Abstract
Individuals with schizophrenia tend to be heavy smokers and are at high risk for tobacco dependence. However, the nature of the comorbidity is not entirely clear. We previously reported evidence for association of schizophrenia with SNPs and SNP haplotypes in a region of chromosome 5q containing the SPEC2, PDZ-GEF2 and ACSL6 genes. In this current study, analysis of the control subjects of the Molecular Genetics of Schizophrenia (MGS) sample showed similar pattern of association with number of cigarettes smoked per day (numCIG) for the same region. To further test if this locus is associated with tobacco smoking as measured by numCIG and FTND, we conducted replication and meta-analysis in 12 independent samples (n>16,000) for two markers in ACSL6 reported in our previous schizophrenia study. In the meta-analysis of the replication samples, we found that rs667437 and rs477084 were significantly associated with numCIG (p = 0.00038 and 0.00136 respectively) but not with FTND scores. We then used in vitro and in vivo techniques to test if nicotine exposure influences the expression of ACSL6 in brain. Primary cortical culture studies showed that chronic (5-day) exposure to nicotine stimulated ACSL6 mRNA expression. Fourteen days of nicotine administration via osmotic mini pump also increased ACSL6 protein levels in the prefrontal cortex and hippocampus of mice. These increases were suppressed by injection of the nicotinic receptor antagonist mecamylamine, suggesting that elevated expression of ACSL6 requires nicotinic receptor activation. These findings suggest that variations in the ACSL6 gene may contribute to the quantity of cigarettes smoked. The independent associations of this locus with schizophrenia and with numCIG in non-schizophrenic subjects suggest that this locus may be a common liability to both conditions.
Introduction
Smoking is an important public health problem, and smoking-related diseases take a heavy toll on society. Many studies have shown that smoking is addictive and both genetic and environmental factors influence smoking behaviors. In recent years, with the application of genome-wide association study (GWAS) approach, genetic studies of smoking have made significant progress. One example is the identification of the CHRNA5/CHRNA3/CHRNB4 locus as a risk factor for smoking quantity [1]–[3]. However, this and other established loci explain only a small proportion of the heritability observed for smoking behaviors. Many more genes remain to be identified for their effects on smoking behaviors.
Smoking is highly prevalent among individuals with a variety of psychiatric disorders [4], [5]. Among them, the comorbidity with schizophrenia is particularly high. Smoking prevalence in those with a schizophrenia diagnosis is 3–4 times higher than that in the general population. Most individuals with schizophrenia smoke more heavily than smokers in the population at large [6]. Several hypotheses have been proposed to explain these phenomena, including a self-medication theory [7], [8]. It is possible that schizophrenic patients smoke to enhance cognition and to inhibit side-effects of neuroleptic drugs, or schizophrenia has an inherited vulnerability to heavy smoking behavior. In a previous study, we found evidence that the SPEC2/PDZ-GEF2/ACSL6 locus is associated with schizophrenia [9]. In the present study, in our analyses of smoking phenotypes for the control subjects of the Molecular Genetics of Schizophrenia (MGS) sample, we found substantial association in the acyl CoA synthetase long chain 6 (ACSL6) gene with a phenotype based on a categorized number of cigarettes smoked per day (numCIG). To verify if this locus is associated with numCIG, we initiated replication studies using 12 other independent samples.
It is unknown how ACSL6 variants might affect tobacco smoking and there are no probes available to assess levels of ACSL6 in the brains of humans. Because nicotine is a major psychoactive ingredient in tobacco that is self-administered in both humans and rodents [10], we used rodent models to determine if chronic nicotine exposure alters ACSL6 expression in brain areas involved in addictive behaviors and cognition. We conducted in vitro and in vivo studies, measuring the effects of chronic nicotine exposure on ACSL6 mRNA expression in rat primary cortical culture and assaying levels of ACSL6 protein in mouse prefrontal cortex (PFC), hippocampus (HIP), ventral tegmental area (VTA) and nucleus accumbens (NAC) following chronic nicotine exposure in the presence or absence of a nicotinic receptor antagonist, mecamylamine. In this article, we report our findings from these experiments.
Results
ACSL6 association with numCIG in the MGS controls
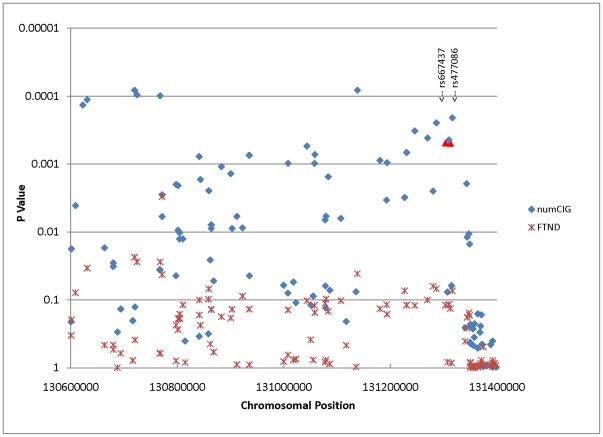
In our analysis of the interval covering the SPEC2, PDZ-GEF2 and ACSL6 genes (about 800 kb), where we reported a long-range haplotype association with schizophrenia [9], 69 of the 145 markers have a p value≤0.05 for numCIG, far in excess of chance expectations. In contrast, only 11 markers reach nominal significance for FTND (Figure 1). The lowest p value (8×10−5) is observed at rs6870930 for numCIG (beta = −0.156, r2 = 0.0096), which is located in the PDZ-GEF2 gene. Of the 24 markers typed in our previous schizophrenia study [9], rs667437 and rs477086 in the ACSL6 gene are amongst the markers with the lowest p values for numCIG (p = 0.0005; see Figure 1 and Table 1). However, the association with FTND is not significant. For both markers, the alleles significantly associated with greater number of daily cigarettes smoked (allele G for rs667437 and allele C for rs477086) reside on the risk haplotypes associated with schizophrenia in our previous study.
Figure 1. Association analyses of the MGS_EA controls for the SPEC2, PDZ-GEF2, FNIP1 and ACSL6 region.
Of the 145 markers tested in this interval, 68 and 12 markers respectively reached nominal significance for numCIG and FTND. The two markers selected for replication were highlighted as red.
Table 1. ACSL6 association with numCIG and FTND in the MGS_EA sample.
| numCIG | FTND | ||||||||
| SNP | Effect allele | BETA | SE | STAT | P | BETA | SE | STAT | P |
| rs667437 | A | −0.1351 | 0.03881 | −3.492 | 0.0005 | −0.1897 | 0.1219 | −1.557 | 0.1197 |
| rs477086 | T | −0.1351 | 0.03883 | −3.491 | 0.0005 | −0.1829 | 0.1219 | −1.501 | 0.1337 |
Replication and meta-analyses of rs667437 and rs477086
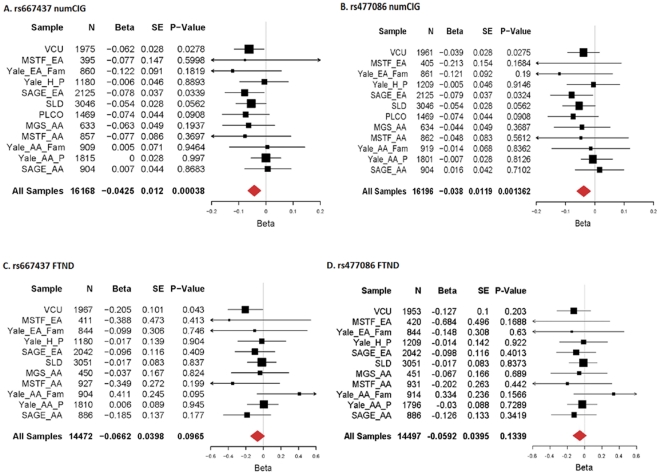
We first initiated replication of the finding using our VCU subjects. For rs667437, both numCIG and FTND reach nominal significance (p = 0.0262 and 0.0427); for rs477086, although no association is found with FTND (p = 0.203), the association with numCIG reaches nominal significance (p = 0.0275) (Figure 2A–2D). Following these results, we extended the replication study to other samples, and conducted meta-analyses for all replication samples. Figure 2 shows the results of the meta-analyses. There is no significant heterogeneity for either phenotype across the replication samples (numCIG: rs667437, Q = 7.938, p = 0.719; rs477086, Q = 7.865, p = 0.725; FTND: rs667437, Q = 9.192, p = 0.513, rs477086, Q = 6.052, p = 0.811). For both markers, numCIG results are significant (rs667437, z = −3.56, p = 3.8×10−4; rs477086, z = −3.20, p = 0.001362). Similar to the results we observed in our VCU subjects, the results for FTND are not significant (rs667437, z = −1.66, p = 0.097; rs477086, z = −1.50, p = 0.134).
Figure 2. Meta-analyses of rs667437 and rs477086 for numCIG and FTND.
A) rs667437, numCIG; B) rs477086, numCIG; C) rs667437, FTND; and D) rs477086, FTND.
Nicotine stimulates ACSL6 mRNA expression in rat cortical culture
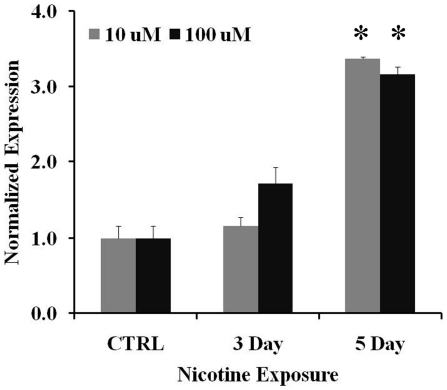
To follow up the finding that ACSL6 is associated with the number of cigarettes smoked, we next questioned if nicotine in tobacco might affect expression of ACSL6 in the central nervous system. We used animal models to determine whether nicotine affects the expression of ACSL6 in comparative rodent brain areas thought to regulate nicotine/tobacco use and involved in schizophrenia. We first conducted chronic nicotine stimulation experiments in primary rat cortical cultures using 10 and 100 µM nicotine, and measured the mRNA expression of ACSL6 by real-time quantitative PCR. There is a main effect of nicotine exposure on mRNA expression of ACSL6 (F 2, 6 = 78.844, p<0.001; Figure 3). A post hoc t-test reveals that on day 5, the mRNA expression in the nicotine-treated samples is significantly higher than that in the saline controls independent of the concentration of nicotine (t 8 = −3.579, p<0.01). The effects on days between the doses are marginal (t 8 = 2.293, p = 0.051 for day 3 and t 8 = 2.278, p = 0.052 for day 5). These results suggest that nicotine stimulates ACSL6 mRNA expression in cortical cells. Rat GAPDH and TATA box binding protein (TBP) primers were used as internal controls. Results from GAPDH and TBP did not differ significantly and therefore, only TBP data was shown (Figure 3).
Figure 3. ACSL6 mRNA expression in rat cortical primary culture after nicotine treatment.
The expression levels of ACSL6 mRNA were significantly increased by 5 days of nicotine exposure for both doses. *p<0.01 compared to saline controls; n = 3. mRNAs were measured by real-time-PCR, and were presented as normalized ΔCt values. Bars represent ± SD.
Nicotine increases ACSL6 protein levels in mouse prefrontal cortex (PFC) and hippocampus (HIP)
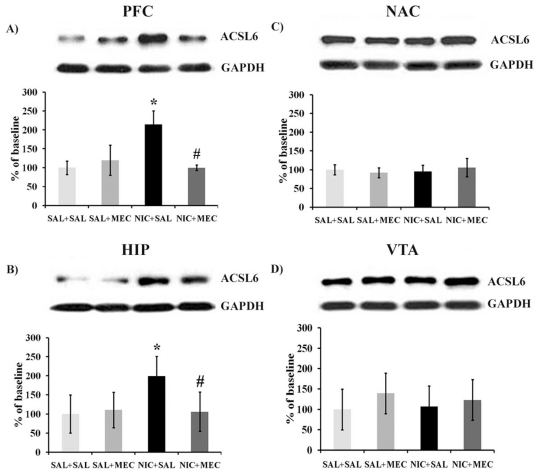
We further tested whether nicotine alters ACSL6 protein expression following in vivo nicotine exposure. We chronically administrated a dosing regimen of 36 mg/kg/day nicotine via mini-pumps for 14 days that has been shown to result in dependence-like behavior in mice [11], [12]. On day 15, mice received a challenge s.c. injection of 1 mg/kg mecamylamine or saline vehicle 30 min prior to brain harvest. Four brain areas (PFC, HIP, NAC and VTA) were dissected and processed for Western blotting to quantify the expression levels of ACSL6 protein. The results are shown in Figure 4. There is a significant interaction of nicotine pretreatment with mecamylamine challenge in the PFC (Figure 4A, F 1,11 = 16.420, p = 0.004) and HIP (Figure 4B, F 1,27 = 8.290, p = 0.008), but not in the NAC (Figure 4C) and VTA (Figure 4D) after chronic nicotine treatment. Post hoc t-tests reveal that animals receiving chronic nicotine show a significant increase in ACSL6 protein expression in the PFC (t 4 = 4.872, p = 0.008) and HIP (t 16 = 4.583, p<0.001) of NIC-SAL compared to SAL-SAL control animals. This was not observed following a withdrawal-precipitating dose of the non-selective nicotinic antagonist, mecamylamine. Mecamylamine treatment alone does not change the expression of ACSL6 protein in the brain regions tested (F's<1.0 in SAL-SAL compared to SAL-MEC mice), but does reverse nicotine associated increases in ACSL6. Mice receiving an injection of mecamylamine following 14 days of chronic nicotine treatment (NIC-MEC) produce significantly less ACSL6 protein than saline-injected controls (NIC-SAL) in both the PFC (t 4 = 5.38, p = 0.006) and HIP (t 12 = 3.221, p = 0.007). Together these data suggest that continued activation of nicotinic acetylcholine receptors is necessary for the increases of ACSL6 protein in these brain regions.
Figure 4. Western blot analyses of ACSL6 protein in mouse brain regions after chronic nicotine exposure and mecamylamine challenge.
Nicotine (NIC+SAL) led to a significant increase in ACSL6 protein levels in A) the prefrontal cortex and B) the hippocampus [*p<0.01 compared to saline controls (SAL+SAL)]. These increases were suppressed by the administration of mecamylamine (NIC+MEC; # p<0.01 compared to nicotine treated NIC-SAL mice). There was no effect of nicotine on levels of ACSL6 in C) the NAC and D) VTA. (n sizes = 3–8 per group).
Discussion
In our analyses of the numCIG and FTND phenotypes in the MGS control subjects, we found that multiple markers in the broad region containing the SPEC2, PDZ-GEF2 and ACSL6 genes showed substantial association with number of cigarettes smoked. While the association signals in these genes did not reach genome-wide significance, the wide spread association signals in the region are consistent with the patterns we observed in our previous schizophrenia studies. Given the high rate of smoking in schizophrenic patients, we sought to test whether this locus is independently associated with tobacco smoking as measured by numCIG and FTND scores in non-schizophrenic subjects. Of the many markers typed in this region in the MGS control sample, we found two overlap markers in our previous study [9], rs667437 and rs477086. The association signals for numCIG from these two markers are among the most significant SNPs identified and are representative of the region. Therefore, we initiated replication of these two ACSL6 markers in a large population twin sample selected from our twin studies (VCU subjects). In our VCU subjects, both markers, rs667437 and rs477086, reached nominal significance and the risk alleles are the same as those observed in the MGS control subjects. Encouraged by these results, we recruited participation by investigators with independent samples. Using standard meta-analyses, we demonstrate that both markers are significantly associated with the numCIG phenotype and the FTND phenotype shows a trend in the same direction. As these studies were performed with non-schizophrenic subjects, the associations observed are independent of schizophrenia diagnosis. The observation that the same alleles in ACSL6 are associated with schizophrenia and with numCIG in non-schizophrenic subjects raises an interesting question as to whether there is a common mechanism underlying these associations. To answer this question, we need a sample with both schizophrenia and cigarettes smoking phenotypes. With such a sample, we could estimate how much of the association with schizophrenia can be accounted for by cigarette smoking, and evaluate the relationship between these two separate associations.
ACSL6 encodes a key enzyme that activates polyunsaturated long chain fatty acids and is involved in lipid metabolism. Its preferred substrates include arachidonic acid, eicosapentaenoic acid and docosahexaenoic acid [13]. Arachidonic acid is a component of arachidonoyl phosphatidyl choline and arachidonic acid-containing inositol phospholipids, which are the major sources of N-arachidonoyl ethanolamine (anandamide) and 2-arachidonoylglycerol (2-AG) [14]. Anandamide and 2-AG are the major endocannabinoids that bind to the cannabinoid receptor 1, a receptor associated with tobacco smoking and other addictive drugs in humans [15]–[18] and shown to promote nicotine reward in animal model studies [19]–[21]. We have reported that the cannabinoid receptor 1 target is associated with tobacco smoking and nicotine dependence [18]. Others have provided evidence that the endogenous cannabinoid system plays a major role in drug reward and addiction, including nicotine [22]–[28]. Intriguingly, it has been proposed that the endocannabinoid system is involved in the etiology of schizophrenia [29] and cannabis use is considered a risk factor for schizophrenia as well [30]. While the exact role of ACSL6 in schizophrenia and tobacco addiction is unclear at this time, these data warrant further investigation into the potential interactions of ACSL6 and the cannibinoid system.
Since nicotine is the major psychoactive ingredient in tobacco and ACSL6 genotype was associated with number of cigarettes smoked, we sought to characterize if nicotine affects expression of ACSL6 in brain areas that regulate cognition, attention, and addiction behaviors. Using primary cell culture of rat cortex, we show that nicotine stimulates ACSL6 mRNA expression after 5 days but not 3 days of nicotine exposure. These data suggest that repeated nicotine exposure is necessary for nicotine-associated changes in ACSL6. In rodents, humans and non-human primates, the cortex is thought to regulate executive functions that support working memory, cognition and inhibitory control [31]–[34]. The PFC communicates with the HIP which is important for declarative memory and integration of higher brain signals with information projected from sensory systems. This circuitry is compromised in those with schizophrenia who are thought to smoke in part to enhance cognition [33]. Utilizing a mouse model of nicotine exposure that we have previously shown to produce nicotine dependence-like behavior as measured by tolerance and withdrawal [11], [35], we find that chronic systemic exposure to nicotine increases ACSL6 protein levels in the PFC and HIP of mice. It is interesting that nicotine does not affect the expression of ACSL6 in the VTA and NAC, areas of the brain that regulate addiction and reward behavior [36]–[38], but the PFC and HIP that have inputs to the VTA and NAC and are also thought to contribute to tobacco addiction [39]–[43]. Interestingly, the ACSL6 gene appeared in the list of amphetamine addiction genes by an independent GWAS [44], suggesting that ACSL6 may be involved in addictive behaviors to drugs other than nicotine. The observation that nicotine-associated increases in ACSL6 are inhibited by the nicotinic receptor antagonist mecamylamine suggests that sustained increases of ACSL6 protein require activation of nicotinic receptors. It is not clear if this effect is due to changes in transcription or protein degradation and since mecamylamine blocks all nicotinic receptors in brain, further studies should question which receptor subtypes mediate these effects. Although the mechanism of these findings has yet to be determined, our finding is consistent with several studies that have implicated nicotinic receptors in the etiology of schizophrenia [45]–[47] and the quantity of daily cigarettes smoked [1]–[3], [48].
It is not clear whether nicotine-associated increases in ACSL6 expression might serve to improve cognition and/or enhance neuroplasticity associated with nicotine dependence in individuals with schizophrenia. Many studies have shown that smoking can improve cognitive function, memory and attention in both normal controls and schizophrenia patients [49]–[52] and the effects of cannabinoids on cognitive function and memory [53], [54] are thought to be mediated in part by the cholinergic system [55], [56]. These findings also warrant future studies to determine if alterations of ACSL6 in brain might support self-medication, cigarette craving, withdrawal or primary or conditioned reinforcing effects of cigarettes.
In our previous fine-mapping study of schizophrenia, we found that haplotypes spanning SPEC2, PDZ-GEF2 and ACSL6 genes were associated with the disease [9]. In the current study, we found that this same genomic interval showed a similar pattern of association with numCIG in the MGS controls, an association that we confirmed in a meta-analysis of 12 independent samples of non-schizophrenic subjects. Since the ACSL6 gene is in high LD with other genes, i.e. SPEC2 and PDZ-GEF2, we cannot exclude the possibility that the observed association signals may reflect the activity of other genes in this region, but our rodent studies suggest that nicotine in tobacco increases expression of ACSL6 message and protein. Given the heavy tobacco use of individuals diagnosed with schizophrenia, our results raise interesting questions in regard to whether schizophrenic smokers may ingest tobacco to regulate ACSL6. In separate studies, several other genes have been associated with both nicotine dependence [18], [20], [57] and schizophrenia [58]–[64]. The SPEC2, PDZ-GEF2 and ACSL6 region may be another locus that contributes to shared susceptibility to schizophrenia and tobacco addiction. Taken together, these studies provide convergent evidence that tobacco use and schizophrenia may share some common underlying mechanisms and that common vulnerability genes such as ACSL6 are regulated by nicotinic receptors.
Materials and Methods
Human studies
This study was conducted according to the principles expressed in the Declaration of Helsinki. For all human studies, all participants provided written informed consent. The study protocol, forms, and procedures were approved by Institutional Review Boards/Ethics Committees at Virginia Commonwealth University (VCU subjects), Yale University School of Medicine (The Yale/UConn subjects), University of Virginia (MSTF study) and all the other participating Institutional Review Boards.
MGS controls
The control subjects for the MGS were a population sample selected for the GWAS of schizophrenia [65]. Subjects were sampled proportionally from 25 major population areas. All subjects completed an online, short, self-report clinical assessment after giving informed consent through an online procedure and prior to venipuncture being arranged. The self-report screen focused on common psychiatric disorders including substance use problems, along with age, sex, height, weight, and the ethnic background of their grandparents. Specific to smoking phenotypes, the subjects were ascertained with the full FTND questionnaire. The information corresponding to the FTND was extracted from the phenotype dataset. FTND scores were obtained based on the answers of the participants to the questionnaires. Subjects who did not smoke a whole cigarette (a “No” answer to the question, “Have you ever smoked a whole cigarette?”) were excluded from FTND score calculation. To be consistent with ascertainment of our VCU subjects, only subjects who answered “Yes” to the question “did you smoke cigarettes on a daily basis” and who smoked ≥5 cigarettes per day were included in the analyses of FTND. The numCIG phenotype was constructed using the raw number of cigarettes smoked per day at the time when the subjects smoked most heavily in their lifetime. Based on the raw data distribution, we divided the subjects into 4 categories using these cut-offs: 0 for those who smoked 1 cigarette per day; 1 for those who smoked 2–14 cigarettes per day; 2 for those who smoked 15–25 cigarettes per day and 3 for those who smoked 26 cigarettes per day or more. For the numCIG phenotype, all subjects reporting smoking were included in the analyses. The MGS controls included both European Americans (MGS_EA) and African Americans (MGS_AA). For this study, there were 1,342 and 1,843 European American (EA) subjects having FTND and numCIG phenotypes respectively, and 454 and 639 AA subjects having FTND and numCIG phenotypes, respectively. The distributions of these phenotypes are shown in Figure S1. The distribution of both phenotypes is normal.
The SAGE subjects
The Study of Addiction: Gene and Environment (SAGE) is part of the Gene Environment Association Studies initiative (GENEVA) funded by the National Human Genome Research Institute aiming at understanding the impact of genes and environments on substance dependence and addiction. The SAGE sample consisted of 3 subsamples: the Collaborative Study on the Genetics of Alcoholism (COGA) [66], the Family Study of Cocaine Dependence (FSCD) [67], and the Collaborative Genetic Study of Nicotine Dependence (COGEND) [1]. Although FTND scores were available for all subjects, the raw number of cigarettes smoked per day was not available. Instead, subjects were assigned to one of the four categories based on the FTND questionnaire that grouped individuals by the number of cigarettes smoked per day (subjects were assigned 0 if they smoked 0–10 cigarettes; 1 if they smoked 11–20 cigarettes; 2 if they smoked 21–30 cigarettes and 3 if they smoked more than 30 cigarettes per day). The SAGE sample included 2,125 subjects with self-reported European ancestry (SAGE_EA) and 904 subjects with self-reported African ancestry. For the FSCD subsample, only one subject from each family was used.
The Lung cancer and smoking study
This is a GWAS to investigate the genetic determinants of lung cancer risk. This study is also part of the GENEVA initiative and consisted of two samples. The first is the Environment and Genetics in Lung Cancer Etiology Study (EAGLE) [68], a population-based, biologically intensive, case-control study from the Lombardy region of Italy including ∼2,000 newly diagnosed lung cancer cases and ∼2,000 age-, gender- and region- matched controls. This study is also referred to as the Smoking and Lung Disease (SLD) study. The second is the Prostate, Lung, Colon and Ovary (PLCO) [69], [70] Cancer Screening Trial, from which ∼850 lung cancer cases and ∼850 controls matched on age and gender were used. The subjects were ascertained for a variety of smoking phenotypes, including smoking status, persistent smoking, quit attempts, and the Fagerström questionnaire. PLCO participants were all EA and EAGLE included subjects from Italy who were all with European ancestry. EAGLE is a case-control study and includes 3,937 phenotyped subjects. PLCO is a screening trial with a cohort design and includes 1,651 phenotyped subjects.
The VCU subjects
The subjects from the Virginia Commonwealth University were selected from the Mid-Atlantic Twin Registry. In this study, we selected regular smokers (defined as those who smoked at least 7 cigarettes per week for a month or more) from our population twin studies [71]–[73]. Tobacco smoking and nicotine dependence were assessed by the Fagerström Tolerance Questionnaire (FTQ) and/or FTND [74], [75]. All regular smokers with DNA samples were included except when self-reported ancestry was not Caucasian. One of the co-twins was selected randomly for inclusion. The final sample included 2,138 individuals, with 1,438 males and 700 females. All subjects were aged 18 to 65 at the time of FTND ascertainment and all self-reported being of European ancestry.
Mid-South Tobacco Family (MSTF) study
The subjects of the MSTF study with either AA or EA origin were recruited primarily during 1,999–2,004 from the Mid-South states of Tennessee, Mississippi, and Arkansas. Proband smokers were required to be at least 21 years old, to have smoked for at least 5 years, and to have consumed at least 20 cigarettes per day for the last 12 months. Siblings and biological parents of a proband smoker were recruited whenever possible, regardless of their smoking status. The study includes 2,037 participants in 602 nuclear families, with 671 subjects in 200 EA families and 1,366 subjects in 402 AA families. The degree of nicotine dependence of each smoker was ascertained by the three most commonly used measures: Smoking Quantity (SQ; defined as the number of cigarettes smoked per day), the Heaviness of Smoking Index (HSI; 0–6 scale), and the FTND score (0–10 scale). All three measures have been used consistently in previous reports on nicotine dependence in this sample [57], [76]–[78]. In this study, only the FTND scores and numCIG phenotypes were used.
The Yale/UConn subjects
The subjects involved in the Yale/UConn study were both families and unrelated individuals. The family samples were recruited from several clinical sites (principally from Yale University School of Medicine and the University of Connecticut Health Center, but also from McLean Hospital and the Medical University of South Carolina) through siblings meeting DSM-IV criteria for cocaine dependence or opioid dependence. The sample of unrelated individuals was ascertained as cases affected for cocaine, opioids, or alcohol dependence and screened controls. The Yale/UConn family sample included 2,129 AAs (including 132 self-reported Hispanics) and 1,706 EAs (including 310 self-reported Hispanics) [79]. Of these, 1,858 subjects from 893 families had genotype and phenotype data. The Yale/UConn case control sample included 1,912 AAs (including 76 Hispanics) and 1,476 EAs (including 176 Hispanics).
Marker Selection, genotyping and genotype imputation
Since our initial goal was to test whether markers associated with schizophrenia were also associated with smoking and nicotine dependence phenotypes, we compared the markers from our schizophrenia study with those included in the Affymetrix 6.0 chipset (the MGS samples were first genotyped with this chipset) in the PDZ-GEF2/ACSL6 region. We found that only 2 of the markers (rs667437 and rs477086) used in our previous schizophrenia study were in the 6.0 chipset. Although the association of these two markers with schizophrenia was not among the strongest observed in the schizophrenia study, they were among the best results in our initial association analyses of the MGS sample. Therefore, only these 2 markers were selected for the present study.
For the GWAS datasets (MGS, SAGE and Lung Cancer), DNA preparation and genotyping have been reported previously. The MGS subjects were typed using an Affymetrix platform (SNP 6.0), and the SAGE and Lung Cancer subjects were typed using an Illumina platform (Human 1 M-Duo). For these datasets, we accepted the quality filtering procedures of each individual study and used the genotypes directly after download. The VCU subjects, the Yale/UConn and MSTF subjects were genotyped by each group using the TaqMan method. For the 2 SNPs used in this study, the assays were designed and synthesized by Applied BioSystems (Foster city, CA, USA). Standardized procedures recommended by the manufacturer were used. The Illumina marker set included only one of the two SNPs used in this study: rs477086. The genotypes for rs667437 were imputed using the fastPHASE program [80] with the MGS_EA or MGS_AA as a reference panel for the EA and AA samples respectively. Imputations were also conducted with HapMap subjects as references, and the imputed genotypes were almost identical to those using MGS samples as references.
Association and meta-analysis
Association analyses were performed for each sample with the PLINK software package [81]. We used two phenotypes, FTND score and numCIG, where numCIG was a categorical phenotype. Both phenotypes were treated as quantitative traits in linear regression. For population samples, we used linear regression with sex, age and ethnicity (Hispanics) as covariates. For family samples, we used the QFAM module and the within-family statistics (MSTF, Yale_EA_Fam and Yale_AA_Fam samples). For GWAS datasets, we adapted the principle components used in the original studies. Meta-analyses were performed with the GWAMA software package [82] for replication samples only. Summary statistics (beta, se and sample size) were extracted from individual analyses and used in the meta-analyses. The package can perform both fixed effect and random effect meta-analyses. In our analyses, since the heterogeneity tests (both Cochran's Q statistics and I2) were non-significant, we reported the results from the fixed effect analyses. Meta-analysis results were plotted with the R package rmeta.
Animal studies
The experimental protocol was approved by the Institutional Animal Care and Use Committee at Virginia Commonwealth University (the University Animal Welfare Assurance Number: A3281-01), and all animals were treated according to the Guidelines for the Care and Use of Laboratory Animals, as set forth by the National Institutes of Health. Animals were maintained on a 12-hour light/dark cycle in a temperature (21°C) and humidity controlled vivarium with ad libitum access to food and water. Experiments were performed during the light cycle.
Rat in vitro expression study
Sprague–Dawley timed-pregnant rats were obtained from Zivic Laboratories (Allison Park, PA, USA). On postnatal day 1, the litter was transferred together to the laboratory where brain harvests took place. Mixed neuronal plus glial cultures were prepared as described previously [83], [84]. Briefly, the cortex of 4–5 pups was dissected in a sterile saline solution (137 mM NaCl, 5.3 mM KCl, 0.17 mM Na2HPO4•7H2O, 0.22 mM KH2PO4 and 0.0012 g/L Phenol Red) under a laminar flow hood then transferred into a sterile-filtered 0.1% porcine trypsin dissection solution, minced and incubated at room temperature for 10 min. Brain sections were rinsed twice in plating medium (DMEM, 10% FBS, 1% L-glutamine and 1% Penicillin/Streptomycin) to stop the trypsin reaction, triterated with a cotton-plugged glass pipette, spun 7 min at 1,000 RPM with medium replaced and then poured through a 70 µm nylon cell filter (BD Falcoln, Bedford, MA, USA) for plating. 1.7×106 cells were seeded into each well of a 6-well plate. Cells were cultured in a 5% CO2 incubator at 37°C. Three days after plating, and every 3 days thereafter, 1 mL medium was removed from each well and replaced with 1 mL of fresh mixed growth medium containing minimal essential medium, 10 mM glucose, penicillin 100 U/mL, streptomycin 100 µg/mL, and 5% horse serum. On day 14, in vitro media was removed and 2.0 mL media was quickly replaced to ensure a consistent stimulating volume. Cells treated with 0 µM, 10 µM, and 100 µM Nicotine ((-)-Nicotine hydrogen tartrate salt, dissolved in 0.9% sodium chloride) were cultured for another 3 and 5 days. At each time point, the media was removed, wells were washed twice with PBS, and cells were harvested in 200 µl PBS. Cells from three wells were pooled into an Eppendorf tube. Total RNA was extracted from cells using TRIzol Reagent (Invitrogen, Eugene, OR, USA) and quantified by Quant-iT™ RNA Assay Kit (Invitrogen, Eugene, OR, USA). cDNA was synthesized using 2 µg of total RNA and 50 ng of random hexamers according to the first-strand cDNA synthesis protocol provided with SuperScript III RNase H-Reverse Transcriptase (Invitrogen, Eugene, OR, USA). For real time-PCR, samples were analyzed in triplicate 20 µl reactions including 25 ng of cDNA, 250 nmol of primer, 1× PCR buffer, 2 mM MgCl2, 0.08 mM dNTPs, 0.01 u/µl Taq polymerase (Invitrogen, Eugene, OR, USA) and 1/10 X SyBR® Green (Sigma, St. Louis, MO, USA). Primers were designed to coding sequence using Primer3 (v. 0.4.0). PCR reactions using rat TATA box binding protein (TBP) and GAPDH primers were used as internal controls. The Primer sequences for the ACSL6, TBP and GAPDH are as follows: ACSL6 forward, 5′-TTTCACGAGCGGTACAACAG-3′, ACSL6 reverse, 5′-GTGTACATCCGCACAAGTGG -3′; TBP forward, 5′-TATAATCCCAAGCGGTTTGC-3′, TBP reverse, 5′-CAGCCTTATGGGGAACTTCA-3′; GAPDH forward, 5′-AAGGGCTCATGACCACAGTC-3′, GAPDH reverse, 5′-CAACGGATACATTGGGGGTA-3′. PCR was conducted and the expression level of each reaction was determined by the CT value. The results from three replicated assays were averaged to produce a single mean CT value for each treatment condition. The relative expression level between ACSL6 and TBP or GAPDH for each condition was calculated by the 2−ΔCT method, where ΔC T = CT ACSL6−CT TBPor GAPDH [85].
Mouse in vivo expression study
Male 129SvJ mice were purchased from Jackson Laboratories (Bar Harbor, ME). Animals were 8–10 weeks of age at the start of the studies. Mice were anesthetized with sodium pentobarbital (45 mg/kg by intraperitoneal injection) and implanted subcutaneously (s.c.) with Alzet osmotic mini pumps [model 2,004, Durect Corporation, Cupertino, CA, USA] filled with (-)-nicotine (NIC) or saline (SAL) solution as described in Jackson et al. [11]. The concentration of nicotine was adjusted according to animal weight and mini pump flow rate so that mice were infused with 36 mg/kg/day for 14 days. The dose and duration of nicotine exposure were chosen based on previous behavioral studies [11], [12] which show that significant tolerance and nicotine withdrawal signs are produced in mice after this treatment regimen. On the morning of Day 15, chronic nicotine- and saline-infused mice were injected s.c. with 1.0 or 2.0 mg/kg of a non-selective nicotinic receptor antagonist, mecamylamine (MEC) or saline vehicle (SAL) followed by a 30 min waiting period. There was no significant difference in ACSL6 protein levels between the 1 and 2 mg/kg doses of mecamylamine used across cohorts of animals, so these data were combined to create 4 experimental treatment groups (SAL+SAL, SAL+MEC, NIC+SAL, NIC+MEC). Mice were then euthanized by cervical dislocation. Brain sections were dissected and placed immediately in cold extraction buffer for dissection of PFC, HIP, NAC and VTA. Brain tissues were dissected and homogenized as described previously [86]. Protein concentrations were determined using the DC protein assay (Bio-Rad Laboratories, Hercules, CA, USA), and 30 µg of protein were mixed with 6× blue gel loading dye (New England Biolabs, Ipswich, MA, USA) and heated for 5 minutes at 95°C. Samples were then separated by SDS-polyacrylamide gel electrophoresis on a 10% Tris-HCL gel and subjected to immunoblotting with anti-ACSL6 antibody (1∶500 from goat; sc-48005, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) or anti-GAPDH (1∶50,000 from mouse; Advanced ImmunoChemical Ins., Long Beach, CA, USA) primary antibodies overnight at 4°C. Blots were rinsed in TBST and then incubated in secondary antibodies (1∶2000 anti-goat or 1∶50,000 mouse, Vector Laboratories, Inc., Burlingame, CA, USA) for 1 hour at room temperature followed by three times of TBST 10 min washes. Specific bands were detected by enhanced chemiluminesence (GE Healthcare Bio-Sciences, Piscataway, NJ, USA), exposed to X-Ray film, and quantified using Image J software (Rasband WS, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/, 1997–2006). ACSL6 protein levels were normalized against GAPDH for loading control and against vehicle-treated control subjects to enable comparisons across blots.
Supporting Information
Distribution of number of cigarettes smoked per day (A) and FTND scores in the MGS control subjects (B).
(DOCX)
Acknowledgments
We thank all the volunteers who had participated in these studies and the investigators who collected and ascertained these samples.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by grants DA019498 to XC and DA011287 to KSK from the National Institutes of Health, by funds from the Virginia Tobacco Settlement Foundation through the Virginia Youth Tobacco Project to Virginia Commonwealth University (subcontracted to KSK, #5100004ST) and by a grant from Virginia Foundation for Healthy Youth 8520667 to DHB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berrettini W, Yuan X, Tozzi F, Song K, Francks C, et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry. 2008;13:368–373. doi: 10.1038/sj.mp.4002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batel P. Addiction and schizophrenia. Eur Psychiatry. 2000;15:115–122. doi: 10.1016/s0924-9338(00)00203-0. [DOI] [PubMed] [Google Scholar]
- 5.Salin-Pascual RJ, Alcocer-Castillejos NV, Alejo-Galarza G. Nicotine dependence and psychiatric disorders. Rev Invest Clin. 2003;55:677–693. [PubMed] [Google Scholar]
- 6.de Leon J, Diaz FJ. A meta-analysis of worldwide studies demonstrates an association between schizophrenia and tobacco smoking behaviors. Schizophr Res. 2005;76:135–157. doi: 10.1016/j.schres.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Kumari V, Postma P. Nicotine use in schizophrenia: the self medication hypotheses. Neurosci Biobehav Rev. 2005;29:1021–1034. doi: 10.1016/j.neubiorev.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 8.Leonard S, Mexal S, Freedman R. Smoking, Genetics and Schizophrenia: Evidence for Self Medication. J Dual Diagn. 2007;3:43–59. doi: 10.1300/J374v03n03_05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Wang X, Hossain S, O'Neill FA, Walsh D, et al. Haplotypes spanning SPEC2, PDZ-G EF2 and ACSL6 genes are associated with schizophrenia. Hum Mol Genet. 2006;15:3329–3342. doi: 10.1093/hmg/ddl409. [DOI] [PubMed] [Google Scholar]
- 10.Le Foll B, Goldberg SR. Effects of nicotine in experimental animals and humans: an update on addictive properties. Handb Exp Pharmacol. 2009;192:335–367. doi: 10.1007/978-3-540-69248-5_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson KJ, Martin BR, Changeux JP, Damaj MI. Differential role of nicotinic acetylcholine receptor subunits in physical and affective nicotine withdrawal signs. J Pharmacol Exp Ther. 2008;325:302–312. doi: 10.1124/jpet.107.132977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson KJ, McIntosh JM, Brunzell DH, Sanjakdar SS, Damaj MI. The role of alpha6-containing nicotinic acetylcholine receptors in nicotine reward and withdrawal. J Pharmacol Exp Ther. 2009;331:547–554. doi: 10.1124/jpet.109.155457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Horn CG, Caviglia JM, Li LO, Wang S, Granger DA, et al. Characterization of recombinant long-chain rat acyl-CoA synthetase isoforms 3 and 6: identification of a novel variant of isoform 6. Biochemistry. 2005;44:1635–1642. doi: 10.1021/bi047721l. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Ueda N. Biology of endocannabinoid synthesis system. Prostaglandins Other Lipid Mediat. 2009;89:112–119. doi: 10.1016/j.prostaglandins.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Comings DE, Muhleman D, Gade R, Johnson P, Verde R, et al. Cannabinoid receptor gene (CNR1): association with i.v. drug use. Mol Psychiatry. 1997;2:161–168. doi: 10.1038/sj.mp.4000247. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt LG, Samochowiec J, Finckh U, Fiszer-Piosik E, Horodnicki J, et al. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug Alcohol Depend. 2002;65:221–224. doi: 10.1016/s0376-8716(01)00164-8. [DOI] [PubMed] [Google Scholar]
- 17.Zuo L, Kranzler HR, Luo X, Covault J, Gelernter J. CNR1 Variation Modulates Risk for Drug and Alcohol Dependence. Biol Psychiatry. 2007;62:616–626. doi: 10.1016/j.biopsych.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Chen X, Williamson VS, An SS, Hettema JM, Aggens HS, et al. The cannabinoid receptor 1 (CNR1) gene association with nicotine dependence. Arch Gen Psychiatry. 2008;65:816–824. doi: 10.1001/archpsyc.65.7.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forget B, Hamon M, Thiebot MH. Cannabinoid CB1 receptors are involved in motivational effects of nicotine in rats. Psychopharmacology (Berl) 2005;181:722–734. doi: 10.1007/s00213-005-0015-6. [DOI] [PubMed] [Google Scholar]
- 20.Merritt LL, Martin BR, Walters C, Lichtman AH, Damaj MI. The endogenous cannabinoid system modulates nicotine reward and dependence. J Pharmacol Exp Ther. 2008;326:483–492. doi: 10.1124/jpet.108.138321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shoaib M. The cannabinoid antagonist AM251 attenuates nicotine self-administration and nicotine-seeking behaviour in rats. Neuropharmacology. 2008;54:438–444. doi: 10.1016/j.neuropharm.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 22.Arnold JC. The role of endocannabinoid transmission in cocaine addiction. Pharmacol Biochem Behav. 2005;81:396–406. doi: 10.1016/j.pbb.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Colombo G, Serra S, Vacca G, Carai MA, Gessa GL. Endocannabinoid system and alcohol addiction: pharmacological studies. Pharmacol Biochem Behav. 2005;81:369–380. doi: 10.1016/j.pbb.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 24.Fattore L, Cossu G, Spano MS, Deiana S, Fadda P, et al. Cannabinoids and reward: interactions with the opioid system. Crit Rev Neurobiol. 2004;16:147–158. doi: 10.1615/critrevneurobiol.v16.i12.160. [DOI] [PubMed] [Google Scholar]
- 25.Lupica CR, Riegel AC, Hoffman AF. Marijuana and cannabinoid regulation of brain reward circuits. Br J Pharmacol. 2004;143:227–234. doi: 10.1038/sj.bjp.0705931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maldonado R, Valverde O, Berrendero F. Involvement of the endocannabinoid system in drug addiction. Trends Neurosci. 2006;29:225–232. doi: 10.1016/j.tins.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Racz I, Bilkei-Gorzo A, Toth ZE, Michel K, Palkovits M, et al. A critical role for the cannabinoid CB1 receptors in alcohol dependence and stress-stimulated ethanol drinking. J Neurosci. 2003;23:2453–2458. doi: 10.1523/JNEUROSCI.23-06-02453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang PW, Ishiguro H, Ohtsuki T, Hess J, Carillo F, et al. Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes and association with polysubstance abuse. Mol Psychiatry. 2004;9:916–931. doi: 10.1038/sj.mp.4001560. [DOI] [PubMed] [Google Scholar]
- 29.Fernandez-Espejo E, Viveros MP, Nunez L, Ellenbroek BA, Rodriguez de FF. Role of cannabis and endocannabinoids in the genesis of schizophrenia. Psychopharmacology (Berl) 2009;206:531–549. doi: 10.1007/s00213-009-1612-6. [DOI] [PubMed] [Google Scholar]
- 30.van OJ, Kenis G, Rutten BP. The environment and schizophrenia. Nature. 2010;468:203–212. doi: 10.1038/nature09563. [DOI] [PubMed] [Google Scholar]
- 31.Goldman-Rakic PS. Working memory dysfunction in schizophrenia. J Neuropsychiatry Clin Neurosci. 1994;6:348–357. doi: 10.1176/jnp.6.4.348. [DOI] [PubMed] [Google Scholar]
- 32.Arnold OH. Schizophrenia - A disturbance of signal interaction between the entorhinal cortex and the dentate gyrus? The contribution of experimental dibenamine psychosis to the pathogenesis of schizophrenia: A hypothesis. Neuropsychobiology. 1999;40:21–32. doi: 10.1159/000026593. [DOI] [PubMed] [Google Scholar]
- 33.Tamminga CA, Stan AD, Wagner AD. The hippocampal formation in schizophrenia. Am J Psychiatry. 2010;167:1178–1193. doi: 10.1176/appi.ajp.2010.09081187. [DOI] [PubMed] [Google Scholar]
- 34.van Erp TG, Therman S, Pirkola T, Tuulio-Henriksson A, Glahn DC, et al. Verbal recall and recognition in twins discordant for schizophrenia. Psychiatry Res. 2008;159:271–280. doi: 10.1016/j.psychres.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson KJ, Walters CL, Miles MF, Martin BR, Damaj MI. Characterization of pharmacological and behavioral differences to nicotine in C57Bl/6 and DBA/2 mice. Neuropharmacology. 2009;57:347–355. doi: 10.1016/j.neuropharm.2009.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fattore L, Fadda P, Spano MS, Pistis M, Fratta W. Neurobiological mechanisms of cannabinoid addiction. Mol Cell Endocrinol. 2008;286:S97–S107. doi: 10.1016/j.mce.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Feltenstein MW, See RE. The neurocircuitry of addiction: an overview. Br J Pharmacol. 2008;154:261–274. doi: 10.1038/bjp.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di MV, Pierucci M, Di GG, Benigno A, Esposito E. The neurobiological bases for the pharmacotherapy of nicotine addiction. Curr Pharm Des. 2007;13:1269–1284. doi: 10.2174/138161207780618920. [DOI] [PubMed] [Google Scholar]
- 39.McClernon FJ, Kozink RV, Rose JE. Individual differences in nicotine dependence, withdrawal symptoms, and sex predict transient fMRI-BOLD responses to smoking cues. Neuropsychopharmacology. 2008;33:2148–2157. doi: 10.1038/sj.npp.1301618. [DOI] [PubMed] [Google Scholar]
- 40.Salas R, Sturm R, Boulter J, De Biasi M. Nicotinic receptors in the habenulo-interpeduncular system are necessary for nicotine withdrawal in mice. J Neurosci. 2009;29:3014–3018. doi: 10.1523/JNEUROSCI.4934-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mansvelder HD, Mertz M, Role LW. Nicotinic modulation of synaptic transmission and plasticity in cortico-limbic circuits. Semin Cell Dev Biol. 2009;20:432–440. doi: 10.1016/j.semcdb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brody AL, Mandelkern MA, Jarvik ME, Lee GS, Smith EC, et al. Differences between smokers and nonsmokers in regional gray matter volumes and densities. Biol Psychiatry. 2004;55:77–84. doi: 10.1016/s0006-3223(03)00610-3. [DOI] [PubMed] [Google Scholar]
- 43.Brody AL, Mandelkern MA, London ED, Childress AR, Lee GS, et al. Brain metabolic changes during cigarette craving. Arch Gen Psychiatry. 2002;59:1162–1172. doi: 10.1001/archpsyc.59.12.1162. [DOI] [PubMed] [Google Scholar]
- 44.Uhl GR, Drgon T, Liu QR, Johnson C, Walther D, et al. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008;65:345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- 45.De LV, Voineskos S, Wong G, Kennedy JL. Genetic interaction between alpha4 and beta2 subunits of high affinity nicotinic receptor: analysis in schizophrenia. Exp Brain Res. 2006;174:292–296. doi: 10.1007/s00221-006-0458-y. [DOI] [PubMed] [Google Scholar]
- 46.De LV, Wong AH, Muller DJ, Wong GW, Tyndale RF, et al. Evidence of association between smoking and alpha7 nicotinic receptor subunit gene in schizophrenia patients. Neuropsychopharmacology. 2004;29:1522–1526. doi: 10.1038/sj.npp.1300466. [DOI] [PubMed] [Google Scholar]
- 47.Voineskos S, De Luca V, Mensah A, Vincent JB, Potapova N, et al. Association of alpha4beta2 nicotinic receptor and heavy smoking in schizophrenia. J Psychiatry Neurosci. 2007;32:412–416. [PMC free article] [PubMed] [Google Scholar]
- 48.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobsen LK, D'Souza DC, Mencl WE, Pugh KR, Skudlarski P, et al. Nicotine effects on brain function and functional connectivity in schizophrenia. Biol Psychiatry. 2004;55:850–858. doi: 10.1016/j.biopsych.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 50.Rezvani AH, Levin ED. Cognitive effects of nicotine. Biol Psychiatry. 2001;49:258–267. doi: 10.1016/s0006-3223(00)01094-5. [DOI] [PubMed] [Google Scholar]
- 51.Sacco KA, Termine A, Seyal A, Dudas MM, Vessicchio JC, et al. Effects of cigarette smoking on spatial working memory and attentional deficits in schizophrenia: involvement of nicotinic receptor mechanisms. Arch Gen Psychiatry. 2005;62:649–659. doi: 10.1001/archpsyc.62.6.649. [DOI] [PubMed] [Google Scholar]
- 52.Moss TG, Sacco KA, Allen TM, Weinberger AH, Vessicchio JC, et al. Prefrontal cognitive dysfunction is associated with tobacco dependence treatment failure in smokers with schizophrenia. Drug Alcohol Depend. 2009;104:94–99. doi: 10.1016/j.drugalcdep.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pattij T, Wiskerke J, Schoffelmeer AN. Cannabinoid modulation of executive functions. Eur J Pharmacol. 2008;585:458–463. doi: 10.1016/j.ejphar.2008.02.099. [DOI] [PubMed] [Google Scholar]
- 54.Solowij N, Battisti R. The chronic effects of cannabis on memory in humans: a review. Curr Drug Abuse Rev. 2008;1:81–98. doi: 10.2174/1874473710801010081. [DOI] [PubMed] [Google Scholar]
- 55.Robinson L, Goonawardena AV, Pertwee R, Hampson RE, Platt B, et al. WIN55,212-2 induced deficits in spatial learning are mediated by cholinergic hypofunction. Behav Brain Res. 2010;208:584–592. doi: 10.1016/j.bbr.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goonawardena AV, Robinson L, Hampson RE, Riedel G. Cannabinoid and cholinergic systems interact during performance of a short-term memory task in the rat. Learn Mem. 2010;17:502–511. doi: 10.1101/lm.1893710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beuten J, Ma JZ, Payne TJ, Dupont RT, Quezada P, et al. Significant association of BDNF haplotypes in European-American male smokers but not in European-American female or African-American smokers. Am J Med Genet B Neuropsychiatr Genet. 2005;139B:73–80. doi: 10.1002/ajmg.b.30231. [DOI] [PubMed] [Google Scholar]
- 58.Ho BC, Andreasen NC, Dawson JD, Wassink TH. Association between brain-derived neurotrophic factor Val66Met gene polymorphism and progressive brain volume changes in schizophrenia. Am J Psychiatry. 2007;164:1890–1899. doi: 10.1176/appi.ajp.2007.05111903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neves-Pereira M, Cheung JK, Pasdar A, Zhang F, Breen G, et al. BDNF gene is a risk factor for schizophrenia in a Scottish population. Mol Psychiatry. 2005;10:208–212. doi: 10.1038/sj.mp.4001575. [DOI] [PubMed] [Google Scholar]
- 60.Qian L, Zhao J, Shi Y, Zhao X, Feng G, et al. Brain-derived neurotrophic factor and risk of schizophrenia: an association study and meta-analysis. Biochem Biophys Res Commun. 2007;353:738–743. doi: 10.1016/j.bbrc.2006.12.121. [DOI] [PubMed] [Google Scholar]
- 61.Zintzaras E. Brain-derived neurotrophic factor gene polymorphisms and schizophrenia: a meta-analysis. Psychiatr Genet. 2007;17:69–75. doi: 10.1097/YPG.0b013e32801119da. [DOI] [PubMed] [Google Scholar]
- 62.Tsai SJ, Wang YC, Hong CJ. Association study of a cannabinoid receptor gene (CNR1) polymorphism and schizophrenia. Psychiatr Genet. 2000;10:149–151. doi: 10.1097/00041444-200010030-00008. [DOI] [PubMed] [Google Scholar]
- 63.Chavarria-Siles I, Contreras-Rojas J, Hare E, Walss-Bass C, Quezada P, et al. Cannabinoid receptor 1 gene (CNR1) and susceptibility to a quantitative phenotype for hebephrenic schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2008;147:279–284. doi: 10.1002/ajmg.b.30592. [DOI] [PubMed] [Google Scholar]
- 64.Martinez-Gras I, Hoenicka J, Ponce G, Rodriguez-Jimenez R, Jimenez-Arriero MA, et al. (AAT)n repeat in the cannabinoid receptor gene, CNR1: association with schizophrenia in a Spanish population. Eur Arch Psychiatry Clin Neurosci. 2006;256:437–441. doi: 10.1007/s00406-006-0665-3. [DOI] [PubMed] [Google Scholar]
- 65.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bierut LJ, Saccone NL, Rice JP, Goate A, Foroud T, et al. Defining alcohol-related phenotypes in humans. The Collaborative Study on the Genetics of Alcoholism. Alcohol Res Health. 2002;26:208–213. [PMC free article] [PubMed] [Google Scholar]
- 67.Bierut LJ, Strickland JR, Thompson JR, Afful SE, Cottler LB. Drug use and dependence in cocaine dependent subjects, community-based individuals, and their siblings. Drug Alcohol Depend. 2008;95:14–22. doi: 10.1016/j.drugalcdep.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Landi MT, Consonni D, Rotunno M, Bergen AW, Goldstein AM, et al. Environment And Genetics in Lung cancer Etiology (EAGLE) study: an integrative population-based case-control study of lung cancer. BMC Public Health. 2008;8:203–212. doi: 10.1186/1471-2458-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Control Clin Trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 70.Simpson NK, Johnson CC, Ogden SL, Gamito E, Trocky N, et al. Recruitment strategies in the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial: the first six years. Control Clin Trials. 2000;21:356S–378S. doi: 10.1016/s0197-2456(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 71.Kendler KS, Neale MC, Sullivan P, Corey LA, Gardner CO, et al. A population-based twin study in women of smoking initiation and nicotine dependence. Psychol Med. 1999;29:299–308. doi: 10.1017/s0033291798008022. [DOI] [PubMed] [Google Scholar]
- 72.Kendler KS, Prescott CA. Genes, Environment, and Psychopathology: Understanding the Causes of Psychiatric and Substance Use Disorders. New York: Guilford Press; 2007. [DOI] [PubMed] [Google Scholar]
- 73.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 74.Fagerstrom KO, Schneider NG. Measuring nicotine dependence: a review of the Fagerstrom Tolerance Questionnaire. J Behav Med. 1989;12:159–182. doi: 10.1007/BF00846549. [DOI] [PubMed] [Google Scholar]
- 75.Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict. 1991;86:1119–1127. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- 76.Huang W, Payne TJ, Ma JZ, Beuten J, Dupont RT, et al. Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African-American sample. Neuropsychopharmacology. 2009;34:319–330. doi: 10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
- 77.Li MD, Sun D, Lou XY, Beuten J, Payne TJ, et al. Linkage and association studies in African- and Caucasian-American populations demonstrate that SHC3 is a novel susceptibility locus for nicotine dependence. Mol Psychiatry. 2007;12:462–473. doi: 10.1038/sj.mp.4001933. [DOI] [PubMed] [Google Scholar]
- 78.Li MD, Beuten J, Ma JZ, Payne TJ, Lou XY, et al. Ethnic- and gender-specific association of the nicotinic acetylcholine receptor alpha4 subunit gene (CHRNA4) with nicotine dependence. Hum Mol Genet. 2005;14:1211–1219. doi: 10.1093/hmg/ddi132. [DOI] [PubMed] [Google Scholar]
- 79.Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, et al. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology. 2010;35:1921–1931. doi: 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stephens M, Sloan JS, Robertson PD, Scheet P, Nickerson DA. Automating sequence-based detection and genotyping of SNPs from diploid samples. Nat Genet. 2006;38:375–381. doi: 10.1038/ng1746. [DOI] [PubMed] [Google Scholar]
- 81.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288–294. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McKinney JS, Willoughby KA, Liang S, Ellis EF. Stretch-induced injury of cultured neuronal, glial, and endothelial cells. Effect of polyethylene glycol-conjugated superoxide dismutase. Stroke. 1996;27:934–940. doi: 10.1161/01.str.27.5.934. [DOI] [PubMed] [Google Scholar]
- 84.Chen T, Willoughby KA, Ellis EF. Group I metabotropic receptor antagonism blocks depletion of calcium stores and reduces potentiated capacitative calcium entry in strain-injured neurons and astrocytes. J Neurotrauma. 2004;21:271–281. doi: 10.1089/089771504322972068. [DOI] [PubMed] [Google Scholar]
- 85.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 86.Jackson KJ, Chen Q, Chen J, Aggen SH, Kendler KS, et al. Association of the histidine-triad nucleotide-binding protein-1 (HINT1) gene variants with nicotine dependence. Pharmacogenomics J. 2010;11:251–257. doi: 10.1038/tpj.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Distribution of number of cigarettes smoked per day (A) and FTND scores in the MGS control subjects (B).
(DOCX)