Abstract
Kinesin-5 motors fulfil essential roles in mitotic spindle morphogenesis and dynamics as slow, processive microtubule (MT) plus-end directed motors. The Saccharomyces cerevisiae kinesin-5 Cin8 was found, surprisingly, to switch directionality. Here, we have examined directionality using single-molecule fluorescence motility assays and live-cell microscopy. On spindles, Cin8 motors mostly moved slowly (∼25 nm/s) towards the midzone, but occasionally also faster (∼55 nm/s) towards the spindle poles. In vitro, individual Cin8 motors could be switched by ionic conditions from rapid (380 nm/s) and processive minus-end to slow plus-end motion on single MTs. At high ionic strength, Cin8 motors rapidly alternated directionalities between antiparallel MTs, while driving steady plus-end relative sliding. Between parallel MTs, plus-end motion was only occasionally observed. Deletion of the uniquely large insert in loop 8 of Cin8 induced bias towards minus-end motility and affected the ionic strength-dependent directional switching of Cin8 in vitro. The deletion mutant cells exhibited reduced midzone-directed motility and efficiency to support spindle elongation, indicating the importance of directionality control for the anaphase function of Cin8.
Keywords: Cin8, kinesin directionality, kinesin-5, microtubules, mitosis
Introduction
Members of the kinesin-5 family are homotetrameric motor proteins, which utilize ATP to slide apart antiparallel spindle microtubules (MTs; Kashina et al, 1997; Kapitein et al, 2005). They are conserved among the eukaryotes and fulfil similar functions in spindle dynamics (Enos and Morris, 1990; Roof et al, 1991; Hagan and Yanagida, 1992; Hoyt et al, 1992; Sawin et al, 1992; Heck et al, 1993; Blangy et al, 1995; Saunders et al, 1995; Walczak and Mitchison, 1996; Straight et al, 1998; Sharp et al, 1999; Touitou et al, 2001; Brust-Mascher et al, 2004; Zhu et al, 2005). S. cerevisiae encodes two kinesin-5 homologues, Cin8 and Kip1, which overlap in function, with Cin8 being more important for mitosis progression (Hoyt et al, 1992; Roof et al, 1992). Before the onset of anaphase, Cin8 and Kip1 function in spindle assembly (Hoyt et al, 1992; Roof et al, 1992; Chee and Haase, 2010) and focus the kinetochore clusters (Tytell and Sorger, 2006; Gardner et al, 2008a; Wargacki et al, 2010). During spindle elongation in anaphase B, their function is important for (i) maintaining the correct spindle elongation rate (Kahana et al, 1995; Saunders et al, 1995; Straight et al, 1998); (ii) the timely transition from the initial fast phase to the final slow phase of anaphase B (Movshovich et al, 2008; Gerson-Gurwitz et al, 2009); (iii) for stabilizing the spindle midzone (Movshovich et al, 2008; Fridman et al, 2009; Gerson-Gurwitz et al, 2009), which consists of an overlapping array of MTs that emanate from the opposite poles (Winey et al, 1995). To guarantee proper mitosis, all components involved, and in particular the motors, have to be tightly regulated. The regulation of kinesin-5 motors is so far poorly understood.
Several kinesin-5 motors were found to be plus-end directed in vitro (Gheber et al, 1999; Kapitein et al, 2005). Independent of our findings, however, a recent report has provided evidence that Cin8 can switch directionality, assumed to be caused by motor–motor coupling (Roostalu et al, 2011). Molecular mechanisms as well as physiological relevance have remained unclear.
To track down the mechanisms of directionality control, we have here studied the motor function of the S. cerevisiae kinesin-5 Cin8 in parallel in-vivo and in-vitro experiments. We provide evidence that during anaphase spindle elongation, Cin8 moves in both plus- and minus-end directions on the spindle MTs. We also show that, in vitro, single molecules of Cin8 can switch directionality from fast processive minus-end directed to slow, processive plus-end directed motility. We provide first insights into the mechanism controlling this switch: (i) change of ionic strength, that is, electrostatic interactions within the motor or between motor and MT, activated the switch in vitro; (ii) deletion of the large insert in loop 8 of the Cin8 motor domain induced bias to minus-end directionality in vitro and reduced Cin8 plus-end directed motility along the anaphase spindles in vivo; (iii) in close to physiological conditions in vitro, individual Cin8 motors switched from fast minus-end motion on single MTs to slow and erratic motion between antiparallel MTs, while they drove plus-end sliding of the linked MTs; (iv) individual motors occasionally moved in the plus-end direction, but mostly maintained fast minus-end motion between parallel MTs.
Results
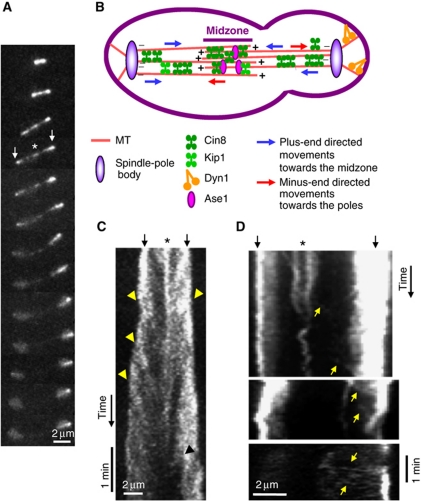
To examine the motile properties of Cin8 in vivo, we imaged live S. cerevisiae cells expressing Cin8–3GFP during anaphase B. As spindles elongated, Cin8 was localized both at the spindle midzone and near the poles, likely near the kinetochores as reported before (Tytell and Sorger, 2006). In late anaphase, Cin8 detached from the spindle and became diffusive (Figure 1A; Supplementary Movie S1), confirming previous reports (Avunie-Masala et al, 2011). Kymograph analysis of fluorescence recordings revealed that Cin8–3GFP moved towards the midzone, in the plus-end direction of MTs, with an average velocity of ∼25 nm/s (Figures 1C and 7A; Table I). This slow plus-end directed motility is consistent with the literature on Cin8 and the Xenopus kinesin-5 Eg5 (Gheber et al, 1999; Kwok et al, 2004; Kapitein et al, 2005). Intriguingly, we also observed occasional movements towards the spindle-pole bodies (SPBs; Figures 1D and 7A; Table I), mainly in long anaphase spindles, when the majority of Cin8 was already detached (Figures 1D and 7A). The scarcity of SPB-directed movements may be due to their rapidity (Table I) and/or to masking by abundant Cin8–3GFP on the spindle. In intermediate anaphase spindles, shorter than 5 μm, interpolar MTs (iMTs) can overlap almost for the entire spindle length (Winey et al, 1995; Gardner et al, 2008b). Therefore, in these spindles, the directionality of Cin8 is difficult to determine. In the long anaphase spindles, however, iMTs overlap only in the middle 10–20% of the spindle length, as determined by Electron Microscopy (Winey et al, 1995), direct imaging of GFP-labelled MTs (Avunie-Masala et al, 2011), or imaging of the midzone-organizing protein Ase1 (Schuyler et al, 2003; Fridman et al, 2009). The fact that Cin8 motility towards the midzone as well as towards the SPBs was observed in spindles longer than 5 μm (Figures 1C, D and 7A), suggests that these movements occur on parallel arrays of iMTs. Moreover, SPB-directed movements were significantly faster and shorter than midzone-directed movements (Table I), indicating that movements in the two directions were genuinely different. Thus, taken together, our data indicate that on the anaphase spindles, Cin8 is a bi-directional motor that moves in both plus-end and minus-end directions of the MTs.
Figure 1.
In vivo, Cin8 occasionally switches directionality. (A) 2D-projection time-lapse confocal fluorescence images of Cin8–3GFP localization in WT cells. Time intervals: 1 min. (B) Model of Cin8 plus-end and minus-end directed movement in the anaphase spindle in S. cerevisiae cells. (C, D) Kymographs of Cin8–3GFP localization to the anaphase spindle. (A, C, D) Top arrows: spindle poles; asterisk: spindle midzone. Arrowheads: plus-end-directed movements towards the midzone; yellow arrows: minus-end directed movements towards the poles.
Table 1. Midzone- and SPB-directed movements of Cin8–3GFP variants in the anaphase spindles.
| Cin8 variant | N a | Midzone-directed movements |
SPB-directed movements |
||||
|---|---|---|---|---|---|---|---|
| % Of spindlesb | Velocity (nm/s) (n)c,d | Run length (nm) (n)c,d | % Of spindlese | Velocity (nm/s) (n)c | Run length (nm) (n)c | ||
| WT Cin8 | 113 | 59 | 26.5±0.7 (120) | 1506±50 (120) | 20 | 56.4±4.2 (29)***f | 845±43 (29)***f |
| Cin8Δ99 | 83 | 29 | 15.3±0.5 (123)***g | 1091±35 (123)***g | 18 | 55.8±2.9 (33)***f | 891±30 (33)*f |
| Cin8-2A | 102 | 41 | 23.0±0.8 (183)*g | 1236±34 (183)***g | 20 | 63.6±3.1 (32)***f | 943±62 (32)**f |
| aN—number of intermediate-long (5–10 μm) spindles analysed. | |||||||
| b% Of spindles with at least one midzone-directed movement spanning from SPB to the midzone (see Figure 5). | |||||||
| cValues represent average±s.e.m., n—number of movements. | |||||||
| dValues determined for movements longer than 0.5 μm, including movements that do not reach the midzone. | |||||||
| e% Of spindles with at least one SPB-directed movement, longer than 0.5 μm. | |||||||
| fCompared with the corresponding movements in cells of the same genotype. | |||||||
| gCompared with cells expressing WT Cin8. | |||||||
| ***P<0.0001; **P<0.001; *P<0.005 (Student's t-test). | |||||||
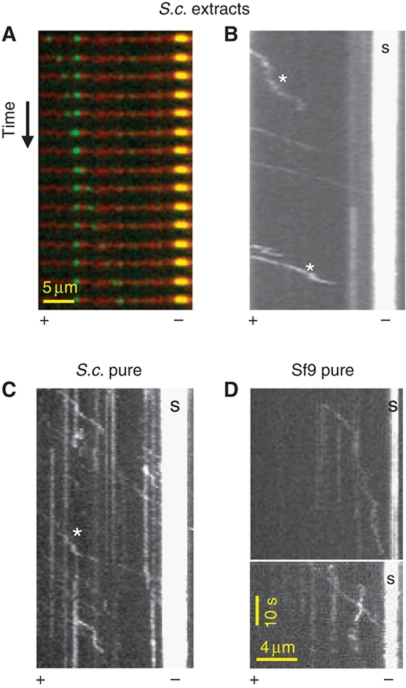
To explore the unconventional minus-end motility of Cin8 on the spindle, we performed single-molecule fluorescence in-vitro motility assays (Vale et al, 1996; Kwok et al, 2006; Kapitein et al, 2008). We used whole-cell extracts of WT and cin8Δ S. cerevisiae cells, in which Cin8–3GFP was expressed from its native promoter, either integrated into the genome or carried on a centromeric (CEN) plasmid (Materials and methods; Supplementary Table S1). We also studied Cin8–GFP purified from Sf9 and yeast cells (Materials and methods; Supplementary Figure S1). Assays were done at saturating ATP (3–5 mM; Lakamper et al, 2010) in a high-salt, close to physiological buffer (175 mM NaCl; Gheber et al, 1999). We found, surprisingly, that in these conditions Cin8 moved unidirectionally and processively towards the minus end of MTs (Figure 2). This is similar to what was recently found in an independent study (Roostalu et al, 2011). Runs were often interrupted by diffusive motion (Figure 2B and C). The average run length between such breaks was ∼2 μm (Figure 3B). In ADP buffer, motors remained attached to MTs, but only moved diffusively without directional bias. We calculated a diffusion coefficient of ∼3 × 103 nm2/s from mean-square-displacement (MSD) analysis of tracks recorded in ADP buffer, comparable to that measured with the closely related Eg5 and other kinesins (Table II; Kwok et al, 2006; Kapitein et al, 2008; Bormuth et al, 2009). At saturating ATP concentration, we determined an average minus-end velocity of Cin8 molecules of ∼360 nm/s by kymograph and mean displacement (MD) analyses (Table II). This velocity is substantially higher than the plus-end velocity seen in vivo and the velocity reported for other kinesin-5 motors (Valentine et al, 2006). In both WT Cin8 batches examined, purified Cin8–GFP and Cin8–3GFP in whole extracts, the distribution of velocities was exceptionally broad in each experiment, ranging from ∼17 to ∼830 nm/s (Supplementary Figure S2A; Figure 4A(I) and B(I)). We ruled out the possibility that the broad distribution of velocities resulted from motor aggregation by analysing fluorescence intensities of individual spots (Supplementary Figure S3).
Figure 2.
At high salt in vitro, Cin8 is a minus-end directed motor. (A) Time-lapse recording showing a single Cin8–3GFP molecule from whole-cell extract (green) moving directionally along the MT (red) towards the bright seed (yellow). Time intervals: 2 s. (B–D) Kymographs of Cin8 movement on polarity-marked MTs. (B) Cin8–3GFP from whole-cell extract (C) Cin8–GFP purified from yeast cells and (D) Cin8–GFP purified from Sf9 cells. S: bright seed indicating the MT minus end. Scale bars in (D) also apply to (B) and (C). * indicates diffusive episotes. See also Supplementary Movies S2 and S3.
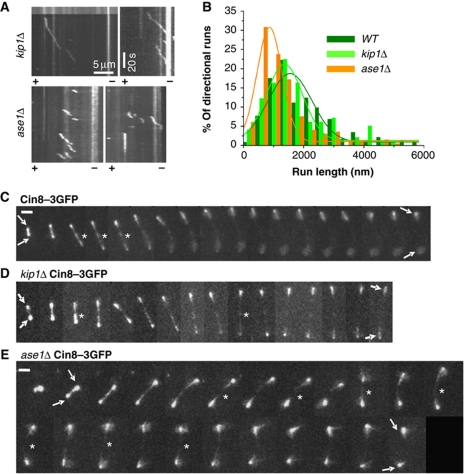
Figure 3.
The effect of spindle proteins on Cin8 in-vitro motile properties and in-vivo localization to anaphase spindles. (A, B) Single-molecule fluorescence motility assay was carried out on polarity-marked MTs with kip1Δ or ase1Δ cell extracts expressing Cin8–3GFP. Extracts were diluted in high-salt (175 mM NaCl) buffer. (A) Representative kymographs of in-vitro runs of Cin8–3GFP in the different extracts. (B) Histograms of run lengths of Cin8–3GFP directional episodes in WT (olive), kip1Δ (light green) or ase1Δ (orange) extracts. Fit: Gaussian distribution. (C–E) Representative 2D-projection time-lapse confocal fluorescence images of Cin8–3GFP localization to anaphase spindles in (C) WT (D) kip1Δ and (E) ase1Δ cells. Asterisk: spindle midzone; Arrows: spindle poles; Time interval between frames: 1 min; Scale bar: 2 μm. Genotypes and Cin8 variants are marked on top.
Table 2. In-vitro motile properties of Cin8 in whole-cell extracts.
| Genotype | VKYMOa (nm/s) | VMDb,c (nm/s) | DADP × 103 c,d (nm2/s) | Stall periodse (%) | Run lengthf (nm) | Interaction timeg (s) |
|---|---|---|---|---|---|---|
| WT (pCIN8-3GFP)h | 353±33 (116) | |||||
| CIN8-3GFP k | 365±27 (178) | 372±7i (52) | 3.7±0.1i (30) | 39 (1645)j | 1700±200 (94) | 8.3±1.0 (65) |
| cin8Δ kip1Δ (pCIN8-3GFP)h | 381±38 (100) | 345±8 (70) | 2.9±0.1***l (24) | 20 (1377)j | 2000±200 (132) | 9.5±0.9 (115) |
| ase1Δ (pCIN8-3GFP)h | 241±17***l (198) | 272±8***l (39) | 4.2±0.2 (23) | 33 (1732)j | 1200±100**l (118) | 8.4±1.0 (66) |
| Values are average±s.e.m. In parentheses, the number of measurements. | ||||||
| aVKYMO—average velocity determined from kymograph analysis. | ||||||
| bVMD—average velocity determined from Mean Displacement analysis. | ||||||
| cFit is done to first 15 s. | ||||||
| dDADP—diffusion coefficient in the presence of 3–5 mM ADP determined from MSD analysis. | ||||||
| ePercentage calculated from total motion time. | ||||||
| fRun length of directional episodes. | ||||||
| gMeasurements applied on kymograph traces of complete directional runs, of which both their beginning and end are apparent. | ||||||
| hCin8–3GFP expressed on CEN plasmid. | ||||||
| iMeasurements carried out on Cin8-3GFP either expressed on CEN plasmid or integrated into the genome. | ||||||
| jTotal time of measurement (s). | ||||||
| kCin8–3GFP integrated into the genome. | ||||||
| lCompared with WT. | ||||||
| ***P<0.0001; **P<0.001 (Student's t-test). | ||||||
| ND—not detected. | ||||||
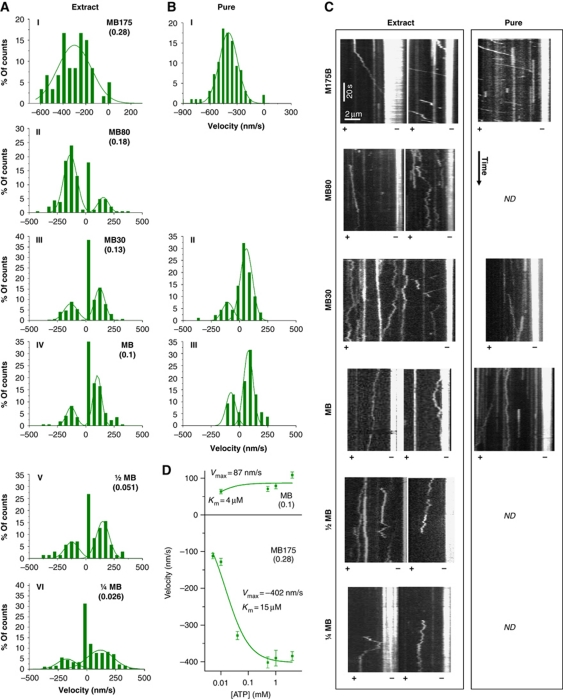
Figure 4.
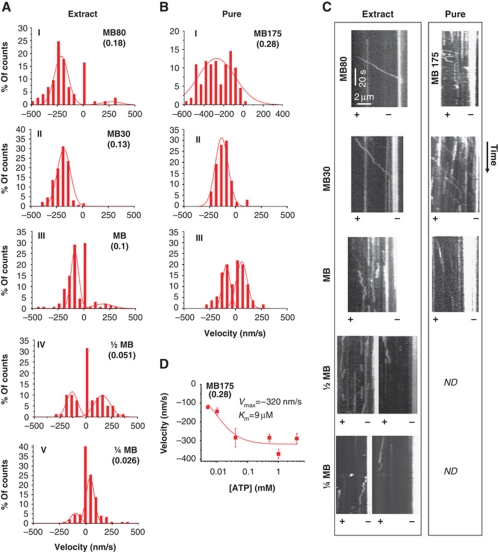
Decrease in ionic strength induces plus-end motility of single molecules of Cin8. (A, B) Histograms of velocities of Cin8 in whole-cell yeast extracts (A) or affinity purified (B) with saturating ATP. Salt and buffer conditions are indicated for each panel: MB—motility buffer; numbers adjacent to ‘MB’ indicate the concentration (mM) of added NaCl. Ionic strength (M) is indicated in parentheses. A control experiment of motility with ADP was carried out using buffer with 30 mM NaCl (see Supplementary Figure S2C). Velocity histograms were assembled by dividing kymograph traces in 3 s segments and piecewise linear fitting. Lines—Gaussian distribution fit. (C) Representative kymographs of Cin8 motility along polarity-marked MTs. Expression/purification conditions are indicated on top. Plus (+) and minus (−) ends of MTs are indicated. See also Supplementary Movie S4. (D) ATP concentration dependence of plus-end (top) and minus-end directed (bottom) velocity of Cin8 (average±s.e.m.). NaCl concentration and ionic strengths (parenthesis) are indicated. Michaelis–Menten parameters Vmax and Km are indicated.
To make sure that Cin8–3GFP was capable of forming functional tetramers, we examined its MT bundling activity. We found that overexpression of Cin8–3GFP caused extensive bundling of MTs in whole-cell extracts (Supplementary Figure S4A, left panel) which was not observed in extracts deleted for CIN8 (Supplementary Figure S4A, right panel) or in extracts expressing Cin8–3GFP from a CEN plasmid (not shown). This excludes the possibility that the bundling was caused by other motors or MT-binding proteins in the extracts. Similar MT bundling was previously reported for biotinylated Cin8 (Cin8-BCP; Gheber et al, 1999), which was shown to be a homotetramer (Hildebrandt et al, 2006). MT bundling is known to occur only with the full-length tetrameric, but not with truncated dimeric Cin8 (Cin8-871) (Gheber et al, 1999; Hildebrandt et al, 2006), suggesting that Cin8–3GFP is a tetramer. Finally, both Cin8–GFP and Cin8–3GFP could maintain the viability of cells deleted for chromosomal copies of CIN8 and KIP1 (Supplementary Figure S4B). Since only tetrameric Cin8 maintains viability of cin8Δkip1Δ cells (Hildebrandt et al, 2006), our findings indicate that Cin8–3GFP is mostly a functional tetramer.
In summary, our data indicate that the motile properties we observed for Cin8, the broad distribution of velocities, the minus-end directionality and the diffusive motion, did not result from altered oligomeric states or aggregation of the examined Cin8 variants, but rather point to an intrinsic regulation of individual Cin8 motors.
The use of whole-cell yeast extracts from deletion mutants in single-molecule motility assays allowed us to examine the influence of other S. cerevisiae cellular factors on Cin8 motility (Table II; Figure 3). We explored the effect, in high-salt buffer, of two major spindle-binding proteins that functionally overlap or interact with Cin8: the kinesin-5 Kip1 (Hoyt et al, 1992; Roof et al, 1992) and the MT-binding and midzone-organizing protein Ase1 (Schuyler et al, 2003). Ase1 was shown to physically interact with Cin8 during mitosis and to recruit Cin8 to the spindle midzone (Khmelinskii et al, 2009).
To examine the influence of Kip1 and Ase1 on Cin8 motor function, we examined Cin8 motility in extracts of kip1Δ or ase1Δ cells, which carried Cin8–3GFP on a CEN plasmid. We found that deletion of Kip1 or Ase1 did not affect the directionality of Cin8, as it remained minus-end directed in high-salt buffer (Figure 3A). In the absence of Kip1, Cin8 was somewhat more processive; its interaction time with MTs was longer, the percentage of runs with stalls was lower, and the diffusion coefficient of Cin8 in the presence of ADP was significantly lower than that of Cin8 in all the other extracts examined (Table II; Figure 3B). Consistently, in kip1Δ cells, the detachment of Cin8 from the spindle MTs in late anaphase was reduced compared with WT cells (Figure 3C and D). In the absence of Ase1, the average run length of Cin8–3GFP directional episodes was significantly decreased, and the average velocity dropped to ∼240 nm/s (Table II; Figure 3A and B). In cells, Cin8–3GFP localized abnormally on the anaphase spindle in the absence of Ase1 and was not detectable in the midzone (Figure 3E). These results suggest that Ase1 regulates Cin8 spindle localization by affecting the motile properties of Cin8.
It remains to understand the circumstances under which the motor switches to plus-end motility, the mode of motion that is believed to drive the poleward relative sliding of antiparallel overlapping MTs in anaphase. Since other kinesins, including the kinesin-5 Eg5, can be turned on and off by varying ionic conditions (Dietrich et al, 2008; Hancock, 2008; Kapitein et al, 2008; Hackney et al, 2009), we tested whether a decrease in ionic strength could affect directionality. We performed single-molecule motility assays with Cin8, either purified or in whole-cell extracts, on polarity-marked MTs at different salt concentrations. We found that with decreasing ionic strength, individual Cin8 motors gradually switched towards plus-end motility (Figure 4A–C). At ionic strengths ⩽0.13, Cin8 moved bi-directionally, moving 60–80% of the time towards the plus ends of MTs (Figure 4A–C). Reduction of ionic strength also significantly decreased the magnitude of the minus-end velocity of Cin8 (Figure 4A–C) from ∼380 nm/s in high ionic strength to ∼130 nm/s in low ionic strength (Supplementary Figure S5). The salt dependence of Cin8 motility in whole-cell extracts was similar to that of affinity-purified Cin8 (Figure 4A and B), although more motors were immobile in the extracts (Figure 4A and B), possibly due to the presence of other MT-binding agents. The velocity of movement in both minus- and plus-end directions was dependent on ATP concentration (Figure 4D; Supplementary Figure S2C), proving that bi-directional motion of single Cin8 molecules is not merely driven by thermal forces in the solution. These results demonstrate that a single Cin8 motor interacting with only one MT can switch directionality. The mechanism for the switch is, therefore, most likely contained in a single motor.
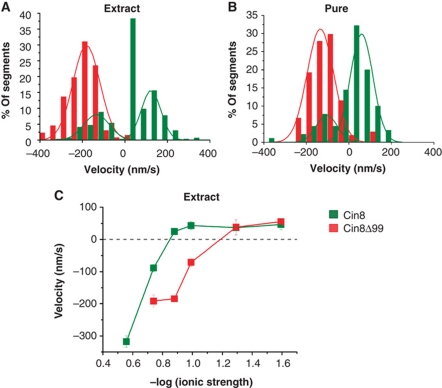
Any switch mechanism is likely to depend, at least in part, on structural elements within the motor's catalytic domains. In comparison with kinesin-5 homologues of higher eukaryotes, several yeast kinesin-5 motors carry inserts of considerable length in loop 8 which is involved in MT binding (Kull et al, 1996; Nitta et al, 2008; Chee and Haase, 2010). Cin8 and the closely related Candida glabrata kinesin-5 carry the largest inserts, of 99 amino-acid length (99aa) (Chua et al, 2007). To probe the role of the large loop 8 in the directionality switch of single Cin8 molecules, we studied a construct in which this segment was replaced with the seven amino acids of loop 8 in the related S. cerevisiae kinesin-5 Kip1 (Cin8Δ99). Yeast cells deleted for the chromosomal copies of CIN8 and KIP1, but expressing Cin8 carrying this deletion were previously shown to be viable, indicating that the mutant Cin8 is, at least partially, functional (Hoyt et al, 1992). We found that with decreasing ionic strength, single Cin8Δ99 molecules also switched from minus-end to plus-end directed motility in whole extracts (Figure 5A) and in purified samples (Figure 5B). However, Cin8Δ99–GFP behaved distinctly differently from WT Cin8–GFP (Figures 4,5,6). In whole-cell extracts, at high salt (175 mM NaCl), Cin8Δ99–GFP did not attach to MTs at all. The same was seen for purified WT Cin8, albeit only at >250 mM NaCl. At 175 mM added NaCl, affinity-purified Cin8Δ99 had a slightly lower average velocity than WT Cin8 (Supplementary Figure S5). With 30 mM added NaCl, the presence or absence of loop 8 in the WT Cin8 correlated with clearly opposite behaviours: in whole extracts and purified samples, Cin8Δ99 remained minus-end directed, while WT Cin8 moved predominantly in the plus-end direction with some bi-directional shuttling (Figure 6A and B). A systematic comparison between Cin8 and Cin8Δ99 motility in different buffers showed that the deletion of the 99aa insert did not eliminate the switch of directionality, but pushed the transition from minus-end to plus-end directionality to lower salt concentrations (Figure 6C), supporting the notion that the directionality switch of Cin8 involves loop 8.
Figure 5.
Decrease in ionic strength induces plus-end motility of single molecules of Cin8Δ99. (A, B) Histograms of velocities of Cin8Δ99 in whole-cell yeast extracts (A) or affinity purified (B) with saturating ATP. Salt and buffer conditions are indicated for each panel: MB—motility buffer; numbers adjacent to ‘MB’ indicate the concentration (mM) of added NaCl. Ionic strength (M) is indicated in parentheses. Velocity histograms were assembled by dividing kymograph traces in 3 s segments and piecewise linear fitting. A control experiment of motility with ADP was carried out using buffer with 30 mM NaCl (see Supplementary Figure S2C). Lines—Gaussian distribution fit. (C) Representative kymographs of Cin8Δ99 motility along polarity-marked MTs. Expression/purification conditions are indicated on top. Plus (+) and minus (−) ends of MTs are indicated. See also Supplementary Movie S5. (D) ATP concentration dependence of minus-end velocity of Cin8Δ99 (average±s.e.m.) at high ionic strength. NaCl concentration and ionic strength, M (parenthesis) are indicated. Michaelis–Menten parameters Vmax and Km are indicated.
Figure 6.
Deletion of the 99aa insert of Cin8 induces bias towards minus-end motility. (A, B) Histograms of velocities of Cin8 (green) and Cin8Δ99 (red) in whole-cell yeast extracts (A) or affinity purified (B) in buffer containing 30 mM NaCl, with saturating ATP. Velocity histograms were assembled by dividing kymograph traces in 3 s segments and piecewise linear fitting. Lines—Gaussian distribution fit. See also Supplementary Movies S4 and S5. (C) Ionic strength dependence of mean velocity of Cin8 (green) and Cin8Δ99 (red) in whole-cell extracts in saturating ATP conditions. Values represent mean s.e.m. of 50–200 velocity values.
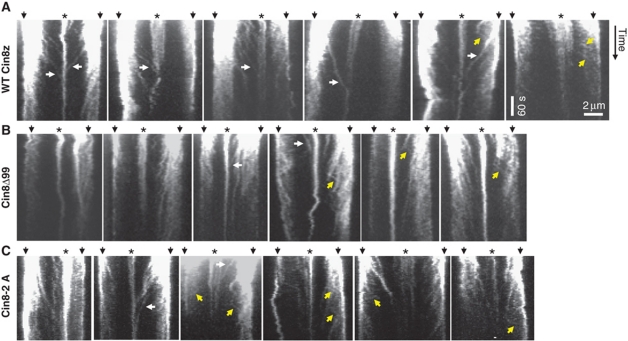
To examine how the bias to minus-end directionality of the mutant Cin8Δ99 affects its function in vivo, we monitored motor localization in cells expressing Cin8Δ99. In contrast to WT Cin8–3GFP, Cin8Δ99–3GFP did not detach from the spindles and seemed to be asymmetrically distributed on the anaphase spindles (Supplementary Figure S6). The plus-end directed movements of Cin8Δ99–3GFP towards the midzone were fewer, significantly slower than that of Cin8–3GFP and spanned shorter distances (Figure 7; Table I). Nonetheless, minus-end directed motility events were also observed (Figure 7B). Finally, the rate of the initial fast phase of anaphase B in Cin8Δ99 cells was 0.61±0.06 μm/min (average±s.e.m., n=9) which is significantly slower than in WT cells (0.85±0.03 μm/min, average±s.e.m., n=8, P<0.005) and is similar to what was observed in cin8Δ cells (Straight et al, 1998). This result indicates that Cin8Δ99 is unable to provide sufficient plus-end directed force for spindle elongation. These in-vivo data are consistent with our in-vitro results of diminished plus-end motility of Cin8Δ99 (Figure 5) and point to a regulatory role of the 99aa insert in loop 8 of Cin8 in promoting its plus-end motility in cells.
Figure 7.
Movements of Cin8–3GFP (A), Cin8Δ99–3GFP (B) and Cin8-2A–3GFP (C) in anaphase spindles. Kymographs of movements of Cin8–3GFP variants (indicated on the left) in intermediate to long anaphase spindles. White arrows: midzone-directed movements that span from the SPBs to the midzone; Yellow arrows: SPB-directed movements; White arrowheads: movements towards the SPB during spindle disassembly; Top arrows: spindle poles; Asterisks: midzone.
It is known that during anaphase, Cin8 localization to the spindle is regulated by phosphorylation of the three Cdk1 sites in its catalytic domain, two of which are located in loop 8 of Cin8 (Avunie-Masala et al, 2011). To examine if phosphorylation of these sites affects Cin8 directionality on the spindle, we examined spindle movements of a Cin8–3GFP mutant that carried phosphorylation-deficient mutations to alanine at the two Cdk1 sites located in loop 8: Cin8-S277A T285A (Cin8-2A). Similarly to a phosphorylation-deficient Cin8 that carried mutations to alanine at all three catalytic-domain Cdk1 sites (Avunie-Masala et al, 2011), Cin8-2A–3GFP remained attached to the anaphase spindles for longer times, compared with cells expressing WT Cin8 (Supplementary Figure S6B, compare with Figures 1A and 3C). This result indicates that the two Cdk1 sites within loop 8 are involved in the regulation of Cin8 spindle localization. Examination of Cin8-2A–3GFP movements on the spindle revealed that this mutant exhibited both midzone (plus)- and SPB-directed (minus) movements (Figure 7C), with SPB-directed movements being shorter and faster than midzone-directed movements (Table I). The movements of Cin8-2A–3GFP were similar to those of Cin8Δ99–3GFP: midzone-directed movements were fewer, and significantly slower and shorter compared with the spindle movements of the WT Cin8–3GFP (Figure 7; Table I), indicating that Cin8-2A is impaired in its midzone (plus-end)-directed motility on the spindle. This result suggests that the bias towards plus-end directed motility by the 99aa insert in loop 8, stems, at least in part, from phosphorylation of the Cdk1 sites located in this insert.
An important part of the switch mechanism might be related to cargo activation of Eg5. The homologous Xenopus kinesin-5 Eg5 is only turned on to move processively when it crosslinks two MTs, that is, when both of its dimeric catalytic domains are engaged (Kapitein et al, 2008). This mechanism was suggested to be related to the cargo activation of dimeric kinesins by straightening of the back-folded stalk and tail (Hackney et al, 1992; Stock et al, 1999; Seiler et al, 2000).
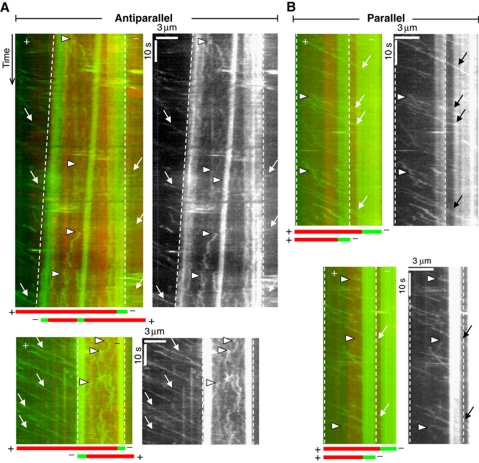
To see if a similar mechanism might be responsible for switching the directionality of Cin8, we performed single-molecule fluorescence experiments on bundled pairs of polarity-marked MTs. Between antiparallel MTs in high-salt buffer (175 mM added NaCl), we saw an immediate switch from fast minus-end directed motion of individual motors to a slow erratic motion without a clear directional bias when motors reached the overlap zone (Figure 8A). In this geometry, the two crosslinked MTs were typically sliding apart with their minus ends leading with a relative velocity of about 30–60 nm/s. This reflects force generation by the motors in the plus-end direction. The fact that in the overlap region, extended fast motion was no longer observed at all implies that motors are attracted to the overlap region, likely due to their ability to bind MTs through both ends of the tetramers. In contrast to the antiparallel case, it was evident from kymographs of single motor motility between parallel MTs (Figure 8B) that most motors kept moving in the minus-end direction at undiminished speed when entering the overlap zone. Occasional short plus-end excursions were observed between parallel MTs (Figure 8B, arrowheads), which were not observed on single MTs. These events were too rare to reliably evaluate details. We thus conclude that one of the major determinants of Cin8 directionality is binding geometry, with binding between two antiparallel MTs, as it occurs in the spindle midzone, switching the motor from minus-end to plus-end motility in near-physiological salt conditions.
Figure 8.
MT orientation changes the motile properties of single Cin8 molecules. Kymographs of movements of purified Cin8–GFP between antiparallel (A) and parallel (B) MTs in high-salt buffer (MB-175). For each event, a merged kymograph in colour (red—MT; green—Cin8 and minus ends of MTs) is shown on the left and a kymograph of the GFP-channel only is shown on the right. Overlapping region between MTs is marked by dashed lines. Cartoon depicting the orientation of overlapping MTs is shown at the bottom of the colour kymographs. Arrows: minus-end directed motility events; arrowheads: plus-end directed motility events. See also Supplementary Movie clips S6,S7,S8,S9,S10.
Discussion
In the kinesin superfamily, the majority of the members are plus-end directed. Until recently (Roostalu et al, 2011), minus-end motion was seen only for kinesin-14 family members, which are structurally distinct from all other kinesin subfamilies in that they carry the conserved motor domain at the C-terminus instead of the N-terminus (McDonald et al, 1990; Walker et al, 1990; deCastro et al, 2000; Block, 2007). No full-length kinesin-14 has been found to be processive so far, that is, these motors produce isolated power strokes and can only produce persistent motion when acting in ensembles. A reversal of power stroke directionality has been reported for mutants of the non-processive kinesin-14 ncd from Drosophila melanogaster (Sablin et al, 1998; Endow and Higuchi, 2000) and could be also generated by swaps of the core and neck domains of ncd and kinesin-1 (Case et al, 1997; Endow and Waligora, 1998). Evidence for active bi-directionality of a given motor construct has been reported for a specific neck-domain mutant of ncd (Endow and Higuchi, 2000) and for cytoplasmic dynein (Dixit et al, 2008). Evidence for bi-directional power strokes of individual ncd motors has also been seen in the analysis of single-molecule recordings (Butterfield et al, 2010).
Here, we show an entirely novel behaviour for a kinesin motor. Individual kinesin-5 Cin8 motors could be switched by varying ionic conditions between processive minus- and plus-directed movements when travelling on single MTs (Figure 4), and they could be switched from processive minus-end motion to plus-end force generation in high-salt conditions by binding and crosslinking two MTs (Figure 8). In vitro, low ionic strength (an unphysiological environment) induced plus-end directed motion of single molecules, while high ionic strength induced minus-end directed motion. Lower ionic strength, in general, reduces electrostatic screening which, in turn, enhances electrostatic interactions between motor subelements or between motor and MT. Thus, the unphysiological change of ionic conditions might mimic the effects of phosphorylation or binding of accessory proteins, which modify electrostatic interactions under constant physiological conditions. A similar phenomenon was reported for cargo regulation of kinesins. Binding of a cargo vesicle to kinesin-1 or of a second MT to kinesin-5 Eg5, respectively, can activate the motors, but the activation also occurs at low ionic strength. In the case of Cin8, a related mechanism might not just turn the motor on or off, but lead to the switching of directionality when the motor tetramer binds between two antiparallel MTs. An alternative model that was recently proposed (Roostalu et al, 2011) relies on a collective effect involving physical load on the motors via the binding between MTs. Based on our findings, it appears that more individual mechanisms such as binding of a single motor between two MTs or phosphorylation in the catalytic domain are able to cause or modify directionality switching.
A case in point is the observed regulatory influence of the large 99aa insert in loop 8 of the Cin8 motor domain, deletion of which did not abolish the shift in directionality, but created a strong bias towards minus-end motility (Figures 5 and 6). The mechanism by which phosphorylation in Cin8 catalytic domain regulates its in-vivo function is likely to be a combination of a number of factors such as interaction with the midzone-organizing Ase1 (Khmelinskii et al, 2009), as was previously suggested (Avunie-Masala et al, 2011), or with kinetohore proteins. The fact that Cin8Δ99 and the phosphorylation-deficient Cin8-2A exhibited reduced motility towards the midzone (Figure 7; Table I) suggests that one of the roles of Cin8 phosphorylation in the 99aa insert is to mediate the switch to plus-end directed motility of Cin8 on the spindle.
The observation that switching of directionality at high salt only occurred between antiparallel MTs is consistent with the reported preference of Drosophila kinesin-5 Klp61f for bundling antiparallel MTs (Kapitein et al, 2008). A preferred orientation was for that motor conferred by the ATP-independent binding sites in the C-terminal tail of the molecules. An ATP-independent binding mechanism appears to also exist for Cin8 because full-length Cin8 also supports diffusive MT attachment in ADP buffer (Table II). Sticky tails with preferred orientation might not prevent parallel crosslinking by the motor, but it was found for Eg5 that all eight binding sites were necessary for motor engagement between MTs (Weinger et al, 2011). It is tempting to speculate that this might also be the case for Cin8, but in this case with the further consequence that directionality is reversed to plus-end motion between the antiparallel MTs. As the spindle midzone is the place where antiparallel overlaps occur and as that is the location where the motors need to exert force, such a regulation appears advantageous.
The discovery of the exceptional properties of Cin8 raises the question how these motile properties aid Cin8 in performing its multiple mitotic roles. The ionic strength in S. cerevisiae cells is high, estimated as ∼300 mM salt (Olz et al, 1993; van Eunen et al, 2010). Under these conditions, Cin8 motors were minus-end directed on single MTs in our in-vitro experiments (Figures 2, 4 and 6). Prior to spindle elongation, Cin8 is known to be important for kinetochore clustering or positioning near the SPBs (Tytell and Sorger, 2006; Gardner et al, 2008a; Wargacki et al, 2010). The proposed mechanism for this function had been the crosslinking of kinetochore MTs (kMTs; Tytell and Sorger, 2006) and the promotion of disassembly of long kMTs (Gardner et al, 2008a). Since in S. cerevisiae cells, each kinetochore is attached to a single MT and since on a single MT Cin8 is minus-end directed, active motion of Cin8 in the minus-end direction of the kMTs may be an alternative/additional mechanism by which Cin8 contributes to kinetochore positioning.
The slow plus-end directed motility that we observed in vivo in anaphase spindles (Figures 1C and 7; Table I) indicates that Cin8 is switched to plus-end directed motility in the cell, even on single MTs or on parallel bundles. During anaphase spindle elongation, bi-directionality is likely to be important to dynamically partition Cin8 motors between different reservoirs, that is, near the poles where they focus the kinetochore clusters (Tytell and Sorger, 2006; Gardner et al, 2008a; Wargacki et al, 2010) and in the midzone where Cin8 promotes plus-end directed MT sliding (Figure 8; Roostalu et al, 2011) and spindle elongation (Saunders et al, 1995; Movshovich et al, 2008). In fact, we observed that until anaphase spindles reach a length of ∼5 μm, Cin8 is localized throughout the spindle, with no preferential accumulation at the midzone or near the spindle poles, nor obvious detachment (Figures 1A and 3C). A way to maintain this even distribution without detachment might be bi-directional motility of Cin8. Interestingly, higher eukaryotes, which show poleward flux in the spindle MTs, appear not to have kinesin-5 motors capable of minus-end motility, possibly because it became unnecessary for motor transport to the poles in fluxing spindles.
A factor that appears to be important for directionality is the geometry of binding and allosteric regulation by two bound antiparallel MTs. Intermediate-long S. cerevisiae anaphase spindles contain a small number of MTs, two emanating from each pole at the end of anaphase (Winey et al, 1995). Motility between antiparallel MTs emanating from opposing poles should move both MTs and keep the motor fixed in the midzone. Therefore, the plus-end motility observed in vivo can only take place on single or between parallel MTs and is likely to utilize a further mode of regulation without which the motors would rapidly converge back to the poles.
In conclusion, Cin8 has turned out to be an exceptional kinesin in that it is truly bi-directional and processive in both directions. This unique feature of Cin8 appears to play a role in cellular function. First, hints about the molecular mechanism indicate a role of charge interactions and possibly phosphorylation, and most importantly binding geometry between pairs of MTs. It remains to be explored in more detail if Cin8 regulation is a variation of the scheme of the regulation of other kinesins, in particular kinesin-5 motors. In general, our findings demonstrate that in order to fulfil their physiological functions, kinesin motors are much less rigidly programmed than was broadly believed so far and that the extent to which their function is regulated in the cell encompasses much more than simple on-off switches.
Materials and methods
Detailed procedures and S. cerevisiae strains used in this study are described in Supplementarydata. In brief, we produced fluorescently labelled Cin8 motors in three ways. We first used whole-cell extracts of S. cerevisiae expressing Cin8 fused with three consecutive C-terminal GFPs (Cin8–3GFP) under its own promoter. Cin8–3GFP was either integrated into the yeast genome or expressed from a CEN plasmid. Cells with integrated Cin8–3GFP were also used for in-vivo imaging (Supplementary Table S1). Second, we expressed Cin8 fused with a single C-terminal GFP and N-terminal 6HIS tag (6HIS–Cin8–GFP) in Sf9 insect cells, and third, we overexpressed Cin8–GFP–6HIS in S. cerevisiae. For yeast strains and plasmids, see Supplementary Table S1 (Supplementary data). We purified motor by HIS tag and MT affinity.
Live-cell imaging was done on a spinning-disc confocal microscope (Zeiss Axiovert 200M, UltraView ESR, Perkin-Elmer, UK; Fridman et al, 2009). Z-stacks of 0.2–0.4 μm separation were acquired in 1-min time intervals (Movshovich et al, 2008). For the Cin8–3GFP spot-motility analysis, images were acquired every 2 s.
In-vitro motility assays were performed following standard procedures (Howard et al, 1993; Gheber et al, 1999; Lakamper et al, 2010) in motility buffer MB-175 (50 mM Tris/HCl, 30 mM PIPES/KOH, final pH 7.2, 175 mM NaCl, 2 mM EDTA, 1 mM EGTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride and 1 mM dithiothreitol) as well as versions of this buffer with less NaCl added. The MTs, polymerized from TMR-labelled porcine tubulin, were polarity marked using Atto-488-labelled seeds marking the minus end of the MTs. Single-molecule fluorescence data were collected on two microscopes, one at Ben-Gurion University (BGU) and one at Göttingen University (GAUG). BGU: Zeiss Axiovert 200M, HBO 100 Mercury Illuminator, cooled CCD (SensiCam, PCO), frame time 0.8 s. Data were processed using ImageJ and MetaMorph (MDS Analytical Technologies) software. GAUG: custom-built total-internal-reflection fluorescence microscope, using a 473-nm Laser (Viasho, USA) for excitation, and a × 100 objective (Nikon, SFluor, NA 1.49, Oil) and a CCD camera (Cascade 512B, Roper Scientific, USA), frame rate 0.5 s. Software was custom written in Labview. Velocity histograms were assembled by drawing lines through consecutive 3 s segments of kymograph traces.
For relative sliding assays, polarity-marked MTs were polymerized as before whereas a solution of shorter MTs was polymerized by incubation at 37°C for only 6 min. First, the long polarity-marked MTs were allowed to bind for 3 min to the DETA-coated surface of the assay chamber. The motility buffer (MB-175) was the same as used for the single-molecule assays but with double ATP and MgCl2 concentration. To this buffer, three times the single-motor concentration and 1 μl of short polarity-marked MTs were added, and the mix was washed into the assay chamber. The custom-built TIRF set-up described before was expanded such that the emission of the TMR-labelled MTs and the GFP-labelled motor proteins could be detected simultaneously. The TMR and the GFP channel were aligned with ImageJ.
Supplementary Material
Acknowledgments
This work was supported in part by the Lower Saxony Grant no. 11-76251-99-26/08 (ZN2440) awarded to LG, SL and CFS. LG was supported by the ISF grant no. 1043/09 and the BSF grant no. 2003141 and CFS was supported by the Center for Molecular Physiology of the Brain (CMPB), funded by the Deutsche Forschungsgemeinschaft (DFG). We thank Yael Nissenkorn, BGU, Israel for providing Cin8-2A plasmids and to Florian Rehfeldt and Marcel Bremerich, GAUG, Germany for data analysis.
Author contributions: AGG, CT, NM, VF, MP, TD, SL and DRK performed the experiments; AGG, CT, CFS and LG analysed the data and wrote the paper; CFS and LG supervised and coordinated the project. All authors read and commented on the draft versions of the manuscript and approved the final version.
Footnotes
The authors declare that they have no conflict of interest.
References
- Avunie-Masala R, Movshovich N, Nissenkorn Y, Gerson-Gurwitz A, Fridman V, Koivomagi M, Loog M, Hoyt MA, Zaritsky A, Gheber L (2011) Phospho-regulation of kinesin-5 during anaphase spindle elongation. J Cell Sci 124: 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d’Herin P, Harper M, Kress M, Nigg EA (1995) Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell 83: 1159–1169 [DOI] [PubMed] [Google Scholar]
- Block SM (2007) Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys J 92: 2986–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormuth V, Varga V, Howard J, Schaffer E (2009) Protein friction limits diffusive and directed movements of kinesin motors on microtubules. Science 325: 870–873 [DOI] [PubMed] [Google Scholar]
- Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM (2004) Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci USA 101: 15938–15943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield AE, Stewart RJ, Schmidt CF, Skliar M (2010) Bidirectional power stroke by ncd kinesin. Biophys J 99: 3905–3915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case RB, Pierce DW, Hom-Booher N, Hart CL, Vale RD (1997) The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell 90: 959–966 [DOI] [PubMed] [Google Scholar]
- Chee MK, Haase SB (2010) B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast. PLoS Genet 6: e1000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Roof DM, Lee Y, Sakowicz R, Clarke D, Pierce D, Stephens T, Hamilton M, Morgan B, Morgans D, Nakai T, Tomasi A, Maxon ME (2007) Effective killing of the human pathogen Candida albicans by a specific inhibitor of non-essential mitotic kinesin Kip1p. Mol Microbiol 65: 347–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCastro MJ, Fondecave RM, Clarke LA, Schmidt CF, Stewart RJ (2000) Working strokes by single molecules of the kinesin-related microtubule motor ncd. Nat Cell Biol 2: 724–729 [DOI] [PubMed] [Google Scholar]
- Dietrich KA, Sindelar CV, Brewer PD, Downing KH, Cremo CR, Rice SE (2008) The kinesin-1 motor protein is regulated by a direct interaction of its head and tail. Proc Natl Acad Sci USA 105: 8938–8943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Ross JL, Goldman YE, Holzbaur EL (2008) Differential regulation of dynein and kinesin motor proteins by tau. Science 319: 1086–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Higuchi H (2000) A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature 406: 913–916 [DOI] [PubMed] [Google Scholar]
- Endow SA, Waligora KW (1998) Determinants of kinesin motor polarity. Science 281: 1200–1202 [DOI] [PubMed] [Google Scholar]
- Enos AP, Morris NR (1990) Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. Cell 60: 1019–1027 [DOI] [PubMed] [Google Scholar]
- Fridman V, Gerson-Gurwitz A, Movshovich N, Kupiec M, Gheber L (2009) Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Rep 10: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O’Toole ET, Haase J, Soubry A, Joglekar AP, Winey M, Salmon ED, Bloom K, Odde DJ (2008a) Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell 135: 894–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Haase J, Mythreye K, Molk JN, Anderson M, Joglekar AP, O’Toole ET, Winey M, Salmon ED, Odde DJ, Bloom K (2008b) The microtubule-based motor Kar3 and plus end-binding protein Bim1 provide structural support for the anaphase spindle. J Cell Biol 180: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A, Movshovich N, Avunie R, Fridman V, Moyal K, Katz B, Hoyt MA, Gheber L (2009) Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cell Mol Life Sci 66: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber L, Kuo SC, Hoyt MA (1999) Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. J Biol Chem 274: 9564–9572 [DOI] [PubMed] [Google Scholar]
- Hackney DD, Baek N, Snyder AC (2009) Half-site inhibition of dimeric kinesin head domains by monomeric tail domains. Biochemistry 48: 3448–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney DD, Levitt JD, Suhan J (1992) Kinesin undergoes a 9 S to 6 S conformational transition. J Biol Chem 267: 8696–8701 [PubMed] [Google Scholar]
- Hagan I, Yanagida M (1992) Kinesin-related cut7 protein associates with mitotic and meiotic spindles in fission yeast. Nature 356: 74–76 [DOI] [PubMed] [Google Scholar]
- Hancock WO (2008) Intracellular transport: kinesins working together. Curr Biol 18: R715–R717 [DOI] [PubMed] [Google Scholar]
- Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS (1993) The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol 123: 665–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Gheber L, Kingsbury T, Hoyt MA (2006) Homotetrameric form of Cin8p, a Saccharomyces cerevisiae kinesin-5 motor, is essential for its in vivo function. J Biol Chem 281: 26004–26013 [DOI] [PubMed] [Google Scholar]
- Howard J, Hunt AJ, Baek S (1993) Assay of microtubule movement driven by single kinesin molecules. Methods Cell Biol 39: 137–147 [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS (1992) Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol 118: 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana JA, Schnapp BJ, Silver PA (1995) Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA 92: 9707–9711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Kwok BH, Weinger JS, Schmidt CF, Kapoor TM, Peterman EJ (2008) Microtubule cross-linking triggers the directional motility of kinesin-5. J Cell Biol 182: 421–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435: 114–118 [DOI] [PubMed] [Google Scholar]
- Kashina AS, Rogers GC, Scholey JM (1997) The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation. Biochim Biophys Acta 1357: 257–271 [DOI] [PubMed] [Google Scholar]
- Khmelinskii A, Roostalu J, Roque H, Antony C, Schiebel E (2009) Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev Cell 17: 244–256 [DOI] [PubMed] [Google Scholar]
- Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD (1996) Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature 380: 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok BH, Kapitein LC, Kim JH, Peterman EJ, Schmidt CF, Kapoor TM (2006) Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat Chem Biol 2: 480–485 [DOI] [PubMed] [Google Scholar]
- Kwok BH, Yang JG, Kapoor TM (2004) The rate of bipolar spindle assembly depends on the microtubule-gliding velocity of the mitotic kinesin Eg5. Curr Biol 14: 1783–1788 [DOI] [PubMed] [Google Scholar]
- Lakamper S, Thiede C, Duselder A, Reiter S, Korneev MJ, Kapitein LC, Peterman EJ, Schmidt CF (2010) The effect of monastrol on the processive motility of a dimeric kinesin-5 head/kinesin-1 stalk chimera. J Mol Biol 399: 1–8 [DOI] [PubMed] [Google Scholar]
- McDonald HB, Stewart RJ, Goldstein LS (1990) The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell 63: 1159–1165 [DOI] [PubMed] [Google Scholar]
- Movshovich N, Fridman V, Gerson-Gurwitz A, Shumacher I, Gertsberg I, Fich A, Hoyt MA, Katz B, Gheber L (2008) Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. J Cell Sci 121: 2529–2539 [DOI] [PubMed] [Google Scholar]
- Nitta R, Okada Y, Hirokawa N (2008) Structural model for strain-dependent microtubule activation of Mg-ADP release from kinesin. Nat Struct Mol Biol 15: 1067–1075 [DOI] [PubMed] [Google Scholar]
- Olz R, Larsson K, Adler L, Gustafsson L (1993) Energy flux and osmoregulation of Saccharomyces cerevisiae grown in chemostats under NaCl stress. J Bacteriol 175: 2205–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD (1991) Multiple kinesin-related proteins in yeast mitosis. Cold Spring Harb Symp Quant Biol 56: 693–703 [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD (1992) Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol 118: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T (2011) Directional switching of the Kinesin cin8 through motor coupling. Science 332: 94–99 [DOI] [PubMed] [Google Scholar]
- Sablin EP, Case RB, Dai SC, Hart CL, Ruby A, Vale RD, Fletterick RJ (1998) Direction determination in the minus-end-directed kinesin motor ncd. Nature 395: 813–816 [DOI] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA (1995) Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol 128: 617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ (1992) Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359: 540–543 [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D (2003) The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol 160: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler S, Kirchner J, Horn C, Kallipolitou A, Woehlke G, Schliwa M (2000) Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat Cell Biol 2: 333–338 [DOI] [PubMed] [Google Scholar]
- Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM (1999) The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol 144: 125–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock MF, Guerrero J, Cobb B, Eggers CT, Huang TG, Li X, Hackney DD (1999) Formation of the compact confomer of kinesin requires a COOH-terminal heavy chain domain and inhibits microtubule-stimulated ATPase activity. J Biol Chem 274: 14617–14623 [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW (1998) Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol 143: 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touitou I, Lhomond G, Pruliere G (2001) Boursin, a sea urchin bimC kinesin protein, plays a role in anaphase and cytokinesis. J Cell Sci 114: 481–491 [DOI] [PubMed] [Google Scholar]
- Tytell JD, Sorger PK (2006) Analysis of kinesin motor function at budding yeast kinetochores. JCB 172: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Funatsu T, Pierce DW, Romberg L, Harada Y, Yanagida T (1996) Direct observation of single kinesin molecules moving along microtubules. Nature 380: 451–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine MT, Fordyce PM, Block SM (2006) Eg5 steps it up!. Cell Div 1: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eunen K, Bouwman J, Daran-Lapujade P, Postmus J, Canelas AB, Mensonides FI, Orij R, Tuzun I, van den Brink J, Smits GJ, van Gulik WM, Brul S, Heijnen JJ, de Winde JH, de Mattos MJ, Kettner C, Nielsen J, Westerhoff HV, Bakker BM (2010) Measuring enzyme activities under standardized in vivo-like conditions for systems biology. FEBS J 277: 749–760 [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ (1996) Kinesin-related proteins at mitotic spindle poles: function and regulation. Cell 85: 943–946 [DOI] [PubMed] [Google Scholar]
- Walker RA, Salmon ED, Endow SA (1990) The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature 347: 780–782 [DOI] [PubMed] [Google Scholar]
- Wargacki MM, Tay JC, Muller EG, Asbury CL, Davis TN (2010) Kip3, the yeast kinesin-8, is required for clustering of kinetochores at metaphase. Cell Cycle 9: 2581–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM (2011) A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol 21: 154–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH Jr, McDonald KL, McIntosh JR (1995) Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol 129: 1601–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan JB, Abraham RT, Jiang W (2005) Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell 16: 3187–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.