55 kDa and a ubiquitin-modified 72 kDa protein product (Figure 7D), presumably due to induced phosphorylation in the linker region. We isolated the modified and unmodified forms of Flag–Smad3SD by immunoprecipitation and carried out GST pull-down assays using GST–Smad4 or phosphorylated GST–Smad3. The results showed that only the unmodified 55 kDa but not the modified 72 kDa Smad3SD was brought down by either GST–Smad4 or phosphorylated GST–Smad3 (Figure 7D).
Smurf2 regulates nuclear accumulation of Smad3 complexes
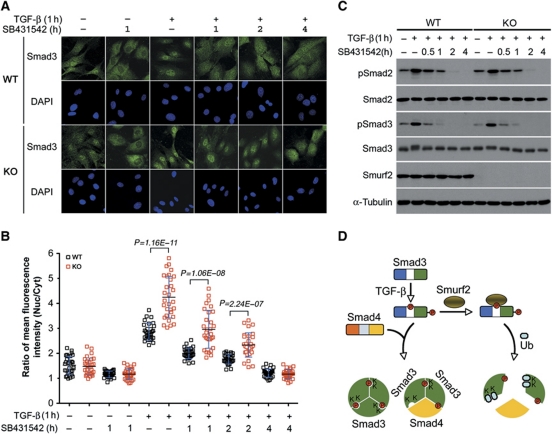
Previous studies indicate that E3 ubiquitin ligases can influence macromolecular trafficking to different cellular compartments through mono-ubiquitin modification (Li et al, 2003; Trotman et al, 2007; Dupont et al, 2009), and complexed form of Smads are imported faster than monomeric Smads (Clarke et al, 2006; Schmierer et al, 2008). Now that we had shown that Smurf2 affects formation of Smad3 complexes, we sought to examine the effect of Smurf2 loss on the subcellular localization of Smad3. By indirect immunofluorescence, we found that the amount of endogenous Smad3 in the nucleus was moderately more than that in the cytoplasm in either WT or Smurf2−/− MEFs prior to adding TGF-β ligand, and the average nuclear to cytoplasmic ratio of indirect immunofluorescence intensity (Nucl/Cyt) was 1.5 in both types of the cells (Figure 8A and B). Blocking background autocrine TGF-β signalling for 1 h with treatment of SB431542, a type I receptor inhibitor, reduced the nuclear content of Smad3 slightly (Figure 8A and B). In contrast, Smad3 accumulated rapidly in the nucleus within 1 h of TGF-β treatment, but the accumulation was much more pronounced in Smurf2−/− MEFs (Nucl/Cyt=4.2) than in control MEFs (Nucl/Cyt=2.9) (Figure 8A and B). Blunting the ligand activation with SB431542 attenuated the phosphorylation of Smad3 (Figure 8C), and caused Smad3 to redistribute more evenly between the nucleus and the cytoplasm (Figure 8A and B). The kinetics of Smad2/3 dephosphorylation was indistinguishable between WT and Smurf2−/− MEFs (Figure 8C); however, the nuclear content of Smad3 was persistently higher in Smurf2−/− MEFs until the pathway activity was completely turned off 4 h after the addition of SB431542 (Figure 8B). Thus, the Smurf2-specific function in attenuating TGF-β signalling can be attributed to its ability to inhibit Smad3 complex formation and decrease Smad3 accumulation in the nucleus, thereby inhibiting TGF-β-mediated transcriptional responses.
Figure 8.
Loss of Smurf2 increases nuclear accumulation of Smad3. (A) Indirect immunofluorescence staining of endogenous Smad3. MEFs were pretreated±TGF-β for 1 h, followed by ligand removal and receptor blocking with SB431542 for indicated duration. (B) Quantification of fluorescence intensity in (A). The results are expressed as ratios of mean fluorescence intensity between nuclear and cytoplasmic staining. Bars indicate mean±s.d., and n=30 in each condition. (C) Western blot analysis of phospho-Smad2/3 showing similar kinetics of dephosphorylation between WT and KO MEFs. TGF-β and SB431542 treatments were as in (A). (D) A model for the inhibitory function of Smurf2 in TGF-β signalling via mono-ubiquitination of Smad3. Upon TGF-β stimulation, Smad3 is phosphorylated at sites in both the linker and the C-terminal regions. Phosphorylation of T179 in the linker region potentiates Smurf2 binding and subsequent mono-ubiquitination in the MH2 domain. This mono-ubiquitination impedes formation of both heterotrimeric and homotrimeric Smad3 complexes, thereby restraining the amplitude of TGF-β signalling. Figure source data can be found with the Supplementary Information.
Discussion
The goal of this study is to define the function of Smurf2 in the regulation of TGF-β signalling. Since the original discovery of Smurfs as ubiquitin ligases of Smads and TβRI, conflicting reports emerged in the literature as to whether Smurf2 promote Smad ubiquitination under physiological conditions. To resolve this issue, we generated mice carrying Smurf2-deficient alleles, and have confirmed that Smurf2 is a physiological inhibitor of TGF-β-induced, Smad-dependent transcriptional responses. However, instead of marking Smad3 for proteasomal degradation by poly-ubiquitination, Smurf2 specifically promotes multiple mono-ubiquitination of Smad3, which blocks formation of both homotrimeric Smad3 and heterotrimeric Smad3–Smad4 complexes.
Smurf1 and Smurf2 share a high degree of amino-acid sequence homology. Mice with a targeted disruption of either the Smurf1 or Smurf2 allele are viable and survive to adulthood; however, disruption of both alleles simultaneously leads to embryonic lethality (Yamashita et al, 2005; Narimatsu et al, 2009). Clearly, some functions of Smurf2 are redundant with or can be compensated by those of Smurf1, but each of these two E3 ligases also has distinct molecular functions. Smurf1-deficient mice display age-dependent bone-mass increases and the corresponding mutant MEFs exhibit a higher sensitivity towards BMP signalling but no significant difference can be ascribed to TGF-β and Smad-specific transcriptional responses in comparison with the WT controls (Yamashita et al, 2005). In contrast, loss of Smurf2 significantly enhances TGF-β/Smad-dependent responses as demonstrated here. Thus, Smurf2 has a more specific role as a negative effector for TGF-β/Smad signalling under physiological conditions than Smurf1.
Previously, the negative regulation of TGF-β signalling by Smurfs had been attributed to proteasomal degradation of Smad2/3 or the type I receptor. However, we did not detect any change in the protein levels of Smad2, Smad3, or the type I receptor in either Smurf1- or Smurf2-deficient cells (Yamashita et al, 2005; this study). The levels of these proteins did not change in Smurf1 and Smurf2 double knockout embryos either (Narimatsu et al, 2009; Supplementary Figure S2B). So, it is questionable whether Smurfs regulate the turnover of Smad2, Smad3, or the type I receptor under physiological conditions. Instead, we found in this study that Smurf2 induces multiple mono-ubiquitination in the Smad3 MH2 domain and, in doing so, inhibits the formation and subsequent nuclear accumulation of Smad3 complexes. These data seamlessly reconcile both the role of Smurf2 as a negative inhibitor of TGF-β signalling, previously revealed by biochemical observations, and the lack of effect on Smad2/3 and the type I receptor turnover.
In normal cells, TGF-β activation of the receptors induces phosphorylation of Smad2 and Smad3 in both the C-terminal tails and the linker regions at T220 and T179, respectively (Alarcon et al, 2009; Millet et al, 2009; Wang et al, 2009). Our biochemical data indicated that phosphorylation in the linker region increased the affinities between Smurf2 and Smad3 by over 10-fold with Kd values in the submicromolar range. This observation is in contrast to a recent study reporting that Smurfs only have a minor role in recognizing the phosphorylated linker regions of Smad2 and Smad3 in comparison to Nedd4L, another HECT-domain E3 ligase with affinities to the phosphorylated pT-PY motifs of Smad2 and Smad3 in the micromolar range (Gao et al, 2009). A possible explanation for this discrepancy may be that the previous study only measured the affinity of Smurf1 to the phosphorylated Smad3 pT-PY motif (Gao et al, 2009). Since Smurf2−/− and Smurf1−/− MEFs display different levels of Smad-dependent transcriptional responses, the affinities of Smurf1 and Smurf2 to the phosphorylated linker regions could be different as well. Another conspicuous difference is that a single WW domain of Smurf1 was used in binding assays of the previous study, whereas we used a recombinant Smurf2 fragment containing both WW2 and WW3 repeats. Recent biochemical and structural studies indicate that although the Smad PY motifs are recognized by the WW2 repeat of Smurf1 or the WW3 of Smurf2, other WW repeats augment this interaction by making auxiliary contacts with S/T-P residues immediately upstream of all PY motifs (Chong et al, 2010).
Although the affinities of Smurf2 to the phosphorylated pT-PY motifs of Smad2 and Smad3 measured in vitro were similar (Figure 5B and C), loss of Smurf2 exhibited a lesser impact on Smad2 ubiquitination than Smad3 in vivo (compare Figure 4A and B). Since Smad2 is ubiquitinated in the absence of Smurf2, other E3 ligases may supply a redundant function. Because Smad2 is more essential for embryonic development than Smad3 (Weinstein et al, 2000), a lack of requirement of Smurf2 in Smad2 ubiquitination is consistent with the mild phenotype of the Smurf2−/− mice. It is possible that in the absence of Smurf2, the majority of Smad2 is still ubiquitinated normally, which could partially compensate for the loss of Smad3 regulation. Alternatively, the regulatory circuitry defined by Smurf2 function is possibly selected for non-developmental physiological conditions, such as response to environmental stress, and has escaped the scrutiny of our current methods of interrogation. Further investigation is required to expose the physiological significance of mono-ubiquitin modification of Smad3 by Smurf2.
Our finding of Smurf2 as a specific E3 ligase for Smad3 mono-ubiquitination is similar in mechanism to studies by Piccolo and colleagues, who showed Ecto/Tif1γ as the E3 ligase for Smad4 mono-ubiquitination (Dupont et al, 2009). Ecto/Tif1γ primarily resides in the nucleus and acts as a Smad complex ‘disruptase’ to turn off signalling by forcing dissociation of the Smad3/Smad4 heteromeric complexes. Since Smurf2 has a more balanced distribution between the nucleus and the cytoplasm (Supplementary Figure S5), it could ubiquitinate Smad3 and inhibit the formation of Smad3 complexes at both cellular compartments, thereby attenuating the Smad3-dependent signalling. We mapped Smurf2-dependent Smad3 ubiquitination sites to lysine residues in the MH2 domain, particularly, K333 and K378, which are conserved among all mammalian R-Smads (Konikoff et al, 2008). Since all R-Smads except Smad8 contain the PY motif for binding by Smurfs and other HECT-domain E3 ligases, it is possible that other R-Smads can be targeted for mono-ubiquitination as well. This implies that mono-ubiquitination at these lysines could be a universal mechanism to inhibit Smad complex formation, hence antagonizing ligand actions of the TGF-β superfamily.
Finally, because K378 in the MH2 domain of Smad3 is also involved in the physical interaction with the GS region of the type I receptor (Chen et al, 1998; Huse et al, 2001), ubiquitin modification of this lysine residue should theoretically prevent the interaction between Smad3 and the type I receptor, thereby blocking Smad3 activation. However, we did not observe any change in ligand-induced C-terminal serine phosphorylation of endogenous Smad3 in the absence of Smurf2. A possible explanation is that in normal cells, binding of Smad3 to Smurf2 requires phosphorylation of T179 in the linker region, which usually occurs slightly later than phosphorylation of C-terminal serines (Sapkota et al, 2007; Alarcon et al, 2009). In cancer cells, the scenario is different as the linker region of Smad3 is phosphorylated prior to any ligand stimulation due to high levels of CDKs or oncogenic Ras. Perhaps, in cancer cells, Smurf2-mediated ubiquitination could abrogate the interaction between R-Smads and the type I receptor, thus preventing activation by C-terminal phosphorylation.
Materials and methods
Cell culture and stimulations
WT and Smurf2−/− MEFs were isolated from E14.5 embryos and primary mouse derma fibroblasts (MDFs) were isolated from newborn mice (Hogan et al, 1994; Takashima, 2001). MEFs and MDFs were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS). Prior to treatment with TGF-β1 (4 ng/ml; PeproTech) or SB431542 (10 μM; Tocris) for indicated times, cells were starved overnight in DMEM containing 0.2% FBS. After the stimulation and wash with PBS, cells were harvested in lysis buffer containing 25 mM Tris–HCl, pH 7.5, 300 mM NaCl, and 1% Triton with protease and phosphatase inhibitors. For ubiquitination analysis, 10 mM N-ethylmaleimide was included in the lysis buffer.
Expression plasmids and transfection
Smad3 T179E, S213A, EPSM, ΔPY, and KR mutations were generated using a PCR-based strategy and subcloned into pCMV5B–Flag. All PCR-amplified regions were verified by sequencing. Plasmids for Flag-tagged full-length Smad3, Smad3N, Smad3NL, Smad3LC, Smad3C (Zhang et al, 1998), Flag-tagged Smad3 T179V, S204A, S208A (Millet et al, 2009), Myc-tagged Smurf2 and Smurf2(CG) (Zhang et al, 2001) have been described previously. pEGFP–Smad3 (Xiao et al, 2000) was provided by Dr Z Xiao. pEGFP–Smad3ΔPY was generated by subcloning the Smad3ΔPY fragment from Flag-tagged Smad3ΔPY into pEGFP-C1. HA–Ubiquitin and HA–Ubiquitin(KO) were provided by Dr C Pickart. HA–Ubiquitin(K48R) or (K63R) were made by PCR-based mutagenesis. Transfection of MEFs and HEK293 cells were carried out with Lipofectamine Plus (Invitrogen) and Fugene 6 (Roche), respectively.
In-vitro ubiquitination assay
Flag–Smad3 was captured from transfected HEK293 cell lysates by anti-Flag agarose. After a thorough wash, Smad3-bound agarose was divided into four aliquots. Empty anti-Flag agarose was used as control. The in-vitro ubiquitination assay was performed by incubating either Smad3-bound agarose or control agarose at 37 °C for 1 h with ubiquitin-activating enzyme UBE1, E2-conjugating enzyme UbcH5c, HA–ubiquitin and ATP (all from Boston Biochem) in the presence or absence of purified His6-Smurf2 or His6-Smurf2CG. The supernatant was removed after assay and agarose was thoroughly washed, and Flag–Smad3 was eluted by the Flag peptide (Sigma). The eluted fraction was subjected to western blot analysis.
Isothermal titration calorimetry
GST–Smurf2 WW2–WW3 fusion protein (amino acids 248–367) (Ying et al, 2003) was expressed in Escherichia coli BL21 cells (Invitrogen), and captured by glutathione sepharose beads (GE Healthcare). After on-beads cleavage by factor Xa (GE Healthcare), Smurf2 WW2–WW3 protein was further purified by Superdex 75 (GE Healthcare). Smad3 peptides were obtained from GenScript, and the Smad2 peptide was from Sigma. Concentrated protein and peptide solutions were dialysed into PBS (pH 7.4) for at least 48 h prior to ITC experiments. Experimental concentrations of the protein and peptide solutions were determined spectrophotometrically at A280 by using molar extinction coefficients (ε280) of 15 595/M/cm for the Smurf2 WW2–WW3 protein, 1490/M/cm for the pT-PY and T-PY Smad3 peptides, and 2980/M/cm for the Smad2 peptide. Diluted solutions were prepared with filtered dialysis buffer and degassed for 8 min under vacuum before each experiment. ITC measurements were carried out using a VP-ITC microcalorimeter equipped with a 315-μl syringe rotating at 300 r.p.m. (MicroCal, Northhampton, MA). All titrations were performed at 25 °C and initiated with a 2-μl injection of 300–440 μM peptide followed by 29 injections of 10 μl into the calorimeter cell (1.4 ml volume) containing 10–13 μM Smurf2 WW2–WW3 protein. Peptide titrations into buffer were performed to measure the heats of dilution. Origin software (Microcal) was used to integrate the raw calorimetric signals and obtain normalized heats of injection by subtracting the heats of titrant dilution from the heats of binding. The resulting data were analysed with a single-site binding model using the program SEDPHAT (Houtman et al, 2007). Active protein concentration was calculated during the analysis based on well-determined peptide concentrations and evidence of single-site binding by NMR studies (Chong et al, 2010). Reported uncertainties represent the 95% confidence interval as determined from 1000 Monte-Carlo simulations using SEDPHAT.
GST pull-down assay
To enrich the amount of ubiquitinated Smad3, HEK293 cells were transfected with Flag–Smad3SD, Myc–Smurf2, and HA–ubiquitin, and total Flag–Smad3SD was captured by anti-Flag agarose. After a thorough wash, Flag–Smad3SD was eluted with 0.8 mg/ml Flag peptide. Flag eluates were subjected to immunoprecipitation with anti-HA agarose followed by elution with 1 mg/ml HA peptide. The HA elution was used for subsequent binding assays. Recombinant GST–Smad3 and GST–Smad4 were purified from E. coli BL21 cells (Zhang et al, 1996). Before the binding experiment, GST–Smad3 was phosphorylated at the C-terminus by an in-vitro kinase assay using a purified TβRI kinase domain as described previously (Zhang et al, 1996). Phosphorylated GST–Smad3 or GST–Smad4 was prebound to the glutathione sepharose 4B, and incubated with HA eluates of Flag–Smad3SD for 1 h. After the incubation, the beads were thoroughly washed with PBS and resuspended in 2 × SDS–PAGE loading buffer (125 mM Tris–HCl, pH 6.8, 4% SDS, 20% (v/v) glycerol, 80 mM DTT, 0.004% bromphenol blue).
In-vitro peptide-binding assay
Smad2 and Smad3 peptides were coupled to an affinity resin using a MicroLink Peptide Coupling Kit (Thermo Scientific). The immobilized peptides were then incubated with cell lysates prepared from Myc–Smurf2-transfected HEK293 cells. Excess protein was removed with a thorough wash before the resin was resuspended in 2 × SDS–PAGE loading buffer.
Transcription reporter assays
Transcription reporter assays in MEFs were performed in 12-well plates by transfecting 0.5 μg of luciferase reporter and 0.2 μg of the pRL-TK (Promega) control reporter per well. Luciferase activity was determined after treating with SB431542 (10 μM) or TGF-β1 (4 ng/ml) for 18–20 h. Results were obtained from at least two independent experiments using cells isolated from different mice with duplicate samples for each data point.
Immunofluorescence
MEFs grown on two-chamber culture slides (BD Falcon) were fixed in 4% paraformaldehyde after the indicated TGF-β and/or SB431542 treatment. After treatment with 0.5% Triton X-100, cells were probed with anti-Smad3 antibody (1:150 dilution; Zymed), followed by Alexa Fluor 488 secondary antibody (1:1000 dilution; Invitrogen). Fluorescence images were acquired with a Zeiss LSM 510 META and analysed using the Imaris x64 v7.0 software (Bitplane). The ratio of mean fluorescence intensity of the nucleus to the cytoplasm was calculated after measuring nuclear and whole-cell green fluorescence intensities and areas.
Supplementary Material
Acknowledgments
We thank Drs F Liu, P ten Dijke, and Z Xiao for various plasmid and antibody reagents, Dr V Barr for assistance with microscope and computer software, N Morris for animal service, and Dr P Randazzo's lab for assistance with protein purification. This research was supported by the intramural research program of the NIH, National Cancer Institute, Center for Cancer Research. MY was partially supported by JSPS Grant #21689053.
Author contributions: SYC and YEZ cloned the Smurf2 mouse genomic DNA and made the targeting construct. CL and CXD transfected the targeting construct into ES cells and generated chimeric mice for the Smurf2-deficient allele. MY picked ES cell colonies, screened for targeted ES clones. MY and YT maintained the mouse colonies and generated primary MEFs and MDFs. NPC performed ITC experiments and analysed the results. LYT, MY, and XW performed all other experiments described in this manuscript. LYT and YEZ analysed the data, conceived the study, and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, Sapkota G, Pan D, Massague J (2009) Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell 139: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Yang C, Hu K, Elly C, Liu YC (2004) Itch E3 ligase-mediated regulation of TGF-beta signaling by modulating smad2 phosphorylation. Mol Cell 15: 825–831 [DOI] [PubMed] [Google Scholar]
- Chacko BM, Qin B, Correia JJ, Lam SS, de Caestecker MP, Lin K (2001) The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat Struct Biol 8: 248–253 [DOI] [PubMed] [Google Scholar]
- Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, De Caestecker M, Lin K (2004) Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol Cell 15: 813–823 [DOI] [PubMed] [Google Scholar]
- Chen YG, Hata A, Lo RS, Wotton D, Shi Y, Pavletich N, Massague J (1998) Determinants of specificity in TGF-beta signal transduction. Genes Dev 12: 2144–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong PA, Lin H, Wrana JL, Forman-Kay JD (2010) Coupling of tandem Smad ubiquitination regulatory factor (Smurf) WW domains modulates target specificity. Proc Natl Acad Sci USA 107: 18404–18409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DC, Betterton MD, Liu X (2006) Systems theory of Smad signalling. Syst Biol (Stevenage) 153: 412–424 [DOI] [PubMed] [Google Scholar]
- Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P (1996) Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell 84: 911–921 [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM (1998) Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 17: 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Miyazono K (2008) TGF-beta and the TGF-beta family. In The TGF-beta Family, Derynck R, Miyazono K (eds), pp 29–43. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S (2009) FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 136: 123–135 [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K (2001) Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem 276: 12477–12480 [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R (2005) Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol 21: 659–693 [DOI] [PubMed] [Google Scholar]
- Feng XH, Zhang Y, Wu RY, Derynck R (1998) The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-beta-induced transcriptional activation. Genes Dev 12: 2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi M, Imamura T, Chiba T, Ebisawa T, Kawabata M, Tanaka K, Miyazono K (2001) Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol Biol Cell 12: 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Alarcon C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massague J (2009) Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell 36: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Dikic I (2005) Ubiquitylation and cell signaling. EMBO J 24: 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E (1994) Manipulating the Mouse Embryos: A Laboratory Manual, 2nd edn, pp 260–261. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Houtman JC, Brown PH, Bowden B, Yamaguchi H, Appella E, Samelson LE, Schuck P (2007) Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci 16: 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massague J (2001) The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell 8: 671–682 [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Hill CS (2002) Nucleocytoplasmic shuttling of Smads 2, 3, and 4 permits sensing of TGF-beta receptor activity. Mol Cell 10: 283–294 [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell 6: 1365–1375 [DOI] [PubMed] [Google Scholar]
- Keeton MR, Curriden SA, van Zonneveld AJ, Loskutoff DJ (1991) Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J Biol Chem 266: 23048–23052 [PubMed] [Google Scholar]
- Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, Miyazawa K (2004) Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23: 6914–6923 [DOI] [PubMed] [Google Scholar]
- Konikoff CE, Wisotzkey RG, Newfeld SJ (2008) Lysine conservation and context in TGFbeta and Wnt signaling suggest new targets and general themes for posttranslational modification. J Mol Evol 67: 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratomi G, Komuro A, Goto K, Shinozaki M, Miyazawa K, Miyazono K, Imamura T (2005) NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem J 386: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302: 1972–1975 [DOI] [PubMed] [Google Scholar]
- Lin X, Duan X, Liang YY, Su Y, Wrighton KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, Shi Y, Chen YG, Meng A, Feng XH (2006) PPM1A functions as a Smad phosphatase to terminate TGFbeta signaling. Cell 125: 915–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem 275: 36818–36822 [DOI] [PubMed] [Google Scholar]
- Lo RS, Massague J (1999) Ubiquitin-dependent degradation of TGF-beta-activated smad2. Nat Cell Biol 1: 472–478 [DOI] [PubMed] [Google Scholar]
- Lonn P, Moren A, Raja E, Dahl M, Moustakas A (2009) Regulating the stability of TGFbeta receptors and Smads. Cell Res 19: 21–35 [DOI] [PubMed] [Google Scholar]
- Millet C, Yamashita M, Heller M, Yu LR, Veenstra TD, Zhang YE (2009) A negative feedback control of transforming growth factor-beta signaling by glycogen synthase kinase 3-mediated Smad3 linker phosphorylation at Ser-204. J Biol Chem 284: 19808–19816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Ishida W, Bhattacharyya S, Li Y, Platanias LC, Varga J (2004) Selective inhibition of activin receptor-like kinase 5 signaling blocks profibrotic transforming growth factor beta responses in skin fibroblasts. Arthritis Rheum 50: 4008–4021 [DOI] [PubMed] [Google Scholar]
- Narimatsu M, Bose R, Pye M, Zhang L, Miller B, Ching P, Sakuma R, Luga V, Roncari L, Attisano L, Wrana JL (2009) Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell 137: 295–307 [DOI] [PubMed] [Google Scholar]
- Sapkota G, Alarcon C, Spagnoli FM, Brivanlou AH, Massague J (2007) Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25: 441–454 [DOI] [PubMed] [Google Scholar]
- Schmierer B, Tournier AL, Bates PA, Hill CS (2008) Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci USA 105: 6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo SR, Lallemand F, Ferrand N, Pessah M, L’Hoste S, Camonis J, Atfi A (2004) The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J 23: 3780–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113: 685–700 [DOI] [PubMed] [Google Scholar]
- Takashima A (2001) Establishment of fibroblast cultures. Curr Protoc Cell Biol, Chapter 2: Unit 2.1 [DOI] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, Pandolfi PP (2007) Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128: 141–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL (2008) TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol 10: 837–848 [DOI] [PubMed] [Google Scholar]
- Wang G, Matsuura I, He D, Liu F (2009) Transforming growth factor-{beta}-inducible phosphorylation of Smad3. J Biol Chem 284: 9663–9673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wau MY, Hill CS (2009) Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell 16: 329–343 [DOI] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Deng C (2000) Functions of mammalian Smad genes as revealed by targeted gene disruption in mice. Cytokine Growth Factor Rev 11: 49–58 [DOI] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Henis YI, Lodish HF (2000) A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc Natl Acad Sci USA 97: 7853–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE (2005) Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Li C, Xu X, Deng C (1998) The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proc Natl Acad Sci USA 95: 3667–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SX, Hussain ZJ, Zhang YE (2003) Smurf1 facilitates myogenic differentiation and antagonizes the bone morphogenetic protein-2-induced osteoblast conversion by targeting Smad5 for degradation. J Biol Chem 278: 39029–39036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE (2009) Non-Smad pathways in TGF-beta signaling. Cell Res 19: 128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA 98: 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R (1998) Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-beta-induced transcription. Nature 394: 909–913 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng X, We R, Derynck R (1996) Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature 383: 168–172 [DOI] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400: 687–693 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.