Abstract
The phosphoinositide 3-kinase (PI3K) pathway has been extensively studied in neuronal function and morphogenesis. However, the precise molecular mechanisms of PI3K activation and its downstream signalling in neurons remain elusive. Here, we report the identification of the Neuronal tYrosine-phosphorylated Adaptor for the PI 3-kinase (NYAP) family of phosphoproteins, which is composed of NYAP1, NYAP2, and Myosin16/NYAP3. The NYAPs are expressed predominantly in developing neurons. Upon stimulation with Contactin5, the NYAPs are tyrosine phosphorylated by Fyn. Phosphorylated NYAPs interact with PI3K p85 and activate PI3K, Akt, and Rac1. Moreover, the NYAPs interact with the WAVE1 complex which mediates remodelling of the actin cytoskeleton after activation by PI3K-produced PIP3 and Rac1. By simultaneously interacting with PI3K and the WAVE1 complex, the NYAPs bridge a PI3K–WAVE1 association. Disruption of the NYAP genes in mice affects brain size and neurite elongation. In conclusion, the NYAPs activate PI3K and concomitantly recruit the downstream effector WAVE complex to the close vicinity of PI3K and regulate neuronal morphogenesis.
Keywords: neuron, NYAP, PI3K, tyrosine phosphorylation, WAVE
Introduction
The phosphoinositide 3-kinase (PI3K) signalling pathway is a fundamental pathway for various cellular events in neurons, including morphogenesis (Rodgers and Theibert, 2002; Shi et al, 2003; Jaworski et al, 2005; Kumar et al, 2005), membrane expansion (Laurino et al, 2005), survival and apoptosis (Brunet et al, 2001), and ion channel regulation (Sanna et al, 2002; Viard et al, 2004). PI3K generates phosphatidylinositol 3,4,5-trisphosphate (PIP3), which then mediates the recruitment and subsequent activation of PH domain-containing effector proteins (Cantrell, 2001). Many different proteins that possess the PH domain, including the Akt and Tec family of kinases, regulators for small G proteins, such as a Rac guanine nucleotide exchange factor, and phospholipase C (PLC) γ isoforms, directly bind to PIP3. Thus, interactions of PIP3 with many different effector proteins enable PI3K to be involved in a number of different cellular responses. Neuronal phenotypes of knockout (KO) mice for PI3K itself include losses of synapses and myelinated axons (Tohda et al, 2006), decreased axonal extension and dampened axonal regeneration (Eickholt et al, 2007), reduction in reproductive hormone secretions in males (Acosta-Martínez et al, 2009), and memory and behavioural phenotypes (Tohda et al, 2006, 2009). Moreover, PTEN, the central negative regulator of the PI3K pathway, is required to obtain proper dendrite morphology, neuronal soma size, and brain size without affecting neuronal cell death during development in mice (Backman et al, 2001; Kwon et al, 2001, 2006). KO mice for Akt3, which is one of the major downstream effectors of PI3K in the brain, demonstrate reduced brain size but do not show aberrant apoptosis (Easton et al, 2005; Tschopp et al, 2005). These reports suggest roles of the PI3K pathway in neuronal morphogenesis rather than regulation of neuronal survival. However, the precise mechanisms leading to the preference of neuronal PI3K for regulation of morphogenesis remain to be identified.
Remodelling of the actin cytoskeleton plays a critical role in altering cellular morphology, and it controls a range of cellular events, such as wound healing, immune defense, embryonic development, and neuronal outgrowth (Suetsugu and Takenawa, 2003). The Rho family of GTPases, including Rho, Cdc42, and Rac, mediates these processes by promoting distinct forms of actin remodelling such as stress fibres, filopodia, and lamellipodia (Hall, 1998). Cdc42 and Rac relay signals to cytoskeletal sites through the WASP and WAVE family of proteins, respectively, which then activate the Arp2/3 complex and stimulate actin polymerization (Stradal and Scita, 2006). In fibroblast cells, PDGF modulates WAVE-mediated cell motility through the activation of PI3K and the production of PIP3, which in turn stimulates the formation of lamellipodia (Suetsugu et al, 2003; Oikawa et al, 2004; Sossey-Alaoui et al, 2005). In this process, the PDGF receptor activates PI3K directly, and PI3K-produced PIP3 is shown to be required for the regulation of Rac activity (Shinohara et al, 2002). The activation of Rac is also mediated by the direct interaction between Rac and PI3K p85 (Hawkins et al, 1995; Tolias et al, 1995). Then, WAVE activity towards the Arp2/3 complex is stimulated by binding with PIP3 (Oikawa et al, 2004) and Rac (Steffen et al, 2004) and phosphorylation by the Abl, Src, and Cdk5 kinases (Leng et al, 2005; Ardern et al, 2006; Kim et al, 2006; Stuart et al, 2006; Sossey-Alaoui et al, 2007; Lebensohn and Kirschner, 2009; Chen et al, 2010). In neurons, however, precise activation mechanisms of the pathway are not fully understood.
Fyn, a member of the Src family of protein tyrosine kinases, is one of the major tyrosine kinases in the brain (Umemori et al, 1992). Substantial genetic evidence suggests that Fyn plays important roles in neuronal morphogenesis (Grant et al, 1992; Sasaki et al, 2002; Morita et al, 2006; Kotani et al, 2007), but the signals that function upstream and downstream of Fyn in neurons are poorly understood. In the course of screening for tyrosine kinase substrates in the brain, we identified a previously uncharacterized family of proteins that we named NYAP—Neuronal tYrosine-phosphorylated Adaptor for the PI 3-kinase. Here, we show that the NYAPs are involved in both the activation of PI3K and the recruitment of the downstream effector WAVE complex to the close vicinity of PI3K, and regulate brain size and neurite outgrowth in mice.
Results
Identification of the NYAP family of proteins as tyrosine kinase substrates in vitro
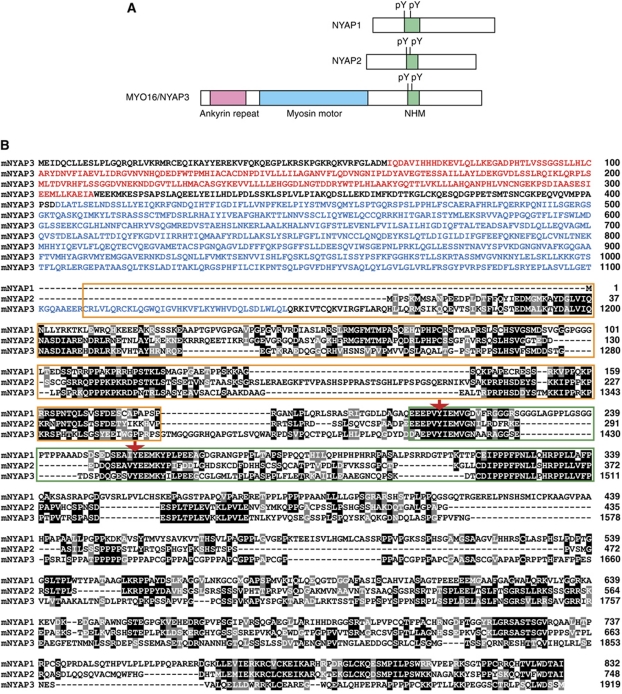
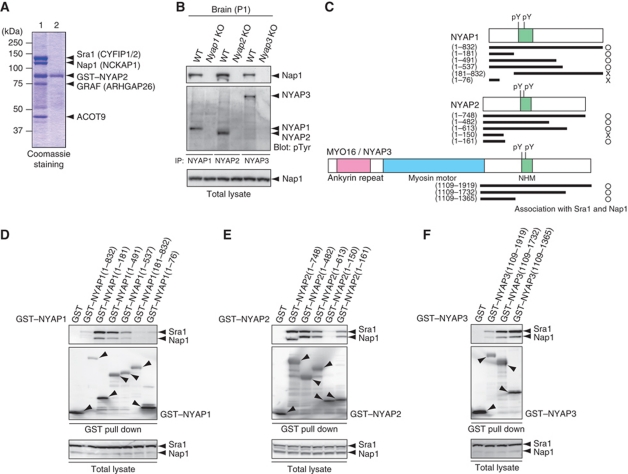
To identify substrates of the Src family of tyrosine kinases in the brain, we performed solid-phase phosphorylation screening (Yokoyama et al, 2002, 2006). This screening yielded a previously uncharacterized protein, which we termed NYAP1. Extensive screening of the protein database identified two proteins with partial similarity to NYAP1: an uncharacterized protein KIAA1486 (termed NYAP2) and an unconventional myosin MYO16 (termed NYAP3 for descriptive purposes). MYO16/NYAP3 is expressed during brain development and associates with F-actin and the protein phosphatase 1 catalytic subunits 1α and 1γ1 (Patel et al, 2001). Moreover, MYO16/NYAP3 regulates S-phase progression when overexpressed in non-neuronal cells (Cameron et al, 2007); however, physiological roles for MYO16/NYAP3 in the brain remain unclear. The short sequence motif present in NYAP1, 2, and 3 contains conserved tyrosine residues and a proline-rich stretch. We termed this the NYAP Homology Motif (NHM). NYAP1 and NYAP2 do not have any identifiable functional domains, whereas MYO16/NYAP3 contains ankyrin repeats and a myosin motor domain (Figure 1). Based on the partial similarity in the NHM, we considered that NYAP1, 2, and MYO16/NYAP3 form a novel family of proteins. It seems likely that no more member of the NYAP family exists in mouse genome sequences. To examine the physiological roles of the NYAPs, we generated Nyap1, 2, and 3 KO mice (Supplementary Figure S1). NYAPs triple knockout (TKO) mice were apparently healthy and fertile and are currently analysed for behavioural abnormalities.
Figure 1.
Identification of the NYAP family of proteins. (A) Schematic representation of NYAP1, NYAP2, and MYO16/NYAP3. (B) Mouse NYAPs amino-acid sequence alignment. Identical or similar residues to the column consensus are printed with black or grey backgrounds, respectively. Ankyrin repeats in MYO16/NYAP3 are indicated in red letters, the MYO16/NYAP3 motor domain is in blue letters, NHM motifs are shown in a green box, regions involved in the interaction with the WAVE complex are shown in an orange box, and phosphorylated tyrosine residues are denoted with red arrows.
The NYAPs are expressed in developing cortical neurons after migration
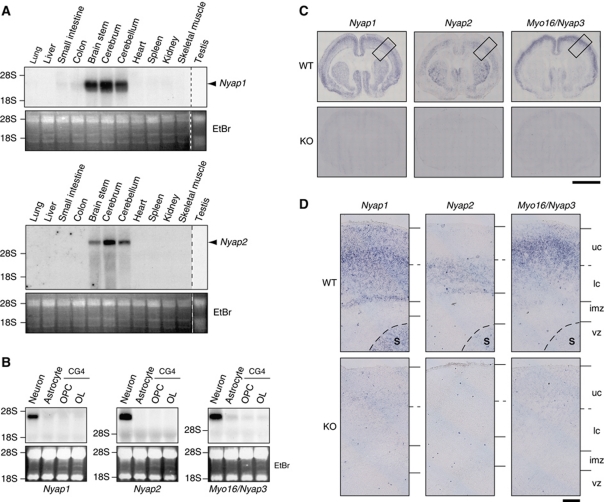
The Myo16/Nyap3 mRNA is specifically expressed in the developing brain (Patel et al, 2001). Northern blot analysis revealed that Nyap1 and Nyap2 mRNA expression was also specific to the brain among the tissues examined (Figure 2A). Furthermore, among three major brain cell types, Nyap1, 2, and 3 mRNAs were all present specifically in neurons but not in astrocytes or oligodendrocytes (Figure 2B).
Figure 2.
Neuron-specific expression of the NYAPs. (A) Northern blot analysis of Nyap1 and Nyap2 mRNA in mouse tissues. Vertical dashed lines represent boundaries between different gels. Ethidium bromide-stained images are shown in the lower panels as loading controls (EtBr). (B) Northern blot analysis of NYAPs mRNA in rat cortical neurons and astrocytes, as well as the rat CG4 oligodendrocyte cell line during precursor (OPC) and mature (OL) stages. (C) In situ hybridization analysis of NYAPs mRNA in postnatal day 1 (P1) mouse brains. Scale bar: 2 mm. (D) Magnified images of the boxed areas in (C). Wild-type (WT) P1 mouse brains (upper panels) and each of the Nyap1, 2, or 3 knockout P1 mouse brains (KO, lower panels) were hybridized with Nyap1, 2, or 3 mRNA probes. S, striatum; uc, upper cortical area; lc, lower cortical area; imz, intermediate zone; and vz, ventricular and subventricular zone. Scale bar: 200 μm.
In situ hybridization of postnatal day 1 (P1) mouse brains revealed that Nyap1 mRNA was present in the neocortex and the striatum but not in the olfactory bulb. Nyap2 mRNA was present in the neocortex, the striatum, and the olfactory bulb; and Myo16/Nyap3 mRNA was present in the neocortex and the olfactory bulb but not in the striatum. No expression of Nyap1, 2, or 3 was found in the ventricular zone or the corpus callosum (Figure 2C). The highest expression of Nyap1, 2, and 3 was observed in the middle, lower, and upper neocortex, respectively, in P1 mouse brains (Figure 2D).
We next examined when neurons began to express the NYAPs (Supplementary Figure S2). Nyap1 expression was detectable in the cortical plate as early as embryonic day 14 (E14); it peaked in the middle neocortex at P1 and then gradually decreased. Nyap2 expression began in the lower neocortex at E18 and decreased thereafter. In contrast to Nyap1, Nyap2 expression persisted in the striatum until at least P7. The expression pattern of Myo16/Nyap3 resembled that of Nyap1 but occurred in slightly upper layers of the neocortex. Because the NYAPs were not expressed in the ventricular zone and intermediate zone during embryonic stages, we concluded that neurons began to express the NYAPs just after their migration into the neocortex.
The NYAPs are tyrosine phosphorylated by Fyn upon Contactin stimulation
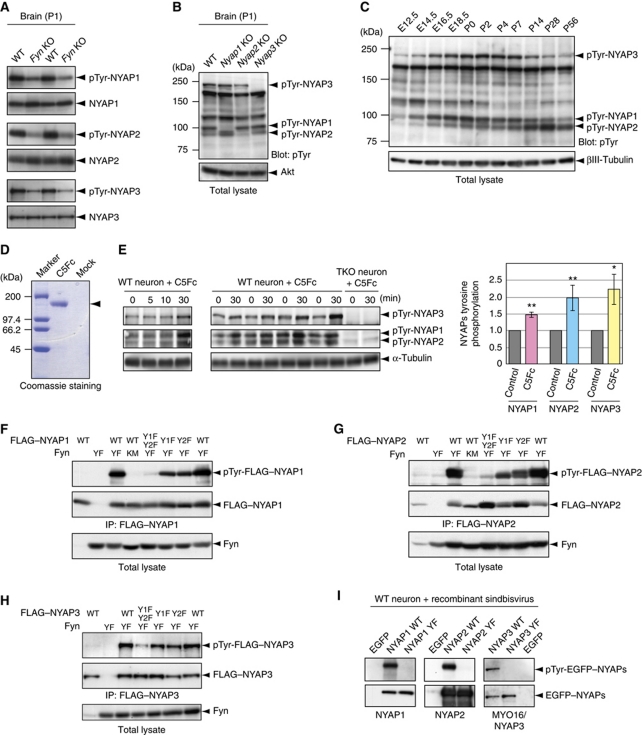
We initially identified NYAP1 as a protein that was directly phosphorylated by Fyn in vitro in the solid-phase phosphorylation screening. To determine whether the NYAPs are genuine tyrosine kinase substrates in vivo, we examined their tyrosine phosphorylation in the brain. When immunoprecipitated from wild-type (WT) P1 mouse brains, the NYAPs were tyrosine phosphorylated (Figure 3A). The phosphorylation levels of the NYAPs were lower in Fyn KO brains (Figure 3A). These data suggest that the Src family of kinases directly phosphorylates the NYAPs in the brain.
Figure 3.
Tyrosine phosphorylation of the NYAPs. (A) Fyn-dependent tyrosine phosphorylation of the NYAPs. Tyrosine phosphorylation of the NYAPs immunoprecipitated from WT and Fyn KO brains was detected with the 4G10 anti-phosphotyrosine antibody. Amounts of the NYAPs in the immunoprecipitates are shown in the lower panels. (B) Overall tyrosine phosphorylation in total lysates of WT and Nyap1, 2, and 3 KO brains. Amounts of Akt are shown as loading controls. (C) Overall tyrosine phosphorylation in total brain lysates obtained from mice of the indicated ages. Amounts of βIII-tubulin are shown as loading controls. E12.5: embryonic day 12.5. (D) Quantification of the recombinant Contactin5–Fc fusion protein. Contactin5–Fc (C5Fc) was captured from the conditioned medium with protein G-sepharose and quantified by Coomassie staining. Protein concentration of C5Fc was estimated by comparing with SDS–PAGE Standards (high range, Bio-Rad). (E) C5Fc-induced tyrosine phosphorylation of the NYAPs. Cultured cortical neurons obtained from WT and NYAPs triple knockout (TKO) brains were stimulated with 6 μg/ml C5Fc for the indicated time periods. Tyrosine phosphorylation of the NYAPs was examined in total cell lysates. TKO neurons were used as negative controls, ensuring that phosphorylation signals in WT neurons were predominantly phosphorylated NYAPs. Upregulation of tyrosine phosphorylation of the NYAPs after 30 min stimulation with C5Fc was quantified (WT, n=8; TKO, n=8). A single asterisk (*) and double asterisk (**) indicate P<0.05 and P<0.01 compared with control neurons, respectively. (F–H) Determination of phosphorylated tyrosine residues in the NYAPs in HEK293T cells. HEK293T cells were transfected with Fyn (YF, the constitutively active mutant; KM, the kinase inactive mutant) and FLAG–NYAP1 (F), FLAG–NYAP2 (G), FLAG–MYO16/NYAP3 (H), or their YF mutants, as indicated. NYAP1 Y1F indicates that Tyr212 was mutated to phenylalanine in mouse NYAP1; NYAP1 Y2F, Tyr257; NYAP2 Y1F, Tyr277; NYAP2 Y2F, Tyr300; NYAP3 Y1F, Tyr1416; and NYAP3 Y2F, Tyr1441. Y1F–Y2F indicates that both tyrosine residues (Y1 and Y2) were mutated. The cell lysates were immunoprecipitated with the M2 anti-FLAG antibody and immunoblotted with the RC20 anti-phosphotyrosine antibody. Although HEK293T cells express Src-family kinases endogenously, the constitutively active mutant of Fyn (FynYF) was introduced to facilitate comparison of phosphorylation levels. (I) Determination of phosphorylated tyrosine residues in the NYAPs in neurons. Cultured cortical neurons were infected with recombinant sindbisviruses expressing EGFP, EGFP-tagged wild-type NYAPs (WT), or their YF mutants. The neurons were lysed, immunoprecipitated with an anti-EGFP antibody, and immunoblotted with the 4G10 anti-phosphotyrosine antibody.
In WT P1 mouse brains, five conspicuous tyrosine-phosphorylated proteins with sizes of 240, 180, 120, 100, and 90 kDa were present (Figure 3B). Interestingly, the sizes of NYAP1, 2, and 3 are 100, 90, and 240 kDa, respectively, which match three of the five phosphorylated proteins. In addition, the bands at 100, 90, and 240 kDa were absent in the lysates of Nyap1, 2, and 3 KO brains, respectively, indicating that these bands represented tyrosine phosphorylation of NYAP1, 2, and 3. The 180- and 120-kDa phosphoproteins have not yet been identified. Next, tyrosine phosphorylation levels of the NYAPs were quantitatively examined during brain development (Figure 3C; see also Supplementary Figure S11 for protein expression levels of the NYAPs). Phosphorylation of NYAP1 and MYO16/NYAP3 began at E14, peaked during perinatal days in the whole brain, and then gradually decreased. Phosphorylation of NYAP2 began at E16; in the adult brain, phosphorylated NYAP2 expression persisted. These results indicated that the NYAPs were heavily phosphorylated on tyrosines, accounting for a large percentage of tyrosine phosphorylation in the brain throughout the entire life of a mouse.
The Src family of kinases is activated in response to various stimuli, so we tested whether these stimuli induced tyrosine phosphorylation of the NYAPs. Recently, members of the Contactin family of GPI-anchored membrane proteins have been demonstrated to activate the Src family of tyrosine kinases directly (Kasahara et al, 2002) or indirectly via PTPα (Ponniah et al, 1999; Su et al, 1999; Ye et al, 2008). Contactins (Contactin1–6) are a subgroup of molecules belonging to the immunoglobulin superfamily that are expressed exclusively in the nervous system. Contactin1 and 2 have been studied extensively in neurite extension and myelination (Shimoda and Watanabe, 2009). We found that the Contactin5–Fc protein induced the phosphorylation of the NYAPs in cortical neurons (NYAP1, 147.7±8.7%; NYAP2, 198.1±23.9%; NYAP3, 223.9±45.9%; Figure 3D and E). Surprisingly, other stimuli we examined (NGF, BDNF, GDNF, EGF, PDGF, IGF, Insulin, Neuregulin, Reelin, Semaphorin-3A, Ephrin-A3, Ephrin-B2, NMDA, KCl, and glutamate) had no effect on the phosphorylation of the NYAPs in our assays.
Next, we determined the phosphorylation sites on the NYAPs. Based on the sequence similarity in the family, we chose to mutate conserved tyrosine residues in the NHM (indicated by red arrows in Figure 1B: NYAP1 Tyr212 and Tyr 257, NYAP2 Tyr277 and Tyr300, and NYAP3 Tyr1416 and Tyr1441) to phenylalanine (YF mutants). Mutation of each tyrosine residues in the NYAPs slightly reduced their phosphorylation in HEK293T cells (Figure 3F–H). When both tyrosine residues were mutated, their phosphorylation was almost abrogated in HEK293T cells (Figure 3F–H) and in neurons (Figure 3I), indicating that most of phosphorylation occurred at these tyrosine residues in the NHM motifs.
The NYAPs are the major binding partners of PI3K p85 in the brain
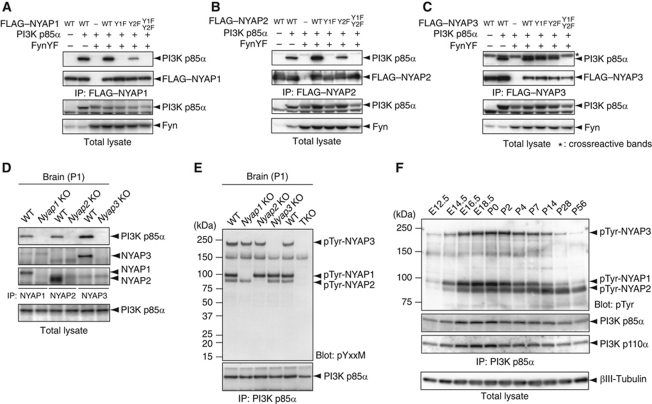
The phosphorylated tyrosine residues in the NYAPs lie in the consensus sequence (YxxM motif) for the binding by SH2 domains of the PI3K p85 subunit (Songyang et al, 1993). Immunoprecipitation analyses revealed that PI3K p85α interacted with the NYAPs in HEK293T cells (Figure 4A–C) and in the brain (Figure 4D). Further, these interactions depended on the phosphorylation state of the NYAPs because the YF mutations abrogated the interactions (Figure 4A–C). Moreover, PI3K p85β interacted with the NYAPs (Supplementary Figure S3). NYAP1 Y1F and NYAP2 Y1F did not bind at all with PI3K p85α, and NYAP1 Y2F and NYAP2 Y2F displayed reduced ability to interact with PI3K p85α (Figure 4A and B). These data suggest that both tyrosine residues participate in the binding with PI3K p85α, but the Y2-mediated binding requires phosphorylation of Y1 and/or the binding of Y1 with PI3K p85α, which might cause conformational changes of the NYAPs or PI3K p85α.
Figure 4.
Interaction between the NYAPs and the PI3K p85. (A–C) Tyrosine phosphorylation-dependent interaction of the NYAPs with the PI3K p85α subunit. HEK293T cells were transfected with Fyn, PI3K p85α, and FLAG-NYAP1 (A), FLAG–NYAP2 (B), FLAG–MYO16/NYAP3 (C), or their YF mutants, as indicated. The cell lysates were immunoprecipitated with the anti-FLAG antibody, immunoblotted with an anti-PI3K p85α antibody, and examined for co-precipitation of PI3K p85α with the NYAPs. The asterisk (*) in (C) represents crossreactive bands, which are faintly apparent in (A) and (B). (D) Interaction between the NYAPs and the PI3K p85 in the brain. WT and Nyap1, 2, and 3 KO brains were immunoprecipitated with anti-NYAP1, 2, or 3 antibodies and immunoblotted with an anti-PI3K p85α antibody. Nyap1, 2, and 3 KO brains were used as negative controls. (E) Phosphoproteins associated with PI3K p85α in the brain. The PI3K p85α immunoprecipitates from WT, Nyap1, 2, 3 KO, and TKO brains were immunoblotted with an anti-phospho(Tyr) p85 PI3K binding YxxM motif antibody (pYxxM). The similar results were obtained when another anti-PI3K p85α antibody (from Millipore) and the 4G10 anti-phosphotyrosine antibody were used. (F) Developmental changes in tyrosine phosphorylated binding partners of PI3K p85α. The brain lysates used in Figure 3C were immunoprecipitated with an anti-PI3K p85α antibody and immunoblotted with the 4G10 anti-phosphotyrosine, anti-PI3K p85α, and anti-p110α antibodies. The bottom panel showing the amounts of βIII-tubulin is the same as that shown in Figure 3C.
Because PI3K is activated when the regulatory p85 subunit binds to tyrosine-phosphorylated proteins, we evaluated all tyrosine-phosphorylated proteins co-immunoprecipitated with PI3K p85. To carry out this experiment, an anti-phospho(Tyr) p85 PI3K binding YxxM motif antibody (Figure 4E) and the anti-phosphotyrosine antibody 4G10 (Figure 4F) were used for detection. Surprisingly, only four phosphoproteins of 240, 150, 100, and 90 kDa were associated with PI3K p85α in the brain. Three of these (the 100, 90, and 240 kDa proteins), which were missing in the immunoprecipitates from Nyap1, 2, and 3 KO brains, corresponded to phosphorylated NYAP1, 2, and 3, respectively (Figure 4E). These proteins accounted for the majority of the PI3K p85-associated tyrosine phosphorylation in the brain throughout the entire life of a mouse (Figure 4F). The 150-kDa phosphoprotein has not been identified. Interestingly, PI3K expression levels in the brain gently peaked around perinatal days, when the expression and phosphorylation of the NYAPs were high (Figure 4F), supporting the cooperative roles of PI3K and the NYAPs.
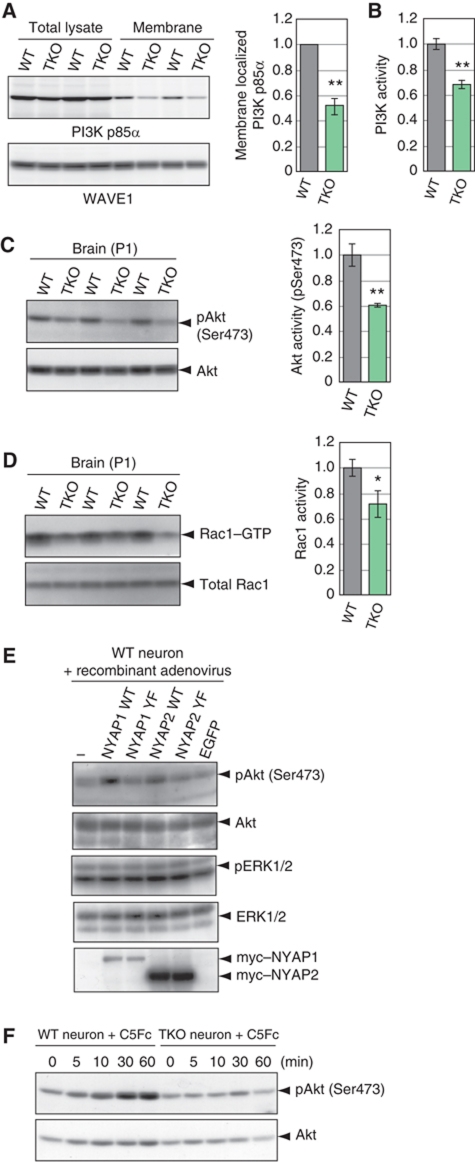
An essential process for PI3K activation is its recruitment to the plasma membrane, whereby PI3K accesses its substrates such as PIP2 (Klippel et al, 1996). To examine this process, we isolated the membrane fraction from WT and TKO P1 mouse brains. In WT brains, a small amount of PI3K p85α was localized in the membrane fraction (27.4±4.5% of total PI3K p85α). However, this recruitment was markedly attenuated in TKO brains (52.5±6.8% of WT; Figure 5A). A certain amount of the NYAPs was also localized in the membrane fraction (Supplementary Figure S4). These results implicated that the NYAPs participate in the recruitment of PI3K to the neuronal plasma membrane. Next, we measured the activity of PI3K and its downstream effector proteins. Activity of PI3K isolated from TKO P1 mouse brains was attenuated (68.1±3.8% of WT; Figure 5B). Activated PI3K produces PIP3, which then activates PH domain-containing effector proteins including the Akt and Tec family of kinases, regulators for small G proteins, and PLCγ (Cantrell, 2001). Akt activation, evaluated as phosphorylation of Akt Ser473, was attenuated in TKO P1 mouse brains (pAkt/Akt, 60.7±1.4% of WT; Figure 5C), whereas expression levels of Akt were unaffected (Supplementary Figure S5). Rac1 activity, which is regulated by PI3K (Hawkins et al, 1995; Tolias et al, 1995; Shinohara et al, 2002), was also attenuated in TKO brains (71.5±10.5% of WT; Figure 5D). Moreover, adenovirus-mediated expression of WT NYAP1 or NYAP2, but not their YF mutants, enhanced Akt activity in neurons (Figure 5E), suggesting that tyrosine phosphorylation of the NYAPs was required for PI3K activation. To further ensure that the NYAPs directly acted in this pathway, we examined activation of the PI3K pathway after induction of NYAPs phosphorylation by Contactin stimulation. Cortical neurons isolated from WT and TKO mice were cultured for 2 days in vitro and stimulated with Contactin5–Fc fusion protein. In WT neurons, Akt was activated after the addition of Contactin5–Fc, but Akt activation did not occur in TKO neurons (Figure 5F). On the other hand, BDNF, which did not induce NYAPs phosphorylation, activated Akt even in the absence of the NYAPs (Supplementary Figure S6). Collectively, these data indicated that the phosphorylated NYAPs activate PI3K and its downstream effectors.
Figure 5.
Activation of the PI3K pathway by the NYAPs. (A) Membrane localization of PI3K p85α and WAVE1 in WT and TKO P1 mouse brains. WT, n=4; TKO, n=4. (B) PI3K activity in WT and TKO P1 mouse brains. PI3K was immunoprecipitated from equal amounts of total lysates obtained from WT and TKO P1 mouse brains with an anti-PI3K p85α antibody, incubated with its substrate phosphatidylinositol and ATP, and subjected to analysis with a PI3K ELISA kit (Echelon). WT, n=6; TKO, n=6. (C) Akt activity in WT and TKO P1 mouse brains. Akt activity was measured in total lysates of WT and TKO brains using an anti-phospho Ser473 Akt antibody. WT, n=3; TKO, n=3. (D) Levels of GTP-bound Rac1 in WT and TKO P1 mouse brains. Due to the low sensitivity of the assay, we pooled four P1 mouse brains per sample. WT, n=3; TKO, n=3. (E) Activation of Akt by phosphorylated NYAPs. Cultured cortical neurons were infected with recombinant adenoviruses expressing EGFP, myc-tagged wild-type NYAP1 and NYAP2, or their YF mutants. Akt activity was measured as in (C). The adenovirus expression system, instead of the sindbisvirus system, was used because of its high expression level. MYO16/NYAP3 was too large to be expressed with the adenovirus system. (F) Contactin5-mediated Akt activation in WT and TKO neurons. Cortical neurons were stimulated with 6 μg/ml C5Fc for the indicated time periods at DIV 2. Akt activation was examined as in (C). A single asterisk (*) and double asterisk (**) indicate P<0.05 and P<0.01 compared with WT, respectively.
The NYAPs interact with the WAVE1 complex
For a comprehensive understanding of the molecular roles of the NYAPs, we searched for NYAPs-interacting proteins using a proteomics approach. Mass spectrometric analysis revealed that NYAP2 tightly interacted with four proteins in the brain (Figure 6A): Sra1 (also called Pir121/CYFIP1/2; Steffen et al, 2004), Nap1 (also called NCKAP1; Yokota et al, 2007), GRAF (also called ARHGAP26; Taylor et al, 1999), and ACOT9 (Poupon et al, 1999). These proteins interacted with NYAP2, as well as NYAP1 and MYO16/NYAP3, in HEK293T cells transfected with the NYAPs (Supplementary Figure S7). WAVE1, Abi2, HSP70, and several other proteins were detected in the NYAP2 proteome. Sra1 and Nap1 are components of the WAVE complex which, consisting of WAVE1/2/3, Sra1, Nap1, Abi1/2, and HSPC300, mediates Arp2/3-dependent actin polymerization (Steffen et al, 2004). HSPC300 was not detected in the proteome analysis, possibly due to its very small molecular weight. Moreover, Sra1 has functions independent of the WAVE complex in the brain (Schenck et al, 2001, 2003; Weiner et al, 2006; Napoli et al, 2008). However, the interaction between NYAP2 and WAVE1 in HEK293T cells (Supplementary Figure S8) and the association of phosphorylated NYAPs in the WAVE1 immunoprecipitates in the brain (Supplementary Figure S9) suggest the function of the NYAPs in the WAVE complex. Because the WAVE complex can be activated by PIP3 and Rac1 (Kobayashi et al, 1998; Eden et al, 2002; Oikawa et al, 2004; Sossey-Alaoui et al, 2005; Lebensohn and Kirschner, 2009; Chen et al, 2010) and because we observed that the NYAPs regulate activities of PI3K and Rac1 (Figure 5), we hypothesized that the NYAPs may be involved in the regulation of the WAVE complex. First, we confirmed the interaction of the NYAPs with the WAVE complex in the brain. We immunoprecipitated the NYAPs from the brain and demonstrated that Nap1 was co-precipitated (Figure 6B). Then, to determine regions of the NYAPs involved in their interaction with the WAVE complex, we constructed various deletion mutants of the NYAPs (Figure 6C) and tested their interactions with Sra1 and Nap1, which were endogenously expressed in HEK293T cells (Figure 6D–F; see also Supplementary Figure S8). GST–NYAP1(1–181), but not GST–NYAP1(1–76), interacted with Sra1 and Nap1 (Figure 6D), suggesting that the region around NYAP1(76–181) is involved in the interaction between NYAP1 and the WAVE complex. Likewise, GST–NYAP2(1–161), but not GST–NYAP2(1–150), interacted with Sra1 and Nap1, suggesting that the region around NYAP2(150–161) is involved in the interaction (Figure 6E). GST–NYAP3(1109–1365), which encompasses a region corresponding to NYAP1(76–181) and NYAP2(150–161), interacted with Sra1 and Nap1 (Figure 6F). These data indicate that the region required for the interaction with the WAVE complex is conserved between the NYAPs just before the NHMs (marked with an orange box in Figure 1B). Thus, the NYAPs utilize distinct regions for interactions with PI3K p85 and the WAVE complex.
Figure 6.
Interaction of the NYAPs with the WAVE complex. (A) Proteomic analysis of NYAP2-interacting proteins. P1 mouse brain lysate was incubated with NYAP2-conjugated sepharose in the presence (lane 2, as a negative control) or absence (lane 1) of 1% SDS. Bound proteins were visualized by Coomassie staining. (B) Interactions of the NYAPs with Nap1 in the brain. P1 mouse brains were immunoprecipitated with anti-NYAP1, 2, or 3 antibodies and immunoblotted with an anti-Nap1 antibody. Immunoprecipitated NYAPs were detected with the RC20 anti-phosphotyrosine antibody. (C) Schematic representation of various deletion mutants of NYAP1, NYAP2, and MYO16/NYAP3. (D–F) Determination of regions of the NYAPs involved in their interaction with the WAVE complex. HEK293T cells were transfected with GST or various deletion mutants of GST-tagged NYAPs as indicated. The cell lysates were incubated with glutathione sepharose (GE Healthcare) and immunoblotted with anti-Sra1 and anti-Nap1 antibodies, and interactions with endogenous Sra1 and Nap1 were tested. Due to relatively low expression levels of GST–NYAP1(1–832) and NYAP3(1109–1919) (D, F), their interactions with Sra1 and Nap1 seemed faint in these assays. See also Supplementary Figure S7A and B for the interactions.
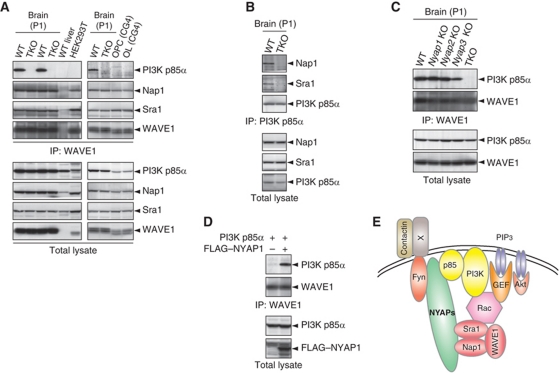
Then, we examined whether these interactions resulted in the ternary complex formation among PI3K, NYAPs, and the WAVE1 complex. Because anti-Nap1 and anti-Sra1 antibodies we used in this study were not applicable for immunoprecipitation, we examined WAVE1 exemplary for the WAVE complex. In WT brains, PI3K p85α was present in the anit-WAVE1 immunoprecipitate, whereas this association completely disappeared in TKO brains (Figure 7A). Further, the association was absent in HEK293T cells and the CG4 oligodendrocyte cell line, none of which express the NYAPs (Figure 7A; WT livers, which do not express WAVE1, were examined as a negative control). On the other hand, in WT brains, Nap1 and Sra1, both of which were components of the WAVE1 complex, were present in the anti-PI3K p85α immunoprecipitates (Figure 7B), ensuring the specificity of the experiment. The association remained in NYAPs single KO brains (Figure 7C), suggesting that all members of the NYAP family were involved in the PI3K–WAVE1 association in neurons. Moreover, exogenous expression of NYAP1 in HEK293T cells resulted in the PI3K–WAVE1 association (Figure 7D). These results indicated that the NYAPs bridge PI3K p85α and the WAVE1 complex, possibly contributing to efficient and/or spatially and temporally controlled conversion of PI3K signals to activation of the WAVE1 complex (Figure 7E).
Figure 7.
The PI3K–WAVE1 association bridged by the NYAPs. (A) The neuron-specific and NYAPs-dependent association between PI3K and WAVE1. WT and TKO brains, WT livers, HEK293T cells, and CG4 cells were lysed and immunoprecipitated with an anti-WAVE1 antibody. PI3K p85α was detected only in the immunoprecipitates from WT brains, in which the NYAPs were expressed. WT livers, which do not express WAVE1, were examined as a negative control. (B) Association of Nap1 and Sra1 in the PI3K p85α immunoprecipitates. WT and TKO brains were immunoprecipitated with an anti-PI3K p85α antibody and immunoblotted with anti-Nap1 and anti-Sra1 antibodies. (C) Involvement of all members of the NYAP family in the PI3K–WAVE1 association in the brain. WT, Nyap1, 2, 3 KO, and TKO brains were immunoprecipitated with an anti-WAVE1 antibody and analysed as in (A). The PI3K–WAVE1 association persisted in NYAPs single KO mouse brains. (D) Association between PI3K and WAVE1 in HEK293T cells expressing the NYAPs. HEK293T cells were transfected with FLAG–NYAP1 and PI3K p85α and tested for the PI3K–WAVE1 association as in (A). (E) Models depicting the roles of the NYAPs. The NYAPs are involved in the activation of PI3K and the recruitment of the WAVE1 complex in neurons. The neuronal receptor(s) that mediates Contactin stimulation is currently unknown.
The NYAPs mediate remodelling of the actin cytoskeleton
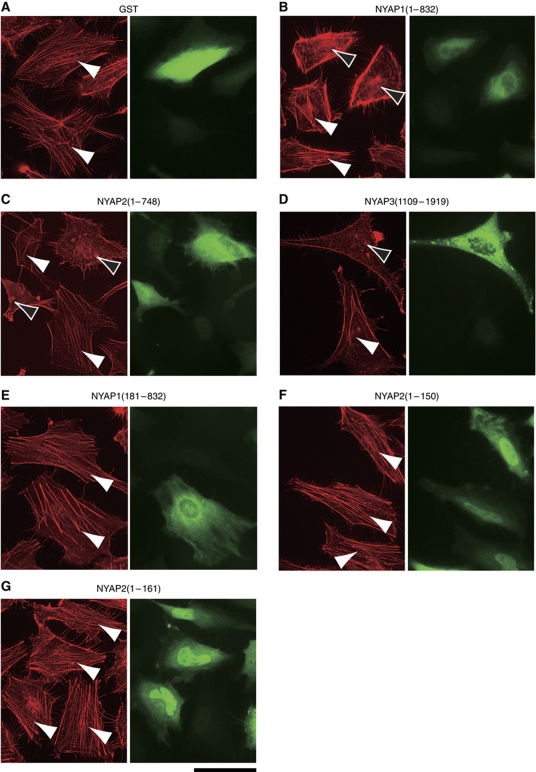
To evaluate whether the NYAPs were involved in regulation of the actin cytoskeleton, we introduced various deletion mutants of the NYAPs and analysed actin fibres in HeLa cells (Figure 8; Supplementary Figure S10). In control cells expressing GST, actin stress fibres were observed (white arrowheads; Figure 8A). When transfected with GST–NYAP1(1–832), GST–NYAP2(1–748), or GST–NYAP3(1109–1919), stress fibres disappeared and, instead, collapsed actin filaments were accumulated in the cytoplasm (black arrowheads; Figure 8B–D). However, when transfected with GST–NYAP1(181–832), which interacted with PI3K but not with the WAVE complex, actin stress fibres were observed (Figure 8E). GST–NYAP2(1–150), which interacted with neither PI3K nor the WAVE complex, and GST–NYAP2(1–161), which interacted with the WAVE complex but not with PI3K, had no effects on actin stress fibres (Figure 8F and G). Some portion of GST–NYAPs appeared to accumulate in the nucleus, which was especially evident for GST–NYAP1(1–181), GST–NYAP2(1–150), and GST–NYAP2(1–161) (Figure 8; Supplementary Figure S10). Because expression levels of cytoplasmic GST–NYAPs seemed similar among the mutants, it was likely that cytoplasmic GST–NYAP1(1–181), GST–NYAP2(1–150), and GST–NYAP2(1–161) have no effect on the actin cytoskeleton. There, however, remains the possibility that nuclear GST–NYAPs negatively regulated remodelling of the actin cytoskeleton. Effects of other deletion mutants were consistent with their interaction with PI3K and the WAVE complex; mutants which interacted with both PI3K and the WAVE complex (NYAP1(1–832), NYAP1(1–491), NYAP2(1–748), NYAP2(1–482), NYAP3(1109–1919), and NYAP3(1109–1732)) induced collapse of actin stress fibres, whereas mutants which did not interacted simultaneously with both PI3K and the WAVE complex (NYAP1(1–181), NYAP1(181–832), NYAP1(1–76), NYAP2(1–150), NYAP2(1–161), and NYAP3(1109–1365)) had no effects on actin stress fibres (Supplementary Figure S10). These data suggest that the association between PI3K and the WAVE complex, which is mediated by the NYAPs, contribute to remodelling of the actin cytoskeleton in this assay.
Figure 8.
NYAPs-mediated remodelling of the actin cytoskeleton. (A–G) Requirement of PI3K- and WAVE1-interacting regions in the NYAPs for remodelling of the actin cytoskeleton in HeLa cells. HeLa cells were transfected with GST (A) or various mutants of GST-tagged NYAPs (B–G) as indicated. Cells were fixed and stained with an anti-GST antibody (green) and phalloidin-rhodamine (red). Actin stress fibres are indicated by white arrowheads. Cytosolic accumulation of collapsed actin fibres is indicated by black arrowheads. Scale bar: 50 μm. See also Supplementary Figure S10.
Neuronal hypotrophy in NYAPs TKO mice
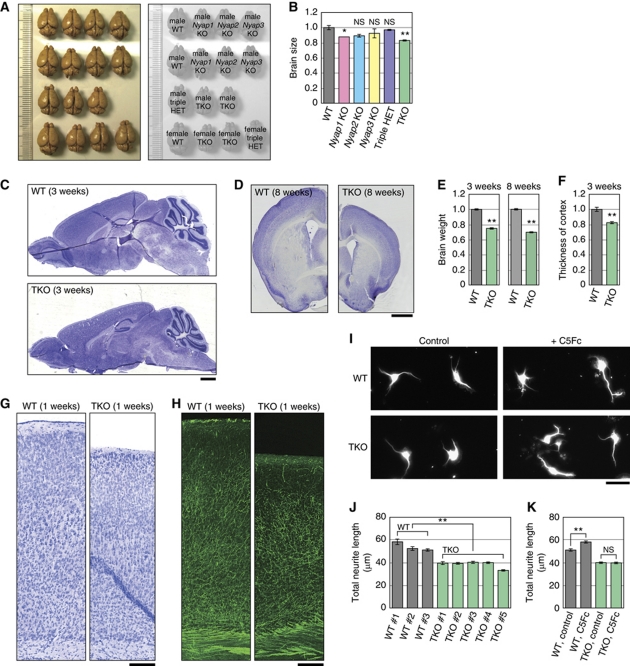
Because the NYAPs controlled remodelling of the actin cytoskeleton (Figure 8), we investigated the involvement of the NYAPs in neuronal morphogenesis. We observed that TKO mice showed a clear reduction in the brain size compared with WT mice (Figure 9A and B; WT cortex area, 100±2.8%, n=3; TKO cortex area, 83.1±0.3%, n=4; P=8.3 × 10−4 in a two-tailed Student's t-test). Nissl-stained TKO mice exhibited a clear reduction in brain size (cortical thickness, 82.9±1.8% of WT, P=4.0 × 10−4; brain weight, 3 weeks, 75.3±1.0% of WT, P=5.3 × 10−10; brain weight, 8 weeks, 70.1±1.1% of WT, P=1.2 × 10−5; Figure 9C–F). Reduction of size was obvious in the cortex and the striatum, but not in the olfactory bulb and the corpus callosum. Because the overall cortical structures of WT and TKO mice appeared to be similar (Figure 9C, D, G, and H), we then examined morphology of individual neurons. Dendrite morphogenesis is regulated by the PI3K-Akt pathway in vitro (Jaworski et al, 2005; Kumar et al, 2005). We isolated cortical neurons from WT and TKO E16 mice and cultured them for 2 days in vitro (Figure 9I). The total neurite length of TKO neurons was significantly shorter than that of WT neurons (71.4±2.5% of WT, P=6.8 × 10−4 in a two-tailed Student's t-test; Figure 9J). Furthermore, WT neurons extended their neurites longer in the presence of Contactin proteins in the culture medium as previously reported (Furley et al, 1990; Ogawa et al, 2001). In contrast, TKO neurons did not respond to Contactin5–Fc (WT control, 51.4±1.3 μm; WT+C5Fc, 58.5±1.2 μm; P=9.5 × 10−5) (TKO control, 40.3±1.0 μm; TKO+C5Fc, 40.1±1.0 μm; P=0.89) (Figure 9K). These data suggested that the Contactin-Fyn-NYAP-PI3K pathway regulates neurite outgrowth. Because the degree of brain size reduction (TKO, brain weight, 8 weeks, 70.1±1.1% of WT; Figure 9E) was comparable to the degree of neurite hypotrophy (TKO neurons, 71.4±2.5% of WT; Figure 9I), reduced brain size in TKO mice may be caused by neurite hypotrophy.
Figure 9.
Neuronal hypotrophy in NYAPs triple knockout mice. (A) WT, Nyap1, 2, 3 KO, and TKO adult mouse brains (8 weeks old) were photographed. The genotypes of each brain are indicated in the right panel. (B) The size of the cortex area (excluding the olfactory bulb and cerebellum) of the brains photographed in (A) was measured using Adobe Photoshop. NS: not significant. (C, D) Nissl-stained sagittal (C) and coronal (D) sections of WT and TKO male mouse brains. Scale bar: 1 mm. (E, F) Brain weight (E) and cortical thickness (F) of WT and TKO male mice (WT, n=7; TKO, n=7). (G) Nissl-stained coronal sections of WT and TKO P7 mouse brains. Scale bar: 50 μm. (H) Neurofilament staining. The sections adjacent to those used in (G) were stained with an anti-68 kDa Neurofilament antibody. Scale bar: 50 μm. (I–K) Neuronal hypotrophy in TKO brains. Cortical neurons were isolated from WT and TKO E16 mouse brains and cultured for 2 days in the presence (C5Fc) or absence (control) of Contactin5–Fc protein (6 μg/ml) in the culture medium. Neurons were stained with the TuJ1 anti-βIII tubulin antibody and total neurite length was measured. For (J), WT, n=1427 neurons (total); TKO, n=3053 neurons (total). For (K), WT control, n=742; WT+C5Fc, n=1037; TKO control, n=859; and TKO+C5Fc, n=853. NS: not significant. Scale bar: 10 μm. A single asterisk (*) and double asterisk (**) indicate P<0.05 and P<0.01 compared with WT, respectively.
Discussion
In this study, we demonstrated that the novel NYAP family of proteins has two roles in the regulation of the PI3K signalling pathway in neurons: the activation of PI3K and the recruitment of the downstream effector WAVE complex to the close vicinity of PI3K. These are achieved by simultaneous interactions with PI3K and the WAVE1 complex, respectively, although precise action of the NYAPs on activity of the WAVE1 complex remains to be elucidated (see the cartoon in Figure 7E). Moreover, we demonstrated that the NYAPs regulate actin remodelling and neurite elongation.
The NYAPs activate PI3K in neurons
We demonstrated that the NYAPs are the major phosphoproteins in the brain. Moreover, they occupied at least three fourths of PI3K-associated tyrosine phosphorylation, and were involved in the activation of PI3K, Akt, and Rac1 in neurons. Tyrosine kinase-PI3K pathways are present in most cell types in mammals, including the brain, testis, liver, muscle, fat, and tissues of the immune system. Commonly expressed adaptor proteins, such as IRS and Gab, and receptor-type tyrosine kinases themselves have been thought to bind to and activate PI3K in these tissues (Cantrell, 2001). In contrast, the repertoires of PI3K p85-associated phosphoproteins vary by cell types (our unpublished observations). Therefore, the molecular mechanisms of PI3K activation may be different across cell types, and the simple application of available data from one cell type to another cell type might be misleading. The present study clearly demonstrated that the NYAPs account for most of PI3K-associated tyrosine phosphorylation in neurons, and would help refinement of previous models of PI3K activation and function in neurons (reviewed in Rodgers and Theibert, 2002) by incorporating the contributions of the NYAPs.
The NYAPs connect PI3K activation to WAVE1 signalling in neurons
We demonstrated that the NYAPs interact with Sra1, Nap1, GRAF, and ACOT9, as well as PI3K and F-actin (for MYO16/NYAP3; Patel et al, 2001). Sra1 and Nap1 are components of the WAVE complex which links PIP3 and Rac1 to the activation of Arp2/3-mediated actin remodelling (Steffen et al, 2004; Chen et al, 2010). Interacting with the NYAPs, the WAVE1 complex was recruited to and associated with PI3K, where PI3K-produced PIP3 and Rac1 may activate WAVE1 efficiently, although precise action of the NYAPs on the WAVE1 complex, as well as the precise mode of interaction between NYAPs and the WAVE complex, remain to be elucidated.
WAVE3 is reported to interact with PI3K p85 directly in the MDA-MB-231 cancer cell line and in yeast cells (Sossey-Alaoui et al, 2005). Moreover, Abi1, a component of the WAVE complex, is also reported to interact with PI3K p85 in MEF cells and the LNCaP prostate adenocarcinoma cell line (Innocenti et al, 2003; Dubielecka et al, 2010). The WAVE3–PI3K direct interaction is mediated by WAVE3 Tyr-151 and the C-terminal SH2 domain of PI3K p85 (Sossey-Alaoui et al, 2005) and the Abi1–PI3K interaction is mediated by Abi1 Tyr-407 and Tyr-213 and the SH2 domain of PI3K p85 (Innocenti et al, 2003; Dubielecka et al, 2010), although WAVE3 Tyr-151 (Tyr151-Phe-Phe-Glu), Abi1 Tyr-407 (Tyr407-Ala-Asp-Gly), and Abi1 Tyr213 (Tyr213-Met-Thr-Ser) do not lie in the consensus sequence for the binding by SH2 domains of the PI3K p85 subunit (YxxM motif; Songyang et al, 1993). As the amino-acid sequence around WAVE3 Tyr-151 is highly conserved among WAVE1, 2, and 3 (Suetsugu et al, 1999), it would be likely that WAVE1 and WAVE2 directly interact with PI3K as well as WAVE3. However, in our semiquantitative assay, tyrosine-phosphorylated WAVE proteins (which will appear at 75 kDa) and Abi proteins (60 kDa) were neither detected with the anti-phosphotyrosine antibody in the PI3K p85α immunoprecipitates (Figure 4F) nor even in the WAVE1-immunoprecipitates (Supplementary Figure S9), suggesting that direct interactions between PI3K and WAVE and between PI3K and Abi would be weak in the brain. In this study, we demonstrated that most of the PI3K–WAVE1 association in the brain was indirect because almost all the interaction was lost in the absence of the NYAPs (Figure 7A) without affecting membrane localization of WAVE1 (Figure 5A), indicating that the direct interaction has minimal, if any, contribution to the PI3K–WAVE1 association in the brain.
Recruitment of downstream effector proteins may be especially important for PI3K signalling because PI3K-produced PIP3 is a lipid signalling molecule that can diffuse across the plasma membrane faster than protein signalling molecules. Other cells may express specific PI3K regulators like the NYAPs, which regulate both the activation and the downstream signal recruitment in the PI3K pathway and contribute to the specificity and/or efficiency of the pathway. Because PI3K is a potential target for anti-cancer therapy, a full understanding of the regulation of the PI3K pathway in many cell types may facilitate the treatment of cancer, as well as obesity and mental disorders.
In addition to the WAVE complex, the NYAPs tightly interact with GRAF and ACOT9. GRAF is a RhoGAP protein with BAR, SH3, and PH domains and regulates Rho activity and actin dynamics (Taylor et al, 1999). Because active RhoA negatively regulates Rac1 activity via GAPs such as ARHGAP22 and ARHGAP24 (Ohta et al, 2006; Sanz-Moreno et al, 2008), local inhibition of RhoA via NYAPs-associated GRAF may contribute to local activation of Rac1. BAR domains induce and stabilize membrane curvature and/or detect curvature in order to recruit cytosolic factors to membranes of a particular shape or size (Frost et al, 2009). ACOT9 is an acyl-CoA thioesterase enzyme that hydrolyses long chain C14 myristoyl-CoA esters (Poupon et al, 1999). By interacting with GRAF and ACOT9, the NYAPs might be involved in regulation of the curvature and composition of the neuronal membrane, as well as in regulation of the actin cytoskeleton.
The NYAPs regulate neuronal morphogenesis
We demonstrated that Contactin5-induced activation of the PI3K-Akt pathway was dependent on the NYAPs (Figure 5F), indicating that Contactin, Fyn, NYAPs, and PI3K function in the same signalling pathway to regulate WAVE1 function (Figure 7E). In addition to similarities in their high expression levels around perinatal days (Figure 4F; Supplementary Figure S11), mice harbouring mutations in this pathway show striking phenotypic similarities with regard to neuronal morphogenesis and brain size. We demonstrated that TKO mice display reduced brain size and neuronal hypotrophy (Figure 9). Previous studies have demonstrated that Fyn KO brains possess reduced cortical thickness (Goto et al, 2008). Mice deficient in PTPα, which links Contactin to Fyn activation (Ponniah et al, 1999; Su et al, 1999; Ye et al, 2008), have reduced brain size (Petrone et al, 2003). Akt3 KO mice also have reduced brain size (Easton et al, 2005; Tschopp et al, 2005). Moreover, a conditional KO of PTEN results in progressive macrocephaly and neuronal hypertrophy (Backman et al, 2001; Kwon et al, 2001, 2006). Cortex-restricted Rac1 KO mice show reduced cortical thickness (Kassai et al, 2008). WAVE1-deficient neurons, as well as Nap1-deficient neurons, display reduced neurite outgrowth (Soderling et al, 2007; Yokota et al, 2007). HSPC300, which is a component of the WAVE complex, is implicated in axonal midline crossing and synaptogenesis in Drosophila (Qurashi et al, 2007). These observations will support our finding that the NYAPs act in the Fyn-NYAP-PI3K pathway to regulate neuronal morphogenesis and brain size.
Human NYAP1 locus (7q22.1) and MYO16/NYAP3 locus (13q33.3) have suggestive association signals with autism spectrum disorders (ASDs; International Molecular Genetic Study of Autism Consortium, 2001a, 2001b; Wang et al, 2009). Intriguingly, PTEN, Contactin3, Contactin4, and CNTNAP2 (Contactin-associated protein-like 2) were mutated in human ASD (Butler et al, 2005; Bourgeron, 2009). Therefore, it would be worthwhile to investigate the molecular links between the NYAP signalling pathway and ASD.
Materials and methods
Substrate screening
Solid-phase phosphorylation screening was performed using a human hippocampus cDNA library (Takara Bio) as described previously (Yokoyama et al, 2002>, 2006) with a purified GST–Lyn fusion protein produced in Sf9 cells and the anti-pTyr antibody PY69 (BD Transduction Laboratories). A fragment of human NYAP1 cDNA (corresponding to amino acids 56–315 in mouse NYAP1) was isolated during the screening.
cDNAs
Human NYAP2 (KIAA1486) and MYO16/NYAP3 (Myosin16, KIAA0865) cDNAs were generously provided by the Kazusa DNA Research Institute. Mouse cDNAs for the NYAPs were obtained via RT–PCR of RNA isolated from the mouse brain on the basis of the sequence information in the NCBI database. Human KIAA1486 possesses a C-terminal truncation based on a comparison with mouse NYAP2. YF mutations were introduced by PCR-based mutagenesis. Deletion mutants were generated by endonuclease digestion. Recombinant adenoviruses and sindbisviruses were generated as described previously (Nakazawa et al, 2008).
Mice
Nyap1, 2, and 3 KO mice were generated from E14.1 ES cells and backcrossed into C57BL/6J for at least 10 generations. Exons 2 and 3 were disrupted in Nyap1 (amino acids 23–643), as was exon 4 in Nyap2 (amino acids 207–567); these regions encode NHM motifs. Exon 13 (amino acids 496–531), which contains a lysine residue in the myosin motor domain that binds ATP, was disrupted in Myo16/Nyap3 (Supplementary Figure S1). Homologous recombination was confirmed by PCR and Southern blotting. NYAPs triple KO mice were generated by successive crossing of Nyap1, 2, and 3 KO mice. All mice were maintained under specific pathogen-free conditions. Experiments were conducted according to the institutional ethical guidelines for animal experiments.
Cell cultures
HEK293T cells and CG4 cells were cultured as previously described (Yokoyama et al, 2006). HeLa cells were cultured in DMEM supplemented with 10% FCS. Cortical neurons were isolated from E18.5 Wistar rats (for Figures 2B, 3I and 5E) or E16.5 mice (for Figures 3E, 5F, and 9I–K and Supplementary Figure S6) and cultured in neurobasal medium with B27 supplement (Invitrogen) at a density of 100 cells/mm2 (for morphological assays) or 1700 cells/mm2 (for biochemical assays) on dishes precoated with 20 μg/ml poly-L-lysine. Astrocytes were isolated from the cortex of E18.5 Wistar rat brains and cultured in DMEM supplemented with 10% FCS.
Antibodies
Polyclonal antibodies against NYAP1, 2, and 3 (designated N1C, N24, and N32, respectively) were raised in rabbits. Monoclonal antibodies against NYAP1, 2, and 3 (designated N1M1, N2M1, and N3RT1, respectively) were generated by immunizing Nyap1 KO mice (for N1M1), Nyap2 KO mice (for N2M1), or Wistar rats (for N3RT1). The antigens were mouse NYAP1 amino acids 536–581 for N1C and 139–318 for N1M1, mouse NYAP2 569–645 for N24 and 207–351 for N2M1, and mouse MYO16/NYAP3 1756–1870 for N32 and 1321–1490 for N3RT1. The monoclonal antibody against mouse Contactin5 was generated essentially as described previously (Toyoshima et al, 2009). Other antibodies used in the experiments were obtained commercially: anti-phosphotyrosine clone 4G10 (Millipore) and RC20 (BD); anti-Akt, anti-phospho(Ser473) Akt, anti-phospho-p44/42 MAPK (Erk1/2)(Thr202/Tyr204), and anti-phospho(Tyr) p85 PI3K binding motif (Cell Signaling); anti-PI3K p85α clone AB6 (MBL) and anti-PI3K p85β (Serotec); anti-GST, anti-Fyn, anti-ERK1, and anti-PI3K p110α (Santa Cruz); anti-α tubulin (Sigma) and anti-βIII tubulin (Babco); anti-Rac1 (BD), anti-WAVE1, anti-Nap1, and anti-Sra1 (Millipore); and anti-68 kDa Neurofilament (Novus Biologicals).
mRNA expression analyses
Northern blotting was performed as described previously (Yokoyama et al, 2002). In situ hybridization with digoxigenin-labelled probes was performed according to the manufacturer's protocol (Roche) using freshly frozen cryosections. WT sections were mounted on the same glass slides containing sections from corresponding Nyap1, 2, or 3 KO mice as negative controls. Sections were counterstained with propidium iodide. The probes were mouse Nyap1 cDNA 1–542, mouse Nyap2 1–453, and mouse Myo16/Nyap3 1488–1593. Overlapping images of the in situ hybridization were captured and merged using a BZ-9000 microscope (Keyence).
Protein analyses
Immunoprecipitation and immunoblotting were performed as previously described (Yokoyama et al, 2002). The images were obtained and quantified with LAS-4000 mini (GE Healthcare). Rac1 activity was measured essentially as previously described (Nakazawa et al, 2008). Briefly, brains were lysed in Rac pulldown buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 10 mM MgCl2, 1% Triton X-100, and 10% glycerol) for 30 min on ice and pulled down with PAK-GST beads (Cytoskeleton Inc.) for 1 h. GTP-bound Rac1 was detected by immunoblotting with an anti-Rac1 antibody. Due to the low sensitivity of the assay, we pooled four P1 mouse brains per sample. To isolate the brain membrane fraction, P1 mouse brains were homogenized in 10 ml of hypotonic buffer (10 mM Tris–HCl (pH 7.4), 2 mM MgCl2, 1 mM KCl, and 1 mM NaF) and centrifuged at 1000 g once for 5 min and twice for 10 min each. One millilitre of the supernatant was centrifuged at 120 000 g for 60 min and then washed twice with hypotonic buffer. The resultant pellet (membrane fraction) was lysed in 150 μl of SDS–PAGE sample buffer.
Immunocytochemistry and immunohistochemistry
HeLa cells were fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and incubated successively with anti-FLAG or anti-GST, rhodamine-phalloidin, and Alexa488-conjugated secondary antibodies. For immunohistochemistry, mouse brains were dissected, immersed in 4% PFA overnight, and dehydrated successively in 10, 20, and 30% sucrose (w/v) solutions in PBS for 24 h each at 4°C. Then, the brains were frozen, cryosectioned at 14 μm thickness, mounted onto MAS-coated glass slides (Matsunami Glass), and air dried. The sections were fixed with 4% PFA, permeabilized with 0.1% Triton X-100, and incubated successively with anti-Neurofilament and Alexa488-conjugated secondary antibodies. For the Nissl staining, cryosections prepared as above were immersed in 0.1% cresyl violet solution overnight and dehydrated successively in 70% ethanol, 80% ethanol, 90% ethanol, absolute ethanol (twice), and xylene (twice) for 15 min each. All images were obtained with a BZ-9000 microscope (Keyence).
Preparation of recombinant Contactin5–Fc fusion protein
HEK293T cell clone stably expressing Contactin5–Fc was washed once with serum-free DMEM and cultured for 3 additional days in serum-free neurobasal medium. The medium was concentrated by ultrafiltration with a Vivaspin 50k MWCO (Sartorius). Then, Contactin5–Fc was captured with protein G-sepharose (GE Healthcare) and quantified by Coomassie staining.
Proteomic analyses
GST-tagged full-length mouse NYAP2 was produced using the baculovirus expression system. Purified GST–NYAP2 was covalently conjugated to the NHS-activated sepharose (GE Healthcare). NYAP2-interacting proteins were purified from three WT P1 mouse brains with 10 μg of NYAP2 sepharose, visualized with Coomassie Brilliant Blue R-250, and identified using QSTAR Elite LC/MS/MS (Applied Biosystems). NYAP2 protein was used because recombinant full-length NYAP1 and MYO16/NYAP3 proteins were less stable in the baculovirus system.
Statistical analyses
All quantified data are expressed as mean values±s.e.m. Significance tests were performed using a two-tailed Student's t-test. A single asterisk (*) and double asterisk (**) indicate P<0.05 and P<0.01 compared with WT, respectively.
Accession numbers
The DDBJ/EMBL/GenBank accession numbers for the NYAPs are AB429289 (mouse Nyap1), AB429290 (mouse Nyap2), and AB429291 (mouse Myo16/Nyap3).
Supplementary Material
Acknowledgments
We thank M Oyama and H Hata for help with the mass spectrometry, S Aizawa for providing Fyn KO mice, and H Umemori and H Bito for critical reading of the manuscript. This work was supported in part by JSPS fellowships for junior scientists, grants-in-aid for young scientists and scientific research, the Global COE program (Integrative life science), and the program for improvement of the research environment for young researchers from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acosta-Martínez M, Luo J, Elias C, Wolfe A, Levine JE (2009) Male-biased effects of gonadotropin-releasing hormone neuron-specific deletion of the phosphoinositide 3-kinase regulatory subunit p85alpha on the reproductive axis. Endocrinology 150: 4203–4212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardern H, Sandilands E, Machesky LM, Timpson P, Frame MC, Brunton VG (2006) Src-dependent phosphorylation of Scar1 promotes its association with the Arp2/3 complex. Cell Motil Cytoskeleton 63: 6–13 [DOI] [PubMed] [Google Scholar]
- Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J, Tsao MS, Shannon P, Bolon B, Ivy GO, Mak TW (2001) Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat Genet 29: 396–403 [DOI] [PubMed] [Google Scholar]
- Bourgeron T (2009) A synaptic trek to autism. Curr Opin Neurobiol 19: 231–234 [DOI] [PubMed] [Google Scholar]
- Brunet A, Datta SR, Greenberg ME (2001) Transcription-dependent and -independent control of neuronal survival by the PI3K-Akt signaling pathway. Curr Opin Neurobiol 11: 297–305 [DOI] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C (2005) Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumor suppressor gene mutations. J Med Genet 42: 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RS, Liu C, Mixon AS, Pihkala JP, Rahn RJ, Cameron PL (2007) Myosin16b: the COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil Cytoskeleton 64: 19–48 [DOI] [PubMed] [Google Scholar]
- Cantrell DA (2001) Phosphoinositide 3-kinase signalling pathways. J Cell Sci 114: 1439–1445 [DOI] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK (2010) Structure and control of the actin regulatory WAVE complex. Nature 468: 533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubielecka PM, Machida K, Xiong X, Hossain S, Ogiue-Ikeda M, Carrera AC, Mayer BJ, Kotula L (2010) Abi1/Hssh3bp1 pY213 links Abl kinase signaling to p85 regulatory subunit of PI-3 kinase in regulation of macropinocytosis in LNCaP cells. FEBS Lett 584: 3279–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton RM, Cho H, Roovers K, Shineman DW, Mizrahi M, Forman MS, Lee VM, Szabolcs M, de Jong R, Oltersdorf T, Ludwig T, Efstratiadis A, Birnbaum MJ (2005) Role for Akt3/protein kinase Bγ in attainment of normal brain size. Mol Cell Biol 25: 1869–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW (2002) Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418: 790–793 [DOI] [PubMed] [Google Scholar]
- Eickholt BJ, Ahmed AI, Davies M, Papakonstanti EA, Pearce W, Starkey ML, Bilancio A, Need AC, Smith AJ, Hall SM, Hamers FP, Giese KP, Bradbury EJ, Vanhaesebroeck B (2007) Control of axonal growth and regeneration of sensory neurons by the p110delta PI 3-kinase. PLoS One 2: e869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P (2009) The BAR domain superfamily: membrane-molding macromolecules. Cell 137: 191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furley AJ, Morton SB, Manalo D, Karagogeos D, Dodd J, Jessell TM (1990) The axonal glycoprotein TAG-1 is an immunoglobulin superfamily member with neurite outgrowth-promoting activity. Cell 61: 157–170 [DOI] [PubMed] [Google Scholar]
- Goto J, Tezuka T, Nakazawa T, Sagara H, Yamamoto T (2008) Loss of Fyn tyrosine kinase on the C57BL/6 genetic background causes hydrocephalus with defects in oligodendrocyte development. Mol Cell Neurosci 38: 203–212 [DOI] [PubMed] [Google Scholar]
- Grant SG, O′Dell TJ, Karl KA, Stein PL, Soriano P, Kandel ER (1992) Impaired long-term potentiation, spatial learning, and hippocampal development in fyn mutant mice. Science 258: 1903–1910 [DOI] [PubMed] [Google Scholar]
- Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279: 509–514 [DOI] [PubMed] [Google Scholar]
- Hawkins PT, Eguinoa A, Qiu RG, Stokoe D, Cooke FT, Walters R, Wennström S, Claesson-Welsh L, Evans T, Symons M, Stephens L (1995) PDGF stimulates an increase in GTP–Rac via activation of phosphoinositide 3-kinase. Curr Biol 5: 393–403 [DOI] [PubMed] [Google Scholar]
- Innocenti M, Frittoli E, Ponzanelli I, Falck JR, Brachmann SM, Di Fiore PP, Scita G (2003) Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J Cell Biol 160: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (2001a) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet 69: 570–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (2001b) Further characterization of the autism susceptibility locus AUTS1 on chromosome 7q. Hum Mol Genet 10: 973–982 [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M (2005) Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of Rapamycin pathway. J Neurosci 25: 11300–11312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K, Watanabe K, Kozutsumi Y, Oohira A, Yamamoto T, Sanai Y (2002) Association of GPI-anchored protein TAG-1 with src-family kinase Lyn in lipid rafts of cerebellar granule cells. Neurochem Res 27: 823–829 [DOI] [PubMed] [Google Scholar]
- Kassai H, Terashima T, Fukaya M, Nakao K, Sakahara M, Watanabe M, Aiba A (2008) Rac1 in cortical projection neurons is selectively required for midline crossing of commissural axonal formation. Eur J Neurosci 28: 257–267 [DOI] [PubMed] [Google Scholar]
- Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, Kim AM, Kwak SP, Park JB, Ho Ryu S, Schenck A, Bardoni B, Scott JD, Nairn AC, Greengard P (2006) Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature 442: 814–817 [DOI] [PubMed] [Google Scholar]
- Klippel A, Reinhard C, Kavanaugh WM, Apell G, Escobedo MA, Williams LT (1996) Membrane localization of phosphatidylinositol 3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol Cell Biol 16: 4117–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K (1998) p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem 273: 291–295 [DOI] [PubMed] [Google Scholar]
- Kotani T, Morone N, Yuasa S, Nada S, Okada M (2007) Constitutive activation of neuronal Src causes aberrant dendritic morphogenesis in mouse cerebellar Purkinje cells. Neurosci Res 57: 210–219 [DOI] [PubMed] [Google Scholar]
- Kumar V, Zhang M, Swank MW, Kunz J, Wu G (2005) Regulation of dendritic morphogenesis by Ras-PI3K-Akt-mTOR and Ras-MAPK signaling pathways. J Neurosci 25: 11288–11299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF (2006) Pten regulates neuronal arborization and social interaction in mice. Neuron 50: 377–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ (2001) Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat Genet 29: 404–411 [DOI] [PubMed] [Google Scholar]
- Laurino L, Wang XX, de la Houssaye BA, Sosa L, Dupraz S, Cáceres A, Pfenninger KH, Quiroga S (2005) PI3K activation by IGF-1 is essential for the regulation of membrane expansion at the nerve growth cone. J Cell Sci 118: 3653–3662 [DOI] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW (2009) Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell 36: 512–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K (2005) Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl Acad Sci USA 102: 1098–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita A, Yamashita N, Sasaki Y, Uchida Y, Nakajima O, Nakamura F, Yagi T, Taniguchi M, Usui H, Katoh-Semba R, Takei K, Goshima Y (2006) Regulation of dendritic branching and spine maturation by semaphorin3A-Fyn signaling. J Neurosci 26: 2971–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T, Kuriu T, Tezuka T, Umemori H, Okabe S, Yamamoto T (2008) Regulation of dendritic spine morphology by an NMDA receptor-associated Rho GTPase-activating protein, p250GAP. J Neurochem 105: 1384–1393 [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C (2008) The fragile × syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134: 1042–1054 [DOI] [PubMed] [Google Scholar]
- Ogawa J, Lee S, Itoh K, Nagata S, Machida T, Takeda Y, Watanabe K (2001) Neural recognition molecule NB-2 of the contactin/F3 subgroup in rat: Specificity in neurite outgrowth-promoting activity and restricted expression in the brain regions. J Neurosci Res 65: 100–110 [DOI] [PubMed] [Google Scholar]
- Ohta Y, Hartwig JH, Stossel TP (2006) FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol 8: 803–814 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T (2004) PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol 6: 420–426 [DOI] [PubMed] [Google Scholar]
- Patel KG, Liu C, Cameron PL, Cameron RS (2001) Myr 8, a novel unconventional myosin expressed during brain development associates with the protein phosphatase catalytic subunits 1α and 1γ1. J Neurosci 21: 7954–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrone A, Battaglia F, Wang C, Dusa A, Su J, Zagzag D, Bianchi R, Casaccia-Bonnefil P, Arancio O, Sap J (2003) Receptor protein tyrosine phosphatase alpha is essential for hippocampal neuronal migration and long-term potentiation. EMBO J 22: 4121–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponniah S, Wang DZ, Lim KL, Pallen CJ (1999) Targeted disruption of the tyrosine phosphatase PTPα leads to constitutive downregulation of the kinases Src and Fyn. Curr Biol 9: 535–538 [DOI] [PubMed] [Google Scholar]
- Poupon V, Bègue B, Gagnon J, Dautry-Varsat A, Cerf-Bensussan N, Benmerah A (1999) Molecular cloning and characterization of MT-ACT48, a novel mitochondrial acyl-CoA thioesterase. J Biol Chem 274: 19188–19194 [DOI] [PubMed] [Google Scholar]
- Qurashi A, Sahin HB, Carrera P, Gautreau A, Schenck A, Giangrande A (2007) HSPC300 and its role in neuronal connectivity. Neural Dev 2: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers EE, Theibert AB (2002) Functions of PI 3-kinase in development of the nervous system. Int J Dev Neurosci 20: 187–197 [DOI] [PubMed] [Google Scholar]
- Sanna PP, Cammalleri M, Berton F, Simpson C, Lutjens R, Bloom FE, Francesconi W (2002) Phosphatidylinositol 3-kinase is required for the expression but not for the induction or the maintenance of long-term potentiation in the hippocampal CA1 region. J Neurosci 22: 3359–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, Sahai E, Marshall CJ (2008) Rac activation and inactivation control plasticity of tumor cell movement. Cell 135: 510–523 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Cheng C, Uchida Y, Nakajima O, Ohshima T, Yagi T, Taniguchi M, Nakayama T, Kishida R, Kudo Y, Ohno S, Nakamura F, Goshima Y (2002) Fyn and Cdk5 mediate semaphorin-3A signaling, which is involved in regulation of dendrite orientation in cerebral cortex. Neuron 35: 907–920 [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Langmann C, Harden N, Mandel JL, Giangrande A (2003) CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile × protein. Neuron 38: 887–898 [DOI] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL (2001) A highly conserved protein family interacting with the fragile × mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc Natl Acad Sci USA 98: 8844–8849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi SH, Jan LY, Jan YN (2003) Hippocampal neuronal polarity specified by spatially localized mPar3/mPar6 and PI 3-kinase activity. Cell 112: 63–75 [DOI] [PubMed] [Google Scholar]
- Shimoda Y, Watanabe K (2009) Contactins: emerging key roles in the development and function of the nervous system. Cell Adh Migr 3: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Terada Y, Iwamatsu A, Shinohara A, Mochizuki N, Higuchi M, Gotoh Y, Ihara S, Nagata S, Itoh H, Fukui Y, Jessberger R (2002) SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature 416: 759–763 [DOI] [PubMed] [Google Scholar]
- Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, Langeberg LK, Banker G, Raber J, Scott JD (2007) A WAVE-1 and WRP signaling complex regulates spine density, synaptic plasticity, and memory. J Neurosci 27: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser G, King F, Roberts T, Ratnofsky S, Lechleider RJ, Neel BG, Birge RB, Fajardo JE, Chou MM, Hanafusa H, Schaffhausen B, Cantley LC (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72: 767–778 [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Li X, Cowell JK (2007) c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J Biol Chem 282: 26257–26265 [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Li X, Ranalli TA, Cowell JK (2005) WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J Biol Chem 280: 21748–21755 [DOI] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TE (2004) Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J 23: 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal TE, Scita G (2006) Protein complexes regulating Arp2/3-mediated actin assembly. Curr Opin Cell Biol 18: 4–10 [DOI] [PubMed] [Google Scholar]
- Stuart JR, Gonzalez FH, Kawai H, Yuan ZM (2006) c-Abl interacts with the WAVE2 signaling complex to induce membrane ruffling and cell spreading. J Biol Chem 281: 31290–31297 [DOI] [PubMed] [Google Scholar]
- Su J, Muranjan M, Sap J (1999) Receptor protein tyrosine phosphatase α activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr Biol 9: 505–511 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T (1999) Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem Biophys Res Commun 260: 296–302 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Takenawa T (2003) Regulation of cortical actin networks in cell migration. Int Rev Cytol 229: 245–286 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Yamazaki D, Kurisu S, Takenawa T (2003) Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell 5: 595–609 [DOI] [PubMed] [Google Scholar]
- Taylor JM, Macklem MM, Parsons JT (1999) Cytoskeletal changes induced by GRAF, the GTPase regulator associated with focal adhesion kinase, are mediated by Rho. J Cell Sci 112: 231–242 [DOI] [PubMed] [Google Scholar]
- Tohda C, Nakanishi R, Kadowaki M (2006) Learning deficits and agenesis of synapses and myelinated axons in phosphoinositide-3 kinase-deficient mice. Neurosignals 15: 293–306 [DOI] [PubMed] [Google Scholar]
- Tohda C, Nakanishi R, Kadowaki M (2009) Hyperactivity, memory deficit and anxiety-related behaviors in mice lacking the p85alpha subunit of phosphoinositide-3 kinase. Brain Dev 31: 69–74 [DOI] [PubMed] [Google Scholar]
- Tolias KF, Cantley LC, Carpenter CL (1995) Rho family GTPases bind to phosphoinositide kinases. J Biol Chem 270: 17656–17659 [DOI] [PubMed] [Google Scholar]
- Toyoshima M, Sakurai K, Shimazaki K, Takeda Y, Nakamoto M, Serizawa S, Shimoda Y, Watanabe K (2009) Preferential localization of neural cell recognition molecule NB-2 in developing glutamatergic neurons in the rat auditory brainstem. J Comp Neurol 513: 349–362 [DOI] [PubMed] [Google Scholar]
- Tschopp O, Yang ZZ, Brodbeck D, Dummler BA, Hemmings-Mieszczak M, Watanabe T, Michaelis T, Frahm J, Hemmings BA (2005) Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132: 2943–2954 [DOI] [PubMed] [Google Scholar]
- Umemori H, Wanaka A, Kato H, Takeuchi M, Tohyama M, Yamamoto T (1992) Specific expressions of Fyn and Lyn, lymphocyte antigen receptor-associated tyrosine kinases, in the central nervous system. Brain Res Mol Brain Res 16: 303–310 [DOI] [PubMed] [Google Scholar]
- Viard P, Butcher AJ, Halet G, Davies A, Nürnberg B, Heblich F, Dolphin AC (2004) PI3K promotes voltage-dependent calcium channel trafficking to the plasma membrane. Nat Neurosci 7: 939–946 [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT et al. (2009) Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW (2006) Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol 4: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Tan YL, Ponniah S, Takeda Y, Wang SQ, Schachner M, Watanabe K, Pallen CJ, Xiao ZC (2008) Neural recognition molecules CHL1 and NB-3 regulate apical dendrite orientation in the neocortex via PTPα. EMBO J 27: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Ring C, Cheung R, Pevny L, Anton ES (2007) Nap1-regulated neuronal cytoskeletal dynamics is essential for the final differentiation of neurons in cerebral cortex. Neuron 54: 429–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Su I-h, Tezuka T, Yasuda T, Mikoshiba K, Tarakhovsky A, Yamamoto T (2002) BANK regulates BCR-induced calcium mobilization by promoting tyrosine phosphorylation of IP3 receptor. EMBO J 21: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama K, Tezuka T, Hoshina N, Nakazawa T, Yamamoto T (2006) Phosphorylation at Tyr-694 of Nogo-A by Src-family kinases. Biochem Biophys Res Commun 349: 1401–1405 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.