Abstract
The p53 tumour suppressor protein is a transcription factor that prevents oncogenic progression by activating the expression of apoptosis and cell-cycle arrest genes in stressed cells. The stability of p53 is tightly regulated by ubiquitin-dependent degradation, driven mainly by the ubiquitin ligase MDM2. In this study, we have identified USP42 as a DUB that interacts with and deubiquitinates p53. USP42 forms a direct complex with p53 and controls level of ubiquitination during the early phase of the response to a range of stress signals. Although we do not find a clear role for USP42 in controlling either the basal or fully activated levels of p53, the function of USP42 is required to allow the rapid activation of p53-dependent transcription and a p53-dependent cell-cycle arrest in response to stress. These functions of USP42 are likely to contribute to the repair and recovery of cells from mild or transient damage.
Keywords: deubiquitination, MDM2, p53, USP42
Introduction
The tumour suppressor protein p53 is activated in response to a range of cellular stresses and functions predominantly as a transcription factor, inducing the expression of genes involved in cell-cycle arrest and DNA repair (e.g., p21) or apoptosis (e.g., PUMA and Bax) (Riley et al, 2008). In addition, p53 can also function in the cytoplasm as a direct activator of apoptosis (Vaseva and Moll, 2009) or as an inhibitor of autophagy (Tasdemir et al, 2008). p53 contributes to tumour suppression in a number of ways. In response to mild or transient stress, p53 can induce a rapid cell-cycle arrest, contributing to the ability of cells to undergo repair and survival. However, under conditions of sustained or irreparable damage, p53 can help to eliminate cells through the induction of apoptosis (Aylon and Oren, 2007). Both of these responses prevent the accumulation of abnormal cells and so help to inhibit malignant progression. Accordingly, loss of p53 activity occurs in most, if not all cancers, with ∼50% of tumours containing inactivating mutations within the p53 gene (Hainaut and Hollstein, 2000).
In the absence of stress, p53 must be maintained at low levels. Aberrant expression of p53 during normal development results in phenotypes ranging from early embryonic lethality to premature ageing (Vousden and Lane, 2007). However, it is also essential that p53 is rapidly stabilized in response to stress, in order to prevent the proliferation of damaged cells. Regulation of p53 stability occurs predominantly through polyubiquitination and degradation by the 26S proteasome and many E3 ubiquitin ligases have now been described for p53 (Horn and Vousden, 2007). However, the best characterized E3 for p53 is MDM2, which is itself a transcriptional target of p53 (Bond et al, 2005). The importance of MDM2-mediated ubiquitination of p53 has been clearly demonstrated in mouse models, where the embryonic lethality resulting from either loss of MDM2 expression, or knockin of a catalytically inactive MDM2, is completely rescued by a simultaneous deletion of p53 (Jones et al, 1995; Montes de Oca Luna et al, 1995; Itahana et al, 2007). Stabilization of p53 in response to stress is believed to result from either a reduction in the affinity of p53 for MDM2 or by inactivation of the E3 ubiquitin ligase activity of MDM2 by proteins such as ribosomal proteins or p14ARF (Horn and Vousden, 2007).
Protein ubiquitination is a reversible process and several families of enzymes have been described which possess deubiquitinating activity, including the ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumour proteases (OTU), Machado-Joseph disease proteins (MJD) and the Jab1/MPN/Mov34 metalloenzymes (JAMM) (Nijman et al, 2005). The deubiquitinating enzymes (or DUBs) have been shown to play a role in the cleavage of ubiquitin from translational precursors and in the maintenance of free ubiquitin levels within the cell. However, DUBs can also remove both monoubiquitin and polyubiquitin chains from proteins, or can trim the distal ubiquitin from polyubiquitin chains. Consequently, these activities can potentially antagonize the functions of ubiquitination within the cell (Komander et al, 2009).
A number of DUBs have been shown to influence p53 stability and activity. The herpes virus-associated USP (HAUSP or USP7) can bind, deubiquitinate and stabilize p53 (Li et al, 2002). However, HAUSP also deubiquitinates MDM2 and reduction of HAUSP levels, either by RNA interference or by gene deletion, produces a complex phenotype (Cummins and Vogelstein, 2004; Li et al, 2004; Meulmeester et al, 2005). Deletion of the HAUSP gene or extensive knockdown by RNAi results in an almost complete loss of MDM2 and consequently significant stabilization of p53 and cell death. In contrast, a more modest reduction in HAUSP causes a decrease in both MDM2 and p53 stability, suggesting that, under these conditions, sufficient MDM2 remains to degrade p53. USP10 is a cytoplasmic DUB that relocalizes to the nucleus in response to DNA damage, where it both stabilizes p53 and prevents nuclear export of p53, so contributing to p53-mediated apoptosis (Yuan et al, 2010). USP29 has been shown to be transcriptionally induced following oxidative stress, when it contributes to the full induction of a p53 response (Liu et al, 2011). Other DUBs involved in the p53 pathway include the MDM2-specific DUB USP2a (Stevenson et al, 2007) and USP5, which degrades K48-linked polyubiquitin chains and so indirectly regulates levels of p53 (Dayal et al, 2009).
We screened a DUB siRNA library and identified the ubiquitin-specific protease USP42 as a novel deubiquitinating enzyme for p53. USP42 is a nuclear protein that is required for the rapid stabilization of p53 and the early activation of a p53 response.
Results
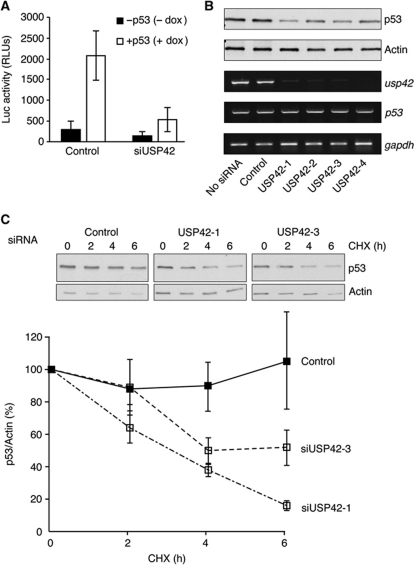
A cell-based screen for modulators of p53 activity was established by stably expressing a p53-dependent luciferase construct (PG13 luciferase) in a p53-inducible Saos-2 cell line. Using this cell line to screen an siRNA library directed against 92 known and predicted deubiquitinating enzymes (DUBs), we identified USP42 as a potential regulator of p53 activity. Pre-treatment of the reporter cells with an siRNA pool directed against USP42 lead to a clear inhibition of p53-induced luciferase expression compared with control siRNA-transfected cells (Figure 1A). Deconvolution of the siRNA pool showed that each individual siRNA could reduce USP42 mRNA levels and dampen the induction of p53 protein in these cells (Figure 1B). While no change in p53 mRNA expression was seen following USP42 knockdown (Figure 1B), a reduction in the half-life of this ectopically expressed p53 protein (which is generally much more stable than endogenous p53) was observed (Figure 1C), suggesting that USP42 can play a role in regulating p53 stability.
Figure 1.
Identification of USP42 as a regulator of p53 stability. (A) p53-inducible Saos-2 cells stably expressing PG13-luciferase reporter were transfected with control or a pool of USP42 siRNA oligonucleotides. Forty-eight hours post transfection, p53 was induced by treatment with 1 μg/ml doxycycline. Cells were lysed 24 h after p53 induction and luciferase activity was determined using a luciferase assay system (Promega). (B) p53-inducible Saos-2 cells were transfected with control or individual USP42 siRNA oligonucleotides. Forty-eight hours post transfection, p53 was induced by treatment with 1 μg/ml doxycycline. Total cell lysates were prepared 24 h after p53 induction. Lysate proteins were resolved by SDS–PAGE and analysed by western blotting with anti-p53 DO1 and Actin antibodies. (C) p53-inducible Saos-2 cells were transfected with control or USP42 siRNA oligonucleotides. Forty-eight hours post transfection, p53 was induced by treatment with 1 μg/ml doxycycline, then 24 h after p53 induction, cells were treated with 100 μg/ml cyclohexamide for the indicated times. Total cell lysates were resolved by SDS–PAGE and analysed by western blotting with anti-p53 and anti-Actin antibodies. Quantification of three independent experiments is shown in the graph below.
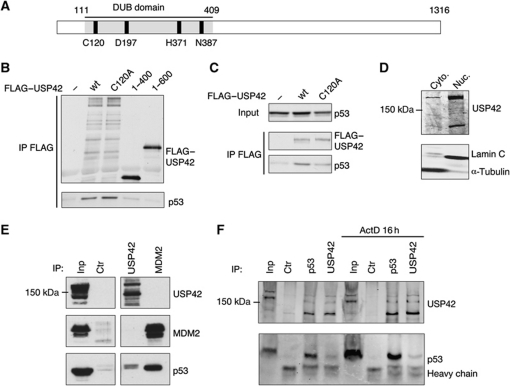
USP42 is a 1316 amino-acid protein (Figure 2A) that has been shown to be expressed in many tissues and can cleave ubiquitin in vitro and in cells (Kim et al, 2007). Expression constructs for full-length USP42, a catalytic domain mutant (C120A) and two C-terminal deletions (1–400 and 1–600) were generated and tested for their ability to bind p53 in vitro (Figure 2B). While both wild-type and the catalytic domain mutant interacted with p53, this interaction was lost in both deletion mutants, suggesting that p53 interacts with the C-terminal region of USP42. An interaction between both wild-type and C120A USP42 was also evident following co-transfection into U2OS cells (Figure 2C). Finally, we examined the interaction of the endogenous proteins. Like p53, USP42 is a predominantly a nuclear protein (Figure 2D), and we were able to detect an interaction between endogenous p53 and endogenous USP42 in U2OS cells (expressing wild-type p53) (Figure 2E). In these experiments, an interaction between MDM2 and p53 could be clearly detected, but MDM2 was not seen associated with the USP42 complex. The interaction of MDM2 and p53 is modulated in response to stress signals, and to determine whether the binding of USP42 with p53 is similarly regulated we carried out co-immunoprecipitation experiments after treatment of cells with actinomycin D, to induce stress (Figure 2F). We were able to detect the p53/USP42 complex following either USP42 or p53 immunoprecipitation. However, although overall p53 levels were increased by treatment with actinomycin D, there was no clear enhancement of the USP42/p53 interaction in response to this stress.
Figure 2.
USP42 binds p53. (A) Schematic showing the DUB domain and amino acids, which are predicted to be important for catalytic activity within USP42. (B) Flag-tagged USP42 and p53 constructs were in-vitro translated and allowed to interact for 1 h. Immunoprecipitations were performed using anti-Flag M2. Precipitated proteins were resolved by SDS–PAGE and analysed by western blotting with anti-Flag M2 and anti-p53 CM1 antibodies. (C) U2OS cells were transfected with p53 and with wt (wild-type) or C120A GFP–Flag-tagged USP42 plasmids where indicated. Cells were lysed and immunoprecipitations were performed using anti-Flag M2. Precipitated and input proteins were resolved by SDS–PAGE and analysed by western blotting with anti-GFP and anti-p53 CM1 antibodies. (D) Western blots of cytoplasmic and nuclear fractions of U2OS cell, using polyclonal antibodies against USP42, Lamin C (a nuclear protein) and α-tubulin (a cytoplasmic protein). (E) Immunoprecipitation of p53, MDM2 and USP42 from U2OS cells, followed by western blotting of the precipitated proteins with anti-USP42, anti-p53 and anti-MDM2 antibodies. (F) Immunoprecipitation of p53 and USP42 from U2OS cells untreated or treated with actinomycin D. Figure source data can be found with the Supplementary data.
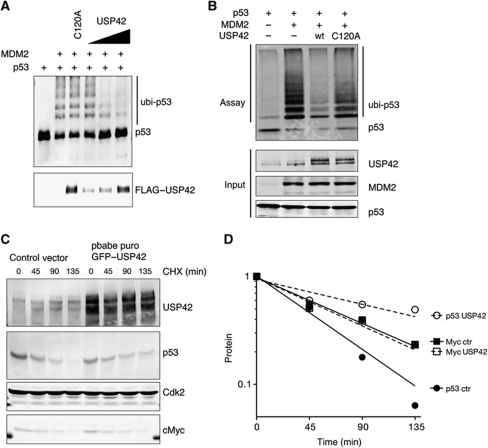
To test whether USP42 can deubiquitinate p53, we examined the effect of adding USP42 to p53 that had been ubiquitinated by MDM2 in vitro (Figure 3A). This assay showed that while wild-type USP42 efficiently removed ubiquitin from p53 in a dose-dependent manner, the catalytic site mutant USP42 C120A did not affect p53 ubiquitination. A similar reduction of MDM2-dependent ubiquitination of p53 was seen following co-transfection of USP42 into U2OS cells (Figure 3B), although again the C120A mutant failed to show this activity. These results show that USP42 can bind to p53 and remove ubiquitin conjugated in response to MDM2. Consistently, overexpression of wild-type USP42 also promoted the stabilization of p53 in U2OS cells (Figure 3C), although the half-life of Myc, another ubiquitinated and rapidly degraded protein, was not affected (Figure 3D).
Figure 3.
USP42 deubiquitininates p53. (A) In vitro translated p53 was pre-incubated for 90 min with a bacterially expressed MDM2 and subsequently immunoprecipitated with an anti-p53 antibody. Immunoprecipitated p53 was then incubated with in vitro translated wild-type (wt) or C120A Flag-tagged USP42, followed by western blotting with anti-p53 and anti-USP42. (B) U2OS cells were co-transfected with p53, MDM2, wild-type or C120A GFP–Flag–USP42 and HIS–ubiquitin, as indicated. Twenty-four hours after transfection, lysates were precipitated with HIS-tag isolation Dynabeads and western blotted for p53. The input level of each protein was assessed by western blotting (below). (C) U2OS cells were infected with wild-type GFP–Flag-tagged USP42 or control vector. Seventy-two hours after transfection, cells were selected with puromycin for 2 weeks to establish USP42 overexpressing or control cell pools. These cells were treated for the indicated time with 100 μg/ml of cyclohexamide to inhibit the translation. Cell lysates were prepared at these times and probed for USP42, p53 and cMyc protein level by western blot. Cdk2 was used as loading control. (D) Quantification of (C). Figure source data can be found with the Supplementary data.
While ectopic expression of USP42 clearly resulted in the deubiquitination and stabilization of p53, we wished to determine the role of endogenous USP42 in the regulation of endogenous p53. Database analysis of expression profiles (e.g., Ensembl and UniProt) suggested that several differentially spliced forms of USP42 can be expressed and our RT-PCR analysis of USP42 expression in U2OS cells confirmed the expression of several isoforms that show variations in the N- and C-terminal regions. This expression pattern is reflected by numerous differently sized proteins detected by western blot in these cells (Supplementary Figure S1A). Preliminary analysis of each USP42 siRNA in the screening pool showed that of the four, siRNA USP42/3 was the most efficient in reducing overall USP42 expression. This siRNAs targets exon 13 (where the epitope for the antibody is located), a region that is preserved in all the isoforms of USP42 detected in these cells. We therefore generated two further independent siRNAs within exon 13 and showed that each of these siRNA reduced USP42 expression (Supplementary Figure S2D). Further analysis was carried out with a pool of the three exon 13-targeting siRNAs, which efficiently depleted all the detectable USP42 proteins (Supplementary Figure S1A).
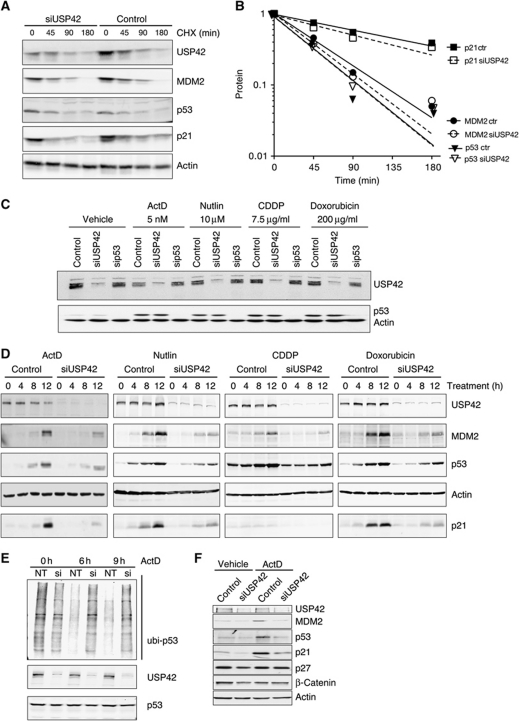
Using the exon 13 siRNA pool, we initially tested the effect of USP42 depletion on p53 levels under unstressed conditions, and found that depletion of endogenous USP42 did not affect the half-life of p53, p21 or MDM2 (Figure 4A and B). These data suggest that USP42 cannot counteract the efficient ubiquitination and degradation of p53 under normal conditions of cell growth and also support the results using USP42 overexpression (Figure 3D), indicating that USP42 does not generally regulate the stability of all ubiquitinated proteins. p53 is stabilized in response to stress, and so we considered a role for USP42 during the activation of p53 by various stress treatments (Figure 4C). We found consistently that after 24 h or longer of treatment of cells with p53 activating signals, the loss of USP42 expression did not obviously impact the levels of p53. The stress signals that activate p53 have been shown to inhibit the ubiquitination of p53 through several mechanisms, each leading to the inactivation of MDM2 (Horn and Vousden, 2007). At these later time points during the stress response, it seems likely that p53 is no longer efficiently targeted for ubiquitination, potentially reducing the effects of a deubiquitinating enzyme like USP42. We therefore considered that the major function of a deubiquitinating enzyme might be at early time points during a stress response, to cooperate with the inhibition of MDM2 and most effectively stabilize p53. We examined the effect of USP42 depletion in U2OS cells during the first 12 h of the response to various forms of p53-inducing stress, and found that in each case overall reduction in USP42 levels led to a defect in the stabilization of p53 protein (Figure 4D). A closer examination of the stress response indicated that within 16 h of actinomycin D treatment, no further effect of loss of USP42 could be detected (Supplementary Figure S2A), indicating that the importance of USP42 in modulating p53 stability is limited to the initial phases of the stress response. Importantly, p53 mRNA levels were not affected by USP42 depletion (Supplementary Figure S2B). The defect in the accumulation of p53 correlated with a delayed activation of the expression of p53-inducible target genes p21 and MDM2 (Figure 4D). This effect of USP42 knockdown was also seen in three further cells lines expressing wild-type p53; RPE, HCT116 and—to a lesser extent—RKO (Supplementary Figure S2C). Deconvolution of the siRNA pool showed that each siRNA reduced USP42 levels and prevented rapid stabilization of p53 in response to actinomycin D (Supplementary Figure S2D). To further provide evidence of specificity of the siRNAs, we showed that the reduction in p53 levels seen following depletion of endogenous USP42 can be rescued by overexpression of ectopic USP42 (Supplementary Figure S2E).
Figure 4.
USP42 regulates the stabilization and activation of the p53 response in response to several stress signals. (A) CHX assay U2OS cell extracts with antibodies against USP42, MDM2 p53, p21 and actin as a loading control. Cells were treated with non-targeting siRNA or a pool of siRNA against USP42 for 48 h before being treated for the indicated time with 100 μg/ml of cyclohexamide to inhibit the translation. Cell lysates were prepared at these times and analysed for the indicated proteins level by western blot. Actin was used as loading control. (B) Quantification of (A). (C) Western blot of U2OS cell extracts with antibodies against USP42, p53 and actin as a loading control. Cells were treated with non-targeting siRNA, a pool of siRNAs against exon 13 of USP42 or an siRNA against p53 and then incubated for 24 h with the indicated drugs to induce p53 expression. (D) Western blot of U2OS cell extracts with antibodies against USP42, MDM2 p53, p21 and actin as a loading control. U2OS were treated as described in (C) for the indicated time. (E) Ubiquitination assay of p53 in U2OS cells after knockdown of USP42. U2OS cells were treated with non-targeting siRNA or a pool of siRNA against USP42 and transfected with HIS–ubiquitin 12 h later. After 36 h, cells were incubated for the indicated times with actinomycin D and MG132 then harvested for HIS pulldown as described in Figure 3A. (F) Western blot of U2OS cell extracts with antibodies against USP42, MDM2 p53, p21, p27, β-catenin and actin as a loading control. U2OS were treated with the indicated siRNAs and then incubated for 10 h with vehicle or actinomycin D. Figure source data can be found with the Supplementary data.
To establish a link between the effects of USP42 depletion on p53 protein levels and ubiquitination, we examined the consequences of USP42 downregulation on p53 ubiquitination during the early stages of the stress response (Figure 4E). While USP42 knockdown had no clear effect on ubiquitination of p53 under unstressed conditions (consistent with the lack of effect on p53 levels or half-life under normal growth conditions), after 6 or 9 h actinomycin D treatment depletion of USP42 clearly prevented the efficient deubiquitination of p53 that is associated with protein accumulation. To further establish whether USP42 depletion generally regulates the stability of ubiquitinated proteins under stressed conditions, we examined the effect of USP42 knockdown on other nuclear proteins that are subject to ubiquitin-dependent degradation. Under circumstances where defects in the accumulation of the p53 transcription targets p21 and MDM2 were observed, we did not see a difference in levels of p27 or β-catenin (Figure 4F), indicating that modulation of USP42 does not generally affect the stability of all ubiquitinated proteins.
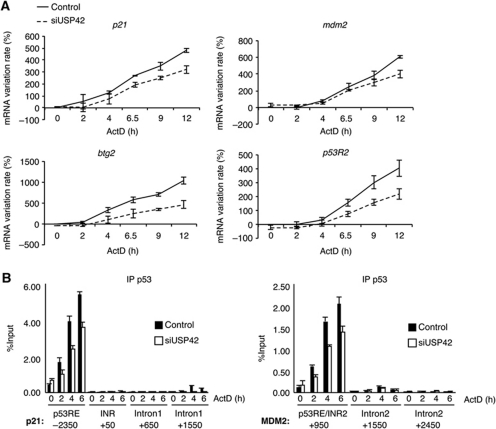
Our data show that USP42 can help in the stabilization and activation of p53 in the initial phases of a stress response. Although we showed that USP42 depletion does not generally affect protein stability (Figure 4A and F), we wished to confirm that the changes in p21 and MDM2 protein levels detected in Figure 4D were the results of decreased p53-mediated transcriptional activity. We, therefore, carried out time course experiments to examine the activation of a number of different p53 target genes by qRT–PCR, examining the changes in gene expression within 12 h of treatment of cells with actinomycin D to induce the p53 response. Most of the genes examined showed a slower induction of mRNA expression following USP42 depletion, correlating with the delayed stabilization of p53 (Figure 5A; Supplementary Figure S3A). We also examined the recruitment of p53 to promoters by chromatin immunoprecipitation and found that consistent with the reduced induction of gene expression, cells depleted of USP42 accumulated less p53 to the p21 and MDM2 promoter during the early phase of the p53 response (Figure 5B). The recruitment of p53 to responsive element regulating p21 expression in response to actinomycin D treatment also correlated with the detection of an elongating form of RNA polymerase II at the coding sequence of p21, which was clearly lower in USP42-depleted cells (consistent with an impaired activation of p21 expression; Supplementary Figure S3B).
Figure 5.
USP42 depletion delays activation of several p53 target genes in response to stress. (A) U2OS cells were transfected with siRNAs against USP42 or non-targeting siRNA. Cells were treated for the indicated time with 5 nM actinomycin D to activate p53. mRNA expression of the indicated genes was then determined by qRT–PCR with specific primers; mRNA variation rate indicates the percentage activation or repression of expression of each gene compared with control. The results were normalized against two different standard genes and the graphs represent the mean of three independent experiments. (B) U2OS cells treated as in (A) were fixed and chromatin was immunoprecipitated with an antibody against p53. p53 bound DNA was then analysed by qPCR with specific primer amplifying the indicated region of the p21 gene. The results are expressed as a percentage of input and the mean of three experiments.
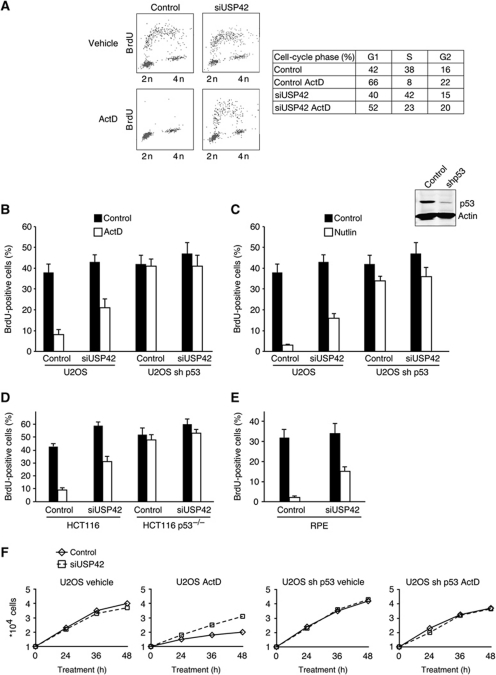
To look more closely at the effect of USP42 depletion on cell growth and survival, we examined cell-cycle progression of wild-type p53 expressing cells in response to actinomycin D treatment. Depletion of USP42 in normally cycling cells had no clear effect on the ability of cells to enter S-phase (as measured by BrdU incorporation), while treatment of control cells with actinomycin D or Nutlin resulted in a clear loss of S-phase and accumulation of cells in G1 (Figure 6A–C) reflecting the activation of p53. This arrest was dependent on p53, as shown by failure to decrease S-phase cells in p53-depleted cells treated with Nutlin and actinomycin D (Figure 6B and C). Knockdown of USP42 clearly impeded the activation of the cell-cycle arrest, significantly increasing the retention of S-phase cells in actinomycin D or Nutlin-treated cells, consistent with a weaker activation of p53 (Figure 6A and B). This USP42-dependent differentiation in cell-cycle progression was lost in p53-depleted cells (Figure 6B and C). Similar effects of USP42 depletion on cell-cycle progression were also seen in two other wild-type p53 expressing cell lines, HCT116 (Figure 6D) and RPE (Figure 6E). To examine the overall effect of USP42 depletion on the growth of cells following p53-activating stress, we examined the proliferation of U2OS cells exposed to low levels of actinomycin D (Figure 6F). In U2OS cells stably expressing an shRNA against p53, depletion of USP42 did not affect the growth rate of the cells. However, while control cells expressing wild-type p53 and exposed to actinomycin D showed almost no proliferation over 48 h, the growth was rescued to some extent by knockdown of USP42, although the growth rate did not revert back to that seen in untreated cells, or cells lacking p53.
Figure 6.
USP42 is required for efficient p53-mediated cell-cycle arrest. (A) Cell-cycle profile of U2OS cells treated with a non-targeting siRNA or a pool of siRNAs against USP42, followed by vehicle or actinomycin D treatment for 16 h. (B) U2OS cells overexpressing a control vector or a vector coding for an shRNA against p53 treated with a non-targeting siRNA or a pool of siRNAs against USP42 were treated 16 h with 5 nM of actinomycin D. During the last 90 min, cells were pulsed with BrdU, then analysed by flow cytometry for BrdU incorporation and DNA content. (B) A graph representative of three independent experiments performed as in (A) showing the percentage of cell in S-phase. (C) Cells depleted for USP42 as in (A) were incubated with Nutlin and analysed for BrdU incorporation by flow cytometry. The graph is representative of three independent experiments and shows the percentage of cells in S-phase. The western blot shows the extent of p53 depletion in the U2OS cells overexpressing the shRNA against p53. (D) HCT116 and HCT116 p53−/− cells were treated as in (B) and analysed for BrdU incorporation by flow cytometry. The graph is representative of three independent experiments and shows the percentage of cells in the subG1 fraction. (E) RPE cells were treated as in (B) and analysed for overall DNA content by flow cytometry. The graph is representative of three independent experiments and shows the percentage of cells in S-phase. (F) Growth curve of same cells as in (B) treated with vehicle or 5 nM of actinomycin D. Cells were counted at the indicated time with an automatic cell counter.
Discussion
One of the principal mechanisms by which p53 activity is regulated is through the control of protein stability. This has been shown to be mediated by proteasome-dependent degradation following the ubiquitination of p53. Many ubiquitin ligases that target p53 have been described, with MDM2 playing a predominant role both in controlling basal levels of p53 in normal unstressed cells and in re-establishing low levels of p53 following the resolution of a stress response. Deubiquitinating enzymes that cleave ubiquitin chains can also control the extent of ubiquitination of p53, and therefore its sensitivity to degradation. Recent studies have identified a number of DUBS that may help to stabilize p53, and here we show that USP42 can also function to deubiquitinate and stabilize p53. Our results show that USP42 does not affect the basal levels of p53 in unstressed cells, but that the activity of USP42 is likely to be most important during the early phase of a stress response, helping to rapidly induce p53 levels to allow recruitment of p53 to its response elements and so mount an efficient p53-dependent cell-cycle arrest. This presents the interesting possibility that USP42 is required for a rapid proliferation arrest, which allows cells to mount a repair and survival response. We did not see any changes in p53/USP42 binding in response to stress, suggesting that this interaction may not be modulated. While it is possible that stress induced modification of USP42 could alter activity towards p53, it may also be the case that the constitutive activity of USP42 plays a role only as the efficiency of MDM2-mediated ubiquitination of p53 declines in response to stress. While the effect of USP42 is somewhat subtle and limited to a small time window immediately following exposure of cells to stress, it is well established that very small changes in the response and activity of p53 can have profound overall effects. The presence of polymorphisms in the promoter of mdm2, for example, subtly alters the efficiency of MDM2 expression but can have clear effects on the cancer susceptibility (Grochola et al, 2010). We would therefore expect that the ability of USP42 to determining the efficacy of the p53 response would be important for tumour suppressor activity, and it will be interesting to determine the consequences of loss or inhibition of USP42 on development and cancer progression in vivo.
Several isoforms of USP42 can be expressed, which may reflect alternative start sites and splicing. In this study, we have focused on using siRNAs that can uniformly deplete the expression of each of these isoforms. However, we have found that siRNAs directed against the N-terminal exons of USP42 target only a subset of the USP42 isoforms, and have variable effects on the ability to impact p53 levels (data not shown). These results suggest that different USP42 isoforms vary in their competence to regulate the ubiquitination of p53, although whether this reflects an inability to bind p53 or loss of DUB activity is not yet clear. We are presently defining the various isoforms of USP42 in more detail, determining where they are expressed and how their activity relates to the stabilization of p53. Furthermore, it seems clear that given the relatively limited number of DUBs available in the cell, each will have several targets. Although we have shown some specificity of USP42 towards p53, with no indication of a clear effect of modulation of USP42 expression on other ubiquitinated proteins such as Myc, p21 and MDM2, it seems most likely that USP42 will have additional p53-independent functions.
Other recently described DUBs that can target p53 have been shown to be active under selective conditions. USP10 is normally cytoplasmic, and is driven to the nucleus where it can affect p53 localization and stability in response to DNA damage. However, unlike USP42, depletion of USP10 was shown to dampen the apoptotic response to p53 activation (Yuan et al, 2010). USP29 is induced by JTV1 and FBP-driven transcription in response to oxidative stress, and was also shown to be important for the full activation of PUMA and apoptosis (Liu et al, 2011). Both USP10 and USP29, therefore, play a role in promoting the ability of p53 to clear irreparably damaged cells through the induction of apoptosis. By contrast, we show here that USP42 is a nuclear protein that is required for the rapid activation of a p53-dependent cell-cycle arrest in response to a broad range of stress signals, including low levels of actinomycin D (functioning through the activation of ribosomal stress and release of ribosomal proteins to inhibit MDM2) (Lohrum et al, 2003), DNA damage inducing drugs (which result in the phosphorylation of p53 and inhibition of MDM2 binding) (Shieh et al, 1997) and even treatment of cells with Nutlin, a direct inhibitor of p53/MDM2 binding (Vassilev et al, 2004). This broad contribution of USP42 to the stabilization of p53 suggests that USP42 activity may be important in the initial response to all stress signals, as MDM2 activity begins to decline. However, in preliminary results we have been unable to detect a contribution of USP42 to the induction of apoptosis in the cell systems studied here, suggesting that USP42 contributes selectively to the ‘protect and repair’ functions of p53. It is, therefore, possible that inhibition of USP42 might drive a more effective p53-mediated apoptotic response in tumour types that have been shown to survive chemotherapy in a p53-dependent manner (Bertheau et al, 2008).
Materials and methods
Plasmids
Plasmids expressing human wild-type p53 and wild-type MDM2 (Chen et al, 1995) glutathione S-transferase (GST)-MDM2 (Fang et al, 2000) were described previously. Plasmids encoding FLAG–USP42 and GFP–FLAG–USP42 were generated by mRNA amplification and restriction digestion followed by ligation into PCDNA 3 FLAG or PEGFP-C1 and verification by DNA sequencing. The USP42 C120A mutation was generated by site-directed mutagenesis (QuikChange site-directed mutagenesis kit; Stratagene, La Jolla, CA) and verified by DNA sequencing.
Cell culture
All cell lines were cultured in DMEM 10% FCS in a 37°C incubator at 5% CO2. The tet-inducible p53 Saos-2 cell line was described previously (Ryan and Vousden, 1998). Actinomycin D, Nutlin, Doxorubicin, Cisplatin and Cycloheximide (all from Sigma) were used at the indicated concentrations.
siRNA and shRNA
siRNA oligonucleotides were transfected using Hiperfect (Qiagen). The siRNA library targeting USPs was obtained from Dharmacon. siRNAs targeting exon 13 were obtained from Eurofins.
USP42-1 CCCUCUUCUACCAUUACCA;
USP42-2 GACAGUGACCCGAAAGAAA;
USP42-3 GUUAAUAGGUCCUCAGUGA.
shRNA against p53 was cloned into pSUPER.retro and transfected in phoenix cell to produce retrovirus.
shRNA p53: GACTCCAGTGGTAATCTAC
In-vivo ubiquitination assays
U2OS cells were transfected with CMV-driven expression plasmids expressing HIS–Ubi, p53 MDM2 and GFP–FLAG–USP42 as indicated. Thirty-six hours after transfection, cells were treated with proteasome inhibitor MG132 (10 μM) for 3 h. Cells were collected in PBS and 5% was kept as input. The rest was centrifuged for 5 min and the cell pellet lysed in 700 μl of UBA buffer (6 M Guanidinium HCl, 300 mM NaCl, 50 mM phosphate pH 8.0, 100 μg/ml N-ethylmaleimide (NEM)) and sonicated for 5 min at 20% amplitude. Lysates were incubated overnight with Invitrogen Dynabeads His-tag matrix, once washed with UBA, UBB, UBC buffer and PBS (UBB:UBA and UBC 1:1; UBC: 300 mM NaCl, 50 mM phosphate pH 8.0, 100 μg/ml NEM) and resolved by 8% SDS–PAGE, followed by immunoblotting with α-p53 (DO-1) antibody.
In-vitro deubiquitination assays
The in-vitro ubiquitination of p53 by MDM2 was carried out as described previously (Uldrijan et al, 2007). Subsequently, in-vitro translated and precipitated FLAG–USP42 was added, followed by incubation at 37°C for 1 h. The reaction was stopped by adding 3 × SDS loading buffer and was analysed by 8% SDS–PAGE, followed by immunoblotting with DO1.
Cell fractionation
3 × 106 cells were washed in PBS and incubated for 5 min at 4°C in Cytoplasmic buffer (10 mM Hepes pH 7.9, 1.5 mM MgCl2, 10 mM KCl) followed by three freeze-thaw cycles in liquid N2. The supernatant was harvested (cytoplasmic fraction). The remaining pellet was washed and the nuclear fraction was harvested in RIPA buffer.
Antibodies and immunoblotting
Cell extracts were prepared in either NP-40 buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP-40) or RIPA buffer (50 mM Tris–HCl, pH 8.0, 120 mM NaCl, 1% NP-40, 1% SDS 0.5% DOC, protease inhibitor cocktail (Roche)). Proteins were resolved by SDS–PAGE and transferred onto nitrocellulose membrane (Millipore). Antibodies were purchased from Sigma (USP42, HPA006752) (Flag, M2) (a tubulin DM1A), Covance (HA, 16B12), Calbiochem (Mdm2 Ab1+Ab2), Santa Cruz (p21 goat C-19, p53 DO1, Lamin (346)), Millipore (Actin C4). Immunoblots were quantitated using ImageJ software or the Licor Odyssey system.
mRNA extraction and qRT–PCR
Analysis of mRNA expression was carried out using qRT-PCR, as previously described (Vigneron et al, 2010). Primer sequences used are available upon request.
Chromatin immunoprecipitation
1 × 106 cells were fixed with 1% formaldehyde and 1.5 mM EGS (Sigma) for 10 min before adding glycine at 250 mM final concentration to stop the reaction. After washing with PBS, cells were resuspended in RIPA buffer supplemented at 1% SDS and sonicated to obtain cell lysate containing nucleosomal fragment of DNA. Lysate was diluted with RIPA buffer to obtain a final concentration of 0.25% SDS, then incubated with the desired antibody (2 μg) overnight and 20 μl of magnetic beads (Dynabeads; Invitrogen). Beads were washed five times with low salt buffer, high salt buffer, LiCl buffer and twice in TE buffer. DNA was eluted from the beads in 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) supplemented with 250 μg/ml of proteinase K for 1 h at 45°C. In all, 8 μl 5 M NaCl was added, followed by overnight incubation at 67°C to reverse the crosslinking. DNA was purified (PCR purification kit; Qiagen) in 150 μl of EB buffer (Qiagen) and used for qRT–PCR with specific primer recognizing indicated region (primer sequence available upon request). Results are expressed as a percentage of input (each PCR was repeated with 0.5% of DNA input treated as the samples) and represent the mean of three experiments.
Cell-cycle analysis
For cell-cycle analysis and S-phase quantification, cells were pulsed with 20 mM BrdU for 90 min, harvested in PBS EDTA (2.5 mM EDTA), and fixed in cold methanol. Cells were then treated 20 min with 2 M HCl, washed twice in PBS containing 0.5% BSA and 0.1% Tween-20, then incubated in the same buffer with an antibody against BrdU (Santa Cruz: sc-51514) and 1 mg/ml of RNAaseA for 30 min. Cells were washed twice, incubated 20 min with an FITC-coupled secondary antibody and 1 mg/ml of RNAaseA and washed once. Cells were then analysed by flow cytometry (FACScan, Becton Dickinson) for BrdU incorporation and DNA content using propidium iodide at 50 mg/ml in PBS 0.1% Tween-20. Cells positive for BrdU with a DNA content between 2 N and 4 N were identified as cell actively replicating their DNA. Results are expressed of a percentage of cells in the whole population.
Statistical analysis
Error bars represent the standard error of the mean between the number of independent experiments detailed in the individual figure legends.
Supplementary Material
Acknowledgments
We would like to acknowledge funding from Cancer Research UK, an EMBO long-term fellowship (to AKH) and the West of Scotland Women's Bowling Association (support for SC).
Author contributions: AKH, AMV, SC and KHV conceived the study, interpreted the experiments and wrote the paper. AKH, AMV, SC and RLL designed and performed the experiments.
The authors declare that they have no conflict of interest.
12/14/2011
This article has had the additional paragraph ‘Statistical analysis’ added at the end of the Materials and methods section since Advance Online Publication.
References
- Aylon Y, Oren M (2007) Living with p53, dying of p53. Cell 130: 597–600 [DOI] [PubMed] [Google Scholar]
- Bertheau P, Espie M, Turpin E, Lehmann J, Plassa L-F, Varna M, Janin A, de The H (2008) TP53 status and response to chemotherapy in breast cancer. Pathobiology 75: 132–139 [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Levine AJ (2005) MDM2 is a central node in the p53 pathway: 12 years and counting. Curr Cancer Drug Targets 5: 3–8 [DOI] [PubMed] [Google Scholar]
- Chen J, Lin J, Levine AJ (1995) Regulation of transcription functions of the p53 tumor suppressor by the mdm-2 oncogene. Mol Med 1: 142–152 [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, Vogelstein B (2004) HAUSP is required for p53 destabilization. Cell Cycle 3: 689–692 [PubMed] [Google Scholar]
- Dayal S, Sparks A, Jacob J, Allende-Vega N, Lane DP, Saville MK (2009) Suppression of the deubiquitinating enzyme USP5 causes the accumulation of unanchored polyubiquitin and the activation of p53. J. Biol Chem 284: 5030–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Grochola LF, Zeron-Medina J, Meriaux S, Bond GL (2010) Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol 2: a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainaut P, Hollstein M (2000) p53 and human cancer; the first ten thousand mutations. Adv Cancer Res 77: 81–137 [DOI] [PubMed] [Google Scholar]
- Horn HF, Vousden KH (2007) Coping with stress: multiple ways to activate p53. Oncogene 26: 1306–1316 [DOI] [PubMed] [Google Scholar]
- Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, Bhat KP, Godfrey VL, Evan GI, Zhang Y (2007) Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell 12: 355–366 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A (1995) Rescue of embyonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208 [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim YS, Yoo KJ, Lee HJ, Lee DR, Yeo CY, Baek KH (2007) The expression of Usp42 during embryogenesis and spermatogenesis in mouse. Gene Expr Patterns 7: 143–148 [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 13: 879–886 [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648–652 [DOI] [PubMed] [Google Scholar]
- Liu J, Chung HJ, Vogt M, Jin Y, Malide D, He L, Dundr M, Levens D (2011) JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J 30: 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum MA, Ludwig RL, Kubbutat MHG, Hanlon M, Vousden KH (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3: 577–587 [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG (2005) Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 18: 565–576 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G (1995) Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206 [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A (2008) Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Ryan KM, Vousden KH (1998) Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol 18: 3692–3698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S-Y, Ikeda M, Taya Y, Prives C (1997) DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91: 325–334 [DOI] [PubMed] [Google Scholar]
- Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK (2007) The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J 26: 976–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D’Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G et al. (2008) Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldrijan S, Pannekoek WJ, Vousden KH (2007) An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J 26: 102–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva AV, Moll UM (2009) The mitochondrial p53 pathway. Biochim Biophys Acta 1787: 414–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, King N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecular antagonists of MDM2. Science 303: 844–848 [DOI] [PubMed] [Google Scholar]
- Vigneron AM, Ludwig RL, Vousden KH (2010) Cytoplasmic ASPP1 inhibits apoptosis through the control of YAP. Genes Dev 24: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z (2010) USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140: 384–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.