Abstract
Autophagic responses are coupled to the activation of the inhibitor of NF-κB kinase (IKK). Here, we report that the essential autophagy mediator Beclin 1 and TGFβ-activated kinase 1 (TAK1)-binding proteins 2 and 3 (TAB2 and TAB3), two upstream activators of the TAK1-IKK signalling axis, constitutively interact with each other via their coiled-coil domains (CCDs). Upon autophagy induction, TAB2 and TAB3 dissociate from Beclin 1 and bind TAK1. Moreover, overexpression of TAB2 and TAB3 suppresses, while their depletion triggers, autophagy. The expression of the C-terminal domain of TAB2 or TAB3 or that of the CCD of Beclin 1 competitively disrupts the interaction between endogenous Beclin 1, TAB2 and TAB3, hence stimulating autophagy through a pathway that requires endogenous Beclin 1, TAK1 and IKK to be optimally efficient. These results point to the existence of an autophagy-stimulatory ‘switch’ whereby TAB2 and TAB3 abandon inhibitory interactions with Beclin 1 to engage in a stimulatory liaison with TAK1.
Keywords: Beclin 1 interactome, mTOR, p53, pifithrin α, rapamycin, stress response
Introduction
Macroautophagy (hereafter referred to as ‘autophagy’) is a catabolic pathway involving the sequestration of cytoplasmic material in double-membraned vesicles, the autophagosomes. Upon fusion with lysosomes, autophagosomes become autophagolysosomes and their content gets degraded by acidic hydrolases, allowing nutrients and macromolecules to fuel cellular metabolism or sustain stress responses (Klionsky, 2004; Mizushima et al, 2008). Multiple distinct perturbations of the cellular physiology can induce autophagy through a complex signalling network that crosstalks with several stress-response pathways (Kroemer et al, 2010; Green et al, 2011), including molecular cascades that are ignited by organellar damage as well as pathways leading to the activation of prominent transcription factors such as p53 (Tasdemir et al, 2008b; Scherz-Shouval et al, 2010) and NF-κB (Herrero-Martin et al, 2009; Criollo et al, 2010; Comb et al, 2011).
Beclin 1 has been the first mammalian protein shown to play a critical role in the initiation of autophagy (Liang et al, 1999). Its complex interactome has a major influence on the positive and negative regulation of autophagy (Kang et al, 2011). One of the decisive events that ignites the autophagic machinery is the activation of the class III phosphatidylinositol 3-kinase (PI3KC3), also called VPS34, to generate phosphatidylinositol-3-phosphate, which is required for the initial steps of vesicle nucleation (Axe et al, 2008). Beclin 1 is an obligatory allosteric activator of VPS34, operating within the so-called ‘Beclin 1 core complex’ that involves Beclin 1, VPS15, VPS34 and, likely, AMBRA1 (He and Levine, 2010). Numerous additional proteins interact with Beclin 1. Pro-autophagic Beclin 1 interactors include ATG14 (also called ATG14L or BARKOR), UV radiation resistance-associated gene (UVRAG) and Bif-1/endophilin B1, which interacts with Beclin 1 via UVRAG. Autophagy-inhibitory interactors of Beclin 1 include RUN domain protein as Beclin 1 interacting and cysteine-rich containing (RUBICON), anti-apoptotic proteins from the BCL-2 family (BCL-2, BCL-XL and MCL-1) and the inositol-1,4,5 trisphosphate receptor, which interacts with Beclin 1 indirectly via BCL-2 (He and Levine, 2010; Kang et al, 2011). Beclin 1 possesses a BH3 domain (residues 114–123) through which it constitutively interacts with BCL-2-like proteins (Maiuri et al, 2007), and autophagy induction requires the dissociation of Beclin 1 from such inhibitory liaisons (He and Levine, 2010; Kang et al, 2011).
The transcription factor NF-κB is activated when the inhibitor of NF-κB (IκB) is phosphorylated by the IκB kinase (IKK), a multiprotein complex that is composed by one regulatory (IKKγ, also known as NEMO) and two catalytic subunits (IKKα and IKKβ). IKK is activated in response to stressors as diverse as reactive oxygen species and DNA damage as well as by the ligation of death receptors (Baud and Karin, 2009). Frequently, the activation of IKK is mediated further upstream by yet another kinase, mitogen-activated protein kinase kinase kinase 7 (MAP3K7), better known as TGFβ-activated kinase 1 (TAK1) (Ninomiya-Tsuji et al, 1999; Takaesu et al, 2003; Landstrom, 2010). The phosphorylation of IκB by IKK stimulates IκB ubiquitination, thus targeting it for proteasomal degradation. In turn, IκB degradation allows NF-κB to translocate from the cytoplasm, where it is usually retained by IκB, to the nucleus, where NF-κB then becomes active as a cytoprotective and pro-inflammatory transcription factor (Baud and Karin, 2009). In both murine and human cells, the genetic inhibition of TAK1 or any of the IKK subunits (but not that of the NF-κB subunit p65) prevents the induction of autophagy in response to a panoply of different stimuli including starvation, rapamycin, p53 inhibition and endoplasmic reticulum stress (Herrero-Martin et al, 2009; Criollo et al, 2010; Comb et al, 2011). Conversely, constitutively active IKK subunits potently stimulate autophagy through a pathway that does not necessarily involve NF-κB, yet relies on the activation of AMPK and JNK1 (Criollo et al, 2010), as well as on several essential autophagy-related proteins including Beclin 1, ATG5 and LC3 (Comb et al, 2011).
Altogether, these results point to the existence of a major crosstalk between autophagy and the TAK1-IKK signalling axis, yet do not reveal the molecular mechanisms through which these pathways intersect. Here, we report the discovery that TAK1-binding protein 2 (TAB2) and TAK1-binding protein 3 (TAB3) function as tonic inhibitors of autophagy. We found that TAB2 and TAB3 constitutively interact with Beclin 1 and that this liaison is lost upon treatment with physiological inducers of autophagy, causing TAB2 and TAB3 to bind TAK1 instead of Beclin 1. Competitive disruption of the Beclin 1/TAB2/TAB3 complex suffices to induce Beclin 1- and TAK1-dependent autophagy, underscoring the importance of the inhibitory interaction between Beclin 1, TAB2 and TAB3.
Results
Identification of TAB2 and TAB3 as novel Beclin 1 interactors
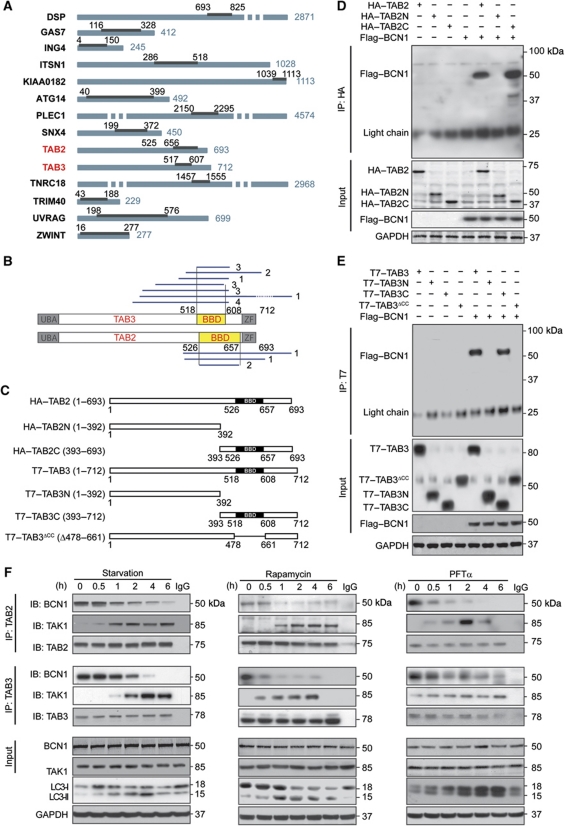
To unveil a possible intersection between autophagy and the TAK1-IKK activation pathway, we identified Beclin 1 interactors in two yeast two-hybrid saturation screens based on a complex human random-primed cDNA library. This approach led us to identify 63 Beclin 1-interacting proteins. Beyond known interactors (such as UVRAG, ATG14 and ZWINT) (Behrends et al, 2010), our screens identified 11 new proteins that would bind Beclin 1 with an elevated Predicted Biological Score (Formstecher et al, 2005) (Figure 1A, also consultable at the following website http://pim.hybrigenics.com/). Among these 11 proteins, 1 (desmoplakin) has previously been described to bind multiple autophagy-relevant proteins including PIK3C3/VPS34 (but not Beclin 1), and two have been reported to interact with one single autophagy-related protein (PLEC1 with SQSTM1/p62 and SNX4 with RABGAP1) (Behrends et al, 2010). Of note, two among the putative Beclin 1 interactors, namely TAB2 and its close homologue TAB3, are known co-activators of TAK1 (Takaesu et al, 2000; Ishitani et al, 2003). Yeast two-hybrid technology allowed us to narrow down the domains that mediate the interaction of TAB2 and TAB3 with Beclin 1 to a C-terminal region that we called ‘Beclin-binding domain’ (BBD). The BBD spans from residues 526 to 657 in TAB2 and from residues 518 to 608 in TAB3 (Figure 1B). Co-immunoprecipitation experiments involving Beclin 1 and multiple TAB2 and TAB3 deletion constructs (Figure 1C) confirmed the interaction of full-length Beclin 1 with full-length TAB2 and TAB3, as well as with TAB2 and TAB3 fragments containing the C-terminal BBD. The binding of Beclin 1 to TAB2 and TAB3 was lost upon deletion of their BBDs (Figure 1D and E). Thus, TAB2 and TAB3 constitute novel bona fide members of the Beclin 1 interactome.
Figure 1.
Identification of novel Beclin 1 interactors. (A) Beclin 1 (BCN1) interactors identified by yeast two-hybrid technology. Proteins binding to BCN1 are listed and their interacting domains are indicated by black bars. Numbers refer to amino-acid positions. (B) Identification of TAB2 and TAB3 fragments interacting with BCN1 in the yeast two-hybrid system. Blue lines depict the fragments of TAB2 or TAB3 that were found to interact with BCN1 (numbers on the right refer to the amount of yeast clones identified for each fragment). The minimal domain required for the interaction is referred to as Beclin-binding domain (BBD). (C) Constructs derived from TAB2 and TAB3 used in this study. (D, E) Co-immunoprecipitation of TAB2 or TAB3 with BCN1. The indicated constructs, namely HA-tagged and T7-tagged TAB2, and TAB3 constructs in (D) and (E), respectively, and Flag-tagged Beclin 1 (Flag–BCN1) were transfected into HeLa cells alone or in combination. Twenty-four hours later, TAB2 and TAB3 were immunoprecipitated with antibodies specific for HA (D) or T7 (E) and the precipitate was separated by SDS–PAGE and revealed with an antibody specific for Flag. (F) Immunoprecipitation of endogenous BCN1 with endogenous TAB2 or TAB3. HeLa cells were subjected to autophagy induction with starvation conditions, 1 μM rapamycin or 30 μM pifithrin α (PFTα) for the indicated time and then processed for TAB2 or TAB3 immunoprecipitation followed by the immunodetection of BCN1, TAK1, TAB2 and TAB3. Results in (E) and (F) are representative for three independent experiments.
TAB2 and TAB3 dissociate from Beclin 1 upon autophagy induction
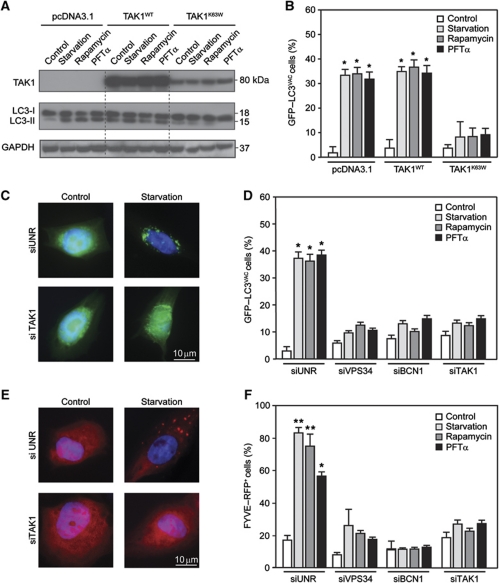
A His-tagged version of Beclin 1 was introduced together with epitope-tagged TAB2 or TAB3 into human cervical carcinoma HeLa cells, which were then driven into autophagy by culture in serum- and nutrient-free conditions (starvation), or by the administration of the mTOR inhibitor rapamycin or of the p53 inhibitor cyclic pifithrin α (PFTα). These three pro-autophagic stimuli all led to a decrease in the amount of TAB2 of TAB3 that co-immunoprecipitated with Beclin 1, as compared with control conditions (Supplementary Figure S1). Consistently, endogenous Beclin 1 co-immunoprecipitated with TAB2 or TAB3 (but not with TAK1) in control conditions, and this interaction was rapidly lost upon autophagy induction in HeLa cells (Figure 1F), as well as in other human cell lines (non small-cell lung cancer A549 cells, colorectal carcinoma HCT 116 cells) and mouse embryonic fibroblasts (MEFs, not shown). While in physiological conditions TAB2 and TAB3 failed to co-immunoprecipitate with TAK1, after the induction of autophagy with starvation, rapamycin or PFTα, both TAB2 and TAB3 were found to engage in such an interaction. Kinetic experiments revealed that TAB2 and TAB3 bind TAK1 as soon as they dissociate from Beclin 1 (Figure 1F). Inhibition of TAK1 by overexpression of a kinase-dead dominant-negative (DN) TAK1 mutant (TAK1K63W) (Ono et al, 2003; Figure 2A and B) or by means of a specific small-interfering RNA (siRNA) (Supplementary Figure S2) prevented the relocalization of a green fluorescence protein (GFP)–LC3 chimera from a diffuse pattern to discrete cytoplasmic puncta (Tasdemir et al, 2008a) by starvation, rapamycin or PFTα (Figure 2C and D). Similarly, TAK1 depletion prevented the autophagy-associated redistribution of a red fluorescent protein (RFP)-tagged FYVE domain (FYVE–RFP) (Zhang et al, 2007) to intracellular membranes containing phosphatidylinositol-3-phosphate (Figure 2E and F).
Figure 2.
Reduced interaction between Beclin 1, TAB and TAB3 in conditions of autophagy induction. (A, B) Inhibition of autophagy by dominant-negative (DN) TAK1. HeLa cells were co-transfected with a GFP–LC3-encoding construct plus pcDNA3.1 (empty vector), or plasmids for the expression of WT TAK1 (TAK1WT) or the DN TAK1K63W mutant. One day later, cells were either left untreated (control) or driven into autophagy by starvation or by the administration of 1 μM rapamycin or 30 μM pifithrin α (PFTα), followed by immunoblotting for the detection of TAK1 and endogenous LC3 (A) or immunofluorescence microscopy for the quantification of cells with cytosolic GFP–LC3 puncta (GFP–LC3VAC cells) (B) (mean values±s.d., n=3; *P<0.01 versus control cells). GAPDH levels were monitored to ensure equal loading. (C, D) Inhibition of autophagy by knockdown of VPS34, Beclin 1 (BCN1) and TAK1. siRNAs that effectively deplete VPS34, BCN1 and TAK1 were co-transfected with a GFP–LC3-encoding plasmid in HeLa cells. Autophagy was then induced as in (A) and the frequency of GFP–LC3VAC cells (mean values±s.d., n=3; *P<0.01 versus control cells) was determined. (E, F) The same setting shown in (C, D) was performed with U2OS cells and FYVE–RFP (mean values±s.d., n=3; *P<0.01, **P<0.001 versus control cells).
Altogether, these results support the idea that TAB2 and TAB3 dissociate from Beclin 1 to engage with TAK1 when autophagy is induced.
TAB2 and TAB3 are endogenous inhibitors of autophagy
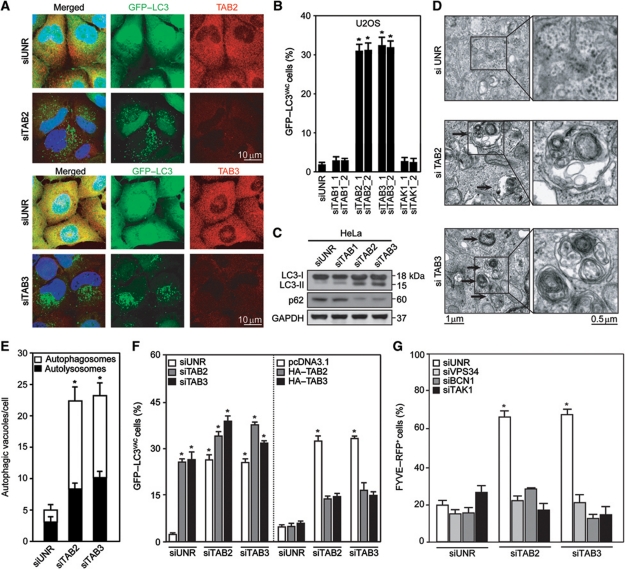
Depletion of TAB2 or TAB3 by specific siRNAs induced the accumulation of GFP–LC3+ dots in HeLa and human osteosarcoma U2OS cells stably expressing GFP–LC3 (Figure 3A and B), in HeLa cells transiently transfected with a GFP–LC3-encoding plasmid as well as in HCT 116 cells (Supplementary Figure S3). Moreover, TAB2 or TAB3 knockdown stimulated the lipidation of endogenous LC3, an autophagy-associated post-translational modification that enhances its electrophoretic mobility (shift from the LC3-I to the LC3-II form), and reduced the abundance of the autophagic substrate SQSTM1/p62 (Figure 3C), suggesting that TAB2 and TAB3 act as endogenous inhibitors of autophagy. Accordingly, the depletion of TAB2 or TAB3 led to the accumulation of bona fide autophagosomes and autophagolysosomes, as determined by transmission electron microscopy (Figure 3D and E). Epistatic experiments revealed that the simultaneous knockdown of TAB2 and TAB3 induced only slightly more GFP–LC3+ puncta than the depletion of each of these proteins alone. Moreover, the induction of autophagy by TAB2 depletion was inhibited by transfection of non-interferable TAB2 or wild-type (WT) TAB3 (and vice versa TAB2 transfection antagonized autophagy induction by TAB3 depletion), suggesting that both proteins inhibit the formation of autophagic puncta in a similar, overlapping manner (Figure 3F). Moreover, the depletion of either TAB induced the formation of FYVE–RFP+ puncta to similar extents (Figure 3G), suggesting that both TAB2 and TAB3 usually restrain the lipid kinase activity of the Beclin 1 complex.
Figure 3.
Induction of autophagosomes by depletion of TAB2 or TAB3. (A, B) Detection of autophagic GFP–LC3+ puncta. HeLa or U2OS cells stably expressing GFP–LC3 were transfected with siRNAs targeting TAK1, TAB1, TAB2 or TAB3 or with a control siRNA (siUNR). One day later, the subcellular localization and abundance of GFP–LC3 or immunostained TAB2 or TAB3 was determined by epifluorescence microscopy. Representative images are shown in (A) (HeLa cells) and quantitative results (mean values±s.d., n=3; *P<0.01 versus siUNR-transfected cells) are depicted in (B) (U2OS cells). (C) Lipidation of LC3 induced by TAB2 or TAB3 knockdown. Representative immunoblots showing the conversion of non-lipidated LC3 (LC3-I) to its lipidated variant (LC3-II) as well as SQSTM1/p62 protein levels are shown. GAPDH levels were monitored to ensure equal loading. (D, E) Quantification of autophagosomes and autophagolysosomes by transmission electron microscopy. Representative images of HeLa cells transfected with siUNR or with TAB2- or TAB3-targeting siRNAs are shown in (D), and quantitative results are depicted in (E) (mean values±s.d., n=3; *P<0.01 versus siUNR-transfected cells). (F) Epistatic analysis of the effects of TAB2 and TAB3 depletion on autophagy. HeLa cells stably expressing GFP–LC3 were transfected with siRNAs specific for TAB2 or TAB3 and/or with cDNAs coding for full-length HA-tagged TAB2 (HA–TAB2) or TAB3 (HA–TAB3). Twenty-four hours later, the frequency of cells exhibiting >5 GFP–LC3+ cytosolic puncta (GFP–LC3VAC cells) was determined. Results are mean values±s.d. (n=3; *P<0.01 versus siUNR-transfected cells). (G) U2OS cells stably expressing FYVE–RFP were transfected with siUNR, or with siRNAs specific for TAB2, TAB3, VPS34, Beclin 1 (BCN1) and TAK1, in the indicated combinations. Forty-eight hours later, the percentage of cells with RFP–FYVE+ puncta cells was determined. Results are mean values±s.d. (n=3; *P<0.01 versus siUNR-transfected cells).
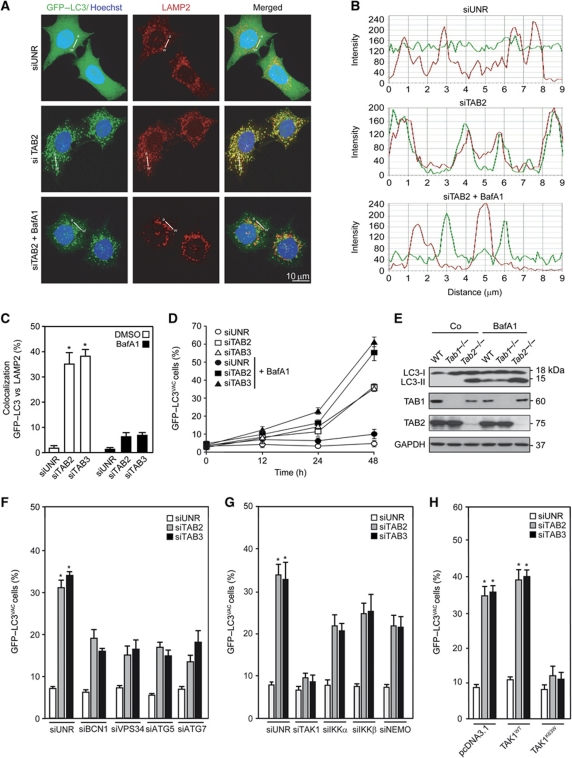
The accumulation of autophagosomes may result from enhanced sequestration of cytoplasmic material (increased on-rate) as well as from reduced removal of autophagosomes by fusion with lysosomes (reduced off-rate). Therefore, we measured autophagosome formation induced by TAB depletion in the absence or presence of bafilomycin A1 (BafA1), which inhibited the colocalization of the autophagic marker GFP–LC3 and the lysosomal marker LAMP2, in accord with its known capacity to block the autophagosome–lysosome fusion (Mizushima et al, 2010) (Figure 4A–C). In the presence of BafA1, the depletion of TAB2 and TAB3 resulted in more GFP–LC3+ puncta than in its absence (Figure 4D). Similar flux determinations were performed in the presence of an alternative lysosomal inhibitor, ammonium chloride, or protease inhibitors (E64d plus pepstatin A) (Supplementary Figure S4). Also, MEFs lacking TAB2 expression due to homologous recombination (Tab2−/−), but neither WT MEFs nor their Tab1−/− counterparts, manifested increased LC3 lipidation, both in the absence and in the presence of BafA1 (Figure 4E). Autophagy induced by TAB2 or TAB3 knockdown followed the canonical pathway, as it was reduced upon depletion of essential autophagy proteins such as Beclin 1, its associated phosphatidylinositol 3-kinase VPS34, ATG5 and ATG7 (Figure 4F). Moreover, it involved the obligatory contribution of TAK1 and that of the three subunits of the IKK complex, as shown by additional experiments of siRNA-mediated knockdown (Figure 4G) or transfection with DN TAK1 (Figure 4H). In conclusion, TAB2 and TAB3 are endogenous inhibitors of the canonical autophagic pathway, which requires the action of kinases from the TAK1-IKK signalling axis.
Figure 4.
Mechanisms of autophagy induced by depletion of TAB2 or TAB3. (A–D) Impact of bafilomycin A1 (BafA1) on the induction of GFP–LC3+ puncta by TAB2 and TAB3 depletion. HeLa cells stably expressing GFP–LC3 were transfected with a control siRNA (siUNR) or with siRNAs targeting TAB2 and TAB3 for 24 h. During the last 12 h of this period, BafA1 was optionally added. After fixation and permeabilization, LAMP2 was detected by immunofluorescence. Representative confocal microphotographs for the TAB2 siRNA are shown (A), together with the profiles of colocalization of fluorescent signals (B) along the indicated direction (α–ω). Columns in (C) represent the percentage of colocalization of GFP–LC3 and LAMP2 (mean values±s.d.; *P<0.01 versus siUNR-transfected cells), as quantified in at least 50 cells for each condition. The frequency (mean±s.d.) of cells with >5 GFP–LC3+ cytosolic puncta (GFP–LC3VAC cells) is plotted in (D). (E) Impact of BafA1 on LC3 lipidation. MEFs with the indicated genotypes were cultured in complete medium supplemented with BafA1 for 12 h and the proportion of LC3-I/LC3-II was determined by immunoblotting. GAPDH levels were monitored to ensure equal loading. (F, G) Impact of autophagy-relevant proteins and of the TAK1-IKK signalling axis on GFP–LC3 aggregation induced by the depletion of TAB2 or TAB3. HeLa cells stably expressing GFP–LC3 were transfected with siUNR or with siRNAs targeting the indicated proteins, alone or in combination, and 48 h later GFP–LC3VAC cells were quantified (mean values±s.d., n=4; *P<0.01 versus siUNR-transfected cells). (H) Inhibition of autophagy by dominant-negative (DN) TAK1. HeLa cells stably expressing GFP–LC3 were co-transfected with pcDNA3.1 (empty vector) or with plasmids encoding WT (TAK1WT) or a DN TAK1 variant (TAK1K63W) together with the indicated siRNAs for 24 h, followed by the quantification of GFP–LC3VAC cells (mean values±s.d., n=3, *P<0.01 versus siUNR-, pcDNA3.1-transfected cells).
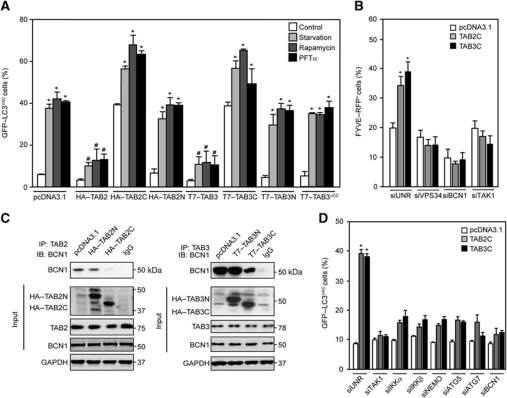
Dissociation of TAB2 and TAB3 from Beclin 1 induces autophagy
Overexpression of full-length TAB2 or TAB3 inhibited starvation-, rapamycin- and PFTα-induced autophagy (Figure 5A), in line with the interpretation that both proteins are negative regulators of autophagic flux. TAB2 or TAB3 variants lacking the BBD (TAB2N, TAB3N, TAB3ΔCC, Figure 1C) failed to modulate autophagy induced by various stimuli (Figure 5A and B). However, TAB2 and TAB3 deletion mutants containing their C-terminal moieties (TAB2C and TAB3C, Figure 1C), which include the BBD, had the striking property to induce the accumulation of GFP–LC3+ and FYVE–RFP+ puncta on their own, as well as to exacerbate autophagy induced by starvation, rapamycin or PFTα (Figure 5A and B). Accordingly, expression of TAB2C or TAB3C (but not that of TAB2N or TAB3N) reduced the interaction between endogenous Beclin 1 and TAB2 or TAB3 (Figure 5C). Moreover, the depletion of Beclin 1, ATG5, ATG7, TAK1, IKKα, IKKβ or NEMO inhibited the generation of GFP–LC3+ puncta by TAB2C or TAB3C, suggesting that TAB2C- and TAB3C-induced autophagy is mediated by the canonical pathway and by TAK1-IKK activity (Figure 5D).
Figure 5.
Inhibition of autophagy by full-length TAB2 and TAB3 but induction by their C-terminal fragments. (A) Effects of full-length TAB2 and TAB3 or their deletion mutants (as in Figure 1C) on autophagy. HeLa cells stably expressing GFP–LC3 were transfected with pcDNA3.1 (empty vector) or with plasmids encoding the indicated TAB2 and TAB3 variants for 24 h, then driven into autophagy by starvation or by the administration of 1 μM rapamycin or 30 μM pifithrin α (PFTα) for 4 h. Finally, the frequency (mean±s.d., n=3) of cells with >5 GFP–LC3+ cytosolic puncta (GFP–LC3VAC cells) was assessed (*P<0.01 versus control cells transfected with the same plasmid; #P<0.01 versus pcDNA3.1-transfected cells treated with the same pro-autophagic trigger). (B) U2OS cells stably expressing FYVE–RFP were co-transfected with pcDNA3.1 or with vectors encoding TAB2C or TAB3C together with a control siRNA (siUNR) or with siRNAs specific for VPS34, Beclin 1 (BCN1) or TAK1 for 24 h, followed by the quantification of cells exhibiting FYVE–RFP+ dots (FYVE–RFP+). Results are mean values±s.d. (n=3; *P<0.01 versus siUNR-, pcDNA3.1-transfected cells). (C) Inhibition of the interaction between endogenous TAB2, TAB3 and BCN1 by C-terminal fragments of TAB2 and TAB3. Forty-eight hours after transfection with pcDNA3.1 or plasmids coding for the indicated proteins, cells were lysed, TAB2 or TAB3 was immunoprecipitated and BCN1 was immunodetected. GAPDH levels were monitored to ensure equal loading. This experiment has been done three times, yielding comparable results. (D) Mechanisms of autophagy induction by the Beclin-binding domain (BBD)-containing C-terminal fragments of TAB2 and TAB3. HeLa cells were transfected with pcDNA3.1, TAB2C- or TAB3C-encoding plasmids in combination with the indicated siRNAs for 24 h, followed by the quantification of GFP–LC3VAC cells (mean values±s.d., n=3; *P<0.01 versus siUNR-, pcDNA3.1-transfected cells).
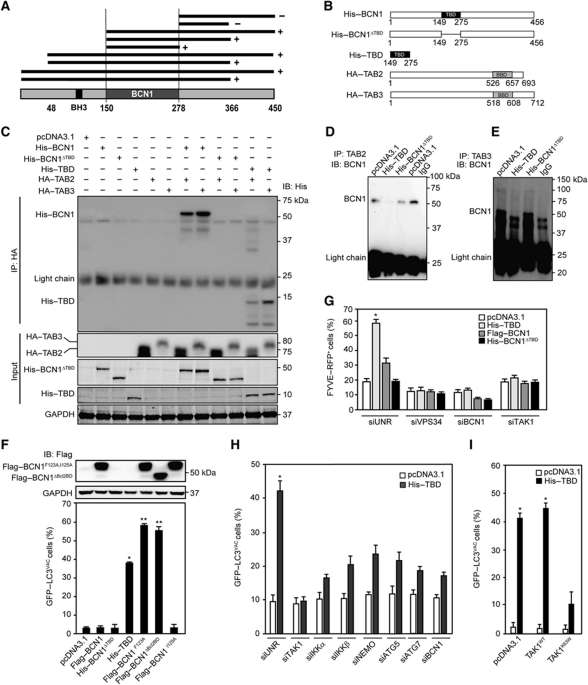
These results suggest that the dissociation of the multiprotein complex including the pro-autophagic protein Beclin 1 and the autophagy inhibitors TAB2 and TAB3 might be sufficient to induce autophagy. To further explore this possibility, we used yeast two-hybrid technology to determine the Beclin 1 domain that interacts with TAB2 and TAB3, finding it comprised between residues 150 and 278 (Figure 6A), a region that overlaps with Beclin 1 central coiled-coil domain (CCD, residues 144–269) (Kang et al, 2011). In control conditions, such a TAB-binding domain (TBD) (Figure 6B) co-immunoprecipitated with TAB2 and TAB3, as did full-length Beclin 1 but not a Beclin 1 deletion mutant lacking the TBD (Beclin 1ΔTBD) (Figure 6C). Moreover, recombinant TAB2 and TAB3 directly interacted with the TBD (but not with Beclin 1ΔTBD) in pull-down experiments (Supplementary Figure S5), and transgene-driven TBD expression caused endogenous Beclin 1 to dissociate from TAB2 and TAB3 (Figure 6D and E). Of note, while WT Beclin 1 failed to induce autophagy upon transfection-enforced expression, TBD did so very efficiently (Figure 6F; Supplementary Figure S6), though slightly less than two gain-of-function mutants of Beclin 1 with a deleted or inactivated inhibitory BH3 domain (Figure 6F). TBD was also highly efficient in stimulating the formation of FYVE–RFP+ puncta (Figure 6G), suggesting that the disruption of the inhibitory interactions between Beclin 1, TAB2 and TAB3 stimulates the lipid kinase activity of the Beclin 1 complex. TBD-induced autophagy depended on endogenous Beclin 1, ATG5 and ATG7 as well as on the TAK1-IKK signalling axis, as demonstrated in knockdown experiments (Figure 6H) or upon transfection with DN TAK1 (Figure 6I). Taken together, these results indicate that the dissociation of multiprotein complex including Beclin 1, TAB2 and TAB3 potently stimulates autophagy.
Figure 6.
Induction of autophagy by a Beclin 1 fragment that disrupts the interaction between endogenous Beclin 1, TAB2 and TAB3. (A) Determination of the TAB-binding domain (TBD) within Beclin 1 (BCN1). Yeast two-hybrid technology was used to screen for positive (+) or negative interactions (−) between BCN1 fragments and full-length TAB2. (B) Schematic presentation of the constructs used in subsequent experiments (BBD, Beclin-binding domain). (C) Confirmation of the TBD by immunoprecipitation. The indicated constructs were transfected into HeLa cells, followed by lysis, immunoprecipitation of HA-tagged full-length TAB2 or TAB3 and detection of His-tagged constructs. (D, E) Inhibition of the interaction between endogenous BCN1, TAB2 and TAB3 by a BCN1 fragment corresponding to the TBD. HeLa cells were transfected with pcDNA3.1 (empty vector) or with plasmids encoding the TBD or a His-tagged Beclin 1 variant lacking the TBD (His–BCN1ΔTBD), followed by co-immunoprecipitation of endogenous TAB2 (D) or TAB3 (E) and detection of BCN1. (F) Induction of autophagy by the TBD. HeLa cells stably expressing GFP–LC3 were transfected with pcDNA3.1 or constructs encoding the indicated BCN1 variants. After 24 h, Flag-tagged proteins were detected by immunoblotting and the frequency of cells with >5 GFP–LC3+ cytosolic puncta (GFP–LC3VAC cells) was assessed (mean values±s.d., n=3; *P<0.01, **P<0.001 versus pcDNA3.1-transfected cells). (G, H) Mechanisms of autophagy induction by the TBD. (G) U2OS cells stably expressing RFP–FYVE were transfected pcDNA3.1 or plasmids for the expression of TBD, BCN1 or BCN1ΔTBD in combination with a control siRNA (siUNR) or siRNAs that effectively deplete VPS4, BCN1 and TAK1. Forty-eight hours later, the percentage of cells exhibiting FYVE–RFP+ dots (FYVE–RFP+) was assessed. (G). Alternatively, HeLa cells stably expressing GFP–LC3 were transfected with pcDNA3.1 or with a TBD-encoding plasmids plus siUNR or siRNAs specific the indicated proteins. Forty-eight hours later, the percentage of GFP–LC3VAC cells was determined (H). Results are mean values±s.d. (n=3, *P<0.01 versus siUNR-, pcDNA3.1-transfected cells). (I) Inhibition of TBD-induced autophagy by dominant-negative (DN) TAK1. Cells were co-transfected pcDNA3.1 or vectors encoding WT TAK1 (TAK1WT) or a DN TAK1 variant (TAK1K63W) alone or together with a TBD-encoding plasmid for 24 h, followed by the quantification of GFP–LC3VAC cells (mean values±s.d., n=3; *P<0.01 versus pcDNA3.1-transfected cells).
Discussion
In this work, we provide evidence in favour of an unsuspected intersection between autophagy regulation and upstream portions of the canonical NF-κB activation pathway. Under physiological conditions, when autophagy is off, Beclin 1 associates with several inhibitory factors that, as we discovered here, include TAB2 and TAB3. However, upon autophagy activation by several distinct stimuli including its most physiological inducer, starvation, as well as pharmacological triggers (rapamycin and PFTα), TAB2 and TAB3 dissociate from Beclin 1 and associate with TAK1. This latter association is well known to stimulate the kinase activity of TAK1 (Takaesu et al, 2000; Ishitani et al, 2003; Besse et al, 2007) and to ignite the IKK pathway (Jin et al, 2004; Kanayama et al, 2004).
TAB2 and TAB3 were found to interact with Beclin 1 in a direct fashion, using several complementary technologies (yeast two-hybrid screens and pull-down assays of recombinant proteins), and their interaction was confirmed by co-immunoprecipitation of tagged as well as endogenous proteins. These data add TAB2 and TAB3 to the ever-expanding list of Beclin 1 interactors that already comprises close to 20 confirmed proteins (including all four anti-apoptotic BCL-2 family proteins, AMBRA1, ATG14, HMGB1, nPIST, RUBICON, SLAM, PINK, Survivin, UVRAG, VMP1, VPS15, VPS34, etc.) (Kroemer et al, 2010; Kang et al, 2011). The detailed mechanisms through which TAB2 and TAB3 dissociate from Beclin 1 remain elusive. TAB2 and TAB3 are known to interact with K63-linked polyubiquitin chains through a highly conserved zinc finger domain (Kanayama et al, 2004). However, the BBD and the zinc finger domain do not overlap, and the latter (aa 684–710 in TAB3) is fully preserved in the TAB3ΔCC mutant, which fails to interact with Beclin 1. Moreover, the Beclin 1 site that can undergo K63-linked polyubiquitination is located in its N-terminal BH3-binding domain (Shi and Kehrl, 2010), and the fraction of Beclin 1 that co-immunoprecipitates with TAB2 or TAB3 does not manifest any shift in electrophoretic mobility that would be compatible with ubiquitination. Rather, TAB2 and TAB3 appear to engage in direct protein-to-protein interactions involving their CCDs (Ishitani et al, 2003), which overlap with their BBD and are required for TAK1 binding (Takaesu et al, 2000; Ishitani et al, 2003), in line with the observation that the binding of TAB2 and TAB3 to Beclin 1 or TAK1 is mutually exclusive.
On the side of Beclin 1, the domain involved in TAB binding (residues 150–278) overlaps with the central CCD (residues 144–269), which is flanked by the BH3 domain on the N-terminal end (residues 114–123) and an evolutionarily conserved domain (ECD, residues 244–337) on the C-terminus. Thus, TAB2 and TAB3 bind to the regulatory region of Beclin 1 that also mediates interactions with multiple autophagy-regulatory proteins including AMBRA1 (Fimia et al, 2007), ATG14 (Sun et al, 2008), nPIST (Yue et al, 2002), RUBICON (Zhong et al, 2009) and UVRAG (Liang et al, 2006). The question how TAB2 and TAB3 affect the binding of these other factors to Beclin 1 will be the subject of future investigations.
Irrespective of the aforementioned molecular details, one fascinating aspect of autophagy regulation by TAB2 and TAB3 resides in its circuitry. Clearly, TAB2 and TAB3 are negative regulators of Beclin 1 (and the Beclin 1-dependent VPS34-mediated production of phosphatidylinositol-3-phosphate), because knockdown of TAB2 or TAB3 induces autophagy, overexpression of TAB2 and TAB3 inhibits autophagy and fragments of Beclin-1 and TAB2 and TAB3 that contain the TBD or the BBD, respectively, induce autophagy as they disrupt the endogenous Beclin 1/TAB2/TAB3 complex. This inhibitory effect of TAB2 and TAB3 on autophagy appears to be dominant, as their deletion (by homologous recombination) or depletion (by siRNAs) induces an increase in the autophagic flux.
Interestingly, the induction of autophagy by TAB2 and TAB3 depletion or TBD transfection is not only inhibited by depletion of Beclin 1 (or VPS34) but also is limited by the knockdown of TAK1 or any IKK subunit. As our paper was under revision, another report was published that confirmed the role of TAB2 and TAB3 as negative regulators of autophagy (Takaesu et al, 2011). siRNA-mediated depletion of TAB2 and TAB3 was found to decrease the abundance of SQSTM1/p62 both in WT MEFs and in MEFs expressing a crippled version of TAK1 that lacks its ATP-binding site (due to the knockout of exon 2) (Takaesu et al, 2011). In sharp contrast, we found that the depletion of TAK1 or the overexpression of a DN TAK1 mutant inhibited autophagy in HeLa or U2OS cells responding to the depletion of TAB2 and TAB3 or to the overexpression of Beclin 1 TBD. Thus, TAK1 may either contribute to autophagy induction in a context-dependent fashion or do so through a mechanism that does not involve its catalytic activity.
The depletion of TAB2 or TAB3 induces autophagy, yet TAK1 appears to be necessary, at least to some extent, for this response, as shown here. We and others found that TAK1 is required for the optimal induction of autophagy by a number of stimuli including starvation, endoplasmic reticulum stress, rapamycin, PFTα, TGFβ and the ligation of the death receptor TRAIL (Herrero-Martin et al, 2009; Criollo et al, 2010; Ding et al, 2010), suggesting a rather broad implication of TAK1 in autophagic signalling. However, it appears counterintuitive that depletion of TAB2 or TAB3 induces autophagy, as TAK1 normally relies on TAB2 and TAB3 as its obligatory cofactors (Takaesu et al, 2000; Ishitani et al, 2003; Besse et al, 2007). This discrepancy may either be explained by the kinetics of depletion (the pool of TAB2 and TAB3 that interacts with Beclin 1 might be less stable than the pool of TAB2 and TAB3 that is free to associate with TAK1) or by the existence of a TAB-independent mode of activation of TAK1 that will have to be explored in future studies.
Irrespective of these incognita, it is tempting to link the present data (which indicate that TAB2 and TAB3 act as negative regulators of the Beclin 1/VPS34 complex) and those published in the literature (showing that TAB2 and TAB3 act to stimulate TAK1) with our observation that TAB2 and TAB3 preferentially interact with Beclin 1 before, and with TAK1 after, the induction of autophagy. In a possible scenario, TAB2 and TAB3 would dissociate from Beclin 1 (thus unleashing the lipid kinase activity of the Beclin 1/VPS34 complex) and associate with TAK1 (thus activating the TAK1-IKK signalling axis), overall functioning as a ‘switch’ in the control of autophagy by molecularly linking a de-inhibitory reaction to a stimulatory one (Supplementary Figure S7).
The existence of such a molecular ‘switch’ (and likely others that remain to be discovered) may explain why autophagy is accompanied by a series of stereotyped alterations in major stress-response signalling pathways, irrespective of the nature of the initiating stimulus. For instance, starvation stimulates well-known pro-autophagic pathways, including the activation of AMPK and of the Beclin 1/VPS34 complex, but also completely unexpected molecular cascades, such as those linked to the activation of IKK and to the depletion of cytoplasmic p53 (Criollo et al, 2010). Conversely, the inhibition, deletion or depletion of p53 as well as the activation of TAK1 or IKK (by transfection of constitutively active IKK subunits) stimulates AMPK-driven autophagy (Tasdemir et al, 2008b; Herrero-Martin et al, 2009; Criollo et al, 2010). Inhibition of p53 also leads to the activation of IKK, which is necessary for autophagy induction, and, vice versa, artificial stimulation of IKK causes p53 depletion (Criollo et al, 2010). Both p53 inhibition and IKK activation cause the dissociation of the BCL-2/Beclin 1 complex (Tasdemir et al, 2008b; Herrero-Martin et al, 2009; Criollo et al, 2010). However, BH3 mimetics that directly disrupt the BCL-2/Beclin 1 complex (Maiuri et al, 2007) also induce p53 depletion and IKK activation (Malik et al, 2011a). Cumulatively, these results suggest that the pathways underlying the regulation of autophagy cannot follow a linear hierarchy but must be interconnected in a complex network. In this setting, the stimulation of autophagy by physiological inducers (such as starvation or organellar stress) or pharmacological agents (such as rapamycin and PFTα) triggers an ensemble of intimately linked changes that are coupled to each other in self-amplificatory loops (Malik et al, 2011b). While the ‘switch’ that we postulate here (and that involves the dissociation of TAB2 and TAB3 from Beclin 1 and their association with TAK1) cannot explain all these coordinated alterations, it unveils some clues explaining the obligatory synchronization of autophagy induction with TAK1-IKK activation.
Materials and methods
Yeast two-hybrid system
Two different Beclin 1 baits have been screened. The first one was constituted by Beclin 1 residues 46–362, as previously identified in a screen for BCL-2-interacting proteins (Maiuri et al, 2007) fused in C-terminal to the lexA DNA-binding domain (pB28) (Formstecher et al, 2005). The second one was constituted by full-length Beclin 1 fused in N-terminal to the lexA DNA-binding domain (pB29) (Formstecher et al, 2005). A random-primed human placenta cDNA library based on the pP6 vector (Fromont-Racine et al, 2002) was transformed into the Y187 Saccharomyces cerevisiae strain. Ten million independent yeast colonies were collected, pooled and stored at −80°C as equivalent aliquot fractions of the same library. In both screens, >80 millions diploid cells have been tested. The first bait (Beclin 1 residues 46–432) has been screened with 1 mM 3-amino-1,2,4-triazole, while the full-length bait required 100 mM 3-amino-1,2,4-triazole, due to consistent autoactivation. Prey fragments from positive clones were PCR amplified and sequenced at their 5′ and 3′ junctions on a PE3700 Capillary DNA Sequencer (PE Applied Biosystems, Foster City, USA) (Fromont-Racine et al, 2002). The results were used to identify the corresponding genes in the GenBank database (NCBI). The baits used to map the Beclin-interacting domain have been obtained by recombination (gap repair) in the C-terminal of the lexA DNA-binding domain, using the following oligonucleotides: OLI6565 GGGCTGGCGGTTGGGGTTATTCGCAACGGCGACTGGCTGGAATTCACCACAGCCCAGGCGAAACA; OLI6566 GGGCTGGCGGTTGGGGTTATTCGCAACGGCGACTGGCTGGAATTCGACACTCAGCTCAACGTCACT; OLI6567 GGGCTGGCGGTTGGGGTTATTCGCAACGGCGACTGGCTGGAATTCACCAACGTCTTTAATGCAACC; OLI6568 AATCATAAATCATAAGAAATTCGCCCGGAATTAGCTTGGCTGCAGGTCTCAAGTGACGTTGAGCTGAGTGTC; OLI6569 AATCATAAATCATAAGAAATTCGCCCGGAATTAGCTTGGCTGCAGGTCTCAGTGCCAGATGTGGAAGGTTG; OLI6570 AATCATAAATCATAAGAAATTCGCCCGGAATTAGCTTGGCTGCAGGTCTCATCAAACTTGTTGTCCCAGAA.
Chemicals, cell lines and culture conditions
Unless otherwise indicated, chemicals were purchased by Sigma-Aldrich (St Louis, USA), and media and supplements for cell culture from Gibco–Invitrogen (Carlsbad, USA). Rapamycin was obtained by Tocris Bioscience (Ellisville, USA). All cells were maintained in standard culture conditions (37°C, 5% CO2). Human cervical carcinoma HeLa and human osteosarcoma U2OS cells (and their GFP–LC3 and FYVE–RFP-expressing derivatives) were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 10 mM HEPES. Human colon carcinoma HCT 116 cells were maintained in McCoy's medium supplemented with 10% FCS, 100 mg/l sodium pyruvate and 10 mM HEPES, while WT, Tab1−/− and Tab2−/− MEFs were cultured in DMEM supplemented with 10% FCS, 10 mM HEPES and 1% non-essential amino acids. For serum and nutrient starvation, cells were cultured in serum-free Earle's balanced salt solution (EBSS) (Boya et al, 2005).
siRNAs, plasmids and transfections
siRNA transfections were performed by means of the Oligofectamine® reagent (Invitrogen), according to the manufacturer's instructions. The following custom-designed siRNA duplexes were purchased from Sigma-Proligo (The Woodlands, USA): siBCN1 (sense 5′-CUCAGGAGAGGAGCCAUUUdTdT-3′) (Boya et al, 2005); siVPS34 (sense 5′-ACGGTGATGAATCATCTCCAAdTdT-3′); siTAB1_1 (sense 5′-GCCUUGGCUGAGAAGGCAAdTdT-3′); siTAB1_2 (sense 5′-GCAUUACAACCAGACAGUUGAUAUUdTdT-3′); siTAB2_1 (sense 5′-GGAACGACUUCAAAGAGAAdTdT-3′); siTAB2_2 (sense 5′-GGAACGACUUCAAAGAGAAdTdT-3′); siTAB3_1 (sense 5′-GGUUGAAGUCUGAAGUUAAdTdT-3′); siTAB3_2 (sense 5′-GCACAUACCUCGGUAUAGUdTdT-3′) siTAK1_1 (sense 5′-GUAGAUCCAUCCAAGACUUTTdTdT-3′); and siTAK1_2 (sense 5′-GAUGGUAUAUACCAAGUUdTdT-3′). An irrelevant siRNA duplex (siUNR, sense 5′-GCCGGUAUGCCGGUUAAGUdTdT-3′) was employed as a negative control. Plasmid transfection was carried out by means of the Lipofectamine 2000® reagent (Invitrogen), as recommended by the manufacturer. A plasmid encoding a GFP–LC3 chimera (Kabeya et al, 2000) was co-transfected with the empty vector pcDNA3.1 (Invitrogen) or with mammalian expression vectors encoding HA–TAK1, Flag–TAK1WT, Flag–TAK1K63W, HA–TAB1, HA–TAB2, HA–TAB3 or T7–TAB3 (Takaesu et al, 2000). TAB2N (the HA-tagged N-terminus of TAB2, residues 1–392), HA–TAB2C (the HA-tagged C-terminus of TAB2, residues 393–693), T7–TAB3N (the T7-tagged N-terminus of TAB3, residues 1–392), T7–TAB3C (the T7-tagged C-terminus of TAB3, residues 393–712) and T7–TAB3ΔCC were generated by PCR as previously described (Ninomiya-Tsuji et al, 1999; Takaesu et al, 2000). Constructs encoding His-tagged Beclin 1 (His–BCN1), Flag-tagged WT Beclin 1 (Flag–BCN1) and Flag-tagged Beclin 1 mutants (Flag–BCN1F123A, Flag–BCN1I125A and Flag–BCN1ΔBcl2BD) (Pattingre et al, 2005) were a generous gift of Dr B Levine (Texas Southwestern University, Dallas, USA). His–BCN1ΔTBD was constructed by first inserting at positions 148 and 276 of the His–BCN1 coding vector two EcoRI restriction sites by site-directed mutagenesis (with the Quickchange© Lightning Site-directed mutagenesis kit, Agilent, Santa Clara, USA) based on the primers 5′-GAATGCACAGATACTCTTTTAGACCAGGAATTCACTCAGCTCAACGTCACTGAAAATGA-3′ and 5′-CAACGTCTTTAATGCAACCTTCCACATCGAATTCAGTGGACAGTTTGGCACAATCAATAAC-3′ and their exact inverse complementary sequences. The resulting plasmid was then digested with EcoRI and religated in the absence of the TBD-coding trait.
Immunoblotting and immunoprecipitation
For immunoblotting, cells were washed with cold PBS at 4°C and lysed as previously described (Criollo et al, 2007a, 2007b). In all experiments, 50 μg of proteins was separated according to molecular weight on 4–12% SDS–PAGE precast gels (Invitrogen) and electrotransferred onto Immobilon membranes (Millipore, Bedford, USA). Unspecific binding sites were blocked by incubating membranes for 30 min in TBS supplemented with 0.05% Tween-20 and 5% w/v non-fat powdered milk. Primary antibodies recognizing the following proteins were employed: Beclin 1, SQSTM1/p62, TAB2, TAB3 (from Santa Cruz Biotechnology, Santa Cruz, USA); LC3, TAB1, TAK1, VPS24, ATG7, His, HA (from Cell Signaling Technology, Danvers, USA); ATG5 (from MBL International, Woburn, USA); Flag (from Sigma-Aldrich); T7 (from Millipore). Upon overnight incubation at 4°C, primary antibodies were detected with the appropriate horseradish peroxidase-labelled secondary antibodies (Southern Biotechnologies Associates; Birmingham; UK) and the SuperSignal West Pico chemoluminescent substrate (Thermo Fisher Scientific Inc., Rockford, USA). A primary antibody that specifically recognizes glyceraldehyde-3-phosphate dehydrogenase (GAPDH, from Millipore) was used to ensure equal lane loading. For immunoprecipitation, 7 × 106 cells were lysed as previously described (Criollo et al, 2007b), and 300 μg of proteins was pre-cleared for 1 h with 30 μl of Pure Proteome™ Protein G Magnetic Beads (Millipore), followed by incubation for 2 h in the presence of 2 μg of specific antibodies or IgG controls. Subsequent immunoblotting was carried out using TrueBlot™-HRP (eBioscience, San Diego, USA) secondary antibodies.
Pull-down experiments with recombinant proteins
Recombinant full-length Beclin 1 (BCN1), the BCN1 TBD, a variant of BCN1 lacking the TBD (BCN1ΔTBD) and the BBDs from TAB2 and TAB3 were all generated as GST-tagged proteins using the PGEX 6P1 vector (GE Healthcare Life Science, Piscataway, USA) and optionally liberated from GST using the PreScission Protease (GE Healthcare Life Science), following the manufacturer's recommendations. Glutathione Sepharose™ 4B beads (GE Healthcare Life Science) were washed in interaction buffer (100 mM Tris pH 7.4, 300 mM NaCl, 0.5% NP-40, 1% glycerol and Complete™ Protease Inhibitor Cocktail Tablet, from Roche Applied Science, Indianapolis, USA) and then GST-tagged recombinant proteins (25–100 μg) were immobilized on 30 μl beads for 2.5 h at 4°C. Thereafter, beads were washed three times and diluted 1:10 in interaction buffer (final volume=500 μl) followed by incubation for 1.5 h at 4°C on a roller with 50 μg of untagged recombinant proteins. Finally, beads were washed three times and proteins were eluted at 95°C for 5 min in the presence of SDS. In all, 5 μl of eluates was analysed by immunoblotting.
Light and immunofluorescence microscopy
Cells were processed for immunofluorescence staining according to established protocols (Criollo et al, 2007b; Boehrer et al, 2008). Fixed and permeabilized cells were incubated with primary antibodies overnight and then revealed with the appropriate AlexaFluor® conjugates (Molecular Probes–Invitrogen). Nuclei were counterstained with 10 μg/ml Hoechst 33342 (Molecular Probes–Invitrogen). Fluorescence and confocal fluorescence microscopy assessments were performed with an IRE2 microscope (Leica Microsystems, Weitzler, USA) equipped with a DC300F camera and with an LSM 510 microscope (Carl Zeiss, Jena, Germany), respectively. To determine the percentage of colocalization of fluorescent signals, the Image J software (freely available from the National Institute of Health, Bethesda, MD, USA at the address http://rsb.info.nih.gov/ij/) was used. Cells exhibiting >5 GFP–LC3+ or FYVE–RFP+ puncta were considered as positive (GFP–LC3VAC and FYVE–RFP+, respectively).
Electron microscopy
Cells were fixed for 1 h at 4°C in 0.1 M Sörensen phosphate buffer (pH 7.3) supplemented with 1.6% glutaraldehyde, washed, fixed again in aqueous 2% osmium tetroxide, stained in 2% uranyl acetate in 30% methanol and finally embedded in Epon. Electron microscopy was performed with a Tecnai 12 Spirit electron microscope (FEI, Eindhoven, The Netherlands), at 80 kV, on ultrathin sections (80 nm) stained with lead citrate and uranyl acetate.
Statistical procedures
All experiments were performed in triplicate parallel instances and repeated at least three times. Unless otherwise specified, figures report the results from one representative experiment (mean values±s.d., n=3). Data were analysed with Microsoft Excel (Microsoft Co., Redmond, USA) and statistical significance was assessed by means of two-tailed unpaired Student's t-tests.
Supplementary Material
Acknowledgments
We thank Drs B Vogelstein (John Hopkins University, Bethesda, MD, USA) and S Sato (Osaka University, Osaka, Japan) for HCT 116 cells and Tab2−/− MEFs, respectively; Drs J Yuan (Harvard Medical School, Boston, MA, USA) and H-B Shu (Wuhan University, Wuhan, China) for the dsRed-FYVE and Flag–TAK1 plasmids, respectively; Dr B Levine (Texas Southwestern University, Dallas, TX, USA) for tagged Beclin 1 variants; Drs O Geneste and JA Hickman (Institut de Recherche Servier, Croissy sur Seine, France) for sharing data on the Beclin 1 interactome; the Hybrigenics team for excellent technical assistance; and the International Collaboration Program ECOS-CONICYT, grant C08S01 (to SL and GK). GK is supported by the Ligue Nationale contre le Cancer (Equipe labellisée), Agence Nationale pour la Recherche (ANR), Fondation Axa (Chair For Longevity Research), European Commission (Apo-Sys, ArtForce, ChemoRes, ApopTrain), Fondation pour la Recherche Médicale (FRM), Institut National du Cancer (INCa), Cancéropôle Ile-de-France, and Fondation Bettencourt-Schueller. MN-S is funded by Junta de Extremadura (Spain). SAM is recipient of a grant from the Higher Education Commission (HEC) of Pakistan. GM and MM are supported by EMBO and Foundation AXA, respectively.
Author contributions: AC, MN-S, SAM, MM, EM, GM, SL, AVA, FH, GP, J-CR and JN-T performed the experiments. JMF, SL, MCM and GK conceived the study. AC, MN-S and LG prepared the figures. LG and GK wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud V, Karin M (2009) Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov 8: 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW (2010) Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse A, Lamothe B, Campos AD, Webster WK, Maddineni U, Lin SC, Wu H, Darnay BG (2007) TAK1-dependent signaling requires functional interaction with TAB2/TAB3. J Biol Chem 282: 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehrer S, Ades L, Braun T, Galluzzi L, Grosjean J, Fabre C, Le Roux G, Gardin C, Martin A, de Botton S, Fenaux P, Kroemer G (2008) Erlotinib exhibits antineoplastic off-target effects in AML and MDS: a preclinical study. Blood 111: 2170–2180 [DOI] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, Larochette N, Metivier D, Meley D, Souquere S, Yoshimori T, Pierron G, Codogno P, Kroemer G (2005) Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comb WC, Cogswell P, Sitcheran R, Baldwin AS (2011) IKK-dependent, NF-kappaB-independent control of autophagic gene expression. Oncogene 30: 1727–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Galluzzi L, Maiuri MC, Tasdemir E, Lavandero S, Kroemer G (2007a) Mitochondrial control of cell death induced by hyperosmotic stress. Apoptosis 12: 3–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criollo A, Maiuri MC, Tasdemir E, Vitale I, Fiebig AA, Andrews D, Molgo J, Diaz J, Lavandero S, Harper F, Pierron G, di Stefano D, Rizzuto R, Szabadkai G, Kroemer G (2007b) Regulation of autophagy by the inositol trisphosphate receptor. Cell Death Differ 14: 1029–1039 [DOI] [PubMed] [Google Scholar]
- Criollo A, Senovilla L, Authier H, Maiuri MC, Morselli E, Vitale I, Kepp O, Tasdemir E, Galluzzi L, Shen S, Tailler M, Delahaye N, Tesniere A, De Stefano D, Younes AB, Harper F, Pierron G, Lavandero S, Zitvogel L, Israel A et al. (2010) The IKK complex contributes to the induction of autophagy. EMBO J 29: 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Kim JK, Kim SI, Na HJ, Jun SY, Lee SJ, Choi ME (2010) TGF-β1 protects against mesangial cell apoptosis via induction of autophagy. J Biol Chem 285: 37909–37919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, Gruss P, Piacentini M, Chowdhury K, Cecconi F (2007) Ambra1 regulates autophagy and development of the nervous system. Nature 447: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, Jacq B, Arpin M, Bellaiche Y, Bellusci S, Benaroch P, Bornens M, Chanet R, Chavrier P, Delattre O, Doye V et al. (2005) Protein interaction mapping: a Drosophila case study. Genome Res 15: 376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Rain JC, Legrain P (2002) Building protein-protein networks by two-hybrid mating strategy. Methods Enzymol 350: 513–524 [DOI] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G (2011) Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science 333: 1109–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B (2010) The Beclin 1 interactome. Curr Opin Cell Biol 22: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, Fumarola C, Farkas T, Lopez-Rivas A, Jaattela M (2009) TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J 28: 677–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani T, Takaesu G, Ninomiya-Tsuji J, Shibuya H, Gaynor RB, Matsumoto K (2003) Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J 22: 6277–6288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Klika A, Callahan M, Faga B, Danzig J, Jiang Z, Li X, Stark GR, Harrington J, Sherf B (2004) Identification of a human NF-kappaB-activating protein, TAB3. Proc Natl Acad Sci USA 101: 2028–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanayama A, Seth RB, Sun L, Ea CK, Hong M, Shaito A, Chiu YH, Deng L, Chen ZJ (2004) TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol Cell 15: 535–548 [DOI] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D (2011) The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 18: 571–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ (2004) Cell biology: regulated self-cannibalism. Nature 431: 31–32 [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B (2010) Autophagy and the integrated stress response. Mol Cell 40: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstrom M (2010) The TAK1-TRAF6 signalling pathway. Int J Biochem Cell Biol 42: 585–589 [DOI] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU (2006) Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 8: 688–699 [DOI] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, Tasdemir E, Pierron G, Troulinaki K, Tavernarakis N, Hickman JA, Geneste O, Kroemer G (2007) Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 26: 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik SA, Orhon I, Morselli E, Criollo A, Shen S, Marino G, Benyounes A, Benit P, Rustin P, Maiuri MC, Kroemer G (2011a) BH3 mimetics activate multiple pro-autophagic pathways. Oncogene 30: 3918–3929 [DOI] [PubMed] [Google Scholar]
- Malik SA, Shen S, Marino G, BenYounes A, Maiuri MC, Kroemer G (2011b) BH3 mimetics reveal the network properties of autophagy-regulatory signaling cascades. Autophagy 7: 914–916 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ (2008) Autophagy fights disease through cellular self-digestion. Nature 451: 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K (1999) The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398: 252–256 [DOI] [PubMed] [Google Scholar]
- Ono K, Ohtomo T, Ninomiya-Tsuji J, Tsuchiya M (2003) A dominant negative TAK1 inhibits cellular fibrotic responses induced by TGF-beta. Biochem Biophys Res Commun 307: 332–337 [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B (2005) Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 122: 927–939 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Weidberg H, Gonen C, Wilder S, Elazar Z, Oren M (2010) p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc Natl Acad Sci USA 107: 18511–18516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH (2010) TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 3: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q (2008) Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 105: 19211–19216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu G, Kishida S, Hiyama A, Yamaguchi K, Shibuya H, Irie K, Ninomiya-Tsuji J, Matsumoto K (2000) TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol Cell 5: 649–658 [DOI] [PubMed] [Google Scholar]
- Takaesu G, Kobayashi T, Yoshimura A (2011) TGFβ-activated kinase 1 (TAK1)-binding proteins (TAB) 2 and 3 negatively regulate autophagy. J Biochem (doi:; DOI: 10.1093/jb/mvr123) [DOI] [PubMed] [Google Scholar]
- Takaesu G, Surabhi RM, Park KJ, Ninomiya-Tsuji J, Matsumoto K, Gaynor RB (2003) TAK1 is critical for IkappaB kinase-mediated activation of the NF-kappaB pathway. J Mol Biol 326: 105–115 [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Galluzzi L, Maiuri MC, Criollo A, Vitale I, Hangen E, Modjtahedi N, Kroemer G (2008a) Methods for assessing autophagy and autophagic cell death. Methods Mol Biol 445: 29–76 [DOI] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G et al. (2008b) Regulation of autophagy by cytoplasmic p53. Nat Cell Biol 10: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Horton A, Bravin M, DeJager PL, Selimi F, Heintz N (2002) A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron 35: 921–933 [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Zhu H, Yu AD, Xie X, Ma D, Yuan J (2007) Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA 104: 19023–19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z (2009) Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 11: 468–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.