Abstract
Centriole duplication involves the growth of a procentriole next to the parental centriole. Mutations in STIL and CPAP/CENPJ cause primary microcephaly (MCPH). Here, we show that human STIL has an asymmetric localization to the daughter centriole and is required for procentriole formation. STIL levels oscillate during the cell cycle. Interestingly, STIL interacts directly with CPAP and forms a complex with hSAS6. A natural mutation of CPAP (E1235V) that causes MCPH in humans leads to significantly lower binding to STIL. Overexpression of STIL induced the formation of multiple procentrioles around the parental centriole. STIL depletion inhibited normal centriole duplication, Plk4-induced centriole amplification, and CPAP-induced centriole elongation, and resulted in a failure to localize hSAS6 and CPAP to the base of the nascent procentriole. Furthermore, hSAS6 depletion hindered STIL targeting to the procentriole, implying that STIL and hSAS6 are mutually dependent for their centriolar localization. Together, our results indicate that the two MCPH-associated proteins STIL and CPAP interact with each other and are required for procentriole formation, implying a central role of centriole biogenesis in MCPH.
Keywords: biogenesis, cell cycle, centrosome duplication, MCPH, microcephaly, procentriole formation
Introduction
The centriole is an essential component of the centrosome, which is required for the formation and maintenance of dynamic arrays of microtubules (MTs), the mitotic spindle, cilia, and flagella (Strnad and Gonczy, 2008; Nigg and Raff, 2009; Azimzadeh and Marshall, 2010). Recent studies reveal an evolutionarily conserved pathway for the formation of centrioles (Delattre et al, 2006; Pelletier et al, 2006; Carvalho-Santos et al, 2010; Hodges et al, 2010) and have identified several key centrosomal proteins (SPD-2, ZYG-1, SAS5/Ana2, SAS6, and SAS4) that are essential for centriole duplication in Caenorhabditis elegans (O’Connell et al, 2001; Kirkham et al, 2003; Leidel and Gonczy, 2003; Kemp et al, 2004; Pelletier et al, 2004; Leidel et al, 2005) and Drosophila melanogaster (Bettencourt-Dias et al, 2005; Peel et al, 2007; Rodrigues-Martins et al, 2007). Human homologues of SPD-2, ZYG-1, SAS4, and SAS6 have been identified as Cep192 (Gomez-Ferreria et al, 2007; Zhu et al, 2008), Plk4/Sak (Bettencourt-Dias et al, 2005; Habedanck et al, 2005), CPAP/CENPJ (Hung et al, 2000), and hSAS6 (Leidel et al, 2005), respectively. Recently, a model for centriole assembly pathway in human cells has been proposed (Kleylein-Sohn et al, 2007). During late G1 and early S phase, a new centriole (procentriole) starts to grow at an orthogonal angle next to each preexisting centriole, a process regulated by the conserved kinase Plk4 (Kleylein-Sohn et al, 2007) and its interacting protein Cep152 (Cizmecioglu et al, 2010; Dzhindzhev et al, 2010; Hatch et al, 2010). Activation of Plk4 induces a cascade including hSAS6, Cep135, CPAP, γ-tubulin, and CP110 that is required for procentriole formation during S and G2 stages (Kleylein-Sohn et al, 2007). Currently, it is not clear whether these proteins are recruited simultaneously or how these proteins cooperate to initiate and promote procentriole assembly.
Primary microcephaly (MCPH) is genetically heterogeneous with eight known loci (MCPH1–8) involved; seven genes have been identified (Thornton and Woods, 2009). Interestingly, all seven known MCPH proteins (MCPH1: Microcephalin; MCPH2: WDR62; MCPH3: CDK5RAP2; MCPH4: Cep152; MCPH5: ASPM; MCPH6: CPAP/CENPJ; MCPH7: STIL/SIL) are ubiquitously expressed and localized to centrosomes for at least part of the cell cycle, implying a central role of the centrosome in this disease (Thornton and Woods, 2009). STIL/SIL was originally identified at the site of a genomic rearrangement in a T-cell acute lymphoblastic leukaemia (Aplan et al, 1991) and has been implicated in regulating centrosome integrity and mitotic spindle organization (Pfaff et al, 2007; Castiel et al, 2011). Mutations in STIL/SIL (MCPH7) cause primary MCPH in humans (Kumar et al, 2009), the left–right asymmetry defects in mouse (Izraeli et al, 1999), and disorganized mitotic spindles in zebrafish (Pfaff et al, 2007). Recently, Drosophila Ana2 (Stevens et al, 2010a) and Caenorhabditis SAS5 (Delattre et al, 2006; Pelletier et al, 2006) were reported to be required for centriole duplication. Human STIL has been proposed to be a likely orthologue of Ana2/SAS5 (Stevens et al, 2010a). However, these three proteins lack obvious sequence homology and share only a short coiled-coil region (∼22 aa) and a conserved STAN motif (∼90 aa) in their C-terminal region (Stevens et al, 2010a). Thus, how human STIL ensures centriole duplication is not known. Here, we provide experimental evidence that human STIL is indeed the functional orthologue of SAS-5/Ana2 and is required for procentriole formation in human cells. Unexpectedly, we found that human STIL (MCPH7) directly interacts with another MCPH protein CPAP (MCPH6), and a natural mutation of CPAP (E1235V) significantly decreased its binding to STIL, implying that interference in centriole biogenesis, particularly at the stage of procentriole formation, may cause MCPH.
Results
STIL is a cell cycle-regulated protein that controls centriole duplication
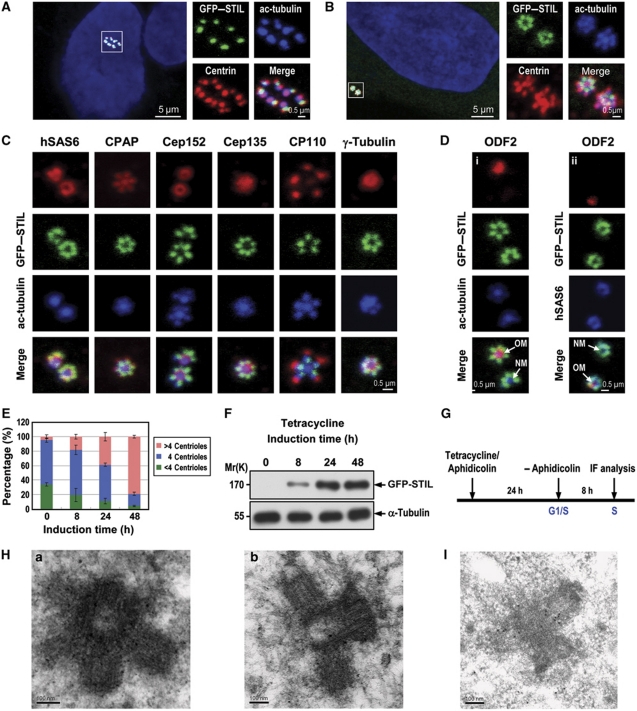
To study the role of STIL in centriole duplication, we generated U2OS-derived stable cell lines that expressed GFP–STIL under tetracycline control. Interestingly, 48 h after STIL induction, ∼80% of asynchronously proliferating cells showed centriole amplification (>4 centrioles; Figure 1A and E). Centrin and acetylated tubulin (ac-tubulin) were used as markers to monitor centriole assembly (Figure 1A and B). A flower-like structure with multiple ‘procentriole-like’ particles surrounding a centriole (Figure 1B), similar to that in Plk4-induced centriole amplification (Kleylein-Sohn et al, 2007), was frequently observed in synchronized GFP–STIL-induced cells (∼8 h after aphidicolin release; S phase). High-resolution immunofluorescence confocal microscopy (Figure 1C) showed a normal incorporation of several centriolar proteins including hSAS6, CPAP, Cep152, Cep135, CP110, and γ-tubulin into these flower-like structures. Further electron microscopic analysis revealed a strong similarity to typical centriolar structures (Figure 1H-a and H-b). We also stained the flower-like structures with an antibody against ODF-2, a component of mature mother centriolar appendages (Ishikawa et al, 2005). Figure 1D shows that only the mature mother (old mother) centriole was stained by the ODF-2 antibody, whereas the surrounding newly assembled procentrioles or the new mother centriole lacked such staining. The centriole amplification effect was also observed in cells transfected with GFP–STIL or Flag–STIL (to eliminate GFP steric effect) in HEK 293T (Supplementary Figure S2A and B) or HeLa lines (Supplementary Figure S2C and D), and with low efficiency in primary bovine aortic endothelial cells (BAECs, <1%, Supplementary Figure S2E).
Figure 1.
Excess STIL induces centriole amplification. Unsynchronized (A) or synchronized (B–D) U2OS-based GFP–STIL-inducible cells treated as shown in (G) were analysed by confocal fluorescence microscopy using indicated antibodies. Procentriole formation was visualized by anti-centrin and anti-ac-tubulin staining (A, B). GFP–STIL was directly visualized by confocal fluorescent microscopy. DNA was counterstained with DAPI. (C, D) Normal incorporation of several centrosomal proteins into GFP–STIL-induced centrioles. (E) The percentage of amplified centrioles induced by excess GFP–STIL at different time points. Error bars represent mean±s.d. of 200 cells from three independent experiments. (F) Immunoblot analysis of the expression levels of exogenously expressed GFP–STIL at indicated times after tetracycline induction. α-Tubulin was used as a loading control. (G) A protocol to analyse GFP–STIL-induced centriole amplification. (H, I) GFP–STIL-inducible cells were treated as described in (G). Four hours after aphidicolin release, the cells were processed for electron microscopy (H-a, transverse section; H-b, longitudinal section) and immunogold EM (I). OM, old mother centriole; NM, new mother centriole.
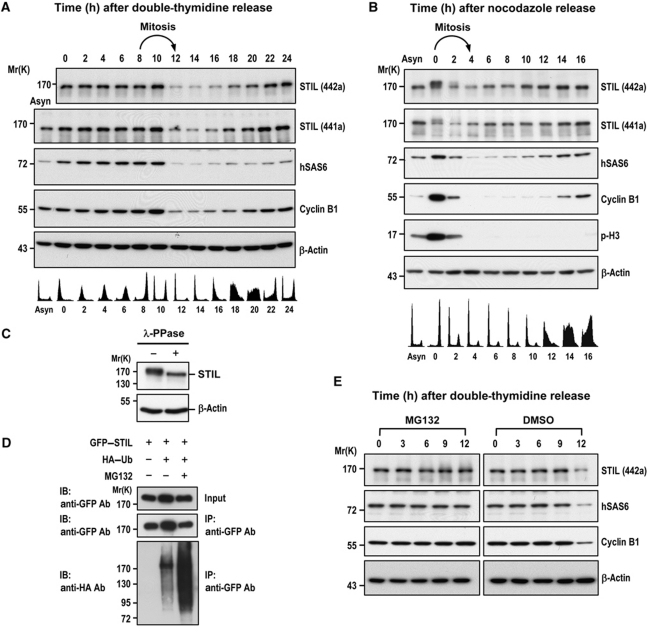
The observation that excess STIL-induced centriole amplification implies that the protein levels of STIL must be carefully regulated to ensure formation of a single procentriole per centriole. We then performed a comparative analysis of the protein expression pattern of STIL and hSAS6 (a known cell cycle-regulated centriole protein; Strnad et al, 2007) in synchronized cells (Figure 2). HeLa cells were released from either a double-thymidine block (Figure 2A) or nocodazole treatment (Figure 2B) that arrested cells at G1/S or G2/M phase, respectively, and analysed by immunoblotting and fluorescence-activated cell sorting (FACS). Two affinity-purified anti-STIL antibodies, #441a and #442a (Bethyl Laboratories Inc.), that specifically recognized the middle portion (residues 450–500) or the C-terminal region (residues 1237–1287) of human STIL/SIL, respectively, were used in this study. Interestingly, both STIL and hSAS6 levels decreased significantly when cells exited mitosis (Figure 2A, ∼12 h after double-thymidine release; Figure 2B, ∼2 h after nocodazole release) and started to accumulate in late G1 and early S phase (Figure 2A, ∼16–18 h after double-thymidine release; Figure 2B, ∼10–12 h after nocodazole release). The high molecular weight bands detected in nocodazole-treated cells (0 h, Figure 2B) may possibly represent hyperphosphorylated STIL proteins, which were lost after λ phosphatase treatment (Figure 2C). Consistent with our findings, Izraeli et al (1997) and Campaner et al (2005) also reported a similar cell-cycle expression pattern of STIL/SIL. Furthermore, we found that GFP–STIL was ubiquitinated in vivo (Figure 2D) and its degradation was significantly inhibited in the presence of the proteasome inhibitor MG132 (∼12 h after double-thymidine release; Figure 2E), implying that STIL degradation may be mediated by the 26S proteasome. Together, our results indicate that STIL is a cell cycle-regulated protein that plays an essential role in controlling centriole duplication.
Figure 2.
STIL is a cell cycle-regulated protein and ubiquitinated in vivo. (A, B) HeLa cells released from a double-thymidine block (A) or nocodazole treatment (B) were analysed at the indicated time points by immunoblotting using indicated antibodies (top) and by FACS (bottom). Two affinity purified antibodies, anti-STIL-441a and anti-STIL-442a, that recognized the middle portion and the C-terminal region of human STIL/SIL, respectively, were used in this study. Asyn, asynchronized. (C) Mitotic HeLa cell extracts, prepared from nocodazole treatment, were incubated with λ phosphatase (λ-PPase) and analysed by immunoblotting using indicated antibodies. (D) HEK 293T cells were transfected with GFP–STIL and HA–ubiquitin constructs. Twenty-two hours after transfection, the cells were treated with MG132 for another 4 h. The cell lysates were immunoprecipitated (IP) with anti-GFP antibody and then immunoblotted (IB) with anti-HA or anti-GFP antibodies. (E) STIL degradation is inhibited by MG132. HeLa cells were arrested at G1/S by double-thymidine treatment and then released into fresh medium with MG132 (2 μM) at indicated times. The cell lysates were analysed by immunoblotting using indicated antibodies. β-Actin was used a loading control.
STIL asymmetrically localizes to the base of procentrioles
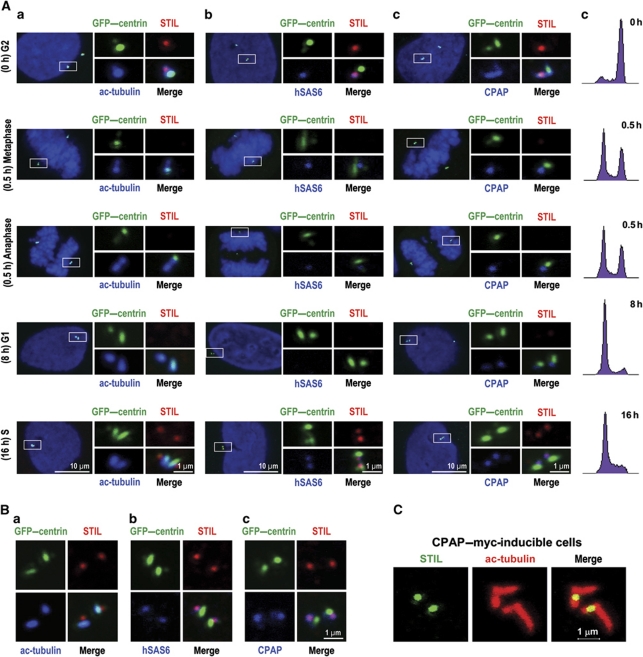
We next examined the pattern of colocalization of endogenous STIL with several known centriolar proteins, including acetylated tubulin (a marker for stabilized centriolar MTs), hSAS6 (a daughter centriole marker localized asymmetrically to the procentriole that disappears in G1 phase; Strnad et al, 2007), and CPAP (located at both mother and daughter centrioles throughout the cell cycle; Tang et al, 2009) in nocodazole-synchronized GFP–centrin expressing cells using confocal immunofluorescence microscopy and FACS (Figure 3). Strikingly, no or very weak STIL signals were detected in mitotic cells (including metaphase and anaphase, 0.5 h after nocodazole release; Figure 3A) and G1-phase cells (8 h after nocodazole release; Figure 3A).
Figure 3.
(A) The subcellular localization of STIL during the cell cycle. U2OS cells stably expressing GFP–centrin were synchronized at G2/M stage by nocodazole treatment for 12 h. (a–d) The mitotic shake-off cells released from nocodazole treatment were replated and cultured on coverslips in the absence of nocodazole and analysed at the indicated times by immunofluorescence confocal microscopy (a–c) or by FACS (d) to monitor their cell-cycle status. The pattern of colocalization of endogenous STIL with ac-tubulin (a), hSAS6 (b), and CPAP (c) in GFP–centrin-labelled centrioles was analysed using indicated specific antibodies. GFP–centrin was directly visualized by confocal fluorescent microscopy. DNA was counterstained with DAPI. (B) The colocalization pattern of STIL with ac-tubulin (a), hSAS6 (b), and CPAP (c) in aphidicolin-synchronized GFP–centrin expressing cells. Twenty-four hours after aphidicolin treatment, cells were analysed by immunofluorescence confocal microscopy using indicated antibodies. (C) CPAP–myc was induced in U2OS-based CPAP–myc-inducible cells in the presence of tetracycline for 2 days and analysed by confocal fluorescence microscopy using indicated antibodies.
When cells exited mitosis, STIL was first detected at the G1/S transition in aphidicolin-synchronized cells, using hSAS6 (a known centriole protein first detected at late G1/early S phase; Kleylein-Sohn et al, 2007; Strnad et al, 2007) as a positive control (Figure 3B-b). Interestingly, STIL colocalized with hSAS6 during S and G2 phases (Figure 3A) and appeared to localize asymmetrically to the proximal end of GFP–centrin/CPAP co-staining centrioles (G2 phase in Figure 3A-c). This asymmetric localization of STIL was clearly visualized in CPAP–myc-inducible cells. In such cells, STIL was frequently detected at the base of elongated centrioles (Figure 3C) that had been induced by CPAP–myc overexpression (Tang et al, 2009). Further immunogold-EM analysis revealed that GFP–STIL signals were clustered around the base of the amplified procentrioles in GFP–STIL-inducible cells (Figure 1I). Together, our results indicate that STIL is recruited to the proximal end of procentrioles at the onset of procentriole formation during late G1 and early S phases.
STIL directly interacts with CPAP and forms a complex with hSAS6
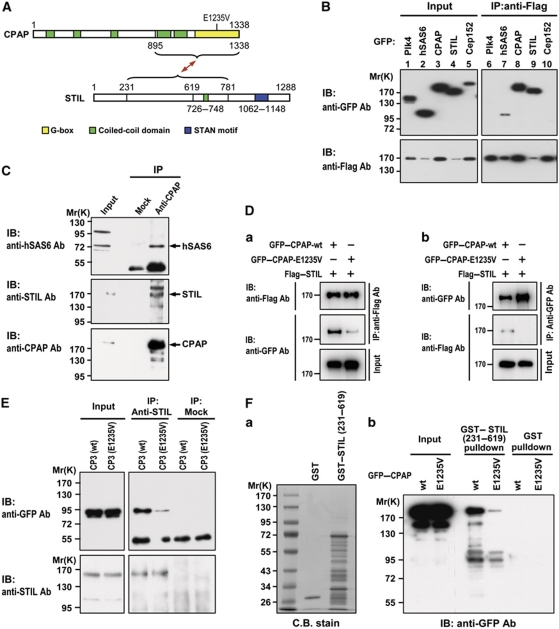
Two putative STIL orthologues, SAS5 and Ana2, were reported to interact with Caenorhabditis SAS6 (Leidel et al, 2005; Boxem et al, 2008) and Drosophila dSas6 (Stevens et al, 2010a), respectively. We next examined whether STIL interacts with human hSAS6 and several other centrosomal proteins including Plk4, CPAP, and Cep152 that are required for procentriole formation. A Flag-tagged STIL was co-transfected with various GFP-tagged Plk4, hSAS6, CPAP, STIL, and Cep152 constructs into HEK 293T cells. Expression of these exogenous proteins was seen in the transfected cell lysates (Figure 4B, lanes 1–5). Flag–STIL was immunoprecipitated (IP) from the transfected cell lysates using anti-Flag antibody and then immunoblotted (IB) with either anti-GFP or anti-Flag antibody. STIL co-precipitated with hSAS6 (Figure 4B, lane 7) and STIL itself (Figure 4B, lane 9), implying that STIL, like SAS5 (Leidel et al, 2005) and Ana2 (Stevens et al, 2010b), may possibly interact with SAS6 (directly or indirectly) and form a homodimer with itself. Surprisingly, we detected no direct interaction between STIL and hSAS6 using either hSAS6 fragments or STIL fragments as baits in yeast two-hybrid assays (Supplementary Figure S3). Similar negative results were also observed in a GST pulldown assay (Supplementary Figure S4A-b), in which no 35S-methionine-labelled full-length STIL or hSAS6 was pulled down by GST–hSAS6 (full length), GST–STIL (residues 1–628), or GST–STIL (residues 620–1288). In contrast, GST–Ana2 (a putative orthologue of human STIL) did bring down 35S-methionine-labelled Drosophila dSAS6 (Supplementary Figure S4B-b, lane 7), consistent with a previous report (Stevens et al, 2010a). Thus, unlike Drosophila Ana2 and dSAS6, human STIL does not directly interact with hSAS6.
Figure 4.
STIL interacts with CPAP and forms a complex with hSAS6. (A) Schematic of interactions between STIL and CPAP. E1235V: a natural mutation of CPAP/CENPJ (E1235V) with glutamate replaced by valine at amino acid 1235 (Bond et al, 2005). (B) STIL interacts with CPAP in transfected cells. HEK 293T cells were co-transfected with Flag–STIL and each GFP-tagged cDNA construct (Plk4, hSAS6, CPAP, STIL, and Cep152). Twenty-four hours after transfection, the cell lysates were first immunoprecipitated (IP) with anti-Flag antibody and then immunoblotted (IB) using anti-GFP and anti-Flag antibodies. (C) Endogenous STIL, CPAP, and hSAS6 form a complex in vivo. Western blot analysis of immunoprecipitated complexes containing endogenous CPAP, STIL, and hSAS6 prepared from HEK 293T cell lysates using anti-CPAP antibody and a non-relevant normal IgG (Mock). (D) The natural E1235V mutation of CPAP reduced its binding to STIL. HEK 293T cells were co-transfected with full-length GFP–CPAP-wt or GFP–CPAP-E1235V mutant with Flag–STIL. Twenty-four hours after transfection, the cell lysates were immunoprecipitated with anti-Flag or anti-GFP antibodies and analysed by immunoblotting using indicated antibodies. (E) Western blot analysis of immunoprecipitated complexes from HEK 293T cells transfected with GFP–CPAP-CP3 (wt, 895–1338) or GFP–CPAP-CP3 (E1235V) mutant. The cell lysates were immunoprecipitated with STIL antibody or a non-relevant normal IgG (Mock) and analysed by western blot using indicated antibodies. (F) GST pulldown assay. GST and GST–STIL (231–619) recombinant proteins were affinity purified and stained with Coomassie blue (C.B. stain, a). Equal amounts of GST and GST–STIL truncated proteins were used to pull down the cell lysates prepared from cells transfected with full-length GFP–CPAP-wt or GFP–CPAP-E1235V mutant constructs. The pulldowns were analysed by western blot using anti-GFP antibody (b).
Unexpectedly, we found that Flag–STIL co-precipitated with CPAP (Figure 4B, lane 8), a protein that regulates centriolar MT assembly (Tang et al, 2009), and associates with neither Plk4 (Figure 4B, lane 6) nor Cep152 (Figure 4B, lane 10). To examine and map the interaction domain between STIL and CPAP, we next performed GST pulldown and yeast two-hybrid assays. Only GST–CPAP-CP3 (residues 895–1338) specifically pulled GFP–STIL down from GFP–STIL transfected cell lysates (Supplementary Figure S5B, lane 14) and interacted directly with 35S-methionine-labelled full-length STIL (Supplementary Figure S6A, lane 6). Consistent with this finding in humans, we also demonstrated that GST–Ana2 can directly interact with 35S-methionine-labelled Drosophila dSAS4, an orthologue of human CPAP (Supplementary Figure S4B-b, lane 5).
The STIL domain that interacts with CPAP-CP3 was then mapped more closely to the central region of STIL (residues 420–781; Supplementary Figure S5C, lane 10). The yeast two-hybrid assay further demonstrated that STIL (residues 231–619) strongly interacts with CPAP-CP3 (residues 895–1338; Supplementary Figure S6B). A schematic diagram of interactions between STIL and CPAP is shown in Figure 4A. We next examined whether STIL, hSAS6, and CPAP are able to form a protein complex in vivo. As shown in Figure 4C, our co-IP experiment using anti-CPAP antibody did detect a protein complex containing endogenous hSAS6, STIL, and CPAP. Further co-IP analyses demonstrated that exogenously expressed GFP–STIL was indeed associated with endogenous CPAP and hSAS6 in transfected cells (Supplementary Figure S7A–C). Together, our results indicate that STIL directly interacts with CPAP and may form a complex with hSAS6 in vivo.
A natural CPAP mutant (E1235V) reduces its binding to STIL
Recently, Bond et al (2005) reported a homozygous missense mutation (E1235V) in the CPAP/CENPJ gene that causes MCPH in humans. Interestingly, this mutation is located at the CPAP-CP3 (895–1338) fragment that binds directly to STIL (Figure 4A). To examine whether this mutation would affect STIL binding, we co-transfected full-length Flag–STIL and GFP–CPAP (both the wild type and the E1235V mutant) into HEK 293T cells and analysed their interaction by co-immunoprecipitation assays. The full-length GFP–CPAP-E1235V had significantly lower binding to STIL, compared with its corresponding wild-type control, using either anti-Flag (Figure 4D-a) or anti-GFP antibody (Figure 4D-b). Similar effects using truncated GFP–CPAP-CP3 (wild type and CP3-E1235V) were also independently demonstrated by co-IP (Figure 4E) and yeast two-hybrid assays (Supplementary Figure S8). This finding is further verified in GST pulldown assay, in which GST–STIL (231–619) interacts only weakly with full-length GFP–CPAP-E1235V (Figure 4F-b). Thus, interference with CPAP binding to STIL could be one of the precipitating causes of MCPH in humans.
STIL is required for the onset of procentriole formation and proper mitotic progression
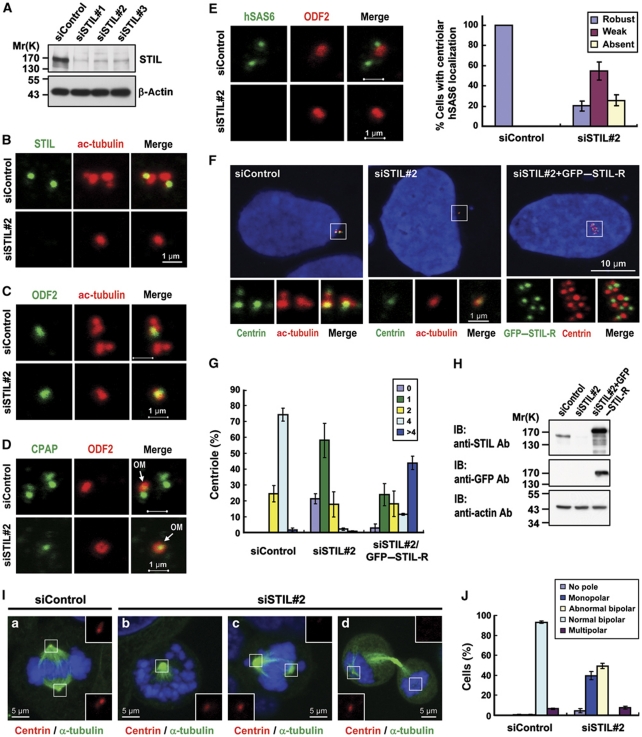
The asymmetric localization of STIL at the base of the procentriole led us to hypothesize that STIL is required for centriole duplication. To test this hypothesis, we depleted STIL protein by specific siRNAs (siSTIL-1, siSTIL-2, and siSTIL-3) in U2OS cells. All three siRNAs showed specific inhibition of STIL expression, as demonstrated by western blotting (Figure 5A) and immunofluorescence staining (Figure 5B) using either anti-STIL-442a (Figure 5; Supplementary Figure S9C) or anti-STIL-441a (Supplementary Figure S9A and B) antibodies. Centriole number was determined by centrin labelling in interphase cells (Figure 5F) and in mitotic cells (Figure 5I). We then selected siSTIL#2 (hereafter referred to as siSTIL) for all other experiments described in this report. As shown in Figure 5F, depletion of STIL significantly suppressed normal centriole duplication; such effects were effectively rescued by expression of a siRNA-resistant GFP–STIL-R construct. Most STIL-depleted cells contained either no centriole (∼21%) or a single centriole (∼58%), while multiple centrioles (>4) were frequently detected in the rescue experiments (∼43%; Figure 5G). Furthermore, STIL depletion also induced mitotic abnormalities including mainly monopolar (40%) and abnormal bipolar spindles (49%) (Figure 5J). At 72 h after STIL depletion, most abnormal bipolar spindles showed only a single centriole at one pole (Figure 5I-c) and monopolar spindles frequently exhibited a single centriole at the centre (Figure 5I-b). In addition, we also observed a single centriole in one of the two dividing cells (Figure 5I-d).
Figure 5.
STIL depletion inhibits centriole duplication and siRNA rescue experiments. Because STIL is absent in G1 cells, we synchronized cells at G1/S by aphidicolin to ensure the detection of STIL after siSTIL treatment. U2OS cells were transfected with control siRNA (siControl) or STIL siRNAs (siSTIL-1, siSTIL-2, and siSTIL-3). Two days after transfection, the cells were treated with aphidicolin for another day and analysed by immunoblotting (A) or by immunofluorescence confocal microscopy using indicated antibodies (B–E). Depletion of STIL significantly inhibits hSAS6 targeting to the procentriole (E). A quantification of the centriolar signals of hSAS6 in siControl and siSTIL-treated cells is shown in (E) (right panel). Values are means of three experiments with s.d. bars (n=100 cells). In siRNA rescue experiments (F–H), U2OS cells were transfected with control siRNA (siControl) or siSTIL#2 for 16 h followed by siRNA-resistant GFP–STIL-R transfection for another 2 days. The cells were then processed for immunofluorescence confocal microscopy (F) or for immunoblotting (H). GFP–STIL-R was directly visualized by confocal fluorescent microscopy. To observe the mitotic effects generated by STIL depletion, U2OS cells were transfected with siControl (a) or siSTIL#2 (b–d). Three days after transfection, cells were analysed by immunofluorescence confocal microscopy using indicated antibodies (I). Histogram illustrating as percentages the numbers of centrioles (G, counted by centrin staining) and abnormal mitotic spindles (J) in siSTIL#2 treated cells. Error bars represent mean±s.d. of 100 cells from three independent experiments. OM, old mother centriole.
We then traced the fate of STIL-knockdown cells. The G2/M (4N) population in siSTIL-depleted cells was significantly increased (∼2 times higher than siControl) at days 2– 3, as evidenced by FACS analysis (Supplementary Figure S10A) and by phospho-histone H3 staining (labelled mitotic cells) (Supplementary Figure S10B). On day 2 and thereafter, nearly 80% of siSTIL-treated cells contained either one or no centriole (Supplementary Figure S10C). Cell proliferation in STIL-knockdown cells was significantly inhibited, and the cell number remained low during siSTIL treatment (Supplementary Figure S10D).
We also analysed siSTIL-treated U2OS-based mCherry–α-tubulin expressing cells by live imaging microscopy, and found that mitosis took thrice as long in siSTIL than in siControl cells (siSTIL, 243±47 min versus siControl 80±20 min) (Supplementary Figure S11). Interestingly, most mitotic cells with monopolar spindles (62.5%, 25/40 tracing cells) still proceeded to assemble a ‘bipolar’ spindle and subsequently divided to produce two separate cells at days 2–3 (Supplementary Figure S11B-a; Supplementary Movie S1). Some mitotic cells (27.5%, 11/40 tracing cells) attempted to divide but failed and resulted in large polyploid cells over a longer period (Supplementary Figure S11B-b; Supplementary Movie S2). A few cells (10%, 4/40 tracing cells) can even divide into three cells (Supplementary Figure S11B-c; Supplementary Movie S3). Thus, STIL appears to be essential for mitotic progression possibly by regulating faithful centriole duplication in mammalian cells.
Intriguingly, depletion of STIL also perturbed the recruitment of hSAS6 and CPAP to the procentriole, mainly hindering the formation of the procentriole (daughter centriole) rather than the parent centriole. Careful immunofluorescence analysis revealed that most STIL-depleted cells displayed no hSAS6 (Figure 5E) or CPAP (Figure 5D) signal on the procentriole, possibly due to complete inhibition of procentriole formation by STIL depletion. The single centriole that remained in siSTIL-treated cells (∼58%) appeared to be derived from the mature mother centriole, and stained positive for ODF-2 (a mature mother centriole marker; Figure 5C) and CPAP (present in both mother and daughter centriole; Figure 5D), but not for hSAS6 (a daughter centriole marker; Figure 5E).
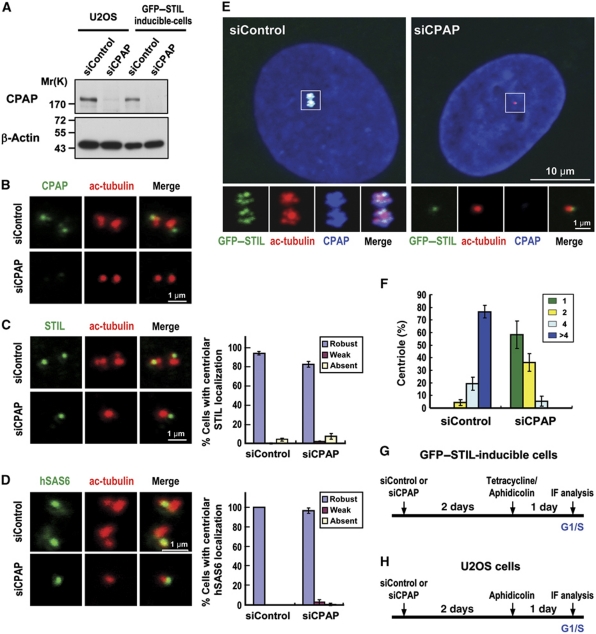
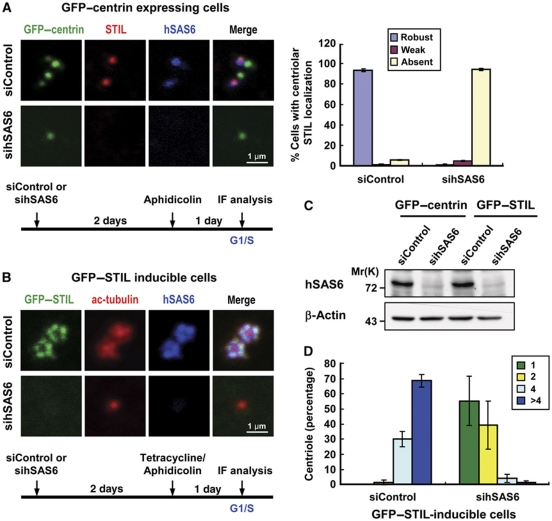
We next examined whether depletion of CPAP or hSAS6 hinders the localization of STIL to the centriole or vice versa. As shown in Figure 6, depletion of CPAP (Figure 6B) in U2OS cells had either no or only minor effects on the targeting of endogenous STIL (Figure 6C) and hSAS6 (Figure 6D) to the procentriole. Intriguingly, CPAP depletion did inhibit GFP–STIL-induced centriole amplification (Figure 6E and F), possibly since the loss of CPAP activity blocked the procentriole MT assembly at a later step. In sharp contrast, hSAS6 depletion not only inhibited GFP–STIL-induced centriole amplification (Figure 7D), but also perturbed both endogenous STIL (Figure 7A) and GFP–STIL (Figure 7B) targeting to the procentriole, implying that STIL and hSAS6 are mutually dependent for their centriolar localization. Thus, during procentriole formation it appears that STIL is required for the correct loading of hSAS6 and CPAP to the base of the procentriole to initiate procentriole assembly.
Figure 6.
CPAP depletion inhibits GFP–STIL-induced centriole amplification, but appears not to affect endogenous STIL or hSAS6 targeting to the procentriole. U2OS (B–D) or GFP–STIL-inducible cells (E, F) were transfected with control siRNA (siControl) or siCPAP and synchronized at G1/S by aphidicolin treatment as depicted in (G) (GFP–STIL-inducible cells) and (H) (U2OS cells). After transfection, cells were analysed by immunoblotting (A) or by immunofluorescence confocal microscopy using indicated antibodies. β-Actin was used as a loading control. A quantification of centriolar signals of STIL (C, right panel) and hSAS6 (D, right panel) in siControl and siCPAP-treated cells. Values are means of three experiments with s.d. bars (n=100 cells). (F) Histogram illustrating as percentages the centriole numbers in siCPAP-treated GFP–STIL-inducible cells. Error bars represent mean±s.d. of 100 cells from three independent experiments. (G, H) A protocol to analyse siCPAP-treated effects in GFP–STIL-inducible cells (G) or U2OS cells (H).
Figure 7.
Depletion of hSAS6 blocks not only GFP–STIL-induced centriole amplification, but also the recruitment of STIL to the procentriole. U2OS-based GFP–centrin expressing cells (A) or GFP–STIL-inducible cells (B) were transfected with control siRNA (siControl) or siRNAs against hSAS6. Forty-eight hours after transfection, cells were synchronized by aphidicolin treatment with or without tetracycline for another day, and analysed by immunofluorescence confocal microscopy using indicated antibodies (A, B) or by immunoblotting (C). Depletion of hSAS6 perturbs the localization of endogenous STIL to the procentriole. Quantification of centriolar signals of STIL (A, right panel) in siControl and sihSAS6-treated cells. Values are means of three experiments with s.d. bars (n=100 cells). (D) Histogram illustrating as percentages the centriole numbers in sihSAS6-treated GFP–STIL-inducible cells. Error bars represent mean±s.d. of 100 cells from three independent experiments.
Delineation of a STIL-mediated centriole duplication pathway
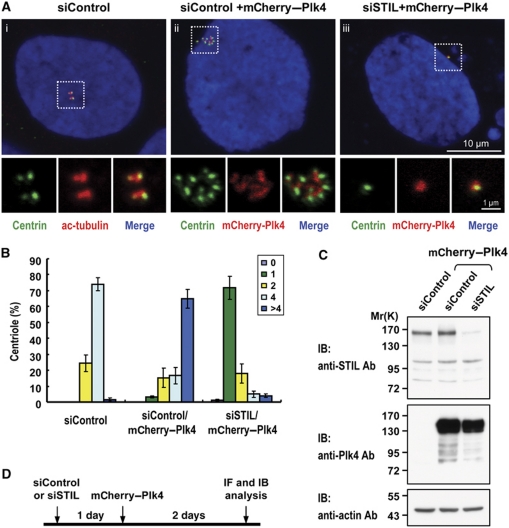
Recently, a composite model for the Plk4-mediated centriole duplication pathway including hSAS6, CPAP (Kleylein-Sohn et al, 2007), and Cep152 (Cizmecioglu et al, 2010; Dzhindzhev et al, 2010; Hatch et al, 2010) in human cells has been proposed. We were interested to see whether STIL is involved in this pathway. Our tests showed that depletion of STIL significantly inhibited Plk4-induced centriole amplification (Figure 8A-iii and 8B). However, STIL depletion did not appear to affect the localization of endogenous Plk4 (Supplementary Figure S12A) and Cep152 (Supplementary Figure S12B) to the centriole, implying that STIL may be a downstream effector in the Plk4-mediated pathway. In addition, we also examined whether depletion of Plk4 affected GFP–STIL-induced centriole amplification and its centriolar localization. Our results showed that depletion of Plk4 not only disrupted STIL targeting to the centriole (Supplementary Figure S13A), but also blocked GFP–STIL-induced centriole amplification (Supplementary Figure S13B), implying that Plk4 is required for STIL-mediated centriole amplification. Furthermore, overexpression of GFP–STIL appears not to affect Plk4 to phosphorylate itself, suggesting that STIL does not influence the kinase activity of Plk4 (Supplementary Figure S14).
Figure 8.
STIL depletion inhibits Plk4-induced centriole amplification. U2OS cells were transfected with siControl or siSTIL for 1 day, followed by mCherry–Plk4 transfection for another 2 days. The cells were then processed for immunofluorescence microscopy (A) and immunoblotting (C). (B) Histogram illustrating the percentages of cells with different centriole numbers (n=100 in triplicate) in siControl or in siSTIL/mCherry–Plk4 transfected cells. (D) A schematic figure showing the timing and the procedures of siRNA-treated cells.
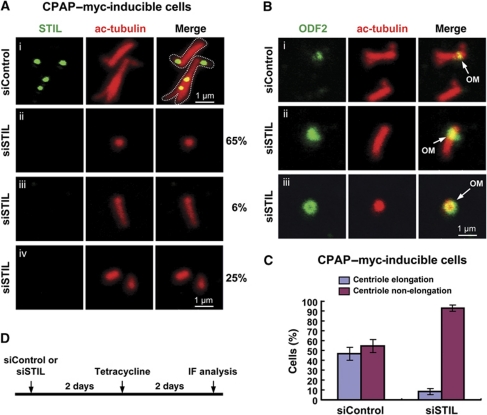
Finally, we observed that depletion of STIL also inhibited CPAP-induced centriole elongation (∼8 versus 46% in control; Figure 9C). Careful immunofluorescence analysis revealed that most STIL-depleted cells in CPAP-inducible clones contained a single mature parent centriole (∼65%, Figure 9A-ii), which showed ODF-2 staining (Figure 9B-iii), whereas the remainder of the cells contained either one longer centriole (∼6%, Figure 9A-iii) or two short centrioles (∼25%, Figure 9A-iv). Thus, STIL depletion mostly affects CPAP-induced centriole elongation from newly assembled procentrioles. Taking all of the data together, our results indicate that STIL is a key player in the early event of procentriole formation in the Plk4-mediated centriole duplication pathway.
Figure 9.
STIL depletion inhibits CPAP-induced procentriole assembly. The CPAP–myc-inducible cells treated as shown in (D) were analysed by confocal fluorescence microscopy using indicated antibodies (A, B). The percentages of CPAP-induced elongated or non-elongated procentriole after siSTIL treatment are shown in (C). Data are shown as mean±s.d. of 200 cells from three independent experiments. OM, old mother centriole.
Discussion
The molecular mechanism of how a single procentriole grows from the base of the preexisting centriole is starting to be revealed. In human cells, the first proteins to be recruited to the site of procentriole formation include Plk4, Cep152, hSAS6, CPAP, Cep135, and γ-tubulin (Kleylein-Sohn et al, 2007; Strnad and Gonczy, 2008; Cizmecioglu et al, 2010; Dzhindzhev et al, 2010; Hatch et al, 2010). Here, we add STIL protein to that list, as one of the first key participants in the early event of procentriole formation in human cells. Based on our current findings and information from other reports (Kleylein-Sohn et al, 2007; Cizmecioglu et al, 2010; Dzhindzhev et al, 2010; Hatch et al, 2010), we propose the possible role of STIL during procentriole formation (Supplementary Figure S1). Following activation of Plk4 and recruitment of Cep152 to the surface of the parent centriole cylinder (Kleylein-Sohn et al, 2007; Cizmecioglu et al, 2010; Dzhindzhev et al, 2010; Hatch et al, 2010), both STIL and hSAS6 are recruited to a primitive site located at the base of the nascent procentriole during late G1 and early S phases. This STIL/hSAS6-containing site may possibly serve as a scaffold for recruitment of CPAP to the base of the procentriole on which CPAP assembles nine triplet MTs during procentriole formation (Kohlmaier et al, 2009; Schmidt et al, 2009; Tang et al, 2009). The observations that depletion of STIL inhibits Plk4-induced centriole amplification (Figure 8), STIL directly interacts with CPAP and forms a complex with hSAS6 (Figure 4), and depletion of STIL (Figure 9) and hSAS6 (Tang et al, 2009) inhibits CPAP-induced procentriole elongation are consistent with this hypothesis. Although Cep152 was also reported to be required for the centrosomal loading of CPAP during procentriole formation in human cells (Cizmecioglu et al, 2010; Dzhindzhev et al, 2010), the relationship between CPAP recruitment by Cep152 and STIL is not clear.
Furthermore, we observed that overexpression of STIL-induced centriole amplification (Figure 1) and that endogenous STIL levels were downregulated during and after mitosis and began to accumulate again at late G1/early S phase (Figure 2). These observations support the notion that the presence of an appropriate amount of STIL, particularly at the G1/S transition, is critical to restrict a single procentriole formation to each parent centriole per cell cycle. Recently, Drosophila Ana2 and dSAS6 were reported to coassemble into the cartwheel-like SAS tubules at the centriole proximal ends that promote centriole duplication (Stevens et al, 2010b). Here, we show that human STIL is only associated indirectly with hSAS6, rather than directly bound. Whether human STIL, like Ana2, coassembles with hSAS6 into this ‘cartwheel’ structure, is an interesting issue yet to be tested.
Finally, mutations in seven known MCPH genes cause primary MCPH in humans (Thornton and Woods, 2009). It is difficult to rationalize why mutations in these genes produce such a specific defect in the human brain. One possibility is that human neuronal progenitors, which divide asymmetrically during normal brain development, are sensitive to centrosome defects (Thornton and Woods, 2009). Recently, asymmetric centrosome inheritance has been observed in mouse neocortex to regulate the differential behaviours of renewing progenitors and their differentiating progeny (Wang et al, 2009). We (this report) and others (Cizmecioglu et al, 2010; Dzhindzhev et al, 2010) have reported that CPAP/CENPJ (MCPH6) interacts not only with STIL/MCPH7, but also with Cep152/MCPH4 (Guernsey et al, 2010), which physically links all three MCPH proteins into a common pathway in the early phase of procentriole assembly. These findings support an intriguing notion that interference in the process of the early asymmetric centrosome inheritance in human embryonic neural progenitors could be one of the major causes of MCPH.
Materials and methods
Plasmids and antibodies
cDNA clones for mCherry–Plk4, CPAP–myc, and GFP–CPAP were described previously (Tang et al, 2009). Plasmid encoding hSAS6 was kindly provided by P Gonczy (Swiss Institute for Experimental Cancer Research). The cDNAs encoding full-length STIL, Cep152, and Cep135 were obtained by RT–PCR using total RNAs from human HEK 293T cells and subcloned in-frame into a pEGFP-C1 (BD Biosciences Clontech) or a pFlag-CMV2 (Sigma-Aldrich) vector. The cDNAs encoding full-length Ana2, dSAS4, and dSAS6 were obtained by RT–PCR using total RNAs from Drosophila larvae and subcloned in-frame into a pcDNA3.0 vector (Invitrogen). For construction of the siRNA-resistant STIL plasmid, the nucleotides targeted by siRNA within the wild-type STIL were partially replaced without changing the amino-acid sequence using a QuickChange kit (Stratagene). The CPAP-CP3 (E1235V) mutant was generated by site-directed mutagenesis using the QuickChange kit as described above. All the resultant plasmids had their sequence confirmed. For generation of GST-fusion constructs, the cDNAs encoding various portions of CPAP, STIL, and Ana2 were fused in-frame to GST in pGEX-2T or pGEX-4T vector as described previously (Tang et al, 2009).
Antibodies against CPAP, SAS-6, centrin 2, and CP110 have been described previously (Tang et al, 2009). Other commercially available antibodies used in this study included anti-STIL-441a, anti-STIL-442a, anti-Plk4, and anti-Cep152 (all from Bethyl Laboratories Inc.); anti-hSAS6 mAb (Abnova); anti-ODF2 (Abcam); anti-β-actin, anti-α-tubulin (DM1a), anti-γ-tubulin (GTU88), anti-Flag (M2), and anti-acetylated tubulin (all from Sigma-Aldrich); anti-cyclin B1 (Santa Cruz Biotechnology); anti-HA (Covance Research Products, Inc.); anti-Myc (4A6, Upstate Biotechnology or NB600-336, Novus Biologicals, Inc); anti-His (Serotec); and anti-GFP (BD Bioscience).
Cell culture, synchronization, immunoblotting, and FACS analysis
Primary BAECs were kindly provided by D-L Wang (Institute of Biomedical Sciences, Academia Sinica, Taiwan). U2OS, HEK 293T, or HeLa cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Cells were transfected with various cDNA constructs by Lipofectamine 2000. U2OS-based GFP–STIL-inducible lines were generated as previously described (Tang et al, 2009). Briefly, the cDNA fragment encoding GFP–STIL was subcloned into pcDNA4/TO/Myc–His-A (Invitrogen), which is under tetracycline control. A stop codon was introduced into the end of GFP–STIL and before the Myc–His sequences of pcDNA4/TO/Myc–His-A. CPAP–myc tetracycline-inducible lines were described previously (Tang et al, 2009). GFP–STIL and CPAP–myc expression was induced by addition of 1 μg/ml tetracycline. U2OS-based GFP–centrin and mCherry–α-tubulin stably expressing lines were generated by transfection of GFP–centrin or mCherry–α-tubulin construct into U2OS cells and selection by G418.
For synchronizing experiments, cells were arrested either at the G1/S phase by double-thymidine block (2 mM) or aphidicolin (2 μg/ml) or at the G2/M phase by nocodazole (100 ng/ml) treatment as previously described (Tang et al, 2009). The synchronized cells were collected at indicated times for immunoblotting or FACS (FACScan, BD Biosciences) analysis as described previously (Tang et al, 2009). To study the degradation of STIL mediated by the 26S proteasome, HeLa cells were arrested at the G1/S phase by double-thymidine block and released at different times in the presence of a 26S proteasome inhibitor MG132 (2 μM, Sigma-Aldrich). The cell lysates were collected at indicated times and analysed by immunoblotting as described previously (Tang et al, 2009).
Live imaging microscopy
U2OS-based mCherry–α-tubulin expressing cells were transfected with either siControl or siSTIL duplexes for 60 h. Time-lapse movies were taken with a Lecia DMI6000 inverted fluorescence microscope equipped with a 37°C incubator and a 5% CO2 supply.
Immunofluorescence confocal microscopy
Cells on coverslips were cold treated and fixed in methanol at −20°C for 10 min as described previously (Tang et al, 2009). After blocking with 10% normal goat serum in PBS, the fixed cells were incubated with the indicated primary antibodies. After washing with PBST (PBS containing 0.1% Tween-20), cells were incubated with Alexa 568-, Alexa 488-, or Alexa 647-conjugated secondary antibodies (Invitrogen). DNA was counterstained with DAPI (4,6-diamidino-2-phenylindole). The samples were observed using a LSM 510 META confocal system (Carl Zeiss, Inc.) with a Plan Apochromat × 100 1.4 NA oil immersion objective. Images were acquired with ZEN or AimImage Browser software (Carl Zeiss, Inc.). All samples were mounted in Vectashield mounting media (Vector Laboratories). Because STIL is absent in G1 cells, in some experiments, cells were synchronized by aphidicolin (2 μg/ml) treatment to insure the detection of STIL at G1/S cells.
Immunogold EM
For electron microscopy, GFP–STIL-inducible cells were fixed and examined as previously described (Tang et al, 2009). For immunogold EM, cells were fixed in 0.1% glutaraldehyde and 4% paraformaldehyde for 2 h at 4°C. The fixed cells were washed three times with PBS, and dehydrated with serial ethanol and embedded in LR white resin. Thin sections were cut (80 nm) and picked up on grids. The samples on grids were incubated in 0.5% BSA for 1 h followed by labelling with rabbit polyclonal anti-GFP antibody (Abcam; ab6556) overnight at 4°C. The samples were then labelled with 12 nm gold conjugated donkey anti-rabbit IgG (H+L) antibody (Jackson ImmunoResearch Laboratories, Inc.) for 30 min. The grids were washed in PBS and distilled water and then stained with uranyl acetate and lead citrate. The grids were air dried and examined in a Tecnai G2 Spirit Twin electron microscope (FEI Company, Hillsboro, OR).
siRNA experiments
siRNAs used for hSAS6 (5′-UUAACUGUUUGGUAACUGCCCAGGG-3′), Plk4 (5′-GACACUGACACAGUCAAGAACACAU-3′), and CPAP were described previously (Tang et al, 2009). siRNAs (siSTIL#1, siSTIL#2, and siSTIL#3) used for STIL are listed below. All siRNAs and a non-targeting siRNA control were from Invitrogen. Transfections were performed using Lipofectamine 2000 or RNAiMAX.
- siSTIL#1:
5′-AAAUCUUCUGACUCACUGGAUGAGG-3′,
- siSTIL#2:
5′-AUUUCCAGCAGAAACUGUUUGGAGC-3′,
- siSTIL#3:
5′-AAACGUUUGUGGAACAGUAAGAUGG-3′.
Immunoprecipitation and ubiquitination experiments
To analyse whether Flag–STIL is associated with other centrosomal proteins, HEK 293T cells were transfected with Flag–STIL and each indicated GFP-tagged constructs including GFP–Plk4, GFP–hSAS6, GFP–CPAP, GFP–STIL, and GFP–Cep152. Twenty-four hours after transfection, cells were lysed in RIPA buffer (50 mM Tris–HCl at pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 20 mM β-glycerophosphate, 20 mM NaF, 1 mM Na3VO4, and protease inhibitors including 1 μg/μl leupeptin, 1 μg/μl pepstatin and 1 μg/μl aprotinin). The cell lysates were immunoprecipitated with anti-Flag antibody for 2 h at 4°C followed by incubation with Protein-G-Sepharose beads for another 2 h. The immunoprecipitated complexes were separated by SDS–PAGE and probed with anti-GFP or anti-Flag antibodies.
To investigate whether exogenous GFP–STIL is associated with endogenous CPAP and hSAS6, HEK 293T cells were transiently transfected with GFP–STIL construct. Twenty-four hours after transfection, cells were lysed in EBR buffer (50 mM Tris–HCl at pH 8.0, 150 mM NaCl, 1% NP-40) for 30 min at 4°C. The cell lysates were immunoprecipitated with anti-hSAS6, anti-CPAP antibody, or a non-relevant rabbit immunoglobulin (as a negative control) as described above. The immunoprecipitates were then separated by SDS–PAGE and analysed by immunoblotting using an anti-hSAS6, anti-STIL(#441a), or anti-GFP antibody. For detection of the immunoprecipitated complex containing endogenous STIL, CPAP, and hSAS6, HEK 293T cells were lysed in EBR buffer. The cell lysates were immunoprecipitated with anti-CPAP antibody or a non-relevant rabbit immunoglobulin and analysed by immunoblotting using indicated antibodies as described above.
For ubiquitination assays, HEK 293T cells were transiently transfected with or without HA–ubiquitin and GFP–STIL constructs for 22 h, followed by MG132 (20 μM) treatment for another 4 h. After treatment, the cell lysates were subjected to immunoprecipitation with anti-GFP antibody, followed by immunoblotting with antibodies against HA or GFP as described (Tang et al, 2009).
GST pulldown assay
For GST pulldown of transfected GFP-tagged proteins, HEK 293T cells were transiently transfected with various GFP-tagged constructs. Twenty-four hours after transfection, cells were lysed in RIPA buffer as described above. Various GST–CPAP-truncated proteins were expressed in Escherichia coli by IPTG induction and affinity purified by glutathione-agarose beads (Sigma-Aldrich; Tang et al, 2009). Cell lysates were incubated with immobilized various GST–CPAP fusion proteins at 4°C for 3 h. After incubation, the samples were washed with RIPA buffer and separated by SDS–PAGE before immunoblot analysis.
To examine any direct interaction between CPAP and STIL, GST–CPAP-CP3 (residues 895–1338) recombinant proteins were used to pull down 35S-methionine labelled full-length STIL and other centrosomal proteins including hSAS6 and Cep135, which had been produced by in-vitro transcription–translation using the TNT T7 Quick Coupled Transcription/Translation System (Promega). GST was used as a negative control. After binding, the beads were boiled in sample buffer, separated by SDS–PAGE and analysed by autoradiography.
Yeast two-hybrid assay
Yeast two-hybrid analysis was used to confirm a direct protein–protein interaction. Pair-wise yeast two-hybrid interaction assays were performed using the Matchmaker Gold yeast two-hybrid system (Clontech) following the manufacturer's instructions. In brief, the indicated cDNA fragments of STIL, CPAP, CPAP-CP3 (E1235V), and hSAS6 were amplified by PCR. The fragments were subcloned either into pGBKT7 as baits or into pGADT7 as preys and subsequently transformed into Y2HGold and Y187 yeast strains, respectively. After mating, the diploids were spread on both DDO (SD minimal medium, −Trp, −Leu) and QDO (SD minimal medium, −Trp, −Leu, −Ade, −His) plates. After 3–4 days incubation, positive colonies growing on QDO or DDO plates were picked and re-streaked to obtain isolated colonies. The activation of β-galactosidase (β-Gal) by protein interactions was assayed using o-nitrophenol-β-galactopyranoside (ONPG) as a substrate with yeast diploids grown in DDO medium, and the values were normalized against the positive control, p53-T (SV40-large-T) pairs.
Kinase assay
HEK 293T cells were transiently transfected with GFP–STIL and Plk4–myc. Twenty-four hours after transfection, Plk4–myc was immunoprecipitated from the transfected cell lysates by anti-myc antibody and added to the kinase reaction buffer (50 mM Tris–HCl at pH 7.6, 10 mM MgCl2, 1 mM NaF, 1 mM DTT, 100 μM ATP) containing 2 μCi [γ-32P]ATP. Kinase reactions were incubated for 30 min at 30°C and stopped in sample buffer followed by SDS–PAGE analysis and autoradiography.
Supplementary Material
Acknowledgments
We thank En-Ju Chou for technical help and Chih-Chieh Chang for generation of U2OS-based mCherry–tubulin expressing cells. This work was supported by an Academia Sinica Investigator Award and partially by a grant from the National Science Council, ROC.
Author contributions: C-JCT conducted cell cycle and siRNA experiments and helped with the data analysis; S-YL generated GFP–STIL-inducible lines, performed GFP–STIL overexpression and EM analysis; Y-NL and C-TW cloned STIL and performed yeast two-hybrid assay; W-BH worked on GST pulldown experiments; C-WC undertook co-IP experiments; Y-CL performed flow analysis and live cell imaging; K-SW performed immunogold EM; TKT wrote the paper, planned and supervised the project, and performed data analysis.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aplan PD, Lombardi DP, Kirsch IR (1991) Structural characterization of SIL, a gene frequently disrupted in T-cell acute lymphoblastic leukemia. Mol Cell Biol 11: 5462–5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Marshall WF (2010) Building the centriole. Curr Biol 20: R816–R825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM (2005) SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15: 2199–2207 [DOI] [PubMed] [Google Scholar]
- Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, Costa SM, Baralle D, Raponi M, Karbani G, Rashid Y, Jafri H, Bennett C, Corry P, Walsh CA, Woods CG (2005) A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 37: 353–355 [DOI] [PubMed] [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C et al. (2008) A protein domain-based interactome network for C. elegans early embryogenesis. Cell 134: 534–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campaner S, Kaldis P, Izraeli S, Kirsch IR (2005) Sil phosphorylation in a Pin1 binding domain affects the duration of the spindle checkpoint. Mol Cell Biol 25: 6660–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M (2010) Stepwise evolution of the centriole-assembly pathway. J Cell Sci 123 (Part 9): 1414–1426 [DOI] [PubMed] [Google Scholar]
- Castiel A, Danieli MM, David A, Moshkovitz S, Aplan PD, Kirsch IR, Brandeis M, Kramer A, Izraeli S (2011) The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J Cell Sci 124: 532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I (2010) Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol 191: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre M, Canard C, Gonczy P (2006) Sequential protein recruitment in C. elegans centriole formation. Curr Biol 16: 1844–1849 [DOI] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM (2010) Asterless is a scaffold for the onset of centriole assembly. Nature 467: 714–718 [DOI] [PubMed] [Google Scholar]
- Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, Sharp DJ (2007) Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol 17: 1960–1966 [DOI] [PubMed] [Google Scholar]
- Guernsey DL, Jiang H, Hussin J, Arnold M, Bouyakdan K, Perry S, Babineau-Sturk T, Beis J, Dumas N, Evans SC, Ferguson M, Matsuoka M, Macgillivray C, Nightingale M, Patry L, Rideout AL, Thomas A, Orr A, Hoffmann I, Michaud JL et al. (2010) Mutations in centrosomal protein CEP152 in primary microcephaly families linked to MCPH4. Am J Hum Genet 87: 40–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA (2005) The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146 [DOI] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T (2010) Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol 191: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K (2010) Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci 123(Part 9): 1407–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LY, Tang CJ, Tang TK (2000) Protein 4.1 R-135 interacts with a novel centrosomal protein (CPAP) which is associated with the gamma-tubulin complex. Mol Cell Biol 20: 7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Kubo A, Tsukita S (2005) Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol 7: 517–524 [DOI] [PubMed] [Google Scholar]
- Izraeli S, Colaizzo-Anas T, Bertness VL, Mani K, Aplan PD, Kirsch IR (1997) Expression of the SIL gene is correlated with growth induction and cellular proliferation. Cell Growth Differ 8: 1171–1179 [PubMed] [Google Scholar]
- Izraeli S, Lowe LA, Bertness VL, Good DJ, Dorward DW, Kirsch IR, Kuehn MR (1999) The SIL gene is required for mouse embryonic axial development and left-right specification. Nature 399: 691–694 [DOI] [PubMed] [Google Scholar]
- Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF (2004) Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell 6: 511–523 [DOI] [PubMed] [Google Scholar]
- Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA (2003) SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell 112: 575–587 [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA (2007) Plk4-induced centriole biogenesis in human cells. Dev Cell 13: 190–202 [DOI] [PubMed] [Google Scholar]
- Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P (2009) Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol 19: 1012–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Girimaji SC, Duvvari MR, Blanton SH (2009) Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet 84: 286–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P (2005) SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol 7: 115–125 [DOI] [PubMed] [Google Scholar]
- Leidel S, Gonczy P (2003) SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell 4: 431–439 [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678 [DOI] [PubMed] [Google Scholar]
- O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG (2001) The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell 105: 547–558 [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW (2007) Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol 17: 834–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T (2006) Centriole assembly in Caenorhabditis elegans. Nature 444: 619–623 [DOI] [PubMed] [Google Scholar]
- Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA (2004) The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol 14: 863–873 [DOI] [PubMed] [Google Scholar]
- Pfaff KL, Straub CT, Chiang K, Bear DM, Zhou Y, Zon LI (2007) The zebra fish cassiopeia mutant reveals that SIL is required for mitotic spindle organization. Mol Cell Biol 27: 5887–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM (2007) DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol 17: 1465–1472 [DOI] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA (2009) Control of centriole length by CPAP and CP110. Curr Biol 19: 1005–1011 [DOI] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW (2010a) Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol 188: 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Roque H, Raff JW (2010b) DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev Cell 19: 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Gonczy P (2008) Mechanisms of procentriole formation. Trends Cell Biol 18: 389–396 [DOI] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P (2007) Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell 13: 203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK (2009) CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol 11: 825–831 [DOI] [PubMed] [Google Scholar]
- Thornton GK, Woods CG (2009) Primary microcephaly: do all roads lead to Rome? Trends Genet 25: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH (2009) Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, Pelletier L (2008) The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol 18: 136–141 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.