Abstract
Eukaryotic chromosomes are replicated from multiple origins that initiate throughout the S-phase of the cell cycle. Why all origins do not fire simultaneously at the beginning of S-phase is not known, but two kinase activities, cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK), are continually required throughout the S-phase for all replication initiation events. Here, we show that the two CDK substrates Sld3 and Sld2 and their binding partner Dpb11, together with the DDK subunit Dbf4 are in low abundance in the budding yeast, Saccharomyces cerevisiae. Over-expression of these factors is sufficient to allow late firing origins of replication to initiate early and together with deletion of the histone deacetylase RPD3, promotes the firing of heterochromatic, dormant origins. We demonstrate that the normal programme of origin firing prevents inappropriate checkpoint activation and controls S-phase length in budding yeast. These results explain how the competition for limiting DDK kinase and CDK targets at origins regulates replication initiation kinetics during S-phase and establishes a unique system with which to investigate the biological roles of the temporal programme of origin firing.
Keywords: CDK, DDK, initiation, replication, timing
Introduction
The first step in eukaryotic replication initiation involves the formation of the pre-replicative complex (pre-RC) at potential origins in G1-phase—a process called ‘licensing’ (Bell and Dutta, 2002). The licensing reaction results in the loading of inactive double hexamers of the replicative MCM2–7 helicase on to double-stranded origin DNA (Evrin et al, 2009; Remus et al, 2009). Replication initiation at ‘licensed’ origins can subsequently only occur in S-phase due to the activation of the S-CDK (cyclin-dependent kinase) and DDK (Dbf4-dependent kinase) kinases. DDK kinase directly phosphorylates the MCM2–7 helicase (Sheu and Stillman, 2006, 2010), while CDK is required to phosphorylate two proteins, Sld2 and Sld3, facilitating their essential interaction with the tandem BRCT repeat protein Dpb11 (Tanaka et al, 2007; Zegerman and Diffley, 2007). The exact functions of these phosphorylation events in the initiation reaction are not fully understood but together DDK and CDK are required to activate the MCM helicase and recruit the necessary proteins for replisome assembly (Labib, 2010). Although all the factors required for replication must be present as eukaryotic cells enter S-phase, initiation events occur as a continuum throughout this period with some origins firing early, others late and some not firing at all (Raghuraman et al, 2001). The purpose of this temporal programme of origin firing is not known.
The time an origin fires is determined at a point in late G1-phase coincident with pre-RC formation (Wu and Gilbert, 1996; Raghuraman et al, 1997). Despite this, pre-RCs form efficiently at all origins in G1-phase in budding yeast, regardless of whether they fire in the subsequent S-phase (Santocanale et al, 1999; Wyrick et al, 2001). On the other hand, CDK and DDK kinase act immediately prior to origin firing and are continually required throughout the S-phase for all initiation events (Bousset and Diffley, 1998; Donaldson et al, 1998a, 1998b). This suggests that although origin timing is determined in G1-phase, the selectivity of the replication initiation apparatus for early origins before late origins likely occurs at a point after pre-RC formation but before or during CDK/DDK function.
The factors that determine the timing of firing of origins are not fully understood, but there is a correlation between chromatin accessibility and the time of replication initiation in eukaryotes (Bell et al, 2010; Mechali, 2010). In budding yeast, changes in the histone acetylation state at origins either by deletion of the deacetylase RPD3 or by artificial recruitment of an acetyltransferase can influence replication timing presumably by increasing origin accessibility (Vogelauer et al, 2002; Aparicio et al, 2004). Therefore, a potential mechanism for the programme of replication initiation is that the chromatin context of a pre-RC generates differential affinities for a limiting replication factor (Supplementary Figure S1; Rhind, 2006). In this study, we set out to identify if any replication factors are limiting for initiation and whether their levels, together with origin accessibility, control the temporal programme of origin firing.
Results
At least four essential replication factors are low abundance in budding yeast
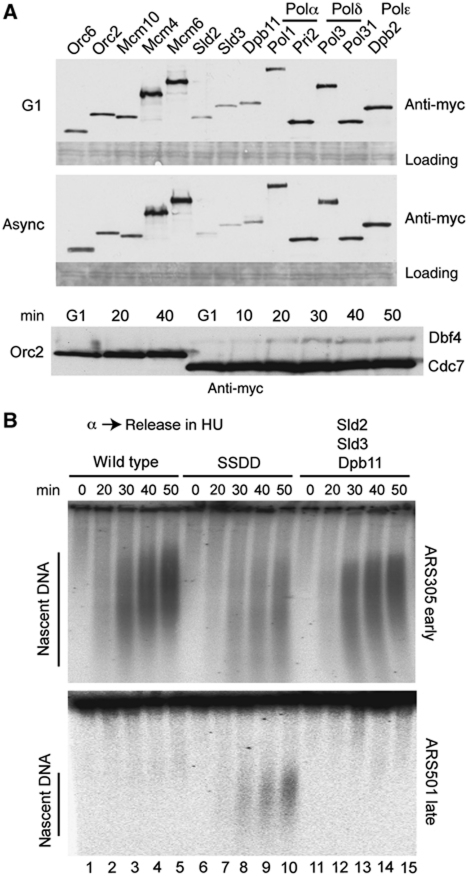
To determine if any factors are indeed limiting for replication initiation in budding yeast, we tagged replication factors at their endogenous loci and compared protein abundance. As expected, we found that components of the Mcm2–7 helicase are more abundant than ORC subunits and the replicative polymerases (Polα, Polδ and Polε; Figure 1A). Importantly, however, the two essential CDK targets Sld2 and Sld3, together with their binding partner Dpb11, are less abundant than ORC (Figure 1A; Supplementary Figure S2A), consistent with a previous proteomic study (Ghaemmaghami et al, 2003). The C-terminal tag has little effect on the levels of these factors (Supplementary Figure S2E). We further confirmed the low abundance of Sld3, Dpb11 and Sld2 in vivo by western blot comparison of a serial dilution of recombinant protein with a lysate from a known number of yeast cells (Supplementary Figure S2B–E). In addition, it has previously been shown that the DDK kinase regulates origin efficiency in fission yeast (Patel et al, 2008; Wu and Nurse, 2009) and must tightly interact with pre-RCs to execute its function (Francis et al, 2009). We show here that while Cdc7 levels are greater than ORC, Dbf4 abundance is extremely low during S-phase (Figure 1A, lower panel). Since previous work has reported that ORC is as abundant as the number of origins in budding yeast (Rowley et al, 1995), the low abundance of Sld2, Sld3, Dpb11 and Dbf4 suggests that they are limiting for replication initiation in budding yeast.
Figure 1.
The low abundance of Sld3, Sld2, Dpb11 and Dbf4 prevents the early firing of late origins in hydroxyurea. (A) Protein abundance measurement by comparative western blotting of tagged protein. Anti-myc western blot of 13myc-tagged protein from α-factor arrested cells (G1) or asynchronous cells (async). All genes were tagged at their endogenous genomic locus. Loading control is a ponceau stain of a section of the membrane. (Bottom) Anti-myc western of Dbf4–13myc/Cdc7–13myc and Orc2–13myc after α-factor arrest (G1) and release for the indicated times. (B) Visualization of nascent DNA from strains arrested in hydroxyurea. Alkaline gel analysis showing replication intermediates after release from α-factor (0) into 200 mM hydroxyurea at 25°C in YP-galactose. SSDD is a strain that expresses a second copy of SLD3, SLD2, DPB11 and DBF4 from the GAL1-10 promoter. The strain used in lanes 11–15 is as SSDD, but without over-expression of Dbf4.
Over-expression of Sld2, Sld3, Dpb11 and Dbf4 allows early firing of late origins in hydroxyurea
To test whether the low levels of Sld2, Sld3, Dpb11 and Dbf4 prevent the early firing of normally late origins, we over-expressed these factors from the galactose-inducible promoter (we hereafter refer to this over-expression strain as SSDD—Supplementary Figure S3) and analysed the timing of replication initiation. As expected, when wild-type cells are released from α-factor into hydroxyurea-containing medium, nascent DNA accumulates at early firing origins, such as ARS305, but late origin firing is inhibited due to the action of the checkpoint kinase Rad53 (Figure 1B, lanes 1–5; Zegerman and Diffley, 2010). However, when the SSDD strain was released into hydroxyurea, we observed early firing of the late origins ARS501 and ARS603 (Figure 1B, lanes 6–10; Supplementary Figure S4), suggesting that Sld3, Sld2, Dpb11 and Dbf4 are limiting for replication initiation in these conditions.
One complication in interpreting the effect of over-expression of these factors in hydroxyurea is that Rad53 inhibits replication initiation by phosphorylating Dbf4 and Sld3 (Lopez-Mosqueda et al, 2010; Zegerman and Diffley, 2010). The activation of late origins upon over-expression of Dbf4 and Sld3 in hydroxyurea could therefore potentially be due to bypass of the checkpoint. Indeed, over-expression of Dbf4 alone causes a low level of late origin firing in budding yeast in hydroxyurea (Supplementary Figure S4), as previously reported in fission yeast (Patel et al, 2008; Wu and Nurse, 2009). Although the near simultaneous firing of early and late origins in the SSDD strain is consistent with an effect on timing (Figure 1B), an effect on the checkpoint is difficult to rule out from experiments in hydroxyurea.
Over-expression of Sld2, Sld3, Dpb11 and Dbf4 allows early firing of late origins in a normal S-phase
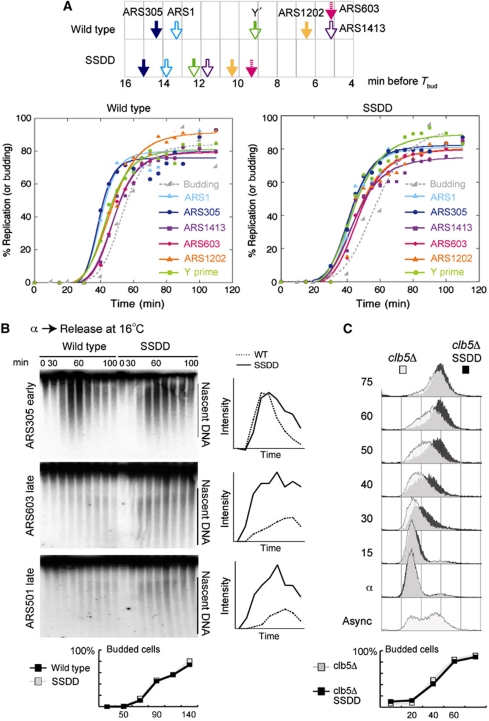
To separate the effects of checkpoint activation from replication timing, we examined S-phase kinetics in a normal cell cycle in the absence of hydroxyurea. We measured the time of replication of origin loci at 30°C using the dense isotope substitution method (McCarroll and Fangman, 1988). For this assay, cells that have been pre-grown in heavy isotope medium are released from G1 into S-phase in light medium and replicated DNA (heavy–light) is separated from unreplicated DNA (heavy–heavy) by CsCl gradient centrifugation and then quantitated. This timing assay shows that several normally late-replicating origin loci are replicated substantially earlier in the SSDD strain (Figure 2A). To determine if this advance in replication timing is due to early initiation of normally late origins, we analysed the appearance of nascent DNA from cells undergoing S-phase at 16°C. At this temperature, the rate of replication elongation is reduced and the delay between early and late origin firing is exacerbated (unpublished observations). In the SSDD strain, the kinetics of bud emergence and the initiation time of an early origin are similar to wild type (Figure 2B), but the late origins ARS501 and ARS603 initiate significantly earlier (Figure 2B). To determine if the early firing of late origins occurs across the genome, we analysed the speed of S-phase in cells lacking the cyclin Clb5. In the absence of Clb5, S-phase is slow because replication initiation only occurs at early origins due to a transient pulse of Clb6-CDK activity (Donaldson et al, 1998b; McCune et al, 2008). Over-expression of Sld3, Sld2, Dpb11 and Dbf4 increases the speed of S-phase in cells lacking Clb5 (Figure 2C) and the late origin ARS501 fires early (Supplementary Figure S5A). This strongly suggests that the SSDD strain allows many origins to initiate in early S-phase when Clb6-CDK is active. This increase in S-phase speed in cells lacking Clb5 is not simply due to over-expression of the essential CDK targets Sld2 and Sld3 (Supplementary Figure S5B). Together these data show that the low abundance of Sld3, Sld2, Dpb11 and Dbf4 delays the firing of late origins relative to early origins in a normal S-phase in budding yeast.
Figure 2.
The low abundance of Sld3, Sld2, Dpb11 and Dbf4 prevents the early firing of late origins in a normal S-phase. (A) Quantitative measurement of replication time using the dense isotope transfer technique. Strains were released into S-phase in light medium plus galactose. The time of replication (Trep) is the point where half of the final level of replication of that locus has been attained. (Top) Replication time (Trep) values (arrows) are shown relative to the time when the strain has attained half of its final level of budding (TBud). (Bottom) Graphs of the data that are represented in the top. (B) Visualization of replication initiation events from strains released from α-factor into YP-galactose at 16°C. Time points were taken every 10 min from 30 to 100 min after release from α-factor (0). (Right) Graphic quantification of nascent DNA using Image J analysis of phosphorimage on the left. The y axis is the intensity of the phosphorimage signal in arbitrary units. (Bottom) Budding index from this experiment. (C) Over-expression of Sld3, Sld2, Dpb11 and Dbf4 increases the speed of S-phase in clb5Δ cells. Flow cytometry after release from α-factor into YP-galactose at 25°C. The flow cytometry results are overlaid to allow direct comparison of S-phase progression. Budding index, lower panel.
Sld3, Sld2, Dpb11 and Dbf4/Cdc7 are not incorporated into replisomes (Gambus et al, 2009) and therefore these proteins are not simply limiting by protein abundance, but also the rate at which they recycle after initiation events. Although the over-expression of all four factors is required for the maximal early firing of late origins observed in our experiments, over-expression of any combination of three of the factors is sufficient for some advance of the firing time (Supplementary Figure S6A). This observation could reflect the fact that when the three factors are over-expressed, replication initiation is restricted by the recycling of only a single protein. Over-expression of Dbf4 alone is not sufficient to advance the time of initiation of late origins in a normal cell cycle, as measured either by detection of replication intermediates or by analysis of the speed of S-phase in a CLB5 mutant (Supplementary Figure S6B and C).
Additional over-expression of Cdc45 and Sld7 affects replication initiation
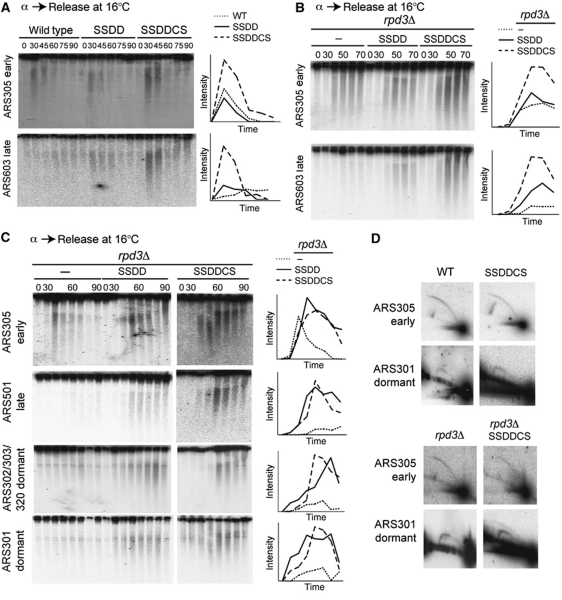
The stoichiometry of proteins at replication origins in vivo is not known and therefore even seemingly abundant factors may be limiting for initiation. It has been shown that the essential replication initiation factor Cdc45, which binds to Sld3 (Kamimura et al, 2001), is preferentially recruited to early origins, but not late origins in G1-phase (Aparicio et al, 1999). An additional Sld3-interacting protein, called Sld7, has recently been identified (Tanaka et al, 2011) and this protein has also been shown to bind to early but not late origins in G1-phase (H Araki, personal communication). Although both Cdc45 and Sld7 are relatively abundant (Ghaemmaghami et al, 2003; Supplementary Figure S2E), the pattern of origin recruitment of these factors might suggest that they are also limiting. When Cdc45 and Sld7 are over-expressed in combination with Sld2, Sld3, Dpb11 and Dbf4 (a strain hereafter referred to as SSDDCS), we observe the early firing of late origins as expected, but also an increase in the amount of nascent DNA at both early and late origins at 16°C (Figure 3A; Supplementary Figure S7), indicative of increased origin efficiency. Importantly, over-expression of Sld2, Sld3, Dpb11 and Dbf4 are all still required for the maximum early firing of late origins in the SSDDCS strain and over-expression of Cdc45 and Sld7 alone does not have this effect (unpublished observations). Cdc45 over-expression affects initiation efficiency in fission yeast (Wu and Nurse, 2009) and since both Cdc45 and Sld7 are Sld3-binding proteins, their preferential recruitment to early origins before late origins may be due to limiting levels of Sld3. Although we cannot exclude that Cdc45 and/or Sld7 are limiting for replication initiation, we do not detect an advance in the timing of origin replication in the SSDDCS strain compared with the SSDD strain using the dense isotope substitution technique at 30°C nor an additional increase in the speed of S-phase in a clb5 null strain (unpublished observations).
Figure 3.
RPD3 deletion combined with Sld3, Sld2, Dpb11 and Dbf4 over-expression allows dormant origin firing. (A) Additional over-expression of Cdc45 and Sld7 increases replication efficiency at 16°C. Replication intermediate assay after release from α-factor (0) into YP-galactose at 16°C for the indicated times as in Figure 2B. SSDDCS corresponds to Sld3, Sld2, Dpb11, Dbf4, Cdc45 and Sld7 expressed from GAL1-10 promoter. (Right) Graphic quantification of nascent DNA using Image J analysis of phosphorimage on the left. The y axis is the intensity of the phosphorimage signal in arbitrary units. (B) Sld3, Sld2, Dbf4 and Dpb11 are limiting in rpd3 null cells. As (A) except time points were taken every 10 min from 30 to 70 min after release from α-factor. (C) Over-expression of limiting factors allows replication initiation of dormant origins in rpd3 null cells. As (A) except time points were taken every 10 min from 30 to 90 min after release from α-factor. The probe for ARS302/303/320 does not distinguish between initiation at ARS302, ARS303 and ARS320. Note that the rpd3Δ SSDDCS strains are on a different gel and so the graphic quantification of this data on the right can only be qualitatively compared with the other two strains. (D) Over-expression of limiting factors allows replication initiation of the dormant origin ARS301 in rpd3 null cells. 2D N/N gel analysis of replication intermediates from S-phase of the indicated strains. S-phase samples were pooled every 5 min from 10 to 30 min after release from α-factor in YP-galactose at 30°C. The genomic DNA digest was Nco1 for ARS305 and Nde1 for ARS301.
Over-expression of limiting factors in the absence of the deacetylase Rpd3 allows firing of heterochromatic dormant origins
Inefficient firing of a particular origin results in passive replication of that origin locus by a replication fork from a neighbouring origin (McCune et al, 2008). In the SSDD and SSDDCS strains that show advanced replication timing, the inefficient origins ARS608 and ARS313, which reside in early-replicating regions of the genome (Raghuraman et al, 2001), initiate more efficiently (Supplementary Figure S8). These data suggest that it is the delay in initiation at these origins, relative to their neighbouring origins, that contributes to their inefficiency. Despite this, we note that the inefficient origins associated with the heterochromatic, late-replicating mating type locus at HML (ARS301 and ARS302/303/320) still do not fire efficiently in the SSDDCS strain (Figure 3D and unpublished observations). Although these origins have a high proportion of pre-RC occupancy across a population of cells (Santocanale and Diffley, 1996), they are normally passively replicated and are referred to as dormant origins. It is possible that pre-RCs at these dormant origins are in a chromatin state that prevents access of these limiting factors even at high concentrations. Previous work has reported that increasing histone acetylation around origins, for example by deletion of the histone deacetylase RPD3, can advance replication timing in budding yeast (Vogelauer et al, 2002; Aparicio et al, 2004; Knott et al, 2009). To test the role of histone acetylation in dormant origin firing, we over-expressed Sld2, Sld3, Dpb11 and Dbf4 in rpd3 null cells. This strain allows the earlier firing of late origins (Figure 3B and C), confirming that these factors are limiting even in the absence of Rpd3 and, as with a wild-type strain, the additional over-expression of Cdc45 and Sld7 also increases the efficiency of both early and late origins (Figure 3B). Importantly, we show by one- (Figure 3C) and two-dimensional electrophoresis (Figure 3D) that the dormant heterochromatic origins ARS302/303/320 and ARS301 initiate replication only when both Rpd3 is absent and the limiting replication initiation factors are over-expressed. Therefore, although increasing the abundance of Sld3, Sld2, Dpb11 and Dbf4 is sufficient for the early initiation of some origins, other origins (such as those at HML) remain refractory to replication initiation, at least in part due to their hypo-acetylated chromatin state (Suka et al, 2001).
The abundance of Sld3, Sld2, Dpb11 and Dbf4 is rate limiting for S-phase length in the absence of Rpd3
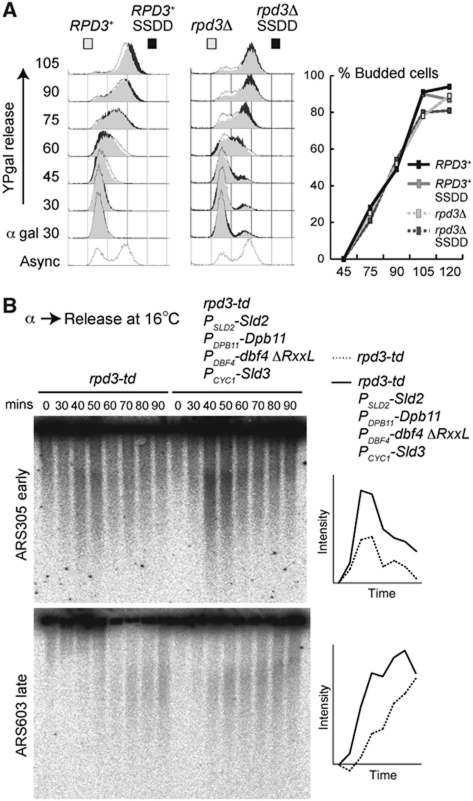
Over-expression of Sld3, Dbf4, Sld2 and Dpb11 in combination with deletion of RPD3 increases the speed of S-phase in budding yeast (Figure 4A; Supplementary Figure S9A—rpd3Δ, 60–90 min time points), without affecting the G1–S transition. S-phase is not faster in the over-expression strain with wild-type RPD3 (Figure 4A; Supplementary Figure S9A), which suggests that it may be the contribution of dormant origin firing that accelerates S-phase in rpd3Δ SSDD cells. The fact that the speed of S-phase is increased by over-expression of Sld3, Dbf4, Sld2 and Dpb11 in combination with deletion of RPD3 strongly suggests that both the levels and accessibility of these proteins at origins is indeed limiting for the rate of replication in budding yeast.
Figure 4.
Limiting factors and chromatin accessibility control replication dynamics. (A) RPD3 inactivation combined with increased expression of Sld3, Sld2, Dpb11 and Dbf4 makes S-phase faster. Flow cytometry of the indicated strains after release from α-factor into YP-galactose at 16°C. The flow cytometry results are overlaid to allow direct comparison of S-phase progression. (Right) Budding index. (B) Increasing origin accessibility allows late origins to fire early with a small increase of the levels of the limiting factors. Replication intermediate assay after release from α-factor (0) into YP-galactose at 16°C for the indicated times as in Figure 2b. The rpd3-td allele corresponds to the temperature degron cassette as an N-terminal fusion at the endogenous RPD3 locus. This allele is driven by the copper-inducible CUP1 promoter. Strains were pre-grown in YP-raffinose + 0.2 mM CuSO4 and released into medium without additional CuSO4. SLD2, SLD3, DPB11 and DBF4 are expressed as second copies from their own promoter, while Sld3 is expressed as a second copy from the Cyc1 promoter. The dbf4 ΔRxxL allele corresponds to the APC/C destruction box mutations R10A, L13A and R62A, L65A. (Right) Graphic quantification of nascent DNA using Image J analysis of phosphorimage on the left. The y axis is the intensity of the phosphorimage signal in arbitrary units.
We reasoned that if the accessibility difference between origins for limiting proteins is reduced in cells lacking Rpd3, then only a small increase in Sld2, Sld3, Dpb11 and Dbf4 levels should be sufficient to advance replication timing in this strain. To slightly increase the levels of these limiting proteins, we integrated second copies of SLD2, DPB11, DBF4 (expressed from their own promoter) and SLD3 (expressed from the Cyc1 promoter) in haploid yeast. A second wild-type copy of DBF4 has little effect on Dbf4 levels, probably because it is an APC/C substrate, so we instead expressed a version of Dbf4 that cannot be degraded by the APC/C (dbf4 ΔRxxL; Ferreira et al, 2000). This approximate doubling of Sld3, Sld2 and Dpb11, together with an increase in Dbf4 levels is on its own not sufficient for the early firing of the late origin ARS603 (Supplementary Figure S9B and C). However, the combination of a modest increase in the levels of these proteins together with a conditional RPD3 mutant allows a late origin to fire early and makes S-phase faster (Figure 4B; Supplementary Figure S9D and E). This experiment strongly suggests that the timing of firing of an origin in budding yeast is a combination of chromatin state and the availability of limiting initiation proteins and proves that the timing effects we are describing in this study are not simply due to high levels of essential replication factors. The rpd3 null allele could not be used in this experiment because in combination with a constitutive increase in SSDD levels this strain grows poorly, consistent with Figure 6B.
dNTPs become limiting when origins fire simultaneously early in S-phase
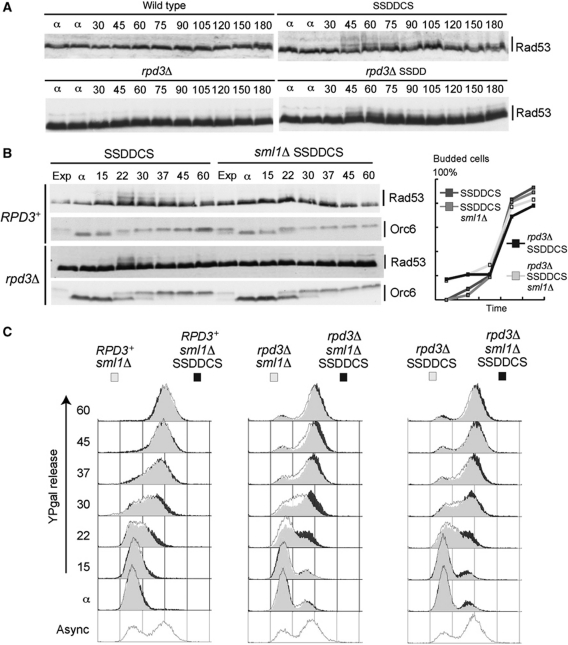
We noticed that when we induce origins to fire simultaneously, the checkpoint kinase Rad53 becomes transiently activated in S-phase (Figure 5A and B), indicative of stochastic replication fork stalling. The increased fork number that results from more origin firing in early S-phase may lead to the depletion of factors that are essential for replication elongation. In support of this hypothesis, in hydroxyurea, when deoxyribonucleotide levels become limiting, the average size of nascent DNA is smaller in strains that allow the early firing of late origins (SSDD and SSDDCS), suggesting that increased replication initiation causes further dNTP depletion (Supplementary Figure S10). Reduction of available dNTPs by the early firing of late origins in an otherwise normal S-phase could potentially increase stochastic fork stalling, resulting in the observed Rad53 activation. To test this hypothesis, we constitutively increased deoxyribonucleotide concentrations by deletion of the ribonucleotide reductase inhibitor SML1. Deletion of SML1 indeed suppresses the activation of Rad53 in these experiments (Figure 5B; Supplementary Figure S11A), suggesting that increased fork number in early S-phase causes nucleotide depletion and subsequent checkpoint activation. Although the G1–S transition is not affected in any of these strains (as determined by Orc6 phosphorylation and budding—Figure 5B), the combination of increased dNTP concentrations and the over-expression of limiting initiation factors further increases the speed of S-phase in cells lacking Rpd3 (Figure 5C; Supplementary Figure S11B; 22–30 min time points). These results show that the temporal programme of origin firing, together with the available supply of nucleotides, controls S-phase length in budding yeast.
Figure 5.
Deoxyribonucleotide levels become limiting when origins fire early. (A) Rad53 is transiently activated in S-phase when origins fire simultaneously early. Rad53 western blot of the indicated strains after release from α-factor into YP-galactose at 16°C. (B) Increasing nucleotide concentrations suppresses this Rad53 activation. Rad53 and Orc6 western blots of indicated strains after release from α-factor into YP-galactose at 25°C. Exp=exponentially growing cells. (Right) Budding index. (C) The additional increase in nucleotide levels makes S-phase even faster when the limiting factors are over-expressed in cells lacking RPD3. Flow cytometry from experiment described in (B). The flow cytometry results are overlaid to allow direct comparison of S-phase progression.
The transient Rad53 activation in the above experiments could restrain S-phase progression directly, for example by the inhibition of replication initiation; however, we found no increase in the speed of S-phase upon over-expression of Sld3 and Dbf4 mutants that can not be inhibited by Rad53 (Zegerman and Diffley, 2010 and unpublished observations). In addition, this low level of replication fork stalling when limiting factors are over-expressed might indirectly increase the firing of late or dormant origins by reducing the passive replication of these loci. To rule out this possibility, we measured late/dormant origin firing in the SSDD strain lacking SML1, where the activation of Rad53 is suppressed. Late origins fire early and dormant origins initiate when both Rpd3 and Sml1 are absent and Sld3, Sld2, Dpb11 and Dbf4 are over-expressed (Supplementary Figure S12), suggesting that the effect on the temporal programme we are describing in this study is independent of fork stalling and checkpoint activation.
Abolition of the temporal programme causes growth defects
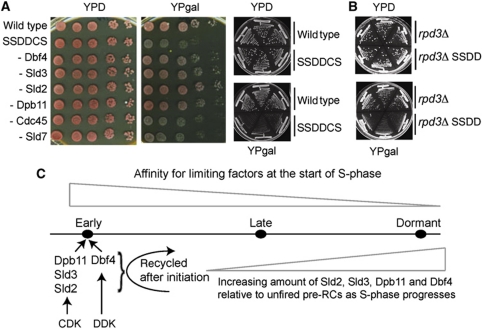
Growth assays showed that the SSDDCS strain is sick in the presence of galactose and this phenotype requires the over-expression of Sld2, Sld3, Dpb11 and Dbf4 (Figure 6A). Restreaking of these strains on a second galactose plate showed that over several cell cycles, the SSDDCS strain and the SSDD strain lacking Rpd3 exhibit a severe growth defect (Figure 6A and B). Constitutively increasing dNTP pools does not suppress the lethality of these strains, suggesting that nucleotide depletion/transient Rad53 activation in S-phase is not likely to be the cause of cell death (Supplementary Figure S13A). To test whether this lethality is due to inappropriate origin firing, we combined the SSDDCS strain with a temperature-sensitive allele of the pre-RC component Cdc6 (cdc6-1). At the semi-permissive temperature, this CDC6 allele reduces pre-RC formation and thus limits initiation events. Importantly, the cdc6-1 mutant suppresses the lethality of the SSDDCS strains (Supplementary Figure S13B), without affecting the expression levels of these factors (Supplementary Figure S13C), strongly suggesting that the lethality is due to increased origin firing. One possible explanation for this lethality is that increased dosage of the CDK targets and the APC/C substrate Dbf4 may allow inappropriate replication initiation outside of S-phase. For example, bypass of CDK phosphorylation of Sld3 and Sld2, combined with over-expression of Dbf4 allows replication in G1-phase (Zegerman and Diffley, 2007). Although we do not detect any replication in G1-phase in the SSDDCS strain by flow cytometry (Supplementary Figure S13D), we cannot rule out that some lethality may be caused by undetectable levels of re-replication. Despite this, the data here show that the low levels of these limiting factors is physiologically important to prevent inappropriate replication initiation in budding yeast.
Figure 6.
Too much replication initiation early in S-phase causes loss of cell viability. (A) Growth assay of wild-type, SSDDCS strain or strains over-expressing five of the six SSDDCS factors (−). (Right) Restreak of three separate colonies onto a second plate. (B) As (A). (C) Model of the temporal regulation of replication initiation.
Discussion
In prokaryotes, the timing of replication initiation is largely determined by the levels and activity of the origin binding protein DnaA (Katayama et al, 2010). In eukaryotes on the other hand, pre-RCs form at all origins, even those that do not initiate in the subsequent S-phase (Santocanale and Diffley, 1996). However, eukaryotic origins do not fire simultaneously at the beginning of S-phase, but initiate according to a temporal programme. One possible explanation for this programme is that the chromatin context of a pre-RC establishes different affinities for limiting replication factors in S-phase (Figure 6C, top triangle). We have shown that the low levels of the two CDK targets Sld3 and Sld2, their binding partner Dpb11 and the DDK subunit Dbf4 are indeed limiting for replication initiation in budding yeast. Importantly, Sld2, Sld3, Dpb11 and DDK are not incorporated into replisomes and therefore after initiation they are free to recycle to other unfired pre-RCs (Figure 6C, curved arrow). The low levels of these factors suggest that this recycling is essential for subsequent replication initiation, which may explain why both CDK and DDK are required throughout the S-phase for all origin firing events (Bousset and Diffley, 1998; Donaldson et al, 1998a, 1998b). The recycling of these factors may also provide a mechanism for the increased probability of origin firing as S-phase progresses (Rhind, 2006), because the concentration of these proteins relative to unfired pre-RCs will increase during S-phase (Figure 6C, bottom triangle). It is noteworthy that it is the downstream targets of CDK, not CDK itself, which is limiting for initiation, as this kinase is vital to inhibit re-replication in S-phase by multiple mechanisms (Nguyen et al, 2001).
In our experiments, additional over-expression of the Sld3-binding proteins Cdc45 and Sld7 seems to increase the efficiency of initiation at early and late origins. We cannot rule out that these factors are themselves limiting although we did not detect any additional effect on timing by density substitution analysis or S-phase length in a CLB5 mutant. Despite this, it is important to note that in any multi-step biological reaction, if the levels of the most limiting component are increased, the second most limiting component becomes rate determining and so on. Recent work has shown that Sld2, Sld3, Dpb11 and Cdc45 are limiting for the in vitro reconstitution of replication initiation from yeast extracts, which is consistent with our findings (Heller et al, 2011). Cdc45 has also recently been shown to be of low abundance in mammalian cells (Wong et al, 2011). Since Cdc45 is incorporated into replisomes, if the levels of this protein are limiting, then replication initiation during S-phase would depend on the release of Cdc45 after termination events. In budding yeast, recycling of essential factors by termination is unlikely to be limiting for initiation because both in checkpoint mutants with stable replication forks (Zegerman and Diffley, 2010) or in our SSDD strain, early and late origins can fire in high levels of hydroxyurea where termination events are virtually zero (Alvino et al, 2007).
Although sufficient to allow earlier initiation of late origins, over-expression of Sld2, Sld3, Dpb11 and Dbf4 does not allow efficient firing of the dormant origins at HML. We found, however, that increased global histone acetylation levels (in cells lacking the histone deacetylase Rpd3) combined with the increased dose of limiting factors allows the firing of such origins. Since dormant origins can initiate if S-phase is sufficiently extended (Vujcic et al, 1999; Doksani et al, 2009), it is possible that the chromatin state of an origin, although established in G1-phase, is actually dynamic during S-phase (Unnikrishnan et al, 2010). Such dynamic modifications are not limited to histone acetylation, but may also include other changes such as histone methylation (Pryde et al, 2009; Rampakakis et al, 2009; Tardat et al, 2010). How such modifications control the access of limiting replication initiation factors at unfired pre-RCs is not known, but the identification of these limiting factors, as described here, will provide the platform for a mechanistic understanding of these affinity differences.
Eukaryotes license many more origins in G1-phase than actually fire in S-phase. While the additional licensing of origins may be an important fail-safe for complete genomic replication (Blow et al, 2011), we show here that too much replication initiation early in S-phase leads to checkpoint activation likely caused by nucleotide depletion. Temporal control of replication initiation therefore allows the number of active forks to be coordinated with the available nucleotide pools, which is important for genome integrity (Chabes and Thelander, 2003). Loss of the temporal order of origin firing may not only disrupt the coordinated replication of the genome (Blow and Ge, 2009) but may also prevent a sufficient window of opportunity for other processes that occur in S-phase, such as transcription, cohesion or chromatin assembly. The system described here will allow these possibilities to be investigated for the first time.
Materials and methods
Yeast strains
All yeast strains are based on W303a ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 and are listed in Supplementary Table SI.
Replication intermediate assays and 2D N/N gels
As previously described (Zegerman and Diffley, 2010), with minor modifications (see Supplementary data).
Total protein extraction
Cells were disrupted with glass beads by bead beating in 20% TCA. The resulting extract was pelleted, resuspended in Laemmli buffer, neutralized with Tris base and boiled.
Density substitution
Replication timing was analysed using the dense isotope transfer procedure at 30°C as previously described (Donaldson et al, 1998b) with the following adjustment: cells were blocked for ∼1 cell cycle time with α-factor, then transferred to YP-2% galactose medium containing α-factor and incubated for 1 h, before release by addition of pronase.
In situ kinase assays
As previously described (Pellicioli et al, 1999).
rpd3-td strain
A conditional allele of RPD3 was made by replacing the endogenous RPD3 with an N-terminal RPD3 temperature degron fusion driven by the copper-inducible CUP1 promoter, as previously described (Sanchez-Diaz et al, 2004). This allele is at least partially inactive because diploids homozygous for the rpd3-td allele are unable to go through meiosis (Burgess et al, 1999).
Supplementary Material
Acknowledgments
We thank Hiroyuki Araki and Seiji Tanaka for sharing unpublished data and Jon Pines, Andrea Brand and members of the Zegerman laboratory for comments on the manuscript. We are grateful to John Diffley and Boris Pfander for anti-Sld3 and anti-Dpb11 serum. PZ is funded by the AICR 10-0908 and DM by the American-Italian Cancer Foundation and EMBO LTF 392-2010. PZ and DM acknowledge the core funding provided by the Wellcome Trust (092096) and CRUK (C6946/A14492). Work by AD and AEM was funded by MRC Grant G0600774 and by a Scottish Universities Life Sciences Alliance studentship.
Author contributions: AM and AD performed and designed the density substitution experiments. DM and PZ performed and designed the remaining experiments. PZ wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alvino GM, Collingwood D, Murphy JM, Delrow J, Brewer BJ, Raghuraman MK (2007) Replication in hydroxyurea: it's a matter of time. Mol Cell Biol 27: 6396–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM (2004) The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Stout AM, Bell SP (1999) Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA 96: 9130–9135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D (2010) Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 17: 894–900 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Ann Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ (2009) A model for DNA replication showing how dormant origins safeguard against replication fork failure. EMBO Rep 10: 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA (2011) How dormant origins promote complete genome replication. Trends Biochem Sci 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousset K, Diffley JF (1998) The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev 12: 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Ajimura M, Kleckner N (1999) GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Natl Acad Sci USA 96: 6835–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Thelander L (2003) DNA building blocks at the foundation of better survival. Cell Cycle 2: 171–173 [DOI] [PubMed] [Google Scholar]
- Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M (2009) Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Cell 137: 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ (1998a) Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev 12: 491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Raghuraman MK, Friedman KL, Cross FR, Brewer BJ, Fangman WL (1998b) CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell 2: 173–182 [DOI] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C (2009) A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA 106: 20240–20245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MF, Santocanale C, Drury LS, Diffley JF (2000) Dbf4p, an essential S phase-promoting factor, is targeted for degradation by the anaphase-promoting complex. Mol Cell Biol 20: 242–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP (2009) Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev 23: 643–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K (2009) A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J 28: 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP (2011) Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell 146: 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamimura Y, Tak YS, Sugino A, Araki H (2001) Sld3, which interacts with Cdc45 (Sld4), functions for chromosomal DNA replication in Saccharomyces cerevisiae. EMBO J 20: 2097–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K (2010) Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8: 163–170 [DOI] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM (2009) Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K (2010) How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev 24: 1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Mosqueda J, Maas NL, Jonsson ZO, Defazio-Eli LG, Wohlschlegel J, Toczyski DP (2010) Damage-induced phosphorylation of Sld3 is important to block late origin firing. Nature 467: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL (1988) Time of replication of yeast centromeres and telomeres. Cell 54: 505–513 [DOI] [PubMed] [Google Scholar]
- McCune HJ, Danielson LS, Alvino GM, Collingwood D, Delrow JJ, Fangman WL, Brewer BJ, Raghuraman MK (2008) The temporal program of chromosome replication: genomewide replication in clb5{Delta} Saccharomyces cerevisiae. Genetics 180: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M (2010) Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ (2001) Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068–1073 [DOI] [PubMed] [Google Scholar]
- Patel PK, Kommajosyula N, Rosebrock A, Bensimon A, Leatherwood J, Bechhoefer J, Rhind N (2008) The Hsk1(Cdc7) replication kinase regulates origin efficiency. Mol Biol Cell 19: 5550–5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J 18: 6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F, Jain D, Kerr A, Curley R, Mariotti FR, Vogelauer M (2009) H3 k36 methylation helps determine the timing of cdc45 association with replication origins. PLoS One 4: e5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Brewer BJ, Fangman WL (1997) Cell cycle-dependent establishment of a late replication program. Science 276: 806–809 [DOI] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Rampakakis E, Di Paola D, Chan MK, Zannis-Hadjopoulos M (2009) Dynamic changes in chromatin structure through post-translational modifications of histone H3 during replication origin activation. J Cell Biochem 108: 400–407 [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF (2009) Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell 139: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N (2006) DNA replication timing: random thoughts about origin firing. Nat Cell Biol 8: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF (1995) Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J 14: 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Diaz A, Kanemaki M, Marchesi V, Labib K (2004) Rapid depletion of budding yeast proteins by fusion to a heat-inducible degron. Sci STKE 2004: PL8. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF (1996) ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J 15: 6671–6679 [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Sharma K, Diffley JF (1999) Activation of dormant origins of DNA replication in budding yeast. Genes Dev 13: 2360–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B (2006) Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell 24: 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B (2010) The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463: 113–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M (2001) Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol Cell 8: 473–479 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Umemori T, Endo S, Muramatsu S, Kanemaki M, Kamimura Y, Obuse C, Araki H (2011) Sld7, an Sld3-associated protein required for efficient chromosomal DNA replication in budding yeast. EMBO J 30: 2019–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E (2010) The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T (2010) Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol 17: 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M (2002) Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Vujcic M, Miller CA, Kowalski D (1999) Activation of silent replication origins at autonomously replicating sequence elements near the HML locus in budding yeast. Mol Cell Biol 19: 6098–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PG, Winter SL, Zaika E, Cao TV, Oguz U, Koomen JM, Hamlin JL, Alexandrow MG (2011) Cdc45 limits replicon usage from a low density of preRCs in mammalian cells. PLoS One 6: e17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JR, Gilbert DM (1996) A distinct G1 step required to specify the Chinese hamster DHFR replication origin. Science 271: 1270–1272 [DOI] [PubMed] [Google Scholar]
- Wu PY, Nurse P (2009) Establishing the program of origin firing during S phase in fission yeast. Cell 136: 852–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD, Jennings EG, Young RA, Bell SP, Aparicio OM (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2010) Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature 467: 474–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.