Abstract
Autophagy controls the quality and quantity of the eukaryotic cytoplasm while performing two evolutionarily highly conserved functions: cell-autonomous provision of energy and nutrients by cytosol autodigestion during starvation, and removal of defunct organelles and large aggregates exceeding the capacity of other cellular degradative systems. In contrast to these autodigestive processes, autophagy in yeast has additional, biogenesis functions. However, no equivalent biosynthetic roles have been described for autophagy in mammals. Here, we show that in mammalian cells, autophagy has a hitherto unappreciated positive contribution to the biogenesis and secretion of the proinflammatory cytokine IL-1β via an export pathway that depends on Atg5, inflammasome, at least one of the two mammalian Golgi reassembly stacking protein (GRASP) paralogues, GRASP55 (GORASP2) and Rab8a. This process, which is a type of unconventional secretion, expands the functional manifestations of autophagy beyond autodigestive and quality control roles in mammals. It enables a subset of cytosolic proteins devoid of signal peptide sequences, and thus unable to access the conventional pathway through the ER, to enter an autophagy-based secretory pathway facilitating their exit from the cytoplasm.
Keywords: autophagy, HMGB1, IL-1, inflammasome, unconventional secretion
Introduction
Autophagy is a fundamental biological process in eukaryotic cells, where it plays a number of roles associated with quality and quantity control of the cytoplasm (Mizushima et al, 2008; Yang et al, 2010). For the most part, autophagy enables cellular viability (Kroemer and Levine, 2008; Kroemer et al, 2010) by removing damaged organelles, for example, depolarized or leaky mitochondria (Youle and Narendra, 2011), digesting potentially toxic protein aggregates (Renna et al, 2010), killing intracellular microbes (Deretic and Levine, 2009), and supplying energy and nutrients to the cell (Kim et al, 2011) and the whole body (Ezaki et al, 2011) by bulk autodigestion of the cytosol at times of starvation. Based on the known and on yet to be uncovered functions, autophagy has broad impact in fundamental biology (Mizushima and Levine, 2010) and in human health (Mizushima et al, 2008) including aging (Alavez et al, 2011), cancer (Guo et al, 2011), neurodegeneration (Renna et al, 2010; Wong and Cuervo, 2010), myopathies (Romanello et al, 2010), metabolic disorders (Gonzalez et al, 2011), infections, immunity, and inflammatory diseases (Deretic, 2011; Levine et al, 2011).
The sensu stricto autophagy, often referred to as macroautophagy, involves Atg factor-driven generation of intracellular membranes that sequester cytoplasmic targets earmarked for autophagic processing (Yang et al, 2010). The principal components of the autophagic signalling and execution machinery are (i) a protein kinase network including Tor, AMPK, and Ulk1/2 (Egan et al, 2011; Kim et al, 2011), (ii) lipid kinases, principally the class III phosphatidylinositol 3-kinase hVPS34 (Kihara et al, 2001) and potentially class I p110β (Dou et al, 2010), in cooperation with Beclin 1 and its multiple modifiers (He and Levine, 2010; Strappazzon et al, 2011); (iii) a protein and protein–lipid conjugation Atg system resulting in a lipo–protein conjugate, LC3-II, which marks the autophagic membranes (Yang et al, 2010); and (iv) Atg8- and LC3-interacting autophagic adaptors that recognize ubiquitinated targets destined for autophagic capture (Johansen and Lamark, 2011) and may be associated, as in the case of p62/sequestosome 1, with autophagosomal nucleation sites (Itakura and Mizushima, 2011). The protein lipidation system resulting in LC3-II is driven by the Atg5–Atg12/Atg16 complex acting as an E3 ligase equivalent that facilitates localized conversion of LC3-I into LC3-II, with the latter consisting of the LC3 moiety conjugated to phosphatidylethanolamine at its C-terminus (Yang et al, 2010). The cargo enveloped by autophagosomes normally follows what is considered to be its principal destiny, that is, degradation of the captured material after conversion of autophagic organelles into degradative autolysosomes (Yang et al, 2010).
Although the central dogma of autophagy is that it is a degradative, disposal pathway, it also participates in several biogenesis processes documented in yeast, for example, the Cvt pathway (Yang et al, 2010) and the unconventional secretion of Acb1 (Duran et al, 2010; Manjithaya et al, 2010). The secretion of proteins devoid of leader peptides differs from the secretion of proteins endowed with this signal that brings them across the ER membrane into the canonical secretory pathway. The secretion of such leaderless peptides may involve unrelated pathways (Giuliani et al, 2011), but some common principles are beginning to emerge that are conserved at least for a subset of unconventional secretion substrates, in particular, a dependence of their secretion on the Golgi reassembly stacking protein (GRASP) (Nickel and Rabouille, 2009; Duran et al, 2010; Manjithaya et al, 2010). The recent findings in yeast with Acb1 (Duran et al, 2010; Manjithaya et al, 2010) suggest that GRASP- and autophagy-dependent pathways may converge, but it is unknown whether these relationships extend to mammalian cells.
One of the promising systems to address the question of the potential role of autophagy in the unconventional secretion in mammalian cells (Keller et al, 2008) is the extracellular delivery of the proinflammatory cytokine IL-1β (Davis et al, 2011; Lamkanfi, 2011). IL-1 family members, IL-1β and IL-18 (IL-1F4), are processed in the cytosol from their precursors upon inflammasome activation and are secreted without transiting through the conventional secretory pathway. However, recent reports have indicated that autophagy plays an inhibitory role in inflammasome and IL-1β activation (Nakahira et al, 2010; Harris et al, 2011; Zhou et al, 2011), thus potentially negating the idea that autophagy may contribute to the unconventional secretion of IL-1β. The mechanisms of autophagy-based suppression of inflammasome activation and IL-1β secretion may be indirect and seem to rely on autophagy-dependent mitochondrial homeostasis that normally suppresses release of reactive oxygen species (ROS; Zhou et al, 2011) and mitochondrial DNA (Nakahira et al, 2010), both of which can stimulate inflammasome. Alternatively, autophagic degradation of the inflammasome components has also been proposed (Harris et al, 2011) but this has not been experimentally tested.
Here, we have addressed the question of whether autophagy plays a direct role in inflammasome and IL-1β activation and secretion. We found that, whereas basal autophagy inhibits IL-1β secretion in concordance with the recent reports (Nakahira et al, 2010; Zhou et al, 2011), induced autophagy augments IL-1β secretion. We show that inflammasome and the autophagy apparatus synergize during IL-1β secretion in cells stimulated to undergo autophagy. We also show that autophagy induction cooperates with GRASP and Rab8a (a GTPase controlling post-Golgi polarized sorting and exocytosis) in driving IL-1β secretion. We, thus, define one of the first biogenesis functions of autophagy in mammalian cells and show that at least one type of unconventional secretion utilizes autophagic machinery in higher vertebrate cells.
Results
Induction of autophagy promotes inflammasome-dependent IL-1β secretion
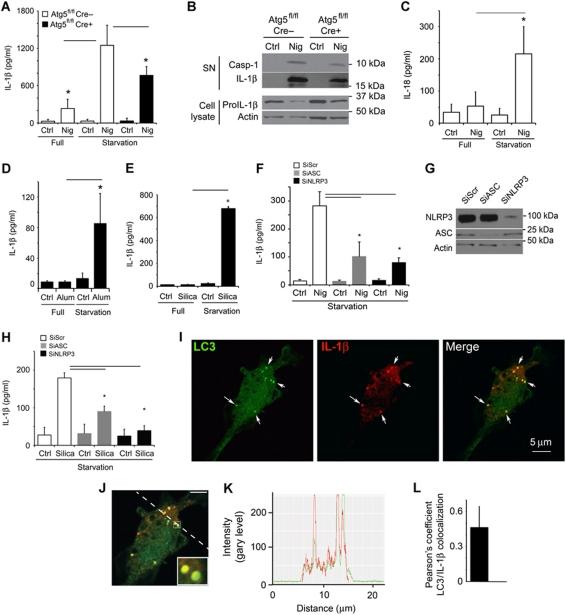
Whereas it has been found that basal autophagy reduces extracellular release of the major proinflammatory cytokine IL-1β (Nakahira et al, 2010; Zhou et al, 2011), we detected the opposite when autophagy was induced in primary murine bone marrow-derived macrophages (BMMs; Figure 1). Stimulation of autophagy by starvation strongly enhanced IL-1β secretion in response to conventional NLRP3 (NALP3) inflammasome agonist nigericin (Figure 1A). This effect was also seen (Figure 1B) in western blots of caspase 1 and mature IL-1β of culture supernatants from cells grown in the absence of serum, as conventionally done when assessing IL-1β secretion by immunoblotting (Martinon et al, 2006). A reduced secretion in BMMs from Atg5fl/fl LyzM-Cre+ mice, compared with BMMs from their Cre− littermates, was accompanied and contrasted by the higher level of cell-associated pro-IL-1β in Cre− versus Cre+ BMMs (Figure 1B). The BMMs derived from Atg5fl/fl LyzM-Cre+ mice for these and other experiments had, as expected, no detectable Atg5 (since the Atg5 gene is excised in Cre+ macrophages; Zhao et al, 2008) and LC3-II, a key marker of autophagy (Supplementary Figure S1A).
Figure 1.
Induction of autophagy enhances IL-1β secretion. (A) Atg5fl/fl Cre− and Atg5fl/fl Cre+ bone marrow-derived macrophages (BMMs), pretreated overnight with 100 ng/ml LPS, were stimulated for 1 h with the inflammasome agonist nigericin (20 μM) with (Starvation; EBSS) or without (Full; full medium) autophagic induction. Cell culture supernatants were assayed for murine IL-1β by ELISA. Data represent mean values±s.d. (n⩾3); *P<0.05. (B) LPS-pretreated Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs were stimulated with 20 μM nigericin for 1 h in OptiMEM and the release of active caspase-1 and IL-1β was determined by immunoblotting. (C) As in (A), assayed for IL-18. Data represent mean values±s.d. (n⩾3); *P<0.05. (D) LPS-pretreated BMMs were exposed to alum (250 μg/ml) for 1 h with or without autophagic induction by starvation. Secreted IL-1β was measured as in (A). Data represent mean values±s.d. (n⩾3); *P<0.05. (E) LPS-pretreated BMMs were exposed to silica (250 μg/ml) for 1 h with or without autophagic induction by starvation. Secreted IL-1β was measured as in (A). Data represent mean values±s.d. (n⩾3); *P<0.05. (F) BMMs were transfected with scramble (Scr) control siRNA or siRNAs against ASC and NLRP3. After 48 h following transfection, cells were treated overnight with LPS and subjected to nigericin (20 μM) and starvation for 1 h. Data represent mean values±s.d. (n⩾3); *P<0.05. (G) Immunoblot analysis of ASC and NLRP3 knockdowns. (H) BMMs were transfected with scramble (Scr) control siRNA or siRNAs against ASC and NLRP3. After 48 h following transfection, cells were treated overnight with LPS and subjected to silica (250 μg/ml) and starvation for 1 h. Data represent mean values±s.d. (n⩾3); *P<0.05. (I) Colocalization of IL-1β with the basal autophagic machinery factor LC3. Fluorescence: LC3 (green, Alexa488); IL-1β (red, Alexa568). BMMs were from GFP–LC3 knock-in mice, treated with LPS then prepared for immunofluorescence microscopy using fluorescently labelled antibodies against GFP and IL-1β. (J, K) A line fluorescence tracing from images in (I). (L) Pearson's colocalization coefficient for IL-1β and LC3. Pearson's coefficient was derived from three independent experiments with five fields per experiment, for a total of 15 fields contributing to the cumulative result. Figure source data can be found in Supplementary data.
The effects of induced autophagy on secretion of inflammasome substrates described above were not limited to IL-1β, since secretion of another inflammasome-dependent cytokine from the IL-1 family, IL-18 (IL-1F4), was enhanced when autophagy was induced (Figure 1C). Pharmacological induction of autophagy by mTOR inhibition with pp242 (Torkinib) increased secretion of IL-1β by BMMs (Supplementary Figure S1B). An enhancement of IL-1β secretion upon induction of autophagy was also detected when particulate inflammasome agonists, alum (Eisenbarth et al, 2008; Figure 1D; Supplementary Figure S1C), silica (Figure 1E; Hornung et al, 2008), and amyloid-β fibrils (Halle et al, 2008; Supplementary Figure S1D), were used as inflammasome inducers. The enhancement of IL-1β secretion associated with autophagy induction was inflammasome dependent, as IL-1β activation was diminished by knockdowns of the inflammasome components ASC and NLRP3, regardless of whether the inflammasome agonist used was nigericin or silica (Figure 1F–H). The knockdowns of ASC and NLRP3 did not change IL-1β expression (Supplementary Figure S1E and F). The increased secretion of IL-1β was not due to the increased cell death or non-specific membrane permeability as LDH release showed a kinetic lag behind release of IL-1β whether the inflammasome agonist used was nigericin or silica (Supplementary Figure S2A–D).
The stimulation of autophagy promoted IL-1β secretion in an Atg5-dependent manner, based on comparisons between BMMs from Atg5fl/fl- LyzM-Cre+ mice and BMMs from their Atg5-competent (Atg5fl/fl Cre−) littermates (Figure 1A). However, the loss was not complete in Cre+ BMMs (Figure 1A). We interpret the incomplete reduction in IL-1β secretion in the absence of Atg5 as a net result of two opposing effects—one described here as a product of positive contribution of induced autophagy on extracellular delivery of IL-1β and the other being the recently reported negative regulation of IL-1β secretion by basal autophagy (Nakahira et al, 2010; Harris et al, 2011; Zhou et al, 2011). In keeping with this interpretation and in contrast to the stimulatory effects of induced autophagy (Figure 1A–E; Supplementary Figure S1B–D), basal autophagy negatively affected IL-1β and IL-18 secretion (Supplementary Figure S2E and F) in agreement with the recent reports in cells not induced for autophagy (Nakahira et al, 2010; Harris et al, 2011; Zhou et al, 2011).
IL-1β and autophagic protein LC3 colocalize in the cytoplasm
How might induced autophagy enhance IL-1β secretion? We considered a model in which autophagy, as a process that can translocate cytosolic proteins and other targets (en masse or specific components) from the cytosol to the inside of vesicular compartments, brought IL-1β into the lumen of autophagic vacuoles followed by exocytosis. When we examined IL-1β and the key marker of autophagosomes LC3 by immunofluorescence confocal microscopy, LC3 and IL-1β colocalized and displayed major similarities in the overall intracellular organellar distribution (Figure 1I–L). The overlap between IL-1β and LC3 remained detectable when cells were treated with nigericin (Supplementary Figure S3A–C). These observations indicate that autophagic organelles and IL-β intersect.
Inhibition of autophagy flux reduces IL-1β secretion
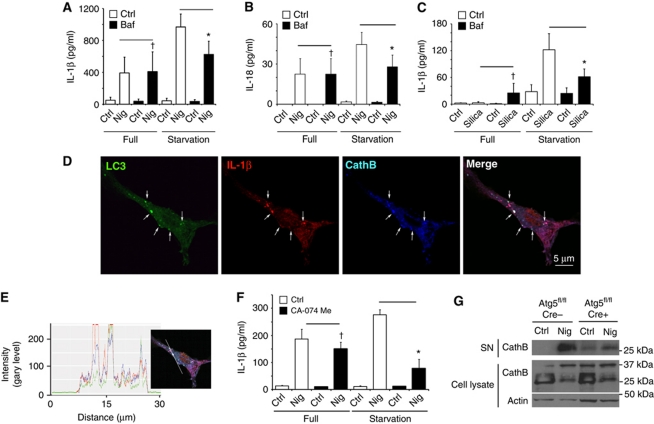
A question arose whether the LC3+ organelles containing IL-1β are on pathway to degradation or facilitated IL-1β secretion. We first tested the effects of bafilomycin A1, a conventional inhibitor of autophagic maturation, which acts as an antagonist of vacuolar H+ ATPase and prevents lumenal acidification and autophagosomal cargo degradation. If induction of autophagy acted to degrade IL-1β, then bafilomycin A1 was expected to increase IL-1β levels. Instead, bafilomycin A1 decreased IL-1β secretion in cells stimulated for autophagy by starvation, whereas no change was observed with bafilomycin A1 in cells undergoing basal autophagy only (Figure 2A). Thus, autophagy flux during autophagy induction was not inhibitory to IL-1β but was instead promoting IL-1β secretion. A similar trend was detected with another inflammasome substrate IL-18 (Figure 2B). Equivalent relationships have been observed for IL-1β secretion whether nigericin (Figure 2A) or silica (Figure 2C) was used as inflammasome agonists. The absence of IL-1β or IL-18 sparing effects of bafilomycin A1 is in keeping with the interpretation that autophagy is not degrading inflammasome components but that an unobstructed autophagy pathway is necessary for inflammasome-dependent IL-1 family members secretion.
Figure 2.
Autophagic pathway progression promotes secretion of inflammasome substrates. (A, B) LPS-pretreated BMMs were treated with 20 μM nigericin (Nig) and 100 nM bafilomycin A1 (Baf) with (Starvation) or without (Full) autophagic induction for 1 h and secreted IL-1β (A) and IL-18 (B) were measured. Data represent mean values±s.d. (n⩾3); *P<0.05. (C) LPS-pretreated BMMs were treated with 250 μg/ml of silica and 100 nM bafilomycin A1 (Baf) with (Starvation) or without (Full) autophagic induction for 1 h and secreted IL-1β were measured. Data represent mean values±s.d. (n⩾3); *P<0.05. (D) Colocalization of cathepsin B with the basal autophagic machinery factor LC3 and IL-1β. Fluorescence; LC3 (green, Alexa488), IL-1β (red, Alexa568), and cathepsin B (blue, Alexa633). BMMs from GFP–LC3 knock-in mice were treated with LPS and then analysed for immunofluorescence. (E) Colocalization line tracing analysis from images in (D). (F) LPS-pretreated BMMs were treated with 20 μM nigericin and cathepsin B inhibitor CA-074 Me (10 μM), with (Starvation) or without (Full) autophagic induction, for 1 h and secreted IL-1β was measured. Data represent mean values±s.d. (n⩾3); *P<0.05. (G) LPS-pretreated Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs were stimulated with 20 μM nigericin for 1 h in OptiMEM and release of cathepsin B was determined by immunoblotting. Figure source data can be found in Supplementary data.
Lysosomal hydrolase cathepsin B is a positive factor in autophagy-driven IL-1β secretion
Next, we investigated the role of lysosomal hydrolases, focusing on cathepsin B. We observed that IL-1β and LC3 colocalized with cathepsin B (Figure 2D and E). However, cathepsin B did not play an inhibitory role. Similarly to bafilomycin A1, cathepsin B inhibitor CA-074 Me suppressed IL-1β production. Instead of protecting IL-1β from potential degradation, CA-074 Me strongly inhibited IL-1β secretion in cells stimulated for autophagy by starvation (Figure 2F). No differences in expression of pro-IL-1β were observed in cells treated with bafilomycin A1 or CA-074 Me (Supplementary Figure S3D). Of further interest was that cathepsin B (mature form) was secreted along with the inflammasome substrates in a manner dependent on an intact autophagic apparatus: loss of Atg5 in BMMs from Cre+ mice (Atg5fl/fl LyzM-Cre) diminished the levels of the secreted mature cathepsin B relative to BMMs from Cre− littermates (Figure 2G). The findings with cathepsin B inhibitor CA-074 Me indicate a positive role for cathepsin B in IL-1β activation and autophagy-driven pathway of extracellular delivery of IL-1β. They can also help explain in part the observations that a lysosomal hydrolase, cathepsin B, assists in inflammasome activation and IL-1β secretion in response to particulate inflammasome agonists (Halle et al, 2008; Hornung et al, 2008).
Rab8a, a regulator of polarized sorting to plasma membrane colocalizes with IL-1β and LC3 and controls IL-1β secretion
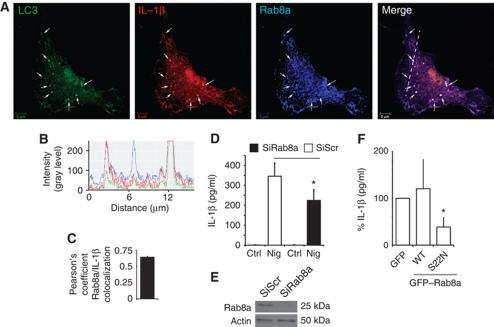
We next addressed the features of the compartment where LC3 and IL-1β colocalized. We observed an overlap between the LC3+ IL-1β+ profiles and Rab8a (Figure 3A–C). Rab8a is a regulator of polarized membrane trafficking, constitutive biosynthetic trafficking, and plasma membrane fusion of insulin-responsive (Sun et al, 2010) and other vesicular carriers (Moritz et al, 2001; Bravo-Cordero et al, 2007; Nachury et al, 2007; Bryant et al, 2010). Rab8a also colocalized with LC3 and IL-1β in cells exposed to nigericin (Supplementary Figure S4A–C). Rab8a was required for enhanced IL-1β secretion caused by starvation-induced autophagy and inflammasome activation with nigericin, since siRNA knockdown of Rab8a diminished IL-1β secretion from BMMs under these conditions (Figure 3D and E). Rab8a knockdown did not change pro-IL-1β mRNA levels (Supplementary Figure S4D). Overexpression of dominant-negative Rab8a mutant (S22N) inhibited IL-1β secretion from RAW264.7 cells, employed in that experiment based on their high efficiency of transfection (Figure 3F) (verified by flow cytometry of GFP–Rab8a for equal yields). Additionally, LC3+ IL-1β+ profiles were positive for subunits of the exocyst complex (Supplementary Figure S4E–J). Exocyst has been shown to cooperate with Rab8a in polarized plasma membrane delivery of vesicular carriers (Mazelova et al, 2009; Bryant et al, 2010). The presence of exocyst components on IL-1β+ autophagic organelles was also in keeping with a recent report implicating exocyst in autophagy (Bodemann et al, 2011). In summary, these experiments indicate that systems involved in vectorial vesicular transport to the plasma membrane participate in autophagy-based unconventional secretion and that Rab8a is required for efficient autophagy-dependent secretion of IL-1β.
Figure 3.
Rab8a is required for autophagy-activated IL-1β secretion. (A) Colocalization of Rab8a with the basal autophagic machinery factor LC3 and IL-1β. Fluorescence; LC3 (green, Alexa488), IL-1β (red, Alexa568), Rab8a (blue, Alexa633). BMMs from GFP–LC3 knock-in mice were pretreated with LPS and analysed by immunofluorescence microscopy. Arrows indicate triple colocalization. (B) Line tracing analysis of fluorescence signal intensity. (C) Pearson's colocalization coefficient for IL-1β and Rab8a. Pearson's coefficients were derived from three completely independent experiments with >5 fields per experiment, for a total of ⩾15 fields contributing to the cumulative result. (D) BMMs were transfected with siRNAs against Rab8a or scramble (Scr) control. At 24 h after the first transfection, cells were transfected again with siRNA, treated with LPS and the day after subjected to nigericin in full medium for 1 h, and IL-1β secretion measured. (E) Immunoblot analysis of Rab8a knockdown in BMMs. (F) RAW 264.7 macrophages were transfected with GFP-tagged Rab8a constructs (WT, wild type; S22N, dominant-negative mutant), treated overnight with LPS and stimulated for 1 h with 20 μM nigericin along with induction of autophagy by starvation. IL-1β secretion was measured by ELISA. Data represent mean values±s.d. (n⩾3); *P<0.05. Figure source data can be found in Supplementary data.
GRASP55 controls secretion of IL-1β
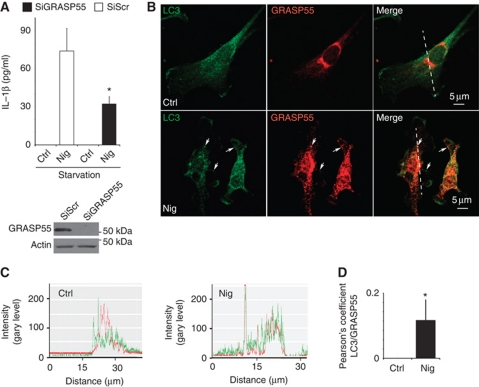
Two studies in yeast (Duran et al, 2010; Manjithaya et al, 2010) have reported that autophagic machinery is required for unconventional secretion of the protein Acb1, and that this pathway depends on the yeast equivalent of a Golgi-associated protein GRASP in mammals (Kinseth et al, 2007; Nickel and Rabouille, 2009). Mammalian cells encode two GRASP paralogues, GRASP55 (GORASP2) and GRASP65 (GORASP1) (Barr et al, 1997; Shorter et al, 1999). We first tested whether any of the mammalian GRASPs were required for IL-1β secretion. We could not obtain a good knockdown of GRASP65 (GORASP1) and thus could not evaluate its involvement. However, a knockdown of GRASP55 diminished IL-1β secretion (Figure 4A; Supplementary Figure S5A). A similar downregulation of IL-18 secretion was observed with GRASP55 knockdown (Supplementary Figure S5B). We next tested whether GRASP55 showed any detectable response to inflammasome stimulation. GRASP55 in resting cells is mostly localized aligned within the perinuclear Golgi (Figure 4B; Supplementary Figure S6A). However, a fraction of it dispersed upon treatment of cells with the inflammasome agonist nigericin (Figure S6B) and was found juxtaposed and partially overlapping with LC3 profiles (Figure 4B and C). Thus, GRASP55 responds to inflammasome stimulation and is important for secretion of the inflammasome substrates IL-1β and IL-18.
Figure 4.
GRASP55 is required for autophagy-activated IL-1β secretion. (A) BMM cells were transfected with scramble (Scr) control siRNA or siRNA against GRASP55. After 48 h of transfection, cells were treated with LPS and the day after subjected to 20 μM nigericin in EBSS, and secreted IL-1β was measured by ELISA. Data represent mean values±s.d. (n⩾3); *P<0.05. Inset: Immunoblot analysis of GRASP55 knockdown. (B) Immunofluorescence confocal microscopy analysis of LC3 and GRASP55 distribution. LC3 (green, Alexa488), GRASP55 (red, Alexa568). BMMs were pretreated overnight with 100 ng/ml LPS and either not stimulated (Ctrl) or stimulated (Nig) for 30 min with the inflammasome agonist nigericin (20 μM) in full medium. (C) Line tracings, analysis of fluorescence signal intensity from images in (B). (D) Pearson's coefficients for LC3 and GRASP55 were quantified using SlideBook morphometric analysis software as a measure of adjacency between GRASP55 and LC3 profiles. Pearson's coefficients were derived from three independent experiments with five fields per experiment, for a total of 15 fields contributing to the cumulative result. Figure source data can be found in Supplementary data.
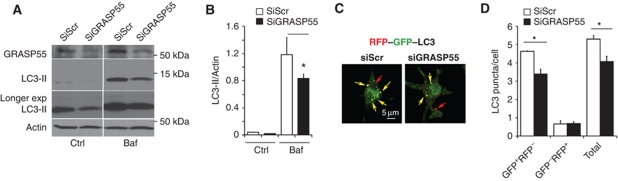
GRASP55 controls autophagy initiation
In addition to being required for IL-1β secretion, GRASP55 showed functional effects on LC3 and autophagy, tested by employing two core assays (Mizushima et al, 2010): LC3-II lipidation and the RFP–GFP–LC3 tandem probe. When GRASP55 was knocked down, autophagy initiation was negatively affected, as LC3-II levels were lower in both untreated and bafilomycin A1-treated cells (Figure 5A and B). A partial downregulation of GRASP65 (to the extent that it could be achieved in BMMs) suggested a minor synergistic effect with GRASP55 on LC3-II levels upon induction of autophagy (Supplementary Figure S5C). Knocking down GRASP55 reduced the total number of autophagic puncta, and selectively reduced the formation of autophagosomes but not their maturation (Figure 5C and D). This was apparent from the data obtained with the RFP–GFP–LC3 probe following published methods (Kimura et al, 2007), which showed reduced GFP+RFP+ LC3 profiles (early autophagosome) and equal number of GFP−RFP+ LC3 profiles (mature autophagic organelles) in cells knocked down for GRASP55 (Figure 5D). Thus, mammalian GRASP55, a paralogue of GRASP from lower organisms that has thus far been the sole definitive molecular factor associated with unconventional secretion (Giuliani et al, 2011), displays important and previously unappreciated positive regulatory effects on autophagy induction. These findings strengthen the connections between autophagy and GRASPs in general, and specifically demonstrate the role of mammalian GRASP55 both in autophagy initiation and in the secretion of leaderless inflammasome substrates such as IL-1β and IL-18.
Figure 5.
GRASP55 controls autophagy initiation. (A, B) Effect of GRASP55 on autophagy induction by measuring LC3-II. BMM cells were transfected with GRASP55 siRNAs or scramble (Scr) control. At 72 h post transfection, cells were induced for autophagy, treated or not with Bafilomycin A1 (Baf) to inhibit autophagic degradation and LC3-II/actin ratios determined by immunoblotting (A) followed by densitometry (B). Data represent mean values±s.d. (n⩾3); *P<0.05. (C, D) RAW 264.7 was transfected with GRASP55 siRNAs or scramble (Scr) siRNA control. Following 48 h of siRNA treatment, cells were transfected with RFP–GFP–LC3 plasmid (GFP is sensitive to acidification, whereas RFP is not), after 24 h induced for autophagy in EBSS for 1 h and autophagic induction and flux quantified (graph in D) by determining the number of early autophagic organelles (GFP+RFP+ puncta) and autolysosomal organelles (GFP−RFP+ puncta) per cell as illustrated in fluorescent images (yellow arrows, GFP+RFP+; red arrows, GFP−RFP+). Total, yellow+red puncta per cell. Data represent mean values±s.d. (n⩾3); *P<0.05. Figure source data can be found in Supplementary data.
Autophagy-based unconventional secretion is not limited to proteolytically processed inflammasome substrates
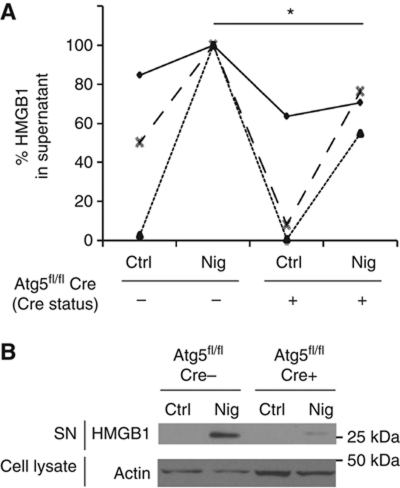
We next wondered whether the unconventional process described above is limited to inflammasome substrates epitomized by IL-1β that are concomitantly with their secretion proteolytically processed from precursor pro-proteins into mature forms. We tested whether induction of autophagy affected other proteins not connected to proteolytic processing in the inflammasome, such as HMGB1 (high mobility group box 1 protein). HMGB1 is a major proinflammatory alarmin or DAMP (damage-associated molecular pattern) normally present in the nucleus (Andersson and Tracey, 2011). This chromatin-associated nuclear protein (with additional intracellular and extracellular signalling roles), upon exposure to inputs including those that induce autophagy (Singh et al, 2010; Tang et al, 2010), undergoes a complex set of biochemical and localization changes. In the process, it first translocates from the nucleus into the cytoplasm and then is released from the cytoplasm to act in tissue remodelling signalling (when acting alone) or as an inflammatory mediator (when combined with bacterial agonists or other alarmins such as IL-1β). When tested, starvation and nigericin co-treatment caused HMGB1 extracellular release in an Atg5-dependent manner (Figure 6A). HMGB1 band was detected by immunoblots in BMMs culture supernatants upon stimulation of cells with nigericin, whereas HMGB1 was largely diminished when BMMs from Atg5fl/fl Cre-LyzM mice were tested (Figure 6B). Nigericin was used in these experiments as an inflammasome agonist based on the reports that HMGB1, along with additional unconventional substrates, depends on inflammasome for secretion although the protein itself is not subjected to proteolytic processing by caspase 1 (Keller et al, 2008; Willingham et al, 2009; Lamkanfi et al, 2010; Lamkanfi, 2011). These experiments show that autophagy-based unconventional secretion affects release of HMGB1 in a manner similar to IL-1β. Our findings broaden the spectrum of autophagy-based unconventional secretion substrates, and establish this type of unconventional secretion as a more general process in extracellular delivery of cytosolic proteins.
Figure 6.
HMGB1 is an autophagy-based alternative secretion substrate. (A) Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs, pretreated overnight with 100 ng/ml LPS, were stimulated for 1 h with 20 μM nigericin (Nig; inflammasome agonist) while incubated in EBSS for induction of autophagy by starvation. Cell culture supernatants were assayed for murine HMGB1 by ELISA. Data (normalized to sample with maximum HMGB1 secretion in each experimental repeat; Cre−and Nig) represent mean values±s.d. (n⩾3); *P<0.05. (B) LPS-pretreated Atg5fl/fl Cre− and Atg5fl/fl Cre+ BMMs were stimulated with 20 μM nigericin for 1 h in OptiMEM and the release of HMGB1 was determined by immunoblotting. Figure source data can be found in Supplementary data.
Discussion
The data presented in this work outline several elements of the autophagy-based unconventional secretory pathway in mammalian cells. This type of unconventional secretion is shown here to support the extracellular delivery of inflammasome substrates, in particular, IL-1β and IL-18 and may potentially have a broader number of clients. A relevant aspect of the process described here is that induction of autophagy is required to observe the manifestations of this type of unconventional secretion. Since basal autophagy suppresses spurious induction of inflammasome (Nakahira et al, 2010; Zhou et al, 2011), autophagy provides both avoidance of unscheduled inflammasome activation and a platform for extracellular delivery of inflammasome substrates. Since a number of hormones, cytokines, pathogen-associated molecular patterns, and danger-associated molecular patterns (Tang et al, 2010; Deretic, 2011) are known to induce or inhibit autophagy, a link between autophagy and secretion of major immunomodulatory cytokines such as IL-1β could significantly influence the extent and duration of inflammation. Connections between metabolic syndrome, high fat diet, and inflammasome activity are now beginning to be appreciated (Vandanmagsar et al, 2011; Wen et al, 2011), and we propose that autophagy-based unconventional secretion may be a key coupler between the metabolism and inflammation. Since a number of genetic links have been found between autophagy and idiopathic inflammatory diseases or infectious diseases with significant inflammatory components (Levine et al, 2011), it is possible that at least in part the genetic associations between autophagy risk loci and disease states may stem from altered autophagy-based unconventional secretion of inflammatory cytokines.
The role of autophagy was represented here by the effects of induction of autophagy and by employing conditional knockout mice with a loss of Atg5 in macrophages. We interpret the incomplete inhibition of IL-1β secretion upon Cre-dependent Atg5fl/fl excision as a result of the composite effects of Atg5-dependent basal autophagy (inhibitory) (Zhou et al, 2011) and induced autophagy (stimulatory) although we cannot exclude the possibility of slight leakiness of the Atg5fl/flLyzM-Cre system or the existence of additional pathways. Importantly, blocking autophagic maturation has not salvaged IL-1β but rather inhibited its secretion. This appears to be in contrast to what has been reported for Acb1 in yeast (Manjithaya et al, 2010). Moreover, cathepsin B activity was needed, suggesting that autophagic organelles here did not function as mere cargo carriers but provided a platform for activation of inflammasome and IL-1β. In keeping with these observations, cathepsin B has been implicated in inflammasome activation in response to particulate inflammasome agonists (Halle et al, 2008; Hornung et al, 2008), such as those (alum, amyloid-β) used here in addition to nigericin, but how the substrate and cathepsin B meet has hitherto not been defined. Our data indicate that induction of autophagy enhances assembly of inflammasome-activating components and suggest that autophagic organelles may be a platform for concentration of components engaged in proteolytic activation of inflammasome components and inflammasome substrates. In keeping with this model of a muster station for inflammasome components, activation and subsequent extracellular delivery, is the translocation of pro-IL-1β to membranous organelles upon stimulation with the inflammasome agonist nigericin, as previously observed by Sitia and colleagues (Rubartelli et al, 1990) who have established early on that this process does not follow the conventional secretory pathway.
The autophagy-based unconventional secretion pathway in mammalian cells includes GRASP, one of the peripheral Golgi proteins involved in lateral organization of Golgi ribbons. Although the role of GRASP in alternative secretory pathway has been studied, its exact mechanism of action has not been elucidated (Kinseth et al, 2007; Nickel and Rabouille, 2009). We observed here a potentially telling connection between GRASP and autophagy, by showing that GRASP affects autophagy induction, which places GRASP upstream of autophagy execution, including the conjugation systems involved in LC3 lipidation. The response of GRASP to nigericin stimulation in terms of its redistribution and juxtaposition to autophagic organelles further links autophagy, inflammasome, and GRASP, although alternative explanations are possible. The finding that Rab8a plays a functional role in autophagy-based unconventional secretion is of significance not just by assigning a trafficking regulator to this pathway but also by providing additional links with exocyst components, implicated to cooperate with Rab8a in other systems (Mazelova et al, 2009; Bryant et al, 2010) and to play a role in autophagy (Bodemann et al, 2011).
In summary, in this study we have uncovered the role of autophagy in the secretion of cytosolic proinflammatory factors that cannot enter the conventional biosynthetic pathway due to the absence of leader peptides that would bring them into the ER and the organelles of the canonical secretory pathway. Both cytosolic IL-1β and IL-18 are processed from their precursor proteins into their active forms via the inflammasome apparatus (Davis et al, 2011; Lamkanfi, 2011); however, the process that delivers the proteolytically activated IL-1β and IL-18 to the extracellular environment has hitherto remained unclear. To be eligible for export outside of the cell without invoking a pore mechanism, cytosolic proteins first need to be brought somehow into the lumen of vesicular carriers, as previously shown by others (Rubartelli et al, 1990). We have elsewhere noted (Deretic, 2005, 2011) that autophagy is a bulk topological inverter for cytosolic proteins and other molecules, ferrying them from the cytosol into the organellar lumen. We now show that induction of autophagy does that with IL-1β in the process of its secretion. In doing so, autophagic machinery cooperates with the Golgi-associated factor GRASP and post-Golgi membrane trafficking and exocytosis regulator Rab8a. Autophagy captures cytosolic IL-1β and brings IL-1β into the organelles of a specialized unconventional secretory pathway.
Broadening the scope of autophagy-based alternative secretion pathway is the observation that it facilitates exit from cells of the alarmin HMGB1. HMGB1 is a DAMP that is actively released from immune cells unlike its passive release from several cell types secondary to cell death (Andersson and Tracey, 2011). It has been recently shown that inflammasome, rather unexpectedly given that HMGB1 is not a known substrate for caspase-1 processing, plays a role in HMGB1 release (Lamkanfi et al, 2010). One explanation that can be offered based on our studies is that a role of inflammasome may not be related solely to proteolytic substrate processing but that it may be hardwired into the secretory pathway studied here. This is in keeping with the requirement for NLRP3 and ASC and not the caspase 1 activity for HMGB1 release as a recently recognized non-canonical inflammasome client (Willingham et al, 2009). We propose here that autophagy-based unconventional secretion may be used for extracellular delivery of a spectrum of cytosolic proteins or processed cytoplasmic substrates, not restricted to the proteins explored here, and possibly including other biological mediators such as the recently discovered cryptides (Deretic, 2005; Ponpuak et al, 2010). A recent study that appeared while this work was in revision suggests that an unconventional secretion process, also dependent on GRASP and autophagic machinery, may facilitate plasma membrane delivery of mutant CFTR, potentially expanding the range of substrates to integral membrane proteins (Gee et al, 2011). Given the capacity for either bulk transport or selectivity when coupled with autophagic adaptors, we predict that autophagy-based unconventional secretion serves a potentially broad spectrum of yet to be uncovered physiological functions.
Materials and methods
Macrophages
Murine BMM cells were prepared from femurs of C57/BL6 mice, Atg5fl/fl LyzM-Cre mice (Zhao et al, 2008) and their Cre-negative Atg5fl/fl littermates, and GFP–LC3 transgene knock-in mice (Mizushima et al, 2004) as previously described (Ponpuak et al, 2010). RAW 264.7 macrophages were maintained and manipulated as previously described (Ponpuak et al, 2010).
Pharmacological agonists, inhibitors, inflammasome, and autophagy
To induce pro-IL-1β expression, cells were pretreated overnight with 100 ng/ml LPS (Sigma). Inflammasome was induced with 20 μM nigericin (Sigma) for 1 h or with 250 μg/ml Alum (Thermoscientific) for 1 h or with 250 μg/ml silica crystals (MIN-U-SIL-15, US Silica) for 1 h or with 5 μM Amyloid-β peptide (Aβ; American Peptide Company) fibrils prepared as described (Moore et al, 2002). Cells were treated with 100 nM bafilomycin A1 (LC Laboratories) or 10 μM cathepsin B inhibitor (CA-074 Me) (Enzo Life Science). Autophagy was induced for 1 h by starvation in EBSS or with pp242 in full medium (Torkinib; Chemidea). Starvation and other treatments (except for macrophage priming with LPS done in advance) were carried out concurrently (i.e., initiated at the same time).
Transfections and siRNA knockdowns
BMM and RAW 264.7 cells were transfected by nucleoporation using Nucleofector Reagent Kit V or Kit Mouse Macrophage (Amaxa/Lonza Biosystems). For murine NLRP3, ASC, Rab8a or GRASP knockdowns, cells were transfected with siGENOME SMARTpool reagents (Dharmacon). Rab8a SMARTpool (GAAUAAGUGUGAUGUGAAU; GAAGACCUGUGUCCUGUUC; GACCUACGAUUACCUGUUC; GAGCAGCCAUGGAGUCAAG), ASC SMARTpool (AUAC AUCCCUACUUGGUGA; GCUUAGAGACAUGGGCUUA; GCAACUGCGAGAAGGCUAU; CUGCAAACGACUAAAGAAG), NLRP3 SMARTpool (GUUCUUCGCUGCUAUGUAC; GCACCCAGGCUGUAACAUU; UGAAGGACCCACAGUGUAA; UCACAUUCCUCUAUGGUAU), GORASP1 SMARTpool (CAUGAAGGUGCGCGAGGUA; CAGAGGACAUUGGUUCUAG; ACUCGAGGCUGAACAAGGA; GCUACGACCUCACAACUUA), and GORASP2 SMARTpool (GAAGACCUGUUCAGCCUUA; UACCAAGUCUGAUGCCUUU; GUAAACCAGUCCCUUGCUU; GAUCAUCACACCAAACUCU). Non-targeting siRNA pool (Scrambled) was used as a control. Plasmid encoding tandem RFP–GFP–LC3 fusion for quantification of autophagic maturation was from T Yoshimori (Osaka, Japan). Plasmids encoding Rab8a wt (wild type) and Rab8a S22N were from Johan Peranen (University of Helsinki, Finland).
Antibodies, immunoblotting, detection assays, and microscopy
Cells extracts were analysed by standard immunoblot techniques with antibodies to pro-IL-1β (Abcam), NLRP3 (AdipoGen), ASC (Enzo Life Sciences), LC3 (Sigma), GRASP65 (Novus), GRASP55 (Abcam), Rab8a (Abcam), and Actin (Sigma); staining was revealed with Super Signal West Dura chemiluminescent substrate (Pierce). For all conditions, cell-free supernatants were assayed by immunoblotting after TCA precipitation for mouse IL-1β p17 (R&D), caspase-1 p10 (Santa Cruz Biotechnology), HMGB1 (Abcam) and Cathepsin B (R&D) or by ELISA for mouse IL-1β (R&D), IL-18 (MBL), and HMGB1 (IBL). Immunofluorescence confocal microscopy was carried out using a Zeiss LSM 510 Meta microscope (laser wavelengths 488, 543, and 633 nm). Antibodies against endogenous proteins IL-1β (Abcam), Sec6 (Shu C Hsu, Rutgers University, NJ, USA), Cathepsin B (R&D), Rab8a (Abcam), GORASP1 (Novus), GORASP2 (ProteinTech Group), GM130 (BD), LC3 (MBL) and GFP (Abcam; for BMMs from GFP–LC3 knock-in transgenic mice) were used for indirect immunofluorescence analysis. Pearson's colocalization coefficients were derived using SLIDEBOOK 5.0 (Intelligent Imaging Innovations) applying the SLIDEBOOK 5 default algorithm command ‘AND’. All Pearson's coefficients were derived from three completely independent experiments with five fields or more per experiment, for a total of ⩾15 fields contributing to the cumulative result.
Statistics
All data were analysed using two-tailed unpaired Student's t-tests. All experiments were performed at least three times, with data representing mean values±s.d. (standard deviation).
Supplementary Material
Acknowledgments
We thank M Delgado-Vargas for help. This work was supported by grants R01 AI069345, RC1AI086845, and R01 AI042999 from National Institutes of Health and a Bill and Melinda Gates Grand Challenge Explorations grant OPP1024376.
Author contributions: ND designed experiments, performed the experiments, analysed the data, and helped write the manuscript. SJ performed the experiments and analysed the data. MP, WO, and DB performed the experiments and analysed the data. VD conceived experiments, analysed the data, and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alavez S, Vantipalli MC, Zucker DJ, Klang IM, Lithgow GJ (2011) Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature 472: 226–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson U, Tracey KJ (2011) HMGB1 is a therapeutic target for sterile inflammation and infection. Annu Rev Immunol 29: 139–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA, Puype M, Vandekerckhove J, Warren G (1997) GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91: 253–262 [DOI] [PubMed] [Google Scholar]
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, Camonis JH, Yeaman C, Levine B, White MA (2011) RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 144: 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Marrero-Diaz R, Megias D, Genis L, Garcia-Grande A, Garcia MA, Arroyo AG, Montoya MC (2007) MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J 26: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DM, Datta A, Rodriguez-Fraticelli AE, Peranen J, Martin-Belmonte F, Mostov KE (2010) A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol 12: 1035–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BK, Wen H, Ting JP (2011) The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 29: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V (2005) Autophagy in innate and adaptive immunity. Trends Immunol 26: 523–528 [DOI] [PubMed] [Google Scholar]
- Deretic V (2011) Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev 240: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Levine B (2009) Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5: 527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, Chattopadhyay M, Pan JA, Guerriero JL, Jiang YP, Ballou LM, Yue Z, Lin RZ, Zong WX (2010) The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol 191: 827–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V (2010) Unconventional secretion of Acb1 is mediated by autophagosomes. J Cell Biol 188: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ (2011) Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 331: 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O′Connor W, Sutterwala FS, Flavell RA (2008) Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature 453: 1122–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezaki J, Matsumoto N, Takeda-Ezaki M, Komatsu M, Takahashi K, Hiraoka Y, Taka H, Fujimura T, Takehana K, Yoshida M, Iwata J, Tanida I, Furuya N, Zheng DM, Tada N, Tanaka K, Kominami E, Ueno T (2011) Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy 7: 727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee HY, Noh SH, Tang BL, Kim KH, Lee MG (2011) Rescue of DeltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell 146: 746–760 [DOI] [PubMed] [Google Scholar]
- Giuliani F, Grieve A, Rabouille C (2011) Unconventional secretion: a stress on GRASP. Curr Opin Cell Biol 23: 498–504 [DOI] [PubMed] [Google Scholar]
- Gonzalez CD, Lee MS, Marchetti P, Pietropaolo M, Towns R, Vaccaro MI, Watada H, Wiley JW (2011) The emerging role of autophagy in the pathophysiology of diabetes mellitus. Autophagy 7: 2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, Coller HA, Dipaola RS, Gelinas C, Rabinowitz JD, White E (2011) Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle A, Hornung V, Petzold GC, Stewart CR, Monks BG, Reinheckel T, Fitzgerald KA, Latz E, Moore KJ, Golenbock DT (2008) The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat Immunol 9: 857–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J, Hartman M, Roche C, Zeng SG, O′Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC (2011) Autophagy controls IL-1{beta} secretion by targeting pro-IL-1{beta} for degradation. J Biol Chem 286: 9587–9597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B (2010) The Beclin 1 interactome. Curr Opin Cell Biol 22: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E (2008) Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol 9: 847–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N (2011) p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol 192: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7: 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Ruegg A, Werner S, Beer HD (2008) Active caspase-1 is a regulator of unconventional protein secretion. Cell 132: 818–831 [DOI] [PubMed] [Google Scholar]
- Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T (2001) Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460 [DOI] [PubMed] [Google Scholar]
- Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, Malhotra V (2007) The Golgi-associated protein GRASP is required for unconventional protein secretion during development. Cell 130: 524–534 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Levine B (2008) Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol 9: 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M (2011) Emerging inflammasome effector mechanisms. Nat Rev Immunol 11: 213–220 [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Sarkar A, Vande Walle L, Vitari AC, Amer AO, Wewers MD, Tracey KJ, Kanneganti TD, Dixit VM (2010) Inflammasome-dependent release of the alarmin HMGB1 in endotoxemia. J Immunol 185: 4385–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW (2011) Autophagy in immunity and inflammation. Nature 469: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjithaya R, Anjard C, Loomis WF, Subramani S (2010) Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol 188: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440: 237–241 [DOI] [PubMed] [Google Scholar]
- Mazelova J, Ransom N, Astuto-Gribble L, Wilson MC, Deretic D (2009) Syntaxin 3 and SNAP-25 pairing, regulated by omega-3 docosahexaenoic acid, controls the delivery of rhodopsin for the biogenesis of cilia-derived sensory organelles, the rod outer segments. J Cell Sci 122: 2003–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B (2010) Autophagy in mammalian development and differentiation. Nat Cell Biol 12: 823–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y (2004) In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell 15: 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, El Khoury J, Medeiros LA, Terada K, Geula C, Luster AD, Freeman MW (2002) A CD36-initiated signaling cascade mediates inflammatory effects of beta-amyloid. J Biol Chem 277: 47373–47379 [DOI] [PubMed] [Google Scholar]
- Moritz OL, Tam BM, Hurd LL, Peranen J, Deretic D, Papermaster DS (2001) Mutant rab8 Impairs docking and fusion of rhodopsin-bearing post-Golgi membranes and causes cell death of transgenic Xenopus rods. Mol Biol Cell 12: 2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peranen J, Merdes A, Slusarski DC, Scheller RH, Bazan JF, Sheffield VC, Jackson PK (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell 129: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM (2010) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 12: 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W, Rabouille C (2009) Mechanisms of regulated unconventional protein secretion. Nat Rev Mol Cell Biol 10: 148–155 [DOI] [PubMed] [Google Scholar]
- Ponpuak M, Davis AS, Roberts EA, Delgado MA, Dinkins C, Zhao Z, Virgin HW 3rd, Kyei GB, Johansen T, Vergne I, Deretic V (2010) Delivery of cytosolic components by autophagic adaptor protein p62 endows autophagosomes with unique antimicrobial properties. Immunity 32: 329–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renna M, Jimenez-Sanchez M, Sarkar S, Rubinsztein DC (2010) Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem 285: 11061–11067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanello V, Guadagnin E, Gomes L, Roder I, Sandri C, Petersen Y, Milan G, Masiero E, Del Piccolo P, Foretz M, Scorrano L, Rudolf R, Sandri M (2010) Mitochondrial fission and remodelling contributes to muscle atrophy. EMBO J 29: 1774–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubartelli A, Cozzolino F, Talio M, Sitia R (1990) A novel secretory pathway for interleukin-1 beta, a protein lacking a signal sequence. EMBO J 9: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Watson R, Giannakou ME, Clarke M, Warren G, Barr FA (1999) GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J 18: 4949–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Ornatowski W, Vergne I, Naylor J, Delgado M, Roberts E, Ponpuak M, Master S, Pilli M, White E, Komatsu M, Deretic V (2010) Human IRGM regulates autophagy and cell-autonomous immunity functions through mitochondria. Nat Cell Biol 12: 1154–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, Piacentini M, Levine B, Cecconi F (2011) Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J 30: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Bilan PJ, Liu Z, Klip A (2010) Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc Natl Acad Sci USA 107: 19909–19914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ 3rd, Lotze MT (2010) Endogenous HMGB1 regulates autophagy. J Cell Biol 190: 881–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD (2011) The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med 17: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, Brickey WJ, Ting JP (2011) Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol 12: 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham SB, Allen IC, Bergstralh DT, Brickey WJ, Huang MT, Taxman DJ, Duncan JA, Ting JP (2009) NLRP3 (NALP3, Cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J Immunol 183: 2008–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM (2010) Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13: 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Geng J, Yen WL, Wang K, Klionsky DJ (2010) Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell 38: 250–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP (2011) Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW (2008) Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4: 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469: 221–225 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.