Abstract
Eukaryotic cells respond to DNA damage by activating checkpoint signalling pathways. Checkpoint signals are transduced by a protein kinase cascade that also requires non-kinase mediator proteins. One such mediator is the Saccharomyces cerevisiae Dpb11 protein, which binds to and activates the apical checkpoint kinase, Mec1. Here, we show that a ternary complex of Dpb11, Mec1 and another key mediator protein Rad9 is required for efficient Rad9 phosphorylation by Mec1 in vitro, and for checkpoint activation in vivo. Phosphorylation of Rad9 by cyclin-dependent kinase (CDK) on two key residues generates a binding site for tandem BRCT repeats of Dpb11, and is thereby required for Rad9 recruitment into the ternary complex. Checkpoint signalling via Dpb11, therefore, does not efficiently occur during G1 phase when CDK is inactive. Thus, Dpb11 coordinates checkpoint signal transduction both temporally and spatially, ensuring the initiator kinase is specifically activated in proximity of one of its critical substrates.
Keywords: cell cycle, checkpoint, DNA damage response
Introduction
Lesions in DNA arising from extrinsic and intrinsic sources can compromise the integrity of genetic information and cause cell death. In eukaryotes, the DNA damage checkpoint modulates many aspects of the cellular program in response to DNA lesions (Melo and Toczyski, 2002; Harrison and Haber, 2006). Checkpoint signalling involves a protein kinase cascade initiated by one of the two apical kinases of the phosphoinositide 3 kinase-related kinases (PIKK) family. In Saccharomyces cerevisiae, these kinases are Mec1 and Tel1 (homologous to vertebrate ATR and ATM, respectively). They phosphorylate and activate the effector kinases Rad53 (in vertebrates, Chk2) and Chk1. Conserved, non-kinase mediator proteins of the DNA damage checkpoint pathway include the BRCT domain-containing Rad9 and Dpb11 proteins and the PCNA-like Ddc1–Mec3–Rad17 (9-1-1) complex (Parrilla-Castellar et al, 2004; Garcia et al, 2005; FitzGerald et al, 2009).
The DNA damage checkpoint must respond to a very wide variety of DNA lesions. The apical kinases, however, do not sense lesions directly, but are recruited via interactions with other proteins that either bind directly to lesions or to processed intermediates. Mec1 is recruited to ssDNA at stalled replication forks or resected DSBs by interactions with RPA (Rouse and Jackson, 2002; Zou and Elledge, 2003; Ball et al, 2005, 2007). Separately, the 9-1-1 complex is loaded at ss-ds-DNA junctions by the Rad24-clamp loader complex (Ellison and Stillman, 2003; Majka and Burgers, 2003; Majka et al, 2006a) where it acts as a co-sensor of DNA damage (Bonilla et al, 2008).
Mediators are recruited to sites of DNA damage by a complex network of interactions. The 9-1-1 complex plays a role in recruiting Dpb11 and its orthologues to sites of DNA damage (Furuya et al, 2004; Delacroix et al, 2007; Lee et al, 2007; Puddu et al, 2008). In budding yeast, this recruitment involves Mec1 phosphorylation of the 9-1-1 complex subunit Ddc1, which generates a binding site for the second pair of phospho-protein binding BRCT repeats (BRCT3&4) of Dpb11 (Wang and Elledge, 2002; Puddu et al, 2008).
Recruitment of Rad9 to sites of DNA damage involves multiple interactions. One pathway depends on histone modifications: Rad9 can bind via its BRCT repeat domain to histone H2A that has been phosphorylated by Mec1 or Tel1 (γH2A) (Hammet et al, 2007). It also binds lysine 79 methylated histone H3 via its TUDOR domain (Grenon et al, 2007). This modification is catalysed by the Dot1 methyltransferase (Ng et al, 2002; van Leeuwen et al, 2002), but currently there is no evidence that this modification is regulated in response to DNA damage. Both unphosphorylatable H2A mutants and dot1Δ mutants show defects in checkpoint activation during G1 phase (Giannattasio et al, 2005; Wysocki et al, 2005; Grenon et al, 2007; Hammet et al, 2007).
In contrast to G1 checkpoint activation, G2/M checkpoint activation occurs in dot1Δ cells (Giannattasio et al, 2005). However, checkpoint activation is abolished in dot1Δ dpb11-1 double mutants suggesting that an additional, alternative mode of Rad9 recruitment may involve Dpb11 (Puddu et al, 2008). This idea is supported by the finding that Crb2, the fission yeast homologue of Rad9, interacts with Cut5/Rad4, the homologue of Dpb11 (Mochida et al, 2004; Du et al, 2006). This interaction is regulated by cyclin-dependent kinase (CDK) phosphorylation of Crb2 and facilitates histone-independent Crb2 recruitment to DNA damage foci (Esashi and Yanagida, 1999; Nakamura et al, 2004; Du et al, 2006). After phosphorylation by Mec1 Rad9 creates a platform for the recruitment of Rad53 (Emili, 1998; Sun et al, 1998; Vialard et al, 1998; Schwartz et al, 2002).
Another feature of checkpoint signalling is the activation of the apical kinase by activator proteins. Mec1 and its homologue ATR are stimulated by binding to Dpb11 and TopBP1, respectively (Kumagai et al, 2006; Mordes et al, 2008; Navadgi-Patil and Burgers, 2008). Mec1 is also stimulated by binding to the Ddc1 subunit of 9-1-1 (Majka et al, 2006b). It is not sufficiently understood how relevant this stimulation is for checkpoint signalling in vivo and especially where in the signalling cascade these activators become important. In this paper, we show that Dpb11 plays an important role in three aspects of checkpoint signalling: cell-cycle regulation, mediator recruitment and Mec1 kinase activation. Dpb11 integrates these functions through formation of a ternary checkpoint complex.
Results
Interactions between Dpb11 and checkpoint proteins
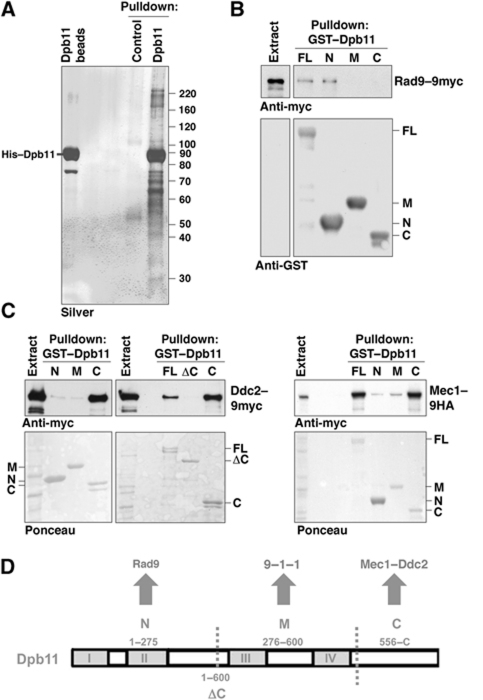
To identify Dpb11 interacting proteins, we purified recombinant, full-length, His-tagged Dpb11 via Ni2+-NTA agarose beads after incubation with whole cell extracts from asynchronous yeast cultures. We identified Mec1, Ddc2 and Rad9 as specific Dpb11 interactors by mass spectrometry (Figure 1A). We next repeated the pulldown experiment with different domains of Dpb11 fused to GST (see Figure 1D). Figure 1B shows that Rad9 from yeast extracts binds to N-terminal BRCT1&2 domain (aa 1–275) but not to BRCT3&4 (aa 276–600) or the C-terminal fragment of Dpb11 (aa 556–764) (Figure 1B). The Rad9 and Dpb11 orthologues in fission yeast have previously been found to interact (Mochida et al, 2004; Du et al, 2006), which suggests evolutionary conservation.
Figure 1.
Dpb11 physically interacts with the DNA damage checkpoint proteins Mec1–Ddc2 and Rad9. (A) Pulldown of Dpb11 protein interactors with purified His–Dpb11 and cell lysates of asynchronously dividing yeast. MS analysis showed the presence of Mec1 (>220 kDa), Rad9 (>160 kDa) and Ddc2 (=90 kDa). (B) Rad9 interacts with the N-terminus of Dpb11. Pulldown experiment with immobilized GST–Dpb11 or GST–Dpb11 fragments (N=aa 1–275, M=aa 276–600, C=556–764; see D) and whole cell extracts of yeast containing Rad9–9myc. (C) The C-terminus of Dpb11 contains an interaction site for Mec1–Ddc2. Pulldown with GST–Dpb11 or GST–Dpb11 fragments (see D) and extracts containing Ddc2–9myc or Mec1–9HA. (D) Schematic diagram of Dpb11 domains and interactors involved in the DNA damage checkpoint. BRCT repeat domains (I–IV) are marked as grey boxes.
Similarly to results published by the Burgers and Cortez laboratories (Mordes et al, 2008; Navadgi-Patil and Burgers, 2008), we observe that the C-terminal domain of Dpb11 is both necessary and sufficient to mediate the binding to Mec1 and Ddc2 (Figure 1C). Consistent with the fact that BRCT repeats are not involved, the interaction does not depend on phosphorylation of Mec1–Ddc2 and appears not to be regulated during the cell cycle or in response to DNA damage (Supplementary Figure S1). Together with previous work showing that Dpb11 also interacts with Mec1-phosphorylated Ddc1 via BRCT3&4 (Wang and Elledge, 2002; Puddu et al, 2008), Figure 1D summarizes interactions between Dpb11 and DNA damage checkpoint proteins.
The C-terminus of Dpb11 affects Mec1 signalling in vitro and in vivo via conserved aromatic residues
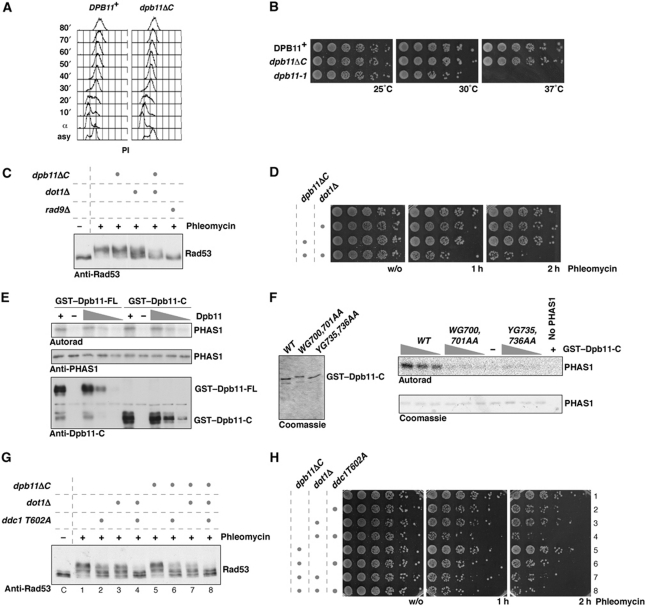
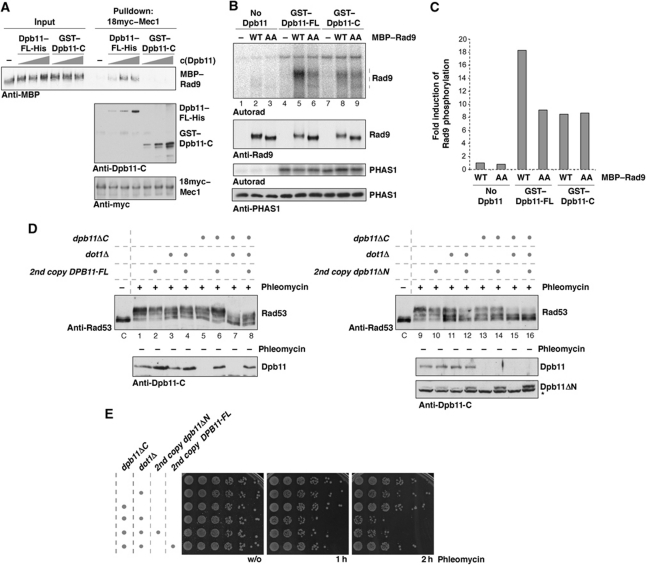
Until now checkpoint studies have generally used the dpb11-1 allele, which introduces a STOP codon in place of W583. However, in addition to defects in interacting with Mec1–Ddc2, this mutant is also thermosensitive and shows strongly reduced binding of the BRCT3&4 interactors Sld2 and Ddc1 (Kamimura et al, 1998; Wang and Elledge, 2002). Since the Sld2–Dpb11 interaction is required for replication initiation and a reduced initiation frequency can mimic a checkpoint defect (Shimada et al, 2002; Tercero et al, 2003), we felt it was important to generate true separation of function mutants. Constructs truncated upstream of W583 were lethal; however, a truncation at amino acid 600 (dpb11ΔC) in the endogenous DPB11 gene resulted in a viable haploid strain. FACS analysis of WT and dpb11ΔC cells released from α-factor arrest showed very similar replication profiles (Figure 2A). Furthermore, in contrast to dpb11-1, we did not observe any temperature sensitivity associated with dpb11ΔC (Figure 2B). Therefore, dpb11ΔC appears to be functional for DNA replication.
Figure 2.
The C-terminal domain of Dpb11 is dispensable for DNA replication, but in the absence of DOT1 is required for the G2/M DNA damage checkpoint in vivo and stimulates the Mec1 kinase via two W/YG motifs in vitro. (A) The C-terminus of Dpb11 is not required for DNA replication, since WT and dpb11ΔC cells synchronously released from G1 arrest (α) show identical replication profiles. (B) The dpb11ΔC mutant, in contrast to dpb11-1, is not temperature sensitive. (C, D) The dpb11ΔC mutation results in a G2/M DNA damage checkpoint defect in the absence of DOT1, as indicated by reduced Rad53 phosphorylation (C) and loss of viability (D) after treatment with phleomycin (50 μg/ml). (C) Samples were taken before (−) or 30′ after (+) addition of phleomycin. (E) The C-terminal domain of Dpb11 is sufficient for stimulation of Mec1 kinase in vitro. Mec1–18myc–Ddc2 phosphorylation towards the model substrate PHAS1 is stimulated by GST–Dpb11 or GST–Dpb11-C (555–764). (F) Stimulation of Mec1 kinase requires two W/YG motifs. GST–Dpb11-C (555–764) and mutant versions WG700,701AA and YG735,736AA of comparable amount and purity (left) were used to activate Mec1 kinase in vitro (right). (G, H) The checkpoint phenotypes of the dpb11ΔC and ddc1-T602A mutants are epistatic. Strains harbouring indicated combinations of mutations were analysed as in (C) and (D).
We next examined checkpoint responses in cells expressing the dpb11ΔC mutant treated with phleomycin. Figure 2C shows that these cells, synchronized in G2/M phase, showed no significant defect in either cell survival or Rad53 activation (Figure 2C). Previous work has shown that dpb11-1 has a G2/M checkpoint defect only when combined with deletion of DOT1 (Puddu et al, 2008). When dpb11ΔC was combined with dot1Δ, we observed a deficient G2/M checkpoint as indicated by reduced phosphorylation of Rad53 and survival after DNA damage treatment (Figure 2C and D). These defects are not as severe as the defect seen in a rad9Δ mutant (Figure 2C; Supplementary Figure S2A). The checkpoint defect in dpb11ΔC dot1Δ is also less severe than that seen in the dpb11-1 dot1Δ double mutant at the permissive temperature for dpb11-1 (Supplementary Figure S2C and D). This may be due to defects in DNA replication in the dpb11-1 mutant that may exacerbate defects in checkpoint activation. Alternatively, this may be because Dpb11-1 is unable to bind both Ddc1 and Mec1–Ddc2 (Wang and Elledge, 2002; Mordes et al, 2008; Navadgi-Patil and Burgers, 2008), while Dpb11ΔC is only deficient in Mec1–Ddc2 interaction (Figure 1C).
Dpb11 and TopBP1 can stimulate the kinase activities of Mec1 and ATR, respectively, in vitro (Kumagai et al, 2006; Mordes et al, 2008; Navadgi-Patil and Burgers, 2008). As shown in Figure 2E, we also observed significant stimulation of immunopurified Mec1–Ddc2 kinase activity by recombinant Dpb11, and the C-terminal domain of Dpb11 is sufficient for this activation (Figure 2E; Mordes et al, 2008). Although it has been reported that Dpb11 does not contain sequences related to the ATR activation domain (AAD) of TopBP1 (Mordes et al, 2008), we noticed that sequence homology among Dpb11 C-termini from Saccharomyces sensu lato was restricted to two patches of amino acids surrounding conserved W/YG motifs (Supplementary Figure S2B). Because a tryptophan residue is critically important in the TopBP1 AAD (Kumagai et al, 2006), we examined the effect of mutating these residues on the interaction with and activation of Mec1–Ddc2. Both Dpb11-WG700,701AA and Dpb11-W700A were unable to interact with Mec1–Ddc2 and Dpb11 YG735,736AA showed a reduced interaction (Supplementary Figure S2E). Moreover, individual mutation of the W/YG motifs strongly reduced the stimulatory effect of Dpb11 on the kinase activity of Mec1–Ddc2 (Figure 2F). Supplementary Figure S2F shows that dpb11 WG700,701AA point mutant is as defective as dpb11ΔC in checkpoint activation and the inability to activate Mec1, therefore, correlates which a deficiency to support checkpoint signalling.
The interaction between the 9-1-1 complex and Dpb11 or their orthologues is thought to recruit Dpb11 orthologues to sites of DNA damage (Furuya et al, 2004; Delacroix et al, 2007; Puddu et al, 2008). We, therefore, tested a specific Mec1-phosphorylation site mutant of Ddc1 (ddc1 T602A), which prevents binding to Dpb11 (Puddu et al, 2008). In ddc1 T602A or ddc1 T602A dot1Δ mutant strains, addition of the dpb11ΔC mutation did not result in increased checkpoint defects as measured by Rad53 phosphorylation and survival after phleomycin treatment (Figure 2G and H compare sample 2 with 6; and 4 with 8). This suggests that the Dpb11–Mec1 interaction is functionally dependent on the Dpb11–Ddc1 interaction, consistent with 9-1-1-dependent recruitment of Dpb11. Defects in the Dpb11–Ddc1 module (i.e., ddc1 T602A) cause more severe phenotypes than defects in the Dpb11–Mec1–Ddc2 module (i.e., dpb11ΔC, Figure 2G and H, compare sample 4 with 7), suggesting that at least in this mutant background Dpb11 has a function in checkpoint signalling independent of its ability to activate Mec1–Ddc2.
CDK phosphorylation of Rad9 regulates binding to Dpb11
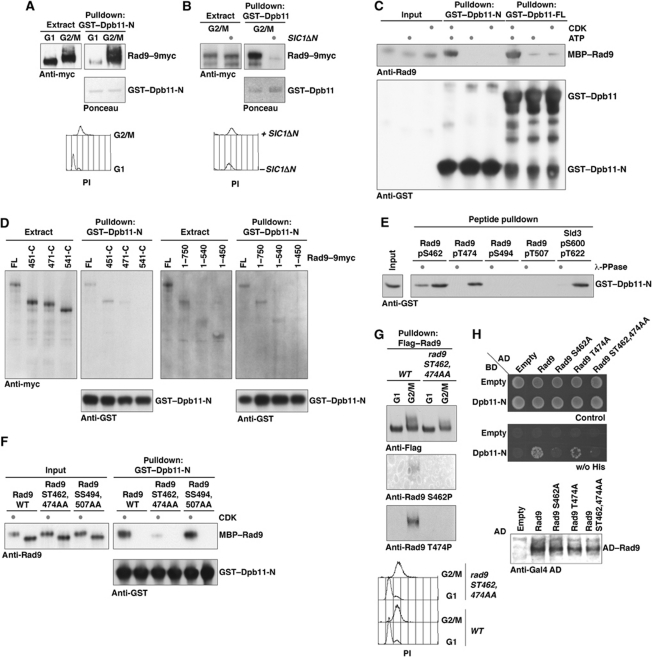
Although the Dpb11–Rad9 interaction could occur in the absence of exogenous DNA damage (Figure 1B), Figure 3A shows that the Rad9–Dpb11 interaction was cell-cycle regulated: it was detected in extracts from G2/M-arrested cells, but not in extracts from G1-arrested cells (Figure 3A). Moreover, the interaction was lost in extracts from G2/M-arrested cells in which a stable version of the CDK inhibitor Sic1 (Sic1ΔN) was overexpressed (Figure 3B). To test whether this cell cycle-regulated interaction was directly mediated by CDK phosphorylation, we expressed and purified recombinant MBP–Rad9 and phosphorylated it with recombinant CDK. Figure 3C shows that CDK phosphorylation strongly stimulated the binding of MBP–Rad9 to either full-length GST–Dpb11 or GST–Dpb11-N, which contains just BRCT1&2.
Figure 3.
The Rad9–Dpb11 interaction is cell-cycle regulated through direct binding of Dpb11 to Rad9 CDK sites S462 and T474. (A) The Rad9–Dpb11 interaction is cell-cycle regulated. Rad9–9myc from G1- or G2/M-arrested cells was tested for binding to GST–Dpb11-N in pulldown. (B) Overexpression of a stable version of Sic1 (Sic1ΔN; Desdouets et al, 1998) inhibits the Rad9–Dpb11 interaction in G2/M-arrested cells. (C) Recombinant, purified MBP–Rad9 specifically interacts with GST–Dpb11 or GST–Dpb11-N after in vitro phosphorylation of Rad9 by CDK (Cyclin AΔN170-Cdk2; Brown et al, 1995). (D) N- and C-terminal truncations of endogenous Rad9 from lysates of G2/M-arrested cells were analysed by GST–Dpb11 pulldowns. A central region of Rad9, which contains a cluster of CDK sites, is required for interaction with Dpb11. (E) Phosphorylated Serine 462 and Threonine 474 peptides of Rad9 pull down GST–Dpb11-N. λ-Phosphatase treatment demonstrated phosphorylation specificity. The Dpb11-N interacting peptide from Sld3 served as a positive control (Zegerman and Diffley, 2007). (F) Rad9 ST462,474AA is deficient for in vitro binding to GST–Dpb11-N after CDK treatment. Experiment as in (B) but with mutant versions of MBP–Rad9. (G) Rad9 is phosphorylated at CDK sites Serine 462 and Threonine 474 in vivo. Phospho-specific antibodies (Supplementary Figure S4) were used to probe pulldowns of Flag–Rad9 or Flag–Rad9 ST462,474AA from G1- or G2/M-arrested cells. (H) Rad9 CDK sites are required for the interaction with Dpb11 in vivo. Gal4–BD–Dpb11-N and Gal4–AD–Rad9 (WT and S462A, T474A, ST462,474AA mutants) fusions were used to test the Rad9–Dpb11 interaction in the two-hybrid system. Expression of Rad9-fusion constructs was confirmed by Gal4–AD–westerns.
Rad9 has previously been shown to be a CDK substrate (Ubersax et al, 2003) and in the fission yeast orthologue Crb2 the CDK site T215 was found to be important for binding of the Dpb11 orthologue Cut5/Rad4 (Esashi and Yanagida, 1999; Du et al, 2006). Homologues of Rad9 that contain conserved C-terminal BRCT and TUDOR domains can be found across the eukaryotic kingdom, but upstream of these domains they differ in length and sequence (Supplementary Figure S5B). Since the majority of putative CDK phosphorylation sites can be found in this region (Supplementary Figure S5B), we decided to unambiguously map the phosphorylation site that regulates the interaction with Dpb11. We generated N- and C-terminal truncations of Rad9 in vivo and tested their ability to bind GST–Dpb11-N in extracts from G2/M-arrested cells. We found that versions of Rad9 harbouring significant N- or C-terminal truncations were still able to interact with Dpb11, but the interaction was abolished when a region between aa 451 and 540 of Rad9 was deleted (Figure 3D). This region of Rad9 contains a cluster of four S/TP sites (S462, T474, S494 and T507; compare Supplementary Figure S3A). In order to test whether any of these are sufficient to mediate phosphorylation-specific binding to Dpb11, we constructed four independent biotinylated 35mer peptides, each harbouring one phosphorylated CDK site at the same position (26) in the peptide, and tested their binding to Dpb11 BRCT1&2 by streptavidin bead pulldown. Figure 3E shows that phospho-S462 and phospho-T474 peptides exhibited phosphorylation-dependent binding to Dpb11, comparable to the Dpb11-binding peptide from Sld3 (Figure 3E; Supplementary Figure S3B). Notably, out of 12 Rad9 peptides tested, which covered 12 out of 16 conserved S/TP sites, only Rad9 pS462 and Rad9 pT474 were able to bind Dpb11 (Supplementary Figure S3A). Among these, we did not see significant Dpb11 binding to peptides containing phosphorylated Ser11, a residue recently implicated in Dpb11–Rad9 interaction (Granata et al, 2010). Sld3 and its mammalian orthologue Treslin/ticrr also utilize two phosphorylated residues to interact with BRCT1&2 of Dpb11/TopBP1 (Tanaka et al, 2007; Zegerman and Diffley, 2007; Boos et al, 2011). We were able to detect limited conservation of sequences surrounding the two phosphorylation sites of Sld3 and Rad9, using the sequences of different Saccharomyces sensu lato species (Supplementary Figure S5A), suggesting that both proteins interact with Dpb11 in a similar fashion.
To assess the importance of S462 and/or T474 phosphorylation to the Dpb11 interaction, we introduced point mutations into full-length Rad9 fused to MBP and examined CDK-dependent binding to Dpb11-N in vitro. Mutation of these two phosphorylation sites, but not mutation of two neighbouring sites (ST494,507AA), greatly reduced the CDK-dependent interaction with Dpb11 in vitro (Figure 3F) indicating that, even in the presence of the other 18 potential CDK sites, S462 and T474 are critical for efficient CDK-dependent Dpb11 interaction. We generated phospho-specific antibodies to these two sites to determine whether S462 and T474 are phosphorylated in vivo (Supplementary Figure S4). Figure 3G shows that both of these antibodies detect wild-type Rad9–3Flag after pulldown of Rad9 with anti-FLAG antibody from G2/M-arrested cells, but not from G1-arrested cells. Moreover, the ST462,474AA mutant was not detected from either G1- or G2/M-arrested cell extracts with these antibodies. Therefore, these sites are phosphorylated in vivo in a cell cycle-dependent manner. Finally, we employed the two-hybrid assay to analyse the requirements of the Dpb11–Rad9 interaction in vivo and found that the T474A mutation reduced the interaction with Dpb11 and S462A or ST462,474AA mutations appeared to abolish it completely (Figure 3H). Taken together, these results show that phosphorylation of S462 and T474 is necessary and sufficient for CDK-dependent, cell cycle-regulated interaction between Dpb11 and Rad9.
Using a degenerate Dpb11 BRCT binding consensus derived from the Rad9 and Sld3 sequences, we were able to detect potential CDK phosphorylation-dependent Dpb11 binding sites in the N-termini of different Rad9 fungal orthologues (Supplementary Figure S5C). The alignment suggests that T215 (Esashi and Yanagida, 1999; Du et al, 2006) and perhaps T235 or T252 of Schizosaccharomyces pombe Crb2 are homologous to S462 and T474. Thus, in addition to the Dpb11–Ddc1 and the Dpb11–Mec1–Ddc2 interactions, the Dpb11–Rad9 interaction is also a conserved feature of Dpb11/TopBP1 function.
CDK regulation of the Rad9–Dpb11 interaction determines cell-cycle regulation of the DNA damage checkpoint
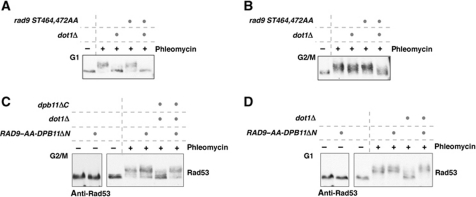
The rad9 ST462,474AA mutant, which is defective in Dpb11 interaction, is fully able to activate the checkpoint in G1 as well as in G2/M in otherwise wild-type cells (Figure 4A and B). However, when this mutant was combined with dot1Δ, it was defective in G2/M checkpoint activation (Figure 4B) similar to the dpb11ΔC mutant (Figure 2C). The rad9 ST462,474AA mutation did not lead to an increase of the phenotype of the ddc1 T602A mutant (Supplementary Figure S6A), which completely abolishes the checkpoint function of Dpb11 providing additional evidence that the rad9 ST462,474AA phenotype is DPB11 dependent. Analysis of the Mec1-dependent phosphorylation of Dpb11 suggests that the Rad9–Dpb11 is not involved in recruitment of Dpb11 to DNA damage sites (Supplementary Figure S6B). We observed a similar checkpoint defect for rad9 ST462,474AA dot1Δ and dpb11ΔC dot1Δ mutants, but the rad9 ST462,474AA dpb11ΔC dot1Δ triple mutant showed a slightly stronger phenotype (Supplementary Figure S6C). This suggests that the Rad9–Dpb11 interaction may be partially independent of the Dpb11–Mec1–Ddc2 interaction at least in these mutant backgrounds.
Figure 4.
CDK regulation of the Rad9–Dpb11 interaction constitutes the essential cell-cycle regulation of checkpoint signalling. (A) The rad9 ST462,474AA allele is proficient for the G1 checkpoint. Rad53 phosphorylation was determined in samples of G1-arrested cells, before (−) or after (+) 30′ treatment with phleomycin (50 μg/ml). (B) rad9 ST462,474AA dot1Δ cells show defects in DNA damage-induced Rad53 phosphorylation in G2/M-arrested cells. Experiment as in (A) but with G2/M-arrested cells. (C) A fusion protein consisting of the two checkpoint deficient alleles rad9 ST462,474AA and dpb11ΔN (276-C) can support checkpoint activation in G2/M. The RAD9–AA–DPB11ΔN fusion was ectopically expressed from the DPB11 promoter as only cellular copy of RAD9 in WT or dot1 dpb11ΔC strains. (D) The RAD9–AA–DPB11ΔN fusion restores checkpoint activation in G1-arrested dot1Δ cells.
The results shown in Figure 4A and B are consistent with the hypothesis that Dot1 and Dpb11 act redundantly in G2/M but the Dpb11 pathway does not function during G1 phase because Rad9 cannot be phosphorylated by CDK. To test whether the Rad9–Dpb11 interaction is sufficient to explain cell cycle-regulated checkpoint signalling, we constructed a covalent fusion of Rad9 ST462,474AA and Dpb11 lacking the N-terminal BRCT1&2 repeat domain (RAD9–AA–DPB11ΔN fusion). This fusion is exactly analogous to the fusion we previously used to show that phosphorylation of Sld3 by CDK generates a binding site for Dpb11 during replication initiation (Zegerman and Diffley, 2007). Expression of RAD9–AA–DPB11ΔN appears not to negatively influence DNA replication (Supplementary Figure S7A). Neither rad9-ST462,474AA nor dpb11ΔN alone is able to support checkpoint signalling in a dot1Δ background (see Figures 4B and 5D). Figure 4C shows that the RAD9–AA–DPB11ΔN fusion was able to restore phleomycin-induced Rad53 activation to WT levels during G2/M phase in a dot1Δ, dpb11ΔC, rad9Δ background. Indeed, the RAD9–AA–DPB11ΔN fusion appears to be a gain-of-function mutant, since the checkpoint was dominantly activated even at lower phleomycin concentrations compared with wild-type cells and also the fusion protein could be recruited in a Ddc1 T602-independent way (Supplementary Figure S7B–D).
Figure 5.
Dpb11 forms a ternary complex with Rad9 and Mec1–Ddc2, which is critical for phosphorylation by Mec1 in vitro and checkpoint activation in vivo. (A) Mec1–18myc can pull down CDK-phosphorylated MBP–Rad9 in the presence of increasing amounts of full-length Dpb11–His but not the isolated C-terminal domain (GST–Dpb11-C), demonstrating formation of a ternary Rad9–Dpb11–Mec1–Ddc2 complex in vitro. (B) In vitro phosphorylation of Rad9 by Mec1 is specifically enhanced by a Rad9–Dpb11–Mec1–Ddc2 complex. Recombinant, purified MBP–Rad9 or MBP–Rad9 ST462,474AA is quantitatively phosphorylated by CDK, repurified (see Supplementary Figure S8A) and used as substrate in Mec1 kinase assays. Mec1 was activated with GST–Dpb11 or GST–Dpb11-C at equimolar concentration as MBP–Rad9. The PIKK substrate PHAS1 was used as a specificity control. (C) Quantification of Dpb11-dependent stimulation of Mec1 phosphorylation of Rad9. Signals are normalized to phosphorylation of WT Rad9 in the absence of Dpb11. (D, E) Dpb11ΔN fails to rescue the checkpoint defect of dot1Δ dpb11ΔC cells. Dpb11 or Dpb11ΔN was ectopically expressed from the endogenous promoter as a second copy of DPB11. G2/M DNA damage checkpoint activation was measured by Rad53 phosphorylation (D) or survival (E) after phleomycin treatment (50 μg/ml). (D) Samples were taken before (−) or 30′ after (+) addition of phleomycin.
Figure 4D shows that the fusion was also able to restore phleomycin-induced Rad53 phosphorylation in G1-arrested cells in the absence of Dot1 and the requirement for Dot1 in the G1 checkpoint is therefore bypassed. Taken together, these results show that CDK phosphorylation of Rad9 is required to induce interaction between Dpb11 and Rad9 and that lack of this interaction in G1 phase is sufficient to explain the cell-cycle regulation of checkpoint signalling.
Dpb11 specifically induces Rad9 phosphorylation by Mec1 in a ternary Rad9–Dpb11–Mec1–Ddc2 complex in vitro
If Dpb11 operates as a molecular scaffold in the DNA damage response, it should be able to simultaneously interact with different checkpoint proteins, for example, Rad9 and Mec1–Ddc2. To test this, we examined the ability of Dpb11 to bridge an interaction between Mec1–Ddc2 and Rad9. Figure 5A shows that, in the presence of full-length Dpb11 but not a C-terminal fragment of Dpb11 (Dpb11-C), CDK-phosphorylated Rad9 was specifically co-immunoprecipitated with Mec1 in a Mec1 pulldown.
To integrate the scaffolding function and the Mec1–Ddc2 activation function of Dpb11 into a mechanistic model, we hypothesized that Dpb11 may work by activating Mec1–Ddc2 and bringing active Mec1–Ddc2 into proximity with one of its key downstream targets, Rad9. We examined the ability of Dpb11 full length and Dpb11-C to activate Mec1–Ddc2 towards the non-specific PHAS1 substrate and towards Rad9. Figure 5B shows that both full-length Dpb11 and Dpb11-C activate Mec1–Ddc2 to a very similar extent when PHAS1 is used as a substrate. However, when we used Rad9 that had been pre-phosphorylated with CDK and repurified as a substrate (Supplementary Figure S8A and B), we found that the full-length Dpb11 promoted ∼2-fold more Rad9 phosphorylation than Dpb11-C (Figure 5B and C). This stimulation was not seen when we used Rad9 ST462,474AA (AA) as substrate, indicating that recruitment of Rad9 to Dpb11 was critical for the stimulation. This stimulation by Dpb11 was maximal at approximately equimolar concentrations of Rad9 and Dpb11, while a further increase of the Rad9 concentration increased overall Rad9 phosphorylation but reduced the stimulatory effect of Dpb11 (Supplementary Figure S8C and D). Taken together, these results support the hypothesis that Dpb11 acts both to activate Mec1–Ddc2 and to bring the active kinase together with CDK-phosphorylated Rad9 protein. When we compared the efficiency of phleomycin-induced Rad9 and Rad53 phosphorylation in vivo, we noticed that in the dpb11ΔC dot1Δ mutant Rad53 phosphorylation was affected more strongly than phosphorylation of Rad9 (Figure 2C; Supplementary Figure S9A). This suggests that activation of Mec1 by Dpb11 may be more important for the efficient phosphorylation of Rad53.
Simultaneous interaction of Dpb11 with Rad9 and Mec1–Ddc2 is required for efficient checkpoint activation in vivo
This model predicts that simultaneous interaction of Dpb11 with Rad9 and Mec1–Ddc2 should be required for checkpoint activation. Alternatively, if the Rad9–Dpb11 and Dpb11–Mec1–Ddc2 interactions were independent of each other, checkpoint signalling should be functional in cells expressing two versions of Dpb11, one able to support the interactions with the 9-1-1 and Mec1–Ddc2 but defective in the Rad9 interaction (dpb11ΔN), and the other able to support interactions with Rad9 and the 9-1-1 complex but not with Mec1–Ddc2 (dpb11ΔC). To address this, we asked whether the dpb11ΔN and dpb11ΔC mutants, when expressed together, could support checkpoint activation. Figure 5D and E show, however, that both Rad53 activation and cell survival are compromised in dpb11ΔC dpb11ΔN dot1Δ cells (compare lanes 15 and 16 in Figure 5D and E). In contrast, if full-length DPB11 is expressed as a second copy in dpb11ΔC dot1Δ cells, the checkpoint defect is rescued to levels comparable to WT or dot1 strains (Figure 5D, compare lanes 7 and 8; Figure 5E). The dpb11ΔC protein is likely to be functional because it can fully support the replicative function of Dpb11 (Figure 2A), which requires functional N-terminal and central BRCT repeat domains. The Dpb11ΔN protein is expressed at a similar level to that of the full-length protein (Figure 5D). Moreover Dpb11ΔN must be able to bind to the 9-1-1 complex and Mec1–Ddc2, because it is able to support residual checkpoint activation in G1, which can be observed under high concentrations of phleomycin (Supplementary Figure S10). This checkpoint activation is independent of the Rad9–Dpb11 interaction, but requires Dpb11 to bind to the 9-1-1 complex and Mec1–Ddc2 (Supplementary Figure S10). As a final test, we examined the effect of a single point mutation in BRCT1, dpb11-T12A. Supplementary Figure S11 shows that dpb11-T12A also fails to complement dpb11ΔC in the checkpoint, similar to dpb11ΔN (Supplementary Figure S11). Taken together, these results indicate that Dpb11 must simultaneously interact with Rad9 and Mec1–Ddc2 for efficient checkpoint signalling.
Discussion
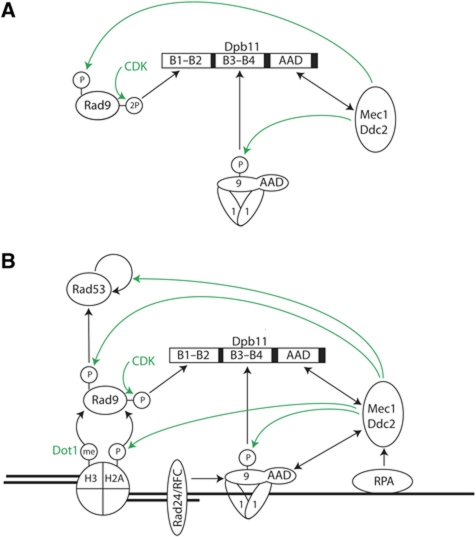
Our work, taken with previous work in budding and fission yeast as well as Xenopus, argues that Dpb11 plays a crucial role in integrating cell cycle and DNA damage signals to ensure correct spatial and temporal checkpoint activation. Dpb11 accomplishes this by providing a scaffolding function for Rad9, 9-1-1 and Mec1–Ddc2 and activating the Mec1–Ddc2 kinase specifically in this context (Figure 6A).
Figure 6.
Model of Dpb11 function in the checkpoint response. (A) The Dpb11 module of checkpoint signalling: Dpb11 engages three protein domains to form a checkpoint signalling complex with Rad9, 9-1-1 and Mec1–Ddc2. In this assembly, Dpb11 coordinates specific activation of Mec1 with cell cycle-regulated recruitment of the Mec1 substrate Rad9. This signalling complex is presumably localized to DNA damage sites by the loaded 9-1-1 complex and RPA-bound Mec1–Ddc2. (B) Holistic view of DNA damage checkpoint signalling upstream of Rad53 activation. The Dpb11 module is integrated with cell cycle- and Dpb11-independent modules: Binding of Rad9 to histones, which are H3-K79 methylated and H2a-S129 phosphorylated can localize Rad9 to DNA damage sites. The Mec1 kinase can be activated Dpb11-independently by the Ddc1 subunit of the 9-1-1 complex. It remains to be clarified, if crosstalk of Dpb11-dependent and -independent modules exists in WT cells.
Our data indicate that Rad9 specifically interacts with Dpb11 after phosphorylation by CDK, thus promoting interaction during S, G2 and M phases of the cell cycle. We identified two key CDK phosphorylation sites on Rad9 (S462 and T474) that bind directly to the N-terminal BRCT repeats 1 and 2 of Dpb11 and are necessary and sufficient for Rad9–Dpb11 interaction. Fusion of Rad9 to Dpb11 bypassed the requirement for CDK phosphorylation of Rad9 and supported Dot1-independent checkpoint signalling even during G1 phase, when CDK is inactive. Thus, CDK phosphorylation of Rad9 is crucial for coordinating DNA damage signalling with the cell cycle. CDK phosphorylation of S11 of Rad9 has recently been implicated in the Dpb11 interaction (Granata et al, 2010). However, peptides containing phospho S11 did not interact with Dpb11 (Supplementary Figure S3) and rad9 mutants in which the N-terminal cluster of CDK sites has been deleted can still interact with Dpb11 (Figure 3D) while full-length Rad9 ST462,474AA cannot interact with Dpb11 (Figure 3F and H). Consequently, phosphorylation of S11 appears to play no direct role in Dpb11 binding. S11 phosphorylation may, of course, contribute in some way indirectly, similar to Sld2 where phosphorylation of a cluster of CDK sites is required for the phosphorylation of T84, which constitutes the direct Dpb11 binding site (Tak et al, 2006). The region around S462 in Rad9 aligns well with T215 of the Schizosaccharomyces pombe Rad9 orthologue, Crb2 (Supplementary Figure S5C), which has previously been implicated in binding of the fission yeast Dpb11 orthologue, Cut5/Rad4 (Du et al, 2006). There are two potential CDK sites in Crb2 downstream of T215 which have some homology with T474 of Rad9 and it will be interesting to know if they also contribute to Cut5/Rad4 binding. Both Rad9 and Sld3 contain two CDK phospho-sites required for efficient interaction with BRCT1&2 of Dpb11. The recent crystal structure of TopBP1 shows that BRCT repeats 1 and 2 each have potential phosphopeptide binding sites (Rappas et al, 2010). We speculate that these two phosphorylation sites may each bind to different BRCT repeats, though further work is required to verify this.
Consistent with previous work, we found a domain of Dpb11 distal to the last BRCT repeat that binds to and activates the Mec1/Ddc2 kinase. While there is very little sequence similarity between this domain and the ATR activating domain (AAD) from TopBP1, we identified two motifs that contain conserved aromatic residues, which are critical for both interaction and activation, similar to the AAD in Xenopus TopBP1. Both Mec1 and Ddc2 are required for interaction with Dpb11, suggesting that either interaction occurs near the interface between Mec1 and Ddc2 or conformation changes that occur when Mec1 and Ddc2 interact are required to generate a Dpb11 binding site on either Mec1 or Ddc2. How the Dpb11 AAD activates Mec1–Ddc2 is unknown, but the presence of conserved aromatic residues in the AADs of Dpb11, TopBP1 and Ddc1 suggests a common mechanism (Kumagai et al, 2006; Navadgi-Patil and Burgers, 2009). Interestingly, a Mec1-phosphorylation site (T731) in Dpb11 has been identified that is required for activation of Mec1 (Mordes et al, 2008) and might be part of the AAD motif. This suggests that AADs may not work constitutively, but may be regulated by post-translational modifications.
Because dpb11ΔC mutants are viable at all temperatures tested, our results demonstrate that the C-terminus of Dpb11 including the AAD is not required for viability. Thus, while Mec1 and Ddc2 are essential, their activation by Dpb11 is not essential. Because deletion of the other AAD-containing protein, Ddc1, is not lethal in combination with dpb11ΔC, either Mec1 activation is not essential or there are other as yet unidentified Mec1-activating proteins in yeast. We favour this latter possibility because the dpb11ΔC ddc1WW352,544AA double mutant is not as sensitive to phleomycin as the rad9Δ mutant and because Rad53 activation is only partly defective in this double mutant (Supplementary Figure S9B). The viability of dpb11ΔC also shows that the C-terminus of Dpb11 does not have an essential role in DNA replication at any temperature.
Our biochemical and genetic data indicate that Dpb11 must be able to interact with both CDK-phosphorylated Rad9 and AAD-activated Mec1–Ddc2 for efficient Rad9 phosphorylation by Mec1–Ddc2 and efficient checkpoint activation. In this regard, Dpb11 appears to act analogously to the archetypal scaffold protein, Ste5, which provides a scaffold for mating type signalling and also specifically activates the bound Fus3 kinase (Choi et al, 1994; Bhattacharyya et al, 2006; Hao et al, 2008; Good et al, 2009). Dpb11 also acts as a scaffold protein in DNA replication, bridging interactions between Sld3 and Sld2 (Tanaka et al, 2007; Zegerman and Diffley, 2007). Although the C-terminal AAD is not required for replication, it is interesting to consider that Dpb11 may, by analogy to its role in checkpoints, play roles in the initiation process beyond simple scaffolding.
Figure 6B considers our results in the broader context of checkpoint activation. After ds break formation and resection, RPA binds to ssDNA and recruits Mec1–Ddc2 (Rouse and Jackson, 2002; Zou and Elledge, 2003; Ball et al, 2007). And separately, the 9-1-1 complex is loaded onto the recessed 5′ end by the Rad24/RFC complex (Ellison and Stillman, 2003; Majka and Burgers, 2003; Zou et al, 2003; Majka et al, 2006a). The phosphorylation of the Ddc1 subunit of the 9-1-1 complex is critical for the function of Dpb11 in checkpoint signalling since the ddc1 T602A mutant is epistatic to both dpb11ΔC and rad9 ST462,474AA. This, however, creates an apparent paradox: Mec1–Ddc2 activation requires the AAD of Dpb11, yet Dpb11 cannot be recruited to ds breaks until Ddc1 is phosphorylated by Mec1–Ddc2. It is possible that direct recruitment of Dpb11 to Mec1/Ddc2 via interaction with the AAD results in sufficient activation of Mec1–Ddc2 to promote Ddc1 phosphorylation, which supports further Dpb11 recruitment and Mec1–Ddc2 activation. Alternatively, the AAD of Ddc1 (or some, as yet unidentified AAD) may play some priming role in Mec1–Ddc2 activation, allowing subsequent Ddc1 phosphorylation and Dpb11 recruitment. Furthermore, the ATM orthologue Tel1 may be involved in this phosphorylation event thus potentially establishing crosstalk between the two apical checkpoint kinases. Finally, Mec1–Ddc2, recruited via RPA, may have sufficient activity prior to AAD interaction to phosphorylate Ddc1, promoting Dpb11 recruitment and complete activation. Further work is required to resolve this issue.
The role of Ddc1 in checkpoint activation is, however, more complex. The Ddc1 AAD mutant (ddc1WW352,544AA) is epistatic to dot1Δ, not to dpb11ΔC (Supplementary Figure S9B), suggesting that the Ddc1 AAD acts primarily within the Dot1 module of the pathway. This suggests that Ddc1 plays different roles in the two pathways: it is required to recruit Dpb11 via Ddc1 phosphorylation in the Dpb11 pathway, but it is required to directly activate Mec1–Ddc2 in the Dot1 pathway. Currently we can only speculate, why Ddc1 cannot simultaneously bind to Dpb11 and activate Mec1: both interactions involve the C-terminal domain of Ddc1 and may be due to steric constraints.
There is evidence for alternative routes to checkpoint activation, at least in mutant backgrounds. For example, at high concentrations of phleomycin, G1 cells can activate Rad53 even in dot1Δ mutants (Supplementary Figure S10). This activation requires the AAD of Dpb11 but not the CDK phosphorylation sites in Rad9, and is therefore most likely mediated by a Dpb11 subcomplex. It is currently unclear whether such subcomplexes (Rad9–Dpb11–9-1-1 or 9-1-1–Dpb11–Mec1–Ddc2) are relevant for checkpoint activation in wild-type cells, since their function can only be observed in mutant backgrounds and/or at high amounts of DNA damage and results only in partial checkpoint activation.
Once Rad9 is recruited, Mec1–Ddc2 phosphorylates it, which promotes both oligomerization of Rad9 via its BRCT domain and recruitment of Rad53 via its FHA domains (Emili, 1998; Sun et al, 1998; Vialard et al, 1998; Soulier and Lowndes, 1999; Sweeney et al, 2005; Usui et al, 2009). Mec1–Ddc2-dependent phosphorylation of Rad53 allows further oligomerization and autoactivation (Sanchez et al, 1996; Sun et al, 1996). Understanding in detail how Rad53 gets activated will ultimately require reconstitution of this complex network with purified proteins. Our partial reconstitution of the Dpb11 module will hopefully contribute towards this goal.
Materials and methods
Strains, plasmids, antibodies and proteins
All yeast strains are based on W303; details are listed in Supplementary Table 1. N-terminal truncations (451–1309, 471–1309 and 541–1309) of Rad9 were created by deletion of the corresponding sequence from the endogenous gene and integration of the RAD9 promoter and new start codon at the site of truncation. Additionally, a 9myc tag was integrated at the C-terminus. C-terminal truncations (1–750, 1–540 and 1–450) of Rad9 were generated through deletion and integration of the 9myc tag at the position of truncation. Unless specifically indicated yeast strains were grown in rich medium at 30°C. Details on plasmid constructs can be found in Supplementary Table 2. Antibodies used in this study are listed in Supplementary Table 3. Purification protocols for proteins used in this study can be found in Supplementary data.
Protein interaction techniques
The initial pulldown was performed with ∼1 nmol Dpb11–His bound to Ni-NTA magnetic beads (Qiagen) and 6 ml of DNAse treated (1500 U, 30′, 4°C) yeast extract (300 mM KOAc, 5 mM Mg2SO4 1 mM CaCl2, 25 mM Hepes pH 7.6, 10% glycerol, 0.02% NP-40, 2 mM β-Me, protease inhibitors) corresponding to 5 × 1010 asynchronously dividing cells. Bound proteins were eluted with 500 mM imidazole and concentrated by TCA precipitation. 10% of the bound sample was loaded on 4–12% NuPAGE (Figure 1A), which was silver stained. 90% of the sample was run under identical conditions and stained with colloidal Coomassie. The gel was cut into 30 slices and after trypsin digestion bound proteins were identified by ESI ion trap MS.
For small-scale pulldowns, GST–Dpb11 or GST-tagged protein fragments were immobilized on glutathione sepharose 4B (GE Healthcare) and incubated with 600 μl ammonium sulphate precipitated (57%) cell extracts (buffer for binding experiments with Ddc2: 300 mM KOAc, 25 mM Hepes pH 7.6, 10% glycerol, 0.02% NP-40, 2 mM β-Me, protease inhibitors; for binding experiments with Rad9: 200 mM KOAc, 100 mM Hepes pH 7.6, 10% glycerol, 0.02% NP-40, 2 mM β-Me, 20 mM β-glycerophosphate, 10 mM NaF, 100 μM okadaic acid, protease inhibitors) corresponding to 1 × 109 cells.
For pulldowns with Dpb11 and purified Rad9, the procedure was as described above, but instead of cell extract GST–Dpb11 beads were incubated with 6 pmol MBP–Rad9, which was quantitatively pre-phosphorylated with btCyclin AΔN170-hsCdk2 (Brown et al, 1995).
In order to test ternary complex formation, immunopurified Mec1–18myc–Ddc2 (protocol in Supplementary data) was used to pull down 0.3 pmol MBP–Rad9 and 0, 1, 3, 10 pmol Dpb11 FL-His or GST–Dpb11-C in buffer containing 500 mM KOAc, 100 mM Hepes pH 7.6, 10% glycerol, 0.02% NP-40, 2 mM β-Me, 20 mM β-glycerophosphate, 10 mM NaF, 100 μM okadaic acid and protease inhibitors.
Peptide binding was investigated with 35mer peptides corresponding to endogenous Rad9 sequence, harbouring a phosphorylated amino acid at position 26, an N-terminal EAhx-linker and biotin tag. 75 μl Streptavidin Dynabeads (Dynal) were saturated with peptide, treated with λ-phosphatase or left untreated, and used to pull down 40 pmol GST–Dpb11-N (aa 1–276) in buffer containing 500 mM KOAc, 100 mM Hepes pH 7.6, 10% glycerol, 0.02% NP-40 and 2 mM β-Me.
In vivo checkpoint assays
To investigate checkpoint activation in vivo, yeast strains were arrested in G1 with α-factor or in G2/M with nocodazole. Phleomycin was added to 50 μg/ml (or the indicated concentration) and samples corresponding to 2 × 107 cells were taken at indicated time points and subjected to TCA precipitation. Rad53 activation was measured by its phosphorylation-dependent shift in electrophoretic mobility. For determination of survival, cells were released from cell-cycle block simultaneously with addition of phleomycin. At indicated time points, cells were diluted to 2 × 106 cells/ml. This solution as well as five 1:5 dilutions were spotted on plates without drugs and incubated for 2 days.
In vitro kinase assays
Mec1 kinase assays were performed as described (Mordes et al, 2008), with small modifications. A protocol of the Mec1–18myc–Ddc2 preparation can be found in Supplementary data. 10 μl of bead-bound Mec1–18myc–Ddc2 was incubated for 30′ at 30°C with ∼0.5 pmol CDK pre-phosphorylated MBP–Rad9 or MBP–Rad9–AA and 0.75 pmol GST–Dpb11 or GST–Dpb11-C (0.75, 0.25 or 0.075 pmol in Figure 2E) and/or 15 pmol PHAS1 (60 pmol in Figure 2F) in 40 μl kinase reactions (100 mM KOAc, 10 mM Hepes pH 7.6, 50 mM β-glycerophosphate, 10 mM MgCl2, 10% glycerol, 2 mM β-Me, 10 mM ATP, 5 μCi γP32-ATP). Phosphorylation of Rad9 and PHAS1 was visualized by autoradiography; equal loading of the substrates was confirmed by western blots (Coomassie stain in Figure 2F).
Supplementary Material
Acknowledgments
Mark Skehel and his Protein Analysis and Proteomics laboratory are thanked for mass spectrometry. We thank P Burgers, J Downs, T Hunt, MP Longhese, N Lowndes and S Jackson for antibodies, constructs and yeast strains; and D Boos for purified Cyclin AΔN170-Cdk2. We thank members of the Diffley laboratory for stimulating discussion and critical reading of the manuscript. BP was supported by a Human Frontiers Science Program Long-Term Fellowship (LT00336/2007-L) and an EMBO Long-Term Fellowship (ALTF 624-2006). Work in the laboratory of JD is supported by Cancer Research UK and by grants from the European Research Council (EUKDNAREP 249883) and the Association for International Cancer Research (10-0270).
Author contributions: BP and JFXD conceived and designed experiments, analysed the data and wrote the manuscript. BP performed all the experiments.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D (2007) Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol 27: 3367–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Myers JS, Cortez D (2005) ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell 16: 2372–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA (2006) The Ste5 scaffold allosterically modulates signaling output of the yeast mating pathway. Science 311: 822–826 [DOI] [PubMed] [Google Scholar]
- Bonilla CY, Melo JA, Toczyski DP (2008) Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell 30: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boos D, Sanchez-Pulido L, Rappas M, Pearl LH, Oliver AW, Ponting CP, Diffley JF (2011) Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr Biol 21: 1152–1157 [DOI] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Garman EF, Wakatsuki S, Mitchell E, Rasmussen B, Hunt T, Johnson LN (1995) The crystal structure of cyclin A. Structure 3: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Choi KY, Satterberg B, Lyons DM, Elion EA (1994) Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell 78: 499–512 [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM (2007) The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 21: 1472–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouets C, Santocanale C, Drury LS, Perkins G, Foiani M, Plevani P, Diffley JF (1998) Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase alpha. EMBO J 17: 4139–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P (2006) Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V, Stillman B (2003) Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol 1: E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A (1998) MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell 2: 183–189 [DOI] [PubMed] [Google Scholar]
- Esashi F, Yanagida M (1999) Cdc2 phosphorylation of Crb2 is required for reestablishing cell cycle progression after the damage checkpoint. Mol Cell 4: 167–174 [DOI] [PubMed] [Google Scholar]
- FitzGerald JE, Grenon M, Lowndes NF (2009) 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans 37: 897–904 [DOI] [PubMed] [Google Scholar]
- Furuya K, Poitelea M, Guo L, Caspari T, Carr AM (2004) Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev 18: 1154–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM (2005) Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst) 4: 1227–1239 [DOI] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280: 9879–9886 [DOI] [PubMed] [Google Scholar]
- Good M, Tang G, Singleton J, Remenyi A, Lim WA (2009) The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell 136: 1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granata M, Lazzaro F, Novarina D, Panigada D, Puddu F, Abreu CM, Kumar R, Grenon M, Lowndes NF, Plevani P, Muzi-Falconi M (2010) Dynamics of Rad9 chromatin binding and checkpoint function are mediated by its dimerization and are cell cycle-regulated by CDK1 activity. PLoS Genet 6: pii: e1001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenon M, Costelloe T, Jimeno S, O’Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF (2007) Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24: 105–119 [DOI] [PubMed] [Google Scholar]
- Hammet A, Magill C, Heierhorst J, Jackson SP (2007) Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep 8: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, Nayak S, Behar M, Shanks RH, Nagiec MJ, Errede B, Hasty J, Elston TC, Dohlman HG (2008) Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell 30: 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, Haber JE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235 [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H (1998) Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol 18: 6102–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG (2006) TopBP1 activates the ATR-ATRIP complex. Cell 124: 943–955 [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG (2007) The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J Biol Chem 282: 28036–28044 [DOI] [PubMed] [Google Scholar]
- Majka J, Binz SK, Wold MS, Burgers PM (2006a) Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem 281: 27855–27861 [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM (2003) Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA 100: 2249–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM (2006b) The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24: 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J, Toczyski D (2002) A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol 14: 237–245 [DOI] [PubMed] [Google Scholar]
- Mochida S, Esashi F, Aono N, Tamai K, O’Connell MJ, Yanagida M (2004) Regulation of checkpoint kinases through dynamic interaction with Crb2. EMBO J 23: 418–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D (2008) Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci USA 105: 18730–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Du LL, Redon C, Russell P (2004) Histone H2A phosphorylation controls Crb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol 24: 6215–6230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM (2008) Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem 283: 35853–35859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM (2009) The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell 36: 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Feng Q, Wang H, Erdjument-Bromage H, Tempst P, Zhang Y, Struhl K (2002) Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev 16: 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrilla-Castellar ER, Arlander SJ, Karnitz L (2004) Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst) 3: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M (2008) Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol 28: 4782–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappas M, Oliver AW, Pearl LH (2010) Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1. Nucleic Acids Res 39: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2002) Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell 9: 857–869 [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360 [DOI] [PubMed] [Google Scholar]
- Schwartz MF, Duong JK, Sun Z, Morrow JS, Pradhan D, Stern DF (2002) Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell 9: 1055–1065 [DOI] [PubMed] [Google Scholar]
- Shimada K, Pasero P, Gasser SM (2002) ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev 16: 3236–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulier J, Lowndes NF (1999) The BRCT domain of the S. cerevisiae checkpoint protein Rad9 mediates a Rad9-Rad9 interaction after DNA damage. Curr Biol 9: 551–554 [DOI] [PubMed] [Google Scholar]
- Sun Z, Fay DS, Marini F, Foiani M, Stern DF (1996) Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev 10: 395–406 [DOI] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF (1998) Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281: 272–274 [DOI] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D (2005) Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 15: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Tak YS, Tanaka Y, Endo S, Kamimura Y, Araki H (2006) A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2-Dpb11. EMBO J 25: 1987–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H (2007) CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445: 328–332 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF (2003) A central role for DNA replication forks in checkpoint activation and response. Mol Cell 11: 1323–1336 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Usui T, Foster SS, Petrini JH (2009) Maintenance of the DNA-damage checkpoint requires DNA-damage-induced mediator protein oligomerization. Mol Cell 33: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Vialard JE, Gilbert CS, Green CM, Lowndes NF (1998) The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J 17: 5679–5688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Elledge SJ (2002) Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics 160: 1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25: 8430–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF (2007) Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445: 281–285 [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ (2003) Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA 100: 13827–13832 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.