Abstract
The light-organ symbiosis of Euprymna scolopes, the Hawaiian bobtail squid, is a useful model for the study of animal–microbe interactions. Recent analyses have demonstrated that chitin breakdown products play a role in communication between E. scolopes and its bacterial symbiont Vibrio fischeri. In this study, we sought to determine the source of chitin in the symbiotic organ. We used a commercially available chitin-binding protein (CBP) conjugated to fluorescein to label the polymeric chitin in host tissues. Confocal microscopy revealed that the only cells in contact with the symbionts that labeled with the probe were the macrophage-like hemocytes, which traffic into the light-organ crypts where the bacteria reside. Labeling of extracted hemocytes by CBP was markedly decreased following treatment with purified chitinase, providing further evidence that the labeled molecule is polymeric chitin. Further, CBP-positive areas co-localized with both a halide peroxidase antibody and Lysotracker, a lysosomal marker, suggesting that the chitin-like biomolecule occurs in the lysosome or acidic vacuoles. Reverse transcriptase polymerase chain reaction (PCR) of hemocytes revealed mRNA coding for a chitin synthase, suggesting that the hemocytes synthesize chitin de novo. Finally, upon surveying blood cells from other invertebrate species, we observed CBP-positive regions in all granular blood cells examined, suggesting that this feature is a shared character among the invertebrates; the vertebrate blood cells that we sampled did not label with CBP. Although the function of the chitin-like material remains undetermined, its presence and subcellular location in invertebrate hemocytes suggests a conserved role for this polysaccharide in the immune system of diverse animals.
Keywords: Host–microbe interactions, Euprymna scolopes, Lysosomes, Squid, Symbiosis
1. Introduction
Chitin, a polymer of β-linked n-acetylglucosamine molecules, is an abundant structural and nutritive polysaccharide. Its synthesis and use is a widespread trait among the invertebrates, from the sponges (Ehrlich et al., 2007) through to the invertebrate chordates (Sannasi and Hermann, 1970). As a structural element, chitin is a principal component of invertebrate endo- and exoskeletons, such as the insect cuticle, and of the pens, beaks, and suckers of squid (Dilly and Nixon, 1976; Hunt and El Sherief, 1990). Chitin-like substances have also been localized to hemocytes in three ecdysozoans (Baikova et al., 1993; Martin et al., 2003), although the precise biochemical identities and functions in the hemocytes are not yet known. In addition, the chitin produced by animals also serves as an important nutrient source for chitinolytic bacteria (Keyhani and Roseman, 1999). One such bacterium is Vibrio fischeri, a luminescent marine organism, specific strains of which form a mutualistic association with sepiolid squid species, including the Hawaiian squid Euprymna scolopes.
In the E. scolopes–V. fischeri light organ mutualism, the symbionts reside throughout the host’s life in deep invaginations, or crypts, that occur within a bi-lobed organ in the center of the body cavity (for review, see Nyholm and McFall-Ngai, 2004). As extracellular partners, the symbionts communicate with the host across the apical surfaces of the crypt epithelial cells. The serum and blood cells, or hemocytes, of the highly vascularized light organ associate with both the host tissues and the symbiont population. The symbionts interact both directly and indirectly with the hemocytes, i.e., these cells not only move through the blood vessels of the organ but also exhibit ‘diapedesis’, a behavior in which they leave the vessels and move between the epithelial cells into the bacteria-filled crypts (Nyholm and McFall-Ngai, 1998).
A recent study of the squid–vibrio symbiosis suggested that the host provides chitin as a nutrient to the symbionts (Wier et al., 2010). In this study, the transcriptomes of both the host and the symbiont population were analyzed at four times over the day–night cycle. These data demonstrated that the host upregulates the expression of one chitinase gene and one chitin synthase gene just before dawn. Simultaneously, the resident bacterial symbionts increase the transcription of genes that have been reported to be important for chitin recognition and breakdown in Vibrio cholerae (Meibom et al., 2004; Li et al., 2007). In addition, V. fischeri is genetically and physiologically capable of chitin breakdown and utilization (Ruby et al., 2005; Sugita and Ito, 2006). The results of this transcriptional study provide evidence that a daily rhythm exists in the presentation of nutritive chitin by the squid host to symbiotic V. fischeri.
To determine whether chitin is provided to the symbiont by the host and to identify host cells responsible for this provision, we sought to define the site or sites of chitin production in the host light-organ tissue. Our analyses provide evidence that all mature host hemocytes, both in the light organ and throughout the body, synthesize chitin-like material that localizes to acidic compartments of these cells. In addition, we analyzed the blood cells of various animal taxa to determine whether the presence of chitin in hemocytes is specific to E. scolopes or more widespread among animals.
2. Materials and methods
2.1. General methods
Adult E. scolopes were collected from the wild in Oahu, Hawaii and then maintained in the lab as previously described (Montgomery and McFall-Ngai, 1993). Juvenile squid used in experiments were collected within 15 min of hatching and then washed three times in filter-sterilized Instant Ocean (FSIO; Aquarium Systems, Mentor, OH, USA) to remove any external bacteria. The animals were then either colonized with V. fischeri by incubation with 5,000 colony-forming units per ml of FSIO overnight or left aposymbiotic by incubating with V. fischeri-free FSIO. Colonization states were confirmed by measuring luminescence levels of the animals in a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA, USA) (Ruby and Asato, 1993).
Animal sources for taxonomic study : A. forbesi, Arbacia sp., C. fornicata, M. mercenaria, M. edulis, N. viridans specimens were purchased from Marine Biological Laboratories (Woods Hole, MA, USA), and S. pharaonis specimens were purchased from the National Resource Center for Cephalopods (Galveston, TX, USA). All confocal experiments were performed on a Zeiss LSM 510 confocal microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany), and all chemicals, except where specifically noted, were purchased from Sigma-Aldrich (St. Louis, MO, USA). Transmission electron microscopy was performed as previously described (Nyholm and McFall-Ngai, 1998).
2.2. RT-PCR analysis
To determine the localization of chitin synthase transcript in E. scolopes, total RNA was isolated from extracted adult E. scolopes tissues (hemocytes, white body, hindgut, and eye). Single-stranded cDNA was then synthesized from the purified RNA using reverse transcriptase (Clontech, Mountain View, CA, USA) and random primers. Using 500 ng of cDNA as a template for each PCR reaction, single gene products were analyzed with specific primers subsequent to cDNA synthesis. All reactions were performed with a no-reverse-transcriptase control to confirm the lack of genomic DNA contamination in the reactions. Two chitin synthases were identified in a 3’ expressed sequence tag database (Chun et al., 2006), called chitin synthase 1 (similar to chitin synthase from nematode Brugia malayi by BLAST analysis with an E-value of 1e-9) and chitin synthase 2 (similar to chitin synthase from oyster Pinctada fucata by BLAST analysis with an E-value of 8e-36). Specific primers to the above transcripts used in this study were: CS1F: 5’-TTGGCGTGTTTGCACTCTCGGCCCT-3’, CS1R: 5’-GACGTGCGTTCATTGCGTTGTTGA-3’ to amplify chitin synthase 1, and CS2F: 5’-TGAATCTGCTGTGGAGTGTGGCTA-3’, CS2R: 5’-AATGCGCCTCTTCTGTTCAACGTC-3’ to amplify chitin synthase 2. As a loading control, we used the primers 40SF: 5’-AATCTCGGCGTCCTTGAGAA-3’, 40SR: 5’-GCATCAATTGCACGACGAGT-3’ to amplify RNA encoding the 40S ribosomal subunit.
2.3. Localization of putative chitin in E. scolopes tissues
To localize chitin-like biomolecules in tissues, whole juvenile E. scolopes and extracted light organs were prepared for immunocytochemistry as previously described (Davidson et al., 2004; Kimbell and McFall-Ngai, 2004). Briefly, whole juvenile squid were anesthetized with 2% ethanol in filter-sterilized Instant Ocean and then fixed in 4% paraformaldehye in marine phosphate-buffered saline (mPBS – 50 mM sodium phosphate buffer with 0.45 M NaCl, pH 7.4) for 18 h at 4 °C. The squid were then washed in mPBS, dissected, and permeabolized in permeabolization buffer (mPBS containing 1% Triton-X) overnight. Samples were then exposed to binding proteins at relevant concentrations: 167 nM for the fluorescein isothiocyanate-conjugated chitin-binding protein (FITC-CBP; New England Biolabs, Ipswich, MA, USA), 0.25 μg/ml rhodamine phalloidin, 20 μg/ml rhodamine-conjugated succinylated wheat-germ agglutinin (Vector Labs, Burlingame, CA, USA), all in permeabolization buffer, for 1–3 days. To visualize nuclei, samples were stained with TOTO-3 (Invitrogen, Carlsbad, CA, USA), a nucleus-specific dye at a concentration of 1 μM. Squid tissues were mounted individually on slides in VectaShield (Vector Labs, Burlingame, CA, USA), a mounting medium that reduces sample autofluorescence and bleaching. The samples were then analyzed by confocal microscopy to determine presence or absence of labeling by FITC-CBP, which only recognizes polymeric chitin (Hardt and Laine, 2004).
To characterize the CBP-labeling of hemocytes, blood cells were extracted from both adult and juvenile samples. Hemocytes were extracted from adult E. scolopes through the cephalic artery using a 1-ml syringe fitted with a 28-gauge needle. To release hemocytes from juvenile host tissues, 10 E. scolopes juveniles were anesthetized with 2% ethanol in FSIO and then the entire cohort was homogenized manually in 1 ml Squid Ringer’s solution (50 mM MgCl2, 10 mM CaCl2, 10 mM KCl, 530 mM NaCl, 10 mM HEPES). The homogenate was applied to coverslips in 12-well tissue culture plates and the hemocytes were allowed to adhere for 30 min. The coverslips were then washed three times for 5 min with Squid Ringer’s solution to remove other tissues. In experiments designed to determine whether CBP-labeling localized to acidic compartments, the hemocytes were exposed to a 1:1000 dilution of Lysotracker Red (Invitrogen, Carlsbad, CA, USA) in Squid Ringer’s solution to label the lysosomes, and then allowed to incubate in the stain for 15 min and rinsed with Squid Ringer’s solution twice for 5 min. The cells were then fixed for 30 min in 4% paraformaldehyde in mPBS. Afterwards, samples were washed three times for 10 min in mPBS and exposed to permeabolization buffer for 1 h. Following permeabolization, samples were washed three times for 10 min with permeabolization buffer and exposed to FITC-CBP for 3 h at room temperature. After another set of three 10-min washes, rhodamine phalloidin at a concentration of 20 μg/ml was added to the samples overnight at 4 °C. After a final set of three 10-min washes with buffer, the coverslips were mounted onto glass slides and processed as for whole tissue mounts.
2.4. Chitinase treatment
To determine whether CBP-labeling of the hemocytes was affected by chitinase treatment, hemocytes were treated with a purified chitinase to disrupt the polymeric structure of any chitin that might have been present. Fixation and permeabolization were performed as described above. Fixed hemocytes were exposed to three different conditions: (i) phosphate-buffered saline (PBS; 50 mM sodium phosphate, 100 mM sodium chloride, pH 7.4) containing 1 mM sodium ethylenediaminetetraacetic acid, pH 6.0, and 40 units of purified Brugia malayi chitinase (New England BioLabs, Ipswich, MA, USA); (ii) buffer without chitinase; or, (iii) as a control for the effects of the treatment conditions, 40 units of chitinase in the same buffer heated to 75 °C for 20 min (heat-inactivated chitinase). Extracted hemocytes were exposed to each of the three treatments for 24 h at 37 °C. The treated hemocytes were washed three times for 10 min with the above buffer and then exposed to CBP and rhodamine phalloidin as described above. Ten hemocytes per condition were then visualized by confocal microscopy and the level of fluorescence in each cell was measured by Zeiss analysis software.
2.5. Assessment of possible protein association of CBP-positive biomolecules
Previous reports of the presence of chitin in invertebrate blood indicated that chitin can occur as a “chitinoprotein”, i.e., with the chitin bound to a protein molecule (Kramerov et al., 1986). Thus, we sought to determine whether the CBP-positive material in the E. scolopes hemocytes was protein-associated. The chitin-like substance in E. scolopes hemocytes is bound by FITC-CBP (this study), so we used the New England BioLabs (Ipswich, MA, USA) SNAP-capture protein purification system to immobilize purified CBP bound to a SNAP tag as bait for the chitin-like substance contained in the squid hemocytes. To bind the SNAP magnetic beads to the purified CBP containing a SNAP-tag, the beads were first resuspended by gentle vortexing and then equilibrated in immobilization buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM DTT and 0.1% Tween 20). Then, 100 μl of a solution of 1 mg/ml SNAP tag-CBP in immobilization buffer was added to the equilibrated SNAP-capture beads in a 1.5 ml Eppendorf tube and incubated with mixing overnight at 4 °C. The beads were then washed four times by adding 500 μl immobilization buffer and vortexing for 1 min, then placed in a magnetic manifold for 10 s and the supernatant was removed. Once the SNAP-CBP was bound to the magnetic SNAP-capture beads, 500 μl of a solution of 1 mg/ml E. scolopes hemocyte lysate in immobilization buffer containing an EDTA-free protease inhibitor cocktail was added to the washed beads and incubated for 3 h at room temperature. The beads were then washed an additional 5 times with 1 ml of immobilization buffer as described above. To remove the bound proteins from the beads, 50 μl of SDS-PAGE loading buffer (Tris-SDS-BME) were added to the beads and heated at 95 °C for 5 min. All fractions were analyzed on SDS-PAGE gels.
Because we found no evidence for the binding of CBP-positive material to protein, we sought to determine the fraction of cell homogenate that contains the putative chitin biomolecules. Two whole gills from adult E. scolopes were homogenized in 1 ml of PBS containing protease inhibitors (Sigma-Aldrich protease inhibitor cocktail III). The homogenate was then centrifuged at 10,000 × g for 20 min at 4 °C to separate the aqueous soluble (supernatant) and insoluble fractions (pellet). The pellet was washed twice with PBS, by resuspension in buffer followed by centrifugation for each wash, to remove residual soluble protein. To extract a subset of the membrane proteins, the pellet was resuspended in PBS containing 2% SDS and centrifuged at 10,000 × g for 20 min at room temperature. The supernatant was removed, and then the resulting pellet was washed twice, as described above, in PBS containing 2% SDS to remove residual membrane proteins soluble in 2% SDS. To solubilize the remainder of the integral membrane proteins and other SDS-soluble proteins, the pellet was resuspended in PBS containing 5% SDS and centrifuged as described above. The supernatant was removed from the sample, and the resulting pellet was washed twice with 5% SDS in PBS to remove any residual membrane-bound proteins. To bring proteins into solution that were insoluble in SDS, the remaining pellet was homogenized in 8 M urea, and then centrifuged as above. The supernatant was then removed and the resulting pellet was retained for subsequent analysis.
To visualize the proteins and determine which fractions contained chitin, 5 μl of each fraction and a positive control of 5 μl of 3.5% colloidal chitin were spotted onto Whatman-1 filter paper in duplicate and allowed to dry and then incubated in ProtoBlue Safe coomassie blue stain (National Diagnostics, Atlanta, GA, USA) for 15 min and destained in deionized water for 30 min and allowed to dry. On a separate piece of filter paper, the same fractions were spotted and allowed to dry, and then the paper was incubated in 0.01% calcofluor white in water for 15 min and then visualized under UV light. Calcofluor white is a dye that binds with polymeric chitin and cellulose.
2.6. Hemocyte preparations for taxonomic survey
Hemolymph containing hemocyte populations was drawn from individuals of other aquatic animals using a 1-ml syringe fitted with a 28-gauge needle as follows: S. pharaonis, from the cephalic artery; Mytilus edulis and Mercenaria mercenaria, from the main hemolymph sinus of the foot; Crepidula fornicata, from the hemocoel of the foot; Eisenia foetida and Nereis viridans, from the dorsal blood vessel; Arbacia sp., through the periosomal membrane; Asterias forbesi, from the tip of an arm; and Ciona intestinalis, from the heart and associated tissues. After extraction, the hemolymph was spotted onto glass coverslips in tissue culture plate wells containing 500 μl of Squid Ringer’s solution for 10 min to allow the hemocytes to adhere to the coverslips. The coverslips were then washed twice for 5 min in Squid Ringer’s solution at room temperature, except for E. foetida samples, which were washed in PBS diluted 1:2 in water. The cells were then fixed on the coverslips by incubation for 30 min at room temperature in mPBS containing 4% paraformaldehyde, except for E. foetida hemocytes, which were fixed in 0.5% PBS containing 4% paraformaldehyde. The fixed cells were then washed three times for 10 min in mPBS and incubated in permabolization buffer for 1 h at room temperature. The buffer was then removed and replaced with permeabolization buffer containing 167 nM FITC-CBP, and then incubated with the samples for 3 h at room temperature. The samples were then washed with permeabolization buffer three times for 5 min at room temperature and then incubated with rhodamine phalloidin in permeabolization buffer for 1 h at room temperature or overnight at 4 °C. The coverslips were then washed three times for 5 min in mPBS and mounted onto glass slides containing VectaShield and visualized by confocal microscopy.
Additional animals used in the taxonomic survey were sampled as follows:
Biomphalaria glabrata: Hemolymph was collected as described previously (Sminia and Barendsen, 1980), then hemocytes were allowed to adhere to a coverslip and processed as described for E. foetida.
Atta colombica and Drosophila melanogaster. Whole animals (3 A. colombica or 10 D. melanogaster) were homogenized as a single cohort manually in 500 μl 0.5 × PBS. The homogenate was then applied to coverslips and the hemocytes were allowed to adhere for 30 min. The samples were then processed as described above for E. foetida. Danio rerio: 48-h juvenile transgenic animals carrying a GFP-tagged myeloperoxidase gene (GFP:MPO), which allows for visualization of neutrophils, were anesthetized in 2% tricaine methanesulfonate and then fixed in 4% paraformaldehyde in 0.5% PBS and processed as whole E. scolopes specimens, except that staining was performed with TRITC-labeled CBP instead of FITC-labeled CBP.
Mus musculus: A whole blood smear was fixed to a microscope slide with methanol and then permeabolized and stained as described above for E. foetida; in addition, the blood sample was stained with 1 μM TOTO-3 in PBS to identify neutrophils. The slides were then analyzed by confocal microscopy for CBP staining.
3. Results
3.1. Localization of chitin to E. scolopes hemocytes
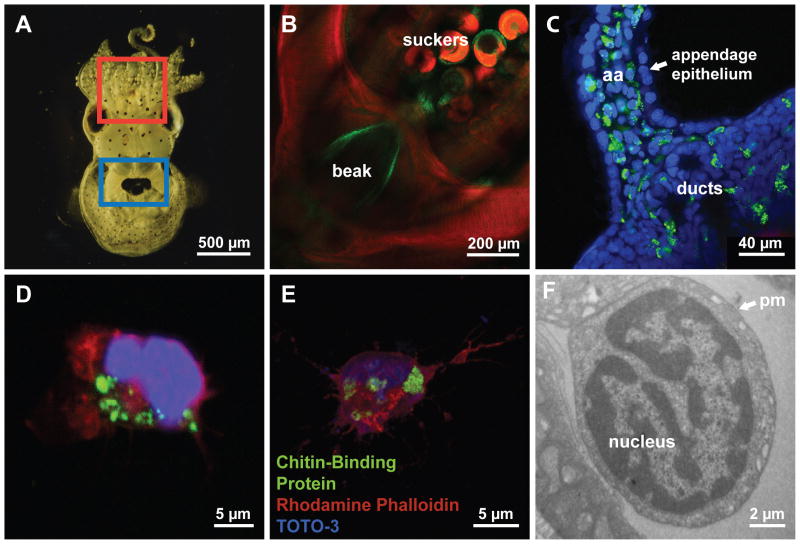
In juvenile E. scolopes (Fig. 1A), the suckers and the surface of the beak, structures that are known to contain chitin (Dilly and Nixon, 1976) labeled strongly with CBP (Fig. 1B). In addition, we observed strong staining in cells inside the body of the light organ, including within appendages of the juvenile-specific ciliated surface (Fig. 1C). These appendages, which comprise a ciliated epithelium overlying a blood sinus, have a population of hemocytes within the sinus (Koropatnick et al., 2007). The CBP staining appeared to localize to the hemocytes. To confirm that E. scolopes hemocytes label with the CBP, we extracted hemocytes, which can be identified by their u-shaped nuclei and adherence to coverslips (Nyholm et al., 2009), from both juvenile (Fig. 1D) and adult (Fig. 1E) animals and incubated them with CBP. The probe bound to vesicle-like areas in all hemocytes that we stained and visualized, (n > 50 for each condition) suggesting that circulating hemocytes contain a molecule recognized by CBP.
Fig. 1.
Localization of chitin in the E. scolopes light organ. (A) Ventral surface of a juvenile E. scolopes resting on the substrate. Boxes represent the position of the buccal area (red) and light organ (blue). (B) Laser-scanning confocal micrograph of the buccal mass (red box in (A)), containing the beak and adjacent suckers, which show chitin-positive regions (green). (C) Confocal micrograph of a portion of one lobe of a juvenile light organ. Chitin-positive labeling (green) occurs in regions near the ducts, as well as in cells in the blood sinus of the anterior appendage (aa). CBP labeling is undetectable in the appendage epithelium that surrounds the blood sinus. (D) Confocal image of a single hemocyte extracted from a juvenile animal. Chitin labeling (green) is localized to intracellular granules surrounding the nucleus (blue). (E) Single confocal image of a three-dimensional reconstruction of a hemocyte from an adult E. scolopes. (F) Transmission electron microscopy image of a hemocyte in the gill of a juvenile E. scolopes. Green: FITC-chitin-binding protein (CBP; chitin-positive areas); red: phalloidin (filamentous actin); blue: TOTO-3 (nuclei). pm, plasma membrane.
3.2. Specificity of the chitin-binding probe
Chitin-binding probes, such as the one used in this study, are widely known to have a high affinity for chains of more than six n-acetyl glucosamine (GlcNAc) residues, generally referred to as polymeric chitin (Hardt and Laine, 2004). To confirm that the substance being stained by the probe is polymeric chitin and not shorter GlcNAc residues, which may theoretically bind the probe in the absence of larger chitin oligomers, we employed two approaches: (1) characterization of the specificity of CBP binding; and (2) determination of the susceptibility of the CBP-positive material to chitinase degradation.
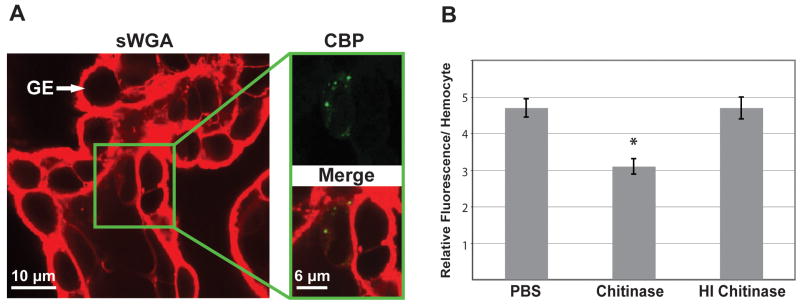
For approach (1), we stained whole juvenile animals with both the CBP and succinylated wheat germ agglutinin (sWGA), a lectin that specifically binds GlcNac residues. If CBP stains polymeric chitin to the exclusion of shorter GlcNac oligomers, we would expect that the CBP would only stain a subset of the areas that sWGA binds. Upon staining of the E. scolopes hemocytes in the gills, only a subset of the areas stained (Fig. 2A), suggesting that the CBP only stains particular types of GlcNac residues, most likely those containing larger numbers of GlcNac groups.
Fig. 2.
Specificity of chitin-binding protein (CBP) labeling. (A) Confocal micrograph of a juvenile E. scolopes gill labeled with succinylated wheat germ agglutinin (sWGA, red) and FITC-CBP (green). A single hemocyte within the gill tissue is shown in the CBP and merged images (right panels). GE, gill epithelium. (B) The effect of chitinase application on the level of fluorescence attributed to FITC-CBP. Fixed E. scolopes hemocytes were treated with a PBS only control (PBS), active chitinase, or heat-inactivated chitinase (HI Chitinase), and the fluorescence of the FITC-CBP was quantified for 10 cells per condition. Asterisk = significant difference from the PBS control at a p-value of < 0.01 as determined by a Student’s t-test with a Bonferroni correction for multiple comparisons.
To determine whether the substance being stained by the probe has properties consistent with polymeric chitin, we applied purified chitinase (see section 2.4) to extracted hemocytes that had been permeabolized and measured the amount of probe binding in comparison with buffer or heat-killed chitinase controls. Active chitinase significantly reduced the fluorescence of CBP-exposed hemocytes (Fig. 2B). In addition, the chitinase treatment did not increase the amount of CBP staining, providing further evidence that CBP does not stain breakdown products of chitin that were produced by the purified chitinase. Taken together, these data suggest that chitinase breaks down polymeric chitin within the hemocytes.
3.3. Expression of chitin synthase mRNA in E. scolopes hemocytes
The 3’ EST library (Chun et al., 2006) of the E. scolopes light organ contains two putative chitin synthases identified by sequence similarity to other known chitin synthase transcripts, which we have named EsCS1 and EsCS2. To determine whether E. scolopes hemocytes expressed either chitin synthase, we performed semi-quantitative reverse transcriptase PCR on cDNA prepared from various E. scolopes tissues, including hemocytes, the hematopoietic gland (white body), hindgut, and eye using primers specific to either chitin synthase or the 40S ribosomal subunit RNA as a control for equal loading. We found that the hemocytes produce transcripts for both chitin synthases. However, the white body, where hemocytes are produced, does not appear to express both chitin synthases to the same degree as the hemocytes did when we carried out end-point PCR using the same amount of cDNA template, suggesting that chitin synthesis only occurs in mature hemocytes (Fig. 3). Upon staining the white body with FITC-CBP, we did not detect any staining of immature hemocytes with the CBP, further supporting this hypothesis (data not shown). The hindgut sample was included as a positive control because it contains high levels of both transcripts, a finding which is consistent with the reported ability of the hindgut to secrete a chitinous layer (Budelmann et al., 1997). These data suggest that E. scolopes hemocytes are capable of producing enzymes that synthesize chitin.
Fig. 3.
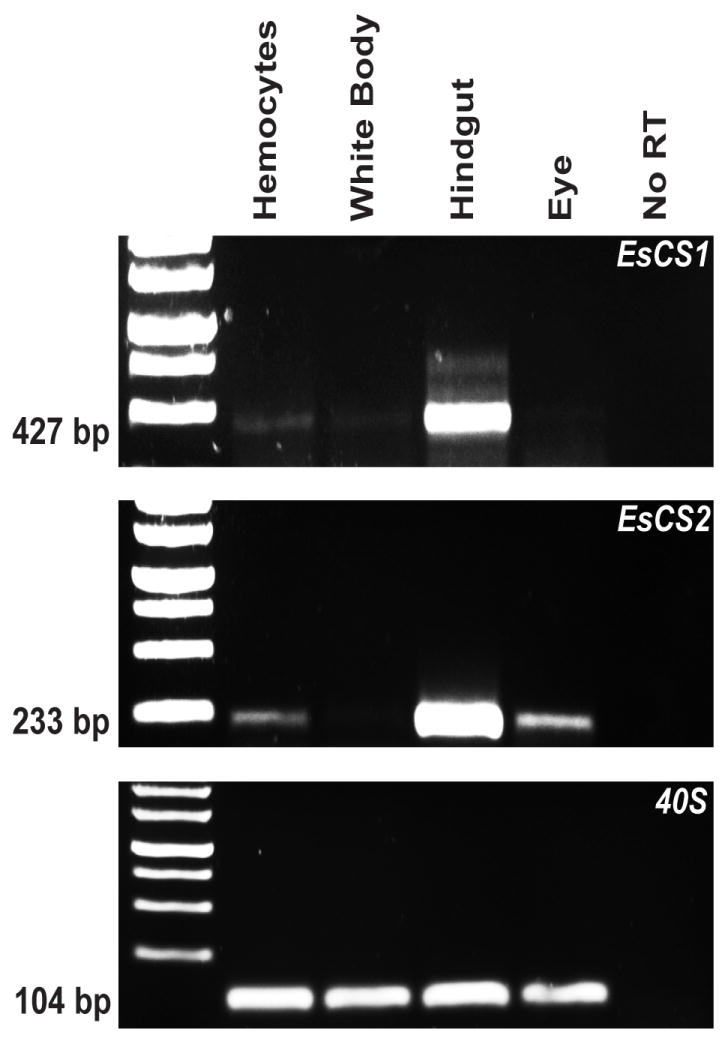
Production of chitin synthase transcript in E. scolopes hemocytes. cDNA was prepared from RNA that had been extracted from four adult squid cell types or tissues (hemocytes, white body, hindgut, and eye). 500 ng of cDNA was used as a template for end-point PCR. CS1 = E. scolopes chitin synthase 1; CS2 = E. scolopes chitin synthase 2, 40S = 40S subunit ribosomal RNA, used as a loading control. Numbers to the left of the gel show the size of each gene product.
3.4. Biochemical characteristics of chitin-containing compartments
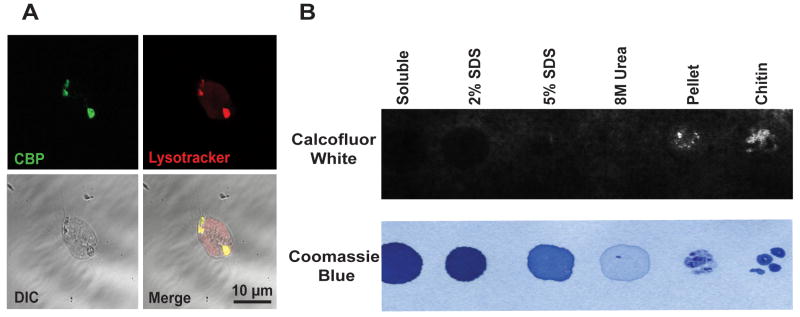
The punctate labeling of the CBP in the hemocytes suggested that the chitin-like molecule localizes to specific subcellular compartments. Extracted hemocytes co-stained with FITC-CBP and Lysotracker Red revealed co-localization of these fluorochromes to intracellular acidic compartments (Fig. 4A). The CBP staining also co-localized with an antibody specific to E. scolopes halide peroxidase (Weis et al., 1996; Small and McFall-Ngai, 1999), a protein that typically localizes to lysosomes (Baggiolini et al., 1969; Nauseef, 1998). Taken together, these data suggest that the intracellular chitin occurs in or is associated with the lysosomes of E. scolopes hemocytes. Because earlier reports of chitin-like molecules in D. melanogaster hemocytes provided evidence that the chitin is bound to protein (Kramerov et al., 1986), we performed two assays to investigate the possibility that chitin in E. scolopes hemocytes also associates with protein. We used calcofluor labeling of protein fractions as well as a pull-down with SNAP-CBP (see Section 2.5) to determine the nature of any potential bound protein. Calcofluor-positive labeling was only present in material pelleted after extraction of hemocyte-rich tissues with aqueous solvents, SDS and urea (Fig. 4B), in which the majority of proteins should be soluble, but chitin is not. A coomassie stain was used as the counterstain for all fractions due to its binding to proteins and chitin (Liau and Lin, 2008). These data, coupled with our inability to capture any material with the CBP fused to a SNAP tag as bait (data not shown), provide evidence that the chitinous material in E. scolopes hemocytes is not a chitinoprotein.
Fig. 4.
Biochemical characterization of CBP-positive compartments. (A) Confocal micrograph of a single E. scolopes hemocyte co-stained with FITC-CBP (green) and Lysotracker Red (red). DIC = image of the same cell by differential interference contrast. (B) Images of protein extracts from adult E. scolopes gill tissue probed with calcofluor white and imaged under UV light (top), or stained with ProtoBlue coomassie blue stain and imaged under visible light (bottom).
3.5. Survey of animal hemocytes for CBP-labeling
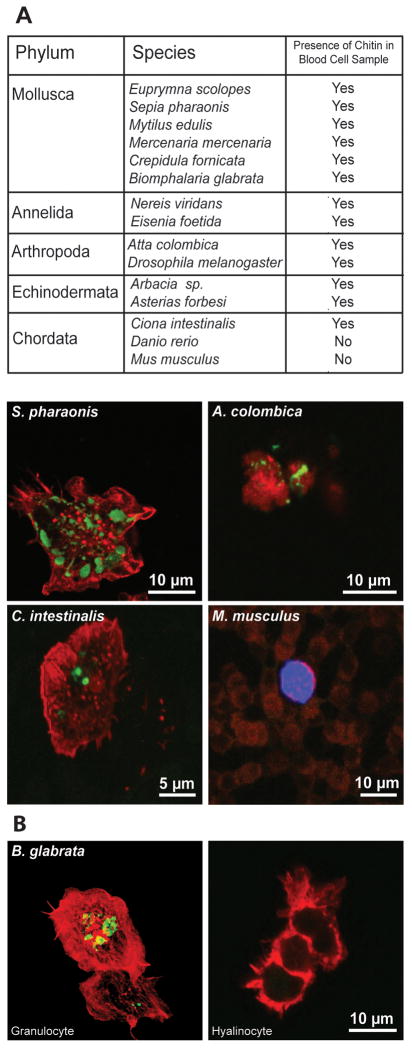
To determine whether the presence of chitin in hemocytes is specific to E. scolopes, we examined circulating blood cells in other animal taxa for CBP-positive labeling. Blood cells from 15 species in five phyla, either from a hemocyte extraction or tissue whole-mount, were labeled with FITC-conjugated CBP and analyzed. Between 15 and 40 cells per preparation were viewed by confocal microscopy (Fig. 5A). Invertebrate granulocytes we surveyed labeled with CBP, regardless of phylum. However, neither of the vertebrates that we sampled (Mus musculus and Danio rerio) had any blood cells that stained positively for chitin, which is consistent with the absence of any known chitin synthetic enzymes in either species.
Fig. 5.
Taxonomic survey of chitin carriage in hemocytes. (A) All organisms surveyed with the presence or absence of chitin as detected by FITC-CBP labeling (green) noted in each sample. Included are confocal micrographs of representative samples from four phyla. TOTO-3-positive blood cell shown for M. musculus is a neutrophil. (B) Confocal micrographs of a granulocyte (left) and a hyalinocyte (right) from B. glabrata. Counterstains: red = rhodamine phalloidin (filamentous actin), blue = TOTO-3 (nuclei).
Blood cell extractions from five species (M. edulis, M. mercenaria, C. fornicata, B. glabrata, and N. viridans) produced the two known distinct blood cell types, identified as granulocytes and hyalinocytes by both size and morphology (Kuchel et al., 2010). In species that produced both of these types of hemocytes, such as Biomphalaria glabrata, we only found CBP staining in the granulocytes (Fig. 5B), a feature that may provide insight into the function of CBP-reactive substances in blood cells.
4. Discussion
The results of this study provide evidence that all mature E. scolopes hemocytes contain endogenously produced chitin in lysosomal compartments. In addition, we demonstrate that the presence of chitin-like molecules in granulocyte-like hemocytes is a widespread character among the invertebrates.
The chitin in E. scolopes hemocytes may be important in the E. scolopes – V. fischeri symbiosis in immune surveillance of the light organ. Hemocytes are known to infiltrate the light organ early in the initiation of the symbiosis, eventually potentiating the development of the light organ into a mature symbiotic structure (Nyholm and McFall-Ngai, 1998; Kimbell et al., 2006; Koropatnick et al., 2007). In addition, studies of the behavior of these cells have demonstrated that the hemocytes sample the crypt spaces and communicate the status of these symbiont-containing regions to the rest of the body of the animal (Nyholm and McFall-Ngai, 1998; Nyholm et al., 2009). As such, they behave similarly to mammalian dendrocytes that sample the gut. To determine whether chitin is important for such immune activity will require further study.
Abundant evidence exists that the symbionts ferment host chitin, which may be provided to the bacteria as a nutrient source (Wier et al., 2010). However, whether this nutrient is provided by chitin synthesized in the abundant population of hemocytes within the bacteria-containing tissue of the light organ, in the crypt epithelial cells themselves, or in both cell types remains to be determined. As the epithelium of the molluscan gut is often lined by chitin (Berkeley, 1935; Moueza and Frenkiel, 1979; Harrison and Kohn, 1997), and the light organ is embryologically derived from the E. scolopes hindgut/ink-sac complex, it would not be surprising to find that the crypt epithelium synthesizes chitin. We did not observe labeling of the crypt epithelium with CBP, but if this tissue synthesizes chitin, the chitin may be produced, transported and consumed immediately by the symbionts, so that any chitin present would be undetectable. This question could be answered in future studies in which the major sites of EsCS1 transcription would be identified using in situ hybridization with probes specific to EsCS1.
The position and size of CBP-positive compartments in E. scolopes hemocytes were consistent with their co-localization within acidic and halide-peroxidase-positive lysosomes of these cells (Nyholm and McFall-Ngai, 1998; Small and McFall-Ngai, 1999). Although their function is unknown, other polysaccharides have been reported to be present in lysosomes in other systems. Abundant polysaccharides, produced in the endoplasmic reticulum of certain mammalian cells, can be targeted to the lysosome (Moore, 1999). In certain lysosomal storage disorders, the polysaccharides that have been targeted to lysosomes for recycling are not broken down, a condition that can impede normal lysosomal function (Meikle et al., 2003). While lysosomal storage disorders have been best characterized in muscle tissue, these defects can also be evident in macrophages (Kieseier et al., 1997). The widespread presence of a chitin-like molecule in invertebrate blood cells is unlikely to be due to a defect in metabolism. However, the presence of polysaccharides in mammalian lysosomes does suggest a possible function for these molecules in the lysosomes of invertebrate granulocytes, i.e., for the storage and recycling of polysaccharides. Since the chitin only localizes to invertebrate granulocytes, it is likely that the compound plays a role that is specific to this cell type. For example, as a nonreactive molecule, chitin may serve as a scaffolding or matrix for the proteins and other biomolecules in the lysosome.
Like other cephalopods, Euprymna scolopes has only been shown to produce one type of hemocyte, which is most similar to invertebrate granulocytes (Cowden and Curtis, 1981; Nyholm and McFall-Ngai, 1998). However, other molluscs surveyed in this study produce both of the major molluscan blood cell types, i.e., granulocytes and hyalinocytes, and only granulocytes stained with the CBP. The presence of chitin in granulocytes but not hyalinocytes may be due to the different functions of these cells; granulocytes are primarily phagocytic and involved in respiratory burst, while the role of hyalinocytes is not yet well defined (Goedken and De Guise, 2004). As such, if the chitin-like molecule is restricted to the lysosome, as suggested in E. scolopes hemocytes, the undetectable labeling of CBP in hyalinocytes may be due to the presence of fewer lysosomes, which is characteristic of this cell type (Mateo et al., 2009).
While ours is the first study to be done on this scale, several previous studies have shown that circulating immune cells in invertebrates may contain chitin-like substances. Drosophila melanogaster cell lines produce a heavily sulfated chitin-like molecule, possibly composed of chitin linked to a protein (Kramerov et al., 1986). An antibody to this molecule labels granules in cell lines of D. melanogaster and other insect species and is present in regions of the imaginal discs of developing D. melanogaster larvae (Kramerov et al., 1990). This same antibody binds to granules in D. melanogaster hemocytes in whole-animal analyses (Baikova et al., 1993). These studies suggested that the chitin-like polysaccharide molecules in Drosophila melanogaster hemocytes are linked to a protein, although the sequence of the hypothetical protein was not determined (Kramerov et al., 1986, 1990; Baikova et al., 1993). In contrast, our analyses suggested that the chitin-like molecules in E. scolopes hemocytes are not linked to protein; specifically, the chitin-positive compound was not soluble in water, SDS, or urea (Fig. 4), and could not be isolated by a CBP-based protein purification protocol. Taken together, the data on chitin-like substances in invertebrate hemocytes suggest that the chitin may occur in different forms in these cells.
Other evidence for possible chitin carriage in invertebrate hemocytes has been suggested by the presence of specific sugar residues. Wheat germ agglutinin, a lectin that binds specifically to n-acetyl-glucosamine and sialic acid residues, recognizes materials in the large granulocytes of the crustaceans Sicyonia ingentis and Homarus americanus (Martin et al., 2003). Our study, probing with fluorescently labeled CBP, suggests that these lectins are labeling chitin within these cells and that this character is common to many invertebrate phyla, not just ecdysozoans (Fig. 5).
While this study provides evidence for the presence of chitin in invertebrate granulocytes, we have not yet determined its function in the immune system. One possible approach would be the study of this chitin-like compound in cell culture of invertebrate granulocytes, where the cells are more easily manipulated directly and can be isolated in higher numbers. Although not present in all blood cells sampled in this study, the conspicuous nature of the chitin in invertebrate hemocytes may help to elucidate the role of polysaccharides in immune cells in general, which is an understudied area. The lack of chitin in vertebrate immune cells, though anticipated due to the lack of chitin synthetic enzymes in most vertebrates, may point to an invertebrate-specific function, and may be tied to the differences between the invertebrate and vertebrate immune systems. Finally, one practical outcome of this research is the development of a tool for the study of invertebrate granulocytes, specifically an easily used, relatively inexpensive, and reliable marker for these cells, i.e., CBP.
Acknowledgments
We thank Dr. G. Boekhoff-Falk, Dr. C. Currie, Dr. A. Huttenlocher, Dr. L. Knoll, Dr. B. Rader, and Dr. T. Yoshino for generously providing blood cell samples. In addition, we thank all members of the McFall-Ngai and E.G. Ruby laboratories for discussion and critique of the manuscript. This work was supported by a NSF grant (IOS 0817232) to M.M.-N., by NIH grants (RO1-AI50661 to M.M.-N. and RR R01-12294 to E.G. Ruby and M.M.-N.), and a NIH Molecular Biosciences Training Grant through UW-Madison to E.H.-H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baggiolini M, Hirsch JG, De Duve C. Resolution of granules from rabbit heterophil leukocytes into distinct populations by zonal sedimentation. J Cell Biol. 1969;40:529–541. doi: 10.1083/jcb.40.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baikova NA, Gvozdev VA, Kramerov AA. The tissue localization of “chitinoprotein”, detectable by using specific antibodies, in the development of Drosophila melanogaster. Ontogenez. 1993;24:33–42. in Russian. [PubMed] [Google Scholar]
- Berkeley C. The chemical composition of the crystalline style and of the gastric shield: with some new observations on the occurrence of the style oxidase. Biol Bull Woods Hole. 1935;68:107–114. [Google Scholar]
- Budelmann BU, Schipp R, Boletzky S, von . Cephalopoda. In: Harrison FW, Kohn AJ, editors. Microscopic Anatomy of Invertebrates, vol 6A Mollusca II. Wiley-Liss; New York, NY: 1997. pp. 119–414. [Google Scholar]
- Chun CK, Scheetz TE, Bonaldo M, de F, Brown B, Clemens A, Crookes-Goodson WJ, Crouch K, DeMartini T, Eyestone M, Goodson MS, Janssens B, Kimbell JL, Koropatnick TA, Kucaba T, Smith C, Stewart JJ, Tong D, Troll JV, Webster S, Winhall-Rice J, Yap C, Casavant TL. An annotated cDNA library of juvenile Euprymna scolopes with and without colonization by the symbiont Vibrio fischeri. BMC Genomics. 2006;7:154. doi: 10.1186/1471-2164-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowden RR, Curtis SK. Cephalopods. In: Ratcliffe NA, Rowley AF, editors. Invertebrate Blood Cells, vol 1. Academic Press; New York: 1981. pp. 301–323. [Google Scholar]
- Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid–vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–1151. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- Dilly PN, Nixon M. The cells that secrete the beaks in octopods and squids (Mollusca, Cephalopoda) Cell Tissue Res. 1976;167:229–241. doi: 10.1007/BF00224330. [DOI] [PubMed] [Google Scholar]
- Ehrlich H, Maldonado M, Spindler KD, Eckert C, Hanke T, Born R, Goebel C, Simon P, Heinemann S, Worch H. First evidence of chitin as a component of the skeletal fibers of marine sponges. Part I. Verongidae (Demospongia: Porifera) J Exp Zool B Mol Dev Evol. 2007;308:347–356. doi: 10.1002/jez.b.21156. [DOI] [PubMed] [Google Scholar]
- Goedken M, De Guise S. Flow cytometry as a tool to quantify oyster defence mechanisms. Fish Shellfish Immunol. 2004;16:539–552. doi: 10.1016/j.fsi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Hardt M, Laine RA. Mutation of active site residues in the chitin-binding domain ChBDChiA1 from chitinase A1 of Bacillus circulans alters substrate specificity: use of a green fluorescent protein binding assay. Arch Biochem Biophys. 2004;426:286–297. doi: 10.1016/j.abb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Harrison FW, Ruppert EE. Microscopic Anatomy of Invertebrates. Wiley-Liss; New York: 1991. [Google Scholar]
- Hunt S, El Sherief A. A periodic structure in the ‘pen’ chitin of the squid Loligo vulgaris. Tissue Cell. 1990;22:191–197. doi: 10.1016/0040-8166(90)90021-z. [DOI] [PubMed] [Google Scholar]
- Keyhani NO, Roseman S. Physiological aspects of chitin catabolism in marine bacteria. Biochim Biophys Acta. 1999;1473:108–122. doi: 10.1016/s0304-4165(99)00172-5. [DOI] [PubMed] [Google Scholar]
- Kieseier BC, Wisniewski KE, Goebel HH. The monocyte macrophage system is affected in lysosomal storage diseases: an immunoelectron microscopic study. Acta Neuropathol. 1997;94:359–362. doi: 10.1007/s004010050719. [DOI] [PubMed] [Google Scholar]
- Kimbell JR, McFall-Ngai MJ. Symbiont-induced changes in host actin during the onset of a beneficial animal–bacterial association. Appl Environ Microbiol. 2004;70:1434–1441. doi: 10.1128/AEM.70.3.1434-1441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimbell JR, Koropatnick TA, McFall-Ngai MJ. Evidence for the participation of the proteasome in symbiont-induced tissue morphogenesis. Biol Bull. 2006;211:1–6. doi: 10.2307/4134572. [DOI] [PubMed] [Google Scholar]
- Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid–Vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- Kramerov AA, Mukha DV, Metakovsky EV, Gvozdev VA. Glycoproteins containing sulfated chitin-like carbohydrate moiety are synthesized in an established Drosophila melanogaster cell line. Insect Biochem. 1986;16:417–432. [Google Scholar]
- Kramerov AA, Rozovsky Ya M, Baikova NA, Gvozdev VA. Cognate chitinoproteins are detected during Drosophila melanogaster development and in cell cultures from different insect species. Insect Biochem. 1990;20:769–775. [Google Scholar]
- Kuchel RP, Raftos DA, Birch D, Vella N. Haemocyte morphology and function in the Akoya pearl oyster, Pinctada imbricata. J Invertebr Pathol. 2010;105:36–48. doi: 10.1016/j.jip.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Li X, Wang LX, Wang X, Roseman S. The chitin catabolic cascade in the marine bacterium Vibrio cholerae: characterization of a unique chitin oligosaccharide deacetylase. Glycobiology. 2007;17:1377–1387. doi: 10.1093/glycob/cwm096. [DOI] [PubMed] [Google Scholar]
- Liau CY, Lin CS. A modified coomassie brilliant blue G 250 staining method for the detection of chitinase activity and molecular weight after polyacrylamide gel electrophoresis. J Biosci Bioeng. 2008;106:111–113. doi: 10.1263/jbb.106.111. [DOI] [PubMed] [Google Scholar]
- Martin GG, Castro C, Moy N, Rubin N. N-acetyl-D-glucosamine in crustacean hemocytes; possible functions and usefulness in classification. Invertebr Biol. 2003;122:265–270. [Google Scholar]
- Mateo DR, Spurmanis A, Siah A, Araya MT, Kulka M, Berthe FC, Johnson GR, Greenwood SJ. Changes induced by two strains of Vibrio splendidus in haemocyte subpopulations of Mya arenaria, detected by flow cytometry with LysoTracker. Dis Aquat Organ. 2009;86:253–262. doi: 10.3354/dao02121. [DOI] [PubMed] [Google Scholar]
- Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci USA. 2004;101:2524–2529. doi: 10.1073/pnas.0308707101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meikle PJ, Fuller M, Hopwood JJ. Mass spectrometry in the study of lysosomal storage disorders. Cell Mol Biol. 2003;49:769–777. [PubMed] [Google Scholar]
- Montgomery MK, McFall-Ngai MJ. Embryonic development of the light organ of the sepiloid squid Euprymna scolopes Berry. Biol Bull. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- Moore SE. Oligosaccharide transport: pumping waste from the ER into lysosomes. Trends Cell Biol. 1999;9:441–446. doi: 10.1016/s0962-8924(99)01648-7. [DOI] [PubMed] [Google Scholar]
- Moueza M, Frenkiel L. Fine structure and histochemistry of the gastric shield in the lamellibranch Donax trunculus L. Z. Mikrosk. Anat Forsch. 1979;93:169–181. [PubMed] [Google Scholar]
- Nauseef WM. Insights into myeloperoxidase biosynthesis from its inherited deficiency. J Mol Med. 1998;76:661–668. doi: 10.1007/s001090050265. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid–vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11:483–493. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–167. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannasi A, Hermann HR. Chitin in the cephalochordata, Branchiostoma floridae. Experientia. 1970;26:351–352. doi: 10.1007/BF01896881. [DOI] [PubMed] [Google Scholar]
- Small AL, McFall-Ngai MJ. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem. 1999;72:445–457. [PubMed] [Google Scholar]
- Sminia T, Barendsen L. A comparative morphological and enzyme histochemical study on blood cells of the freshwater snails Lymnaea stagnalis, Biomphalaria glabrata, and Bulinus truncatus. J Morphol. 1980;165:31–39. doi: 10.1002/jmor.1051650104. [DOI] [PubMed] [Google Scholar]
- Sugita H, Ito Y. Identification of intestinal bacteria from Japanese flounder (Paralichthys olivaceus) and their ability to digest chitin. Lett Appl Microbiol. 2006;43:336–342. doi: 10.1111/j.1472-765X.2006.01943.x. [DOI] [PubMed] [Google Scholar]
- Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna–Vibrio mutualism. Proc Natl Acad Sci USA. 1996;93:13683–13688. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, Splinter-Bondurant S, Brown B, Manzella L, Snir E, Almabrazi H, Scheetz TE, Bonaldo M, de F, Casavant TL, Soares MB, Cronan JE, Reed JL, Ruby EG, McFall-Ngai MJ. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci USA. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]