Abstract
The use of fossil evidence to calibrate divergence time estimation has a long history. More recently, Bayesian Markov chain Monte Carlo has become the dominant method of divergence time estimation, and fossil evidence has been reinterpreted as the specification of prior distributions on the divergence times of calibration nodes. These so-called “soft calibrations” have become widely used but the statistical properties of calibrated tree priors in a Bayesian setting hashave not been carefully investigated. Here, we clarify that calibration densities, such as those defined in BEAST 1.5, do not represent the marginal prior distribution of the calibration node. We illustrate this with a number of analytical results on small trees. We also describe an alternative construction for a calibrated Yule prior on trees that allows direct specification of the marginal prior distribution of the calibrated divergence time, with or without the restriction of monophyly. This method requires the computation of the Yule prior conditional on the height of the divergence being calibrated. Unfortunately, a practical solution for multiple calibrations remains elusive. Our results suggest that direct estimation of the prior induced by specifying multiple calibration densities should be a prerequisite of any divergence time dating analysis.
Keywords: calibration, divergence time estimation, tree priors
In addition to observed sequence data, a Bayesian phylogenetic analysis can incorporate other sources of knowledge through the application of informative priors. The use of so-called “soft calibrations” (Rannala and Yang 2005) in the form of informative prior distributions on the divergence times of internal nodes during a phylogenetic analysis, havehas become increasingly common. This is especially the case in Bayesian phylogenetic models that support “relaxed phylogenetics,” in which genetic distances are partitioned into divergence times and lineage-specific substitution rates using a relaxed molecular clock (Drummond et al. 2006).
Although these methods are now quite widely used, the statistical properties of prior distributions subject to calibration densities have not been carefully investigated, when the ranked tree topology is a random variable. In the relaxed phylogenetic models implemented in BEAST (Drummond and Rambaut 2007), calibration is achieved by one of three means: (i) calibration of the rate of evolution through an informative prior on the substitution rate, (ii) calibration through heterochronous data (Drummond et al. 2002; Drummond et al. 2003) (iii) calibration by specification of an informative prior distribution on the divergence time(s) of one or more internal nodes. Whereas the first two methods are relatively straightforward and have been well studied, the statistical properties of the third option in a Bayesian setting have not been well studied when the tree is a random variable.
Here we aim to highlight some of the statistical properties of calibration on internal nodes in BEAST 1.5, a commonly used Bayesian MCMC implementation, and give a new method of constructing a calibration prior that has more intuitive statistical properties when the tree is a random variable. Below we give two examples illustrating how the current implementation of calibration in BEAST may induce non-uniform prior distribution on the tree topology, and how the marginal prior distribution of the calibrated nodes may differ from the calibration density used to construct the tree prior. While the form of the calibrated tree prior can be computed directly for simple cases, in general, the precise relationship between the calibration densities used to construct the tree prior and the actual marginal priors on the calibrated nodes can only be investigated by direct simulation of the tree prior using Markov chain Monte Carlo (MCMC). When there is only a single calibration density, we introduce an alternative method of specifying the marginal prior distribution of the calibrated node directly. However, when using the existing calibration method, we recommend direction simulation of the tree prior as a standard precursor to all relaxed phylogenetic analyses involving internal node calibration densities.
The Construction of a Calibrated Tree Prior in BEAST
When calibrating the divergence times of some internal nodes, the tree prior is constructed in BEAST using three main ingredients:
One or more “calibration densities,” each applied to the divergence time of the most recent common ancestor of a subset of the taxa.
A parametric “tree prior” and associated hyperparameters and hyperpriors that specifiesspecify a density on the topology and all the divergence times of the tree.
Zero or more additional constraints on the topology in the form of subsets of taxa that are constrained to be monophyletic.
In BEAST, these ingredients are combined in a particular way to construct a prior distribution on time trees. The combination of the latter two ingredients is quite unproblematic from the point of view of interpretation. The resulting distribution is simply the relevant parametric “tree prior” conditional on the topological constraints. Although this interpretation is simple, it is worth noting that the resulting distribution of both the divergence times and (obviously) the tree topology will differ from the unconstrained distribution.
However, the first of these ingredients can be incorporated into the model in a number of ways. A general method for computing a conditional birth–death sampling prior for a tree with a fixed ranked topology has been described (Rannala and Yang 2005), but this is not suitable when we wish to infer the topology. For the birth–death model, the special case where the node in question is the root is given in Gernhard (2008), theorem 4.1 with k = 1. In BEAST, the calibration density is combined with the tree prior by simply taking their product. This ignores the overlap in state space of the two densities and we will call this the multiplicative construction. In papers applying BEAST, the calibration density is often known as the “calibration prior” or the “prior on the divergence times,” but we will avoid using the term prior, and use “calibration density” instead, since in the multiplicative construction this distribution does not correspond to the marginal prior distribution of the associated divergence time. If the birth rate and the calibration density are really independent sources of information about the phylogeny, then this may not be a bad method to construct the calibrated tree prior, although this construction certainly does not follow the rules of probability calculus. Specifically, the multiplicative construction is problematic in situations where the researcher expects the calibration prior to represent the marginal distribution of the calibrated node, and can lead to unexpected results.
For example, consider associating a calibration density on TAB, the time of the most recent common ancestor (MRCA) of A and B in a three-taxon tree A,B,C. A Yule prior with a birth rate λ = 1 is used for the tree and an exponential density with mean 2 is used to calibrate TAB. The density of TAB obtained by running BEAST using only the prior is shown in Figure 1a. The same setup with a gamma (γ) calibration density is shown in Figure 1b.
FIGURE 1.

a) Density of TAB from a BEAST run with a Yule prior (λ = 1) and exponential calibration density with mean 2 (black line). The induced density (gray) matches the theoretical density (black dashed line). b) is the same as a) with a gamma (γ(k = 2,θ = 1)) calibration density.
Note that those are the expected outputs. Although difficult in general, here we can analytically compute the marginal prior density (shown as a dashed line), which exactly matches the distribution sampled by MCMC with BEAST. Also note that in case (a) the tree ((A,B),C) is preferred over the other two possible trees for any value of μ. This may seem counterintuitive at first because one might expect the pairing of A and B to be less probable when the mean of TAB is larger than the expected height of the tree. See Appendix 1 for more details and other examples.
A Multiple Calibration Data Set of Marsupials
As the number of calibration densities used to construct the tree prior grows, it becomes hard to describe the joint prior on the calibrated node heights or their individual marginal priors analytically. However, it is always possible to examine the mismatch between the specified calibration densities and marginal densities that result from the multiplicative construction. In Phillips et al. (2009), sequences of seven nuclear genes and the complete mitochondrial (mt) genome protein-coding and RNA-coding DNA sequences for seven placental mammals, three marsupials, two momotremes, and two sauropsids were analyzed, aimed at dating the echidna–platypus divergence. Here, we have rerun the MCMC analysis without the sequence data to show the marginal distributions that result from the multiplicative construction (Figs 2 and 3) on the eight calibrated nodes, alongside the calibration densities specified. We follow the authors by constraining mammals and sauropsids to be monophyletic. Because no specific prior was placed on the birth rate (implied improper prior between zero and infinity), it can be seen that the root height distribution almost matches the calibration density, but most of the others show strong modality, due to the interaction between the calibration densities and the topological constraints. When all of the calibrated nodes have monophyly enforced, the marginal prior distributions are much better matches to the calibration densities, but there are still small discrepancies evident (Fig3).
FIGURE 2.

The marginal prior distributions that result from the multiplicative construction (gray) versus calibration densities (black line) specified for the calibrated nodes from Phillips et al. (2009). The marginal prior distributions were obtained from an MCMC run using the prior only. The calibration densities are as defined by the authors.
FIGURE 3.

The marginal prior distributions that result from the multiplicative construction (gray) versus calibration densities (black line) specified for the calibrated nodes from Phillips et al. (2009), when monophyly is enforced for all calibrated nodes. The marginal prior distributions were obtained from an MCMC run using the prior only. The calibration densities are as defined by the authors.
CONSTRUCTING A TREE PRIOR WITH AN ARBITRARY MARGINAL DISTRIBUTION ON THE TIME OF AN INTERNAL NODE
What are the desired properties of a calibrated tree prior? First, we would like the marginal density of the calibrated node to match the calibration density; and second, conditional on the calibrated node height, we want two trees to have relative prior densities proportional to some sensible generative process like the Yule, birth–death sampling (Stadler 2009, Stadler:2010kx) or coalescent (Kingman 1982; Griffiths and Tavare 1994) tree prior.
Let τ(g) be the time of the most recent common ancestor (TMRCA) for calibrated taxa on genealogy g from the space of all genealogies G. Consider the function ρG(g), a candidate for a calibrated tree prior density on the space of genealogies and ρT(·), the desired marginal calibration density on τ(g). The following properties are desired: I
 |
(1) |
The marginal density on the calibrated node is equal to the calibration density:
In words, the total density of all trees for which the TMRCA of calibrated taxa is x equals the calibration density at x. The integral is written first informally, then formally using the indicator function 1(·), which is equal to 1 when the argument is true, 0 otherwise. Note that this is an integral over all genealogies, or time trees. When the genealogy is represented as g = {ψ,h}, where ψ is the ranked topology and h is a vector of internal node heights in order, the integral can be written as
 |
(2) |
II When restricted to a subset of trees with equal calibrated node height, the density is proportional to the uncalibrated target density fG:
 |
(3) |
Constructing ρG:G→ℝ, which satisfies (I) and (II) is quite easy:
 |
(4) |
where fT(·) is the marginal distribution of τ under fG. We call this the conditional construction. Informally, equation 4 can be written as
 |
It is easy to see that ρG(·) satisfies (I) and (II), and integrates to 1. For genealogies with equal calibration, the calibration and marginal are equal, so their ratio is fG as the other two terms cancel (II). And when integrating over trees with equal calibration, the calibration and marginal can be moved out of the integral, which leaves only the fG term inside, which then cancels with the marginal, leaving the calibration (I).
The conditional construction is useful in practice only if the marginal density fT(·) can be computed efficiently
 |
(5) |
Note that the domain of fG(·) may depend on the conditions imposed on g. If taxa φ are not required to be monophyletic, the domain is all genealogies (G). When φ is required to be monophyletic, the domain is all genealogies that have φ as a monophyletic clade (Gφ).
Yule Tree Prior on Four Taxa with OneMonophyletic Calibration
We now describe in detail how to compute the marginal prior (equation 5) for the Yule tree prior with calibration on a 2-taxa2-taxon monophyletic clade (A,B) in a 4-taxa4-taxon tree. We then show how the same can be done in the general case for Yule tree prior on n taxa and one calibration. In the following sections we shall use fY for the uncalibrated density instead of fG to make it clear that the results involving fY are specific to the Yule prior.
There are four ranked trees (Fig. 4). One, in which TCD is lower than TAB (Case 1), and three ranked trees where TAB has the most recent divergence time (Case 2).
FIGURE 4.

The four ranked trees with four taxa and (A,B) monophyletic. τAB is the same in all cases.
Let T = (T1,T2,T3) be the intracoalescent time intervals. For example, in Case 1, the interval between the leaves and T3 = TCD, T2 = TAB − TCD, and so on. Under the Yule prior, each interval is distributed exponentially, Ti − 1∼ E x p (iλ), and the joint density for T is
 |
Because for Case 1 we have TAB = T2 + T3, the marginal density is given by
 |
(6) |
Note the range [0,tAB] in the integral of t3, which keeps the branch length positive. For Case 2, we obtain
 |
(7) |
Because there are three ranked trees with density u(2) and one with u(1), the marginal Yule distribution is given by
 |
(8) |
Yule tree on four taxa with one calibration prior, no monophyly.—
The construction for the monophyletic clade can be adapted to placing a calibration on TAB without enforcing monophyly. Instead of two cases we have three: A,B is monophyletic (Case I), the common ancestor of A,B has three descendants (Case II), and the common ancestor of A,B is the root (Case III). We already have the densities when A,B is monophyletic, and the density for Case II is given by equation (6). We still need a density for Case III:
 |
The three densities are combined by weighting them according to the number of ranked topologies to which they apply. For Case I we have, as before, 1 and 3 ranked topologies with densities u(1) and u(2). For Case II, there are 4 ranked topologies with density u(1), and for Case III there are 10 with density u(3). Together we get
 |
Yule tree prior with one monophyletic calibration prior
The four taxa case can be generalized to any monophyletic clade φ of size nc in an n = nc + no taxa tree. Formally, the genealogy g is a pair g = {ψ,h}, where ψ is the ranked topology and h is a vector of the internal node heights in reverse order, h = (h1,h2,…,hn − 1). Since φ is monophyletic it is one of the ranked topologies in Ψφ, the set of ranked topologies containing that clade.Because φ is monophyletic, ψ∈ψφ, where ψφ is the set of ranked topologies that contain the monophyletic clade φ.
Now, the Yule density for the heights (Gernhard (2008) section 6.1) is equally divided between all ranked trees having those heights. Because there are |ψφ| of them, the density for the genealogy g is,
 |
(9) |
Define i(ψ) as the rank of the MRCA of φ. The marginal Yule density is given by
 |
(10) |
Surprisingly, this multiple integral evaluates to a simple expression that depends only on the size of φ (nc) and does not depend on n (Appendix 3)
 |
(11) |
Yule tree prior with one calibration prior, no monophyly.—
Deriving the conditional density for the age of the most recent common ancestor of a subset of taxa φ, without the constraint of monophyly of φ is more involved. Again, assume a subset φ of size nc in an n = nc + no taxa tree. The conditional density is broken into no + 1 cases:
Case 0: taxa set φ is monophyletic,
Case 1: the most recent common ancestor of taxa φ has nc + 1 descendants,
Case 2: the most recent common ancestor of taxa φ has nc + 2 descendants,
⋮
Case no: the most recent common ancestor of taxa φ is the root.
Because in Case k we have a monophyletic clade of size k + nc, the density for that case is given by fTφk(x) (11), where φk is of size k + nc. Note that in (11) only the size of the clade matters. To combine the densities from all cases into an overall density we need the number of ranked topologies for each case. Those counts, when scaled to add to 1, act as the coefficients wk in the final equation
 |
(12) |
For the derivation of wk, see (15) in Appendix 2.
Note that the formula works for the special case nc = 1, that is when we wish to condition on the time a particular taxon “attaches” to the tree. In that case, the marginal density has the simple form 2λe − 2λx.
REVISITING AN ANALYSIS OF THE CHACMABABOON (PAPIO URSINUS)
Sithaldeen et al. (2009) provide a phylogenetic analysis of the Chacma Baboon sequences sampled across the entire range of the species. The authors analyze 52 mtDNA samples, using a Yule prior coupled with calibration densities on the root and two nested monophyletic clades. Although this calibrated tree prior has multiple calibration densities and therefore does not fall under the cases previously described, we can derive the marginal density in this particular case using the same methods (Appendix 4).
Before applying the new prior, we run both the original analysis and a prior-only version. Figure 5a shows the calibration density as specified in the BEAST analysis for the three nodes, together with the induced density from the prior-only run and the posterior values from the full analysis. We can clearly see that the posterior values match the induced prior almost to perfection, and that the induced prior is shifted in varying degrees from the calibration priors due to the interaction with the Yule prior. It is not really surprising that the analysis was able to “match” all three marginal divergence time priors because it used a relaxed clock with a wide and flat prior on the rate mean and variance, accommodating branch rate/time combinations whose products satisfy the desired branch length in substitutions while also producing branch times that closely match the marginal tree prior on the calibrated nodes.
FIGURE 5.

a) Induced marginal prior distributions (gray) versus calibration densities (black line) versus marginal posterior distributions (dark gray) for the calibrated nodes from (Sithaldeen et al. 2009) using the multiplicative construction. The induced marginal prior distributions were obtained from an MCMC run using the prior only. The calibration densities are as defined by the authors. b) Similar to (a) but using the conditional-construction. The marginal distributions match the calibration densities as expected. c) Similar to (b), using the conditional construction and a strict molecular clock.
When using the conditional construction, the calibrated density matches the MCMC-sampled prior as expected, and the posterior from the full analysis is almost identical to the prior, presumably because of the relaxed clock (Fig. 5b). However, with the use of a strict clock and the conditional construction we can see that, whereas the sampled prior matches the specified marginal calibration densities, the posterior distribution shows how this prior knowledge has been updated by the data (Fig 5c). That we did not see this with the relaxed clock suggests we may have overparameterized the model in that case.
Given that the data had no visible effect on the posterior distribution of the calibrated divergence times, it is reasonable to assume that in the relaxed clock analysis the prior plays a significant role in the divergence times of the noncalibrated nodes as well. This is indeed the case. Figure 6 shows the trees with the key divergence times from the original, multiplicative construction run (Fig. 6a), and the tree from the conditional construction run (Fig. 6b). We can see that the lowest calibration node in the conditional construction run matches the expected mean of the calibration prior, and as a result all the divergence times in the subtree below have earlier times as well.
FIGURE 6.

a) Posterior estimates for divergence times using the data from Sithaldeen et al. (2009) and the multiplicative construction. b) Posterior estimates for the same data using the conditional construction and relaxed molecular clock as for (a).
DISCUSSIONAND CONCLUSIONS
It is sometimes possible to construct a calibrated tree prior that factorizes precisely into a standard process-based tree prior conditional on the divergence times of the calibrated nodes and an independent marginal prior on those divergence times. We have demonstrated this for one calibrated node in the Yule prior. In order to produce such a conditional construction, one simply needs to be able to efficiently calculate the marginal distribution of the calibrated nodes under the uncalibrated tree prior of choice.
Other conditions on tree priors are also possible. For example, conditioning on the root height of the tree is fairly straightforward for both the Yule model and the more general birth–death model of speciation (Stadler, T. personal communication). In fact, the original description of both the Yule and birth–death models in a phylogenetic context were in the form of a conditioning on the root height. However, those formulations did not condition on the number of taxa, which is also required. Nevertheless, arriving at a Yule probability density conditional on both the root height and the number of taxa is straightforward from that earlier work.
We are fairly confident that the methods presented here can be extended to handle multiple marginal prior distributions on internal nodes. However, the formulas for the conditional densities grow rapidly in size as a function of the number of conditions and taxa. As a result, the determination and evaluation of those conditional priors may become a practical problem.
The method that BEAST implements for constructing calibrated tree priors can lead to marginal distributions on calibrated nodes that are very different than the calibration densities chosen, as seen in Figure 2. In practice, any multiple calibration analysis should always involve direct computation of the calibrated tree prior (by MCMC), and preferably report the actual marginal calibration prior for nodes of interest. Finally, in general, both multiplicative construction and the conditional construction produce nonuniform distributions on the (ranked) topology.
FUNDING
This work was supported by Marsden grant UOA0502.
Acknowledgments
We would like to thank David Bryant, Ron DeBry, Laura Kubatko, and an anonymous reviewer for helpful comments.
APPENDIX 1
Examples of Calibrated Tree Priors Usingthe multiplicative Construction
In the multiplicative construction used by BEAST 1.5, the tree topology and divergence times are influenced both by the calibration density and by the birth rate (λ) of the Yule model of tree branching. These two sources of information are combined to construct a prior density on the tree.
For our first example, we consider associating a calibration density with TAB, the time of the MRCA of A and B in a three-taxon tree A,B,C. A Yule prior with a birth rate λ is used for the tree and an exponential density with mean 1/μ is used to calibrate TAB. Label with T2 is the time of the youngest internal node and with T1, the time between the root and the youngest internal node. Obviously, because TAB is a function of T1,T2, the Yule model already implies a marginal density on TAB, so the Yule density and the calibration density share state space and are not independent.
In BEAST 1.5, this is ignored and the density of the Yule model is simply multiplied by the calibration density to form the multiplicative construction. This product gives the following density on the spaces of trees:
 |
where the terms involving λ come from the Yule prior and the terms involving μ from the calibration. Since the product of two densities is not in general a proper density, the expressions have to be normalized by the constant factor 1/z. The resulting probabilities of the three possible topologies are,
 |
The constant factor z is easily computed because the three integrals have to sum to 1, giving  . So, the relative ratio of the three topologies is 2λ + μ:2λ:2λ and tree ((A,B),C) is preferred for any value of μ. Furthermore, it can be shown that under the multiplicative construction E[T2] = 1/μ + 3λ instead of 1/3λ under the Yule, whereas
. So, the relative ratio of the three topologies is 2λ + μ:2λ:2λ and tree ((A,B),C) is preferred for any value of μ. Furthermore, it can be shown that under the multiplicative construction E[T2] = 1/μ + 3λ instead of 1/3λ under the Yule, whereas  instead of 1/2λ.
instead of 1/2λ.
One may think that placing a calibration density on the nonmonophyletic clade is the cause of the problem. However, we can repeat the calculation for a four-taxon tree while enforcing monophyly of A,B. In one of the four possible topologies (first left in (Fig. 4), the TMRCA of (A,B) is the larger of the two internal nodes (TAB = T2 + T3) and is the smaller of the two in the other three cases (TAB = T3). The total densities for those two cases are
 |
Now, because there are two ranked topologies with the unranked topology ((A,B),(C,D)), the ratio is
 |
So, a ratio of μ + 6λ:μ + 3λ:μ + 3λ is obtained for the three topologies ((A,B),(C,D)), (((A,B),C),D) and (((A,B),D),C). Again, the first topology is preferred regardless of μ.
Even when restricting the three-taxon tree to a single topology by enforcing monophyly, the induced prior on divergence times depends on the specific interaction between the tree prior and the calibration density. Consider a Yule prior with birth rate λ and a gamma prior with shape 2 and scale θ (γ(2,θ)) on TAB. The expected divergence time under this combination can be shown to be 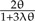 ; which would always be less than the mean of the calibration density, 2θ. Finally, instead of fixing λ we can assume a hyperprior on λ—a very common practice in BEAST. This results in an increase in the dimensionality of the state space, and when deriving expectations or clade probabilities, we need to integrate over the divergence times and λ to obtain the constant normalizing factor. To compute the expected divergence time of (A,B), where λ has a uniform hyperprior of [0,N], we first derive the constant normalization factor,
; which would always be less than the mean of the calibration density, 2θ. Finally, instead of fixing λ we can assume a hyperprior on λ—a very common practice in BEAST. This results in an increase in the dimensionality of the state space, and when deriving expectations or clade probabilities, we need to integrate over the divergence times and λ to obtain the constant normalizing factor. To compute the expected divergence time of (A,B), where λ has a uniform hyperprior of [0,N], we first derive the constant normalization factor,
 |
The expectation under the multiplicative construction is
 |
with N = 100, the average divergence time is approximately  , which is less than θ for any θ > 0.006.
, which is less than θ for any θ > 0.006.
APPENDIX 2
Number of Ranked Topologies for a Nonmonophyletic Clade
Here we derive the coefficients wk used in the formula for calculating the conditional density for the time of the MRCA of the nonmonophyletic taxa set φ. The coefficient wk is the ratio of rk, the number of ranked topologies for case k, to the total number ℛn of ranked topologies for an n = nc + no taxa tree.
 |
Here, rk is the number of ranked topologies, where nc taxa are part of a nc + k taxa subtree. The number is the product of
(i) the number of ways to choose k taxa from no to be part of the clade φ;
(ii) 𝒞k,nc, the number of ranked trees with nc + k taxa whose common ancestor of φ is the root; and
(iii) 𝒟no − k,nc + k, the number of ways to combine the remaining no − k taxa with the clade in(i).
(i) is simply . For (ii), we start with the ℛnc ways to coalesce nc lineages. For each of those, we can add the remaining k in some fixed order. The first lineage can attach itself to places to create a different ranked topology, the second to , and so on, giving
 |
Let 𝒟l,m be the number of ways to combine l lineages and a fixed subtree with m lineages. Examine the possible choices for the first coalescent: Either two of the l lineages are joined ( ways), or this is a coalescent in the subtree. This observation leads to the following recursive formula:
 |
With the initial condition 𝒟0,m = 1 and 𝒟l,1 = ℛl + 1. It is easy to show the above has the solution
 |
(13) |
Substituting no − k for l and nc + k for m gives the required count for (iii). All three put together give
 |
(14) |
Computational Note
The counts rk are large and we need to evaluate wk directly. Some tedious manipulations result in an expression that does not involve large numbers
 |
APPENDIX 3
Conditional Density of the TMRCA fora Monophyletic Clade φ
Here we derive the simple form of the marginal Yule density when the genealogy has a single monophyletic clade φ of size nc in a tree with n taxa.
First note that the total number of those genealogies can be obtained from equation (13)
 |
Partition all ranked topologies according to iφ(ψ) = k + 1, that is group together topologies having k heights above the root of φ.
 |
Under both conditions, the multiintegral has the same value in each case. The integrals can be separated into two independent groups, the n − k − 2 heights below x (nc − 2 from φ, no − k from outside), and the k heights above x. The first group integrates to  , the second to
, the second to 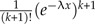 . Both from the simple observation that the integral on k unrestricted heights is k! times the integral on the order statistic. The root of φ contributes λe − λx, giving
. Both from the simple observation that the integral on k unrestricted heights is k! times the integral on the order statistic. The root of φ contributes λe − λx, giving
 |
The last step is possible because none of the terms depend on the specific topology. The number of ranked topologies under our criteria is,
 |
where  , the number of ranked ways to reduce n lineages to k.
, the number of ranked ways to reduce n lineages to k.
Now it is straightforward (but tedious) to show that
 |
After replacing the above and factoring out,
 |
APPENDIX 4
Conditional Density of Three Nested Clades withThree Taxa Outside the Main Clade
Here we derive the marginal density for three nested calibration points in a n + 3 taxon tree. The first (lowest) calibration point is on the root of an n-taxon monophyletic clade φ, the second on the n + 1 clade containing φ and one additional taxon, and the third is on the root of the tree, which includes the remaining two taxa. Let the heights of the calibration points be x0, x1 and x2, where x0 is the height of the root.
Of the n + 2 heights in the tree, three are calibrated and n − 2 are below x2, which leaves just one height, x, outside φ. This gives us three cases, the first where the two outside taxa coalesce before x2 (x < x2), the second where they coalesce between x1 and x2 (x1 ≤ x < x2), and the third where x1 ≤ x < x0. The marginal densities for the three cases are as follows:
 |
For each of the possible ℛn ways to coalesce φ, there are n − 1 ways to place h between the n − 2 heights of φ (Case 1), only one way in Case 2 (no other heights between x2 and x1), and again only one way in Case 3, but here there are ℛ3 ways to coalesce the three lineages between x1 and the root. So, the ratios of the three cases are n − 1:1:3, which let us combine the f1,f2 and f3 into the required density:
 |
(15) |
References
- Drummond A, Ho S, Phillips M, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006:4–e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond A, Rambaut A. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics. 2002;161:1307–1320. doi: 10.1093/genetics/161.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. Genealogies from time-stamped sequence data. In: Buck CE, Millard AR, editors. Tools for constructing chronologies, crossing disciplinary boundaries Lecture Notes in Statistics. Chapter 7. Vol. 177. London: Springer; 2003. pp. 149–174. [Google Scholar]
- Gernhard T. The conditioned reconstructed process. J. Theor. Biol. 2008;253:769–778. doi: 10.1016/j.jtbi.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Griffiths R, Tavare S. Sampling theory for neutral alleles in a varying environment. R. Soc. Lond. Philos. Trans. B. 1994;344:403–410. doi: 10.1098/rstb.1994.0079. [DOI] [PubMed] [Google Scholar]
- Kingman J. The coalescent. Stoch. Process.Appl. 1982;13:235–248. [Google Scholar]
- Phillips M, Bennett T, Lee M. Molecules, morphology, and ecology indicate a recent, amphibious ancestry for echidnas. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17089–17094. doi: 10.1073/pnas.0904649106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 2005;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Sithaldeen R, Bishop J, Ackermann R. Mitochondrial DNA analysis reveals Plio-Pleistocene diversification within the chacma baboon. Mol. Phylogenet. Evol. 2009;53:1042–1048. doi: 10.1016/j.ympev.2009.07.038. [DOI] [PubMed] [Google Scholar]
- Stadler T. On incomplete sampling under birth-death models and connections to the sampling-based coalescent. J. Theor. Biol. 2009;261:58–66. doi: 10.1016/j.jtbi.2009.07.018. [DOI] [PubMed] [Google Scholar]
- Stadler T. Sampling-through-time in birth-death trees. J. Theor. Biol. 2010;267:396–404. doi: 10.1016/j.jtbi.2010.09.010. [DOI] [PubMed] [Google Scholar]


