Abstract
Ritonavir-boosted protease inhibitor regimens are widely used for HIV chemotherapy. However, ritonavir causes multiple side effects, and the mechanisms are not fully understood. The current study was designed to explore the metabolic pathways of ritonavir that may be related to its toxicity. Metabolomic analysis screened out 26 ritonavir metabolites in mice, and half of them are novel. These novel ritonavir metabolites include two glycine conjugated, two N-acetylcysteine conjugated and three ring-open products. Accompanied with the generation of ritonavir ring-open metabolites, the formation of methanethioamide and 2-methylpropanethioamide were expected. Based upon the structures of these novel metabolites, five bioactivation pathways are proposed, which may be associated with sulfation and epoxidation. By using Cyp3a-null mice, we confirmed that CYP3A is involved in four pathways of RTV bioactivation. In addition, all these five bioactivation pathways were recapitulated in the incubation of ritonavir in human liver microsomes. Further studies are suggested to determine the role of CYP3A and these bioactivation pathways in ritonavir toxicity.
INTRODUCTION
Ritonavir (RTV) was originally developed as an HIV protease inhibitor (PI) (1). It is now rarely used for its antiviral activity, but is used as a booster of other PIs (2). The mechanism of RTV as a booster relies upon RTV to inhibit CYP3A-mediated metabolism of other PIs, thus increasing their plasma concentrations (3). RTV-boosted PI regimens have drastically improved the efficacy of PI and highly active antiretroviral therapy. However, RTV-boosted PI regimens may cause toxicity. For example, RTV-boosted PI regimens have been considered as a risk factor of HIV drug-induced liver injury (4, 5). In addition, RTV-boosted PI regimens may cause kidney injury (6, 7). The exact mechanisms of the adverse effects of RTV-boosted PI regimens remain unknown.
Bioactivation of a drug may be associated with drug toxicity (8, 9). RTV metabolism has been studied extensively, and the metabolic pathways, including hydroxylation of the isopropyl side chain, N-demethylation, and cleavage of the terminal thiazole and isopropylthiazole group have been reported (10–14). However, the pathways of RTV bioactivation remain unclear. RTV is a CYP3A inhibitor and increased inhibitory potency of RTV after pre-incubation with microsomes was observed, suggesting that RTV is a mechanism-based inhibitor which converts into reactive intermediate(s) and inactivates CYP3A by irreversibly attaching to heme and/or active site(s) (12, 15, 16). These previous reports suggest the existence of RTV bioactivation.
The current study was designed to explore the pathways of RTV bioactivation. A metabolomic approach was used to re-profile RTV metabolism in mice. RTV-D6 (Supplemental Scheme 1) was used to illustrate the structures of novel RTV metabolites. Cyp3a-null mice were used to determine the contribution of CYP3A in RTV bioactivation. Overall, 13 novel RTV metabolites were identified and 5 bioactivation pathways are proposed. CYP3A is involved in 4 pathways of RTV bioactivation.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
RTV was supplied by the National Institutes of Health AIDS Research and Reference Reagent Program. RTV-D6 was obtained from Toronto Research Chemicals Inc. (North York, ON, Canada). Human liver microsomes (HLM) were purchased from XenoTech (Lenexa, KS, US). 2-Isopropylthiazole-4-carbaldehyde, thiazole-5-carbaldehyde, and 5-(hydroxymethyl)-thiazole were obtained from Synthonix, Inc. (Wake Forest, NC, US). Methoxylamine, semicarbazide and NADPH were purchased from Sigma-Aldrich (St. Louis, MO, US). All the solvents for liquid chromatography and mass spectrometry were of the highest grade commercially available.
Animals and Treatments
Wild-type (WT) and Cyp3a-null mice (17) were used in the current study. All mice (2–4 months old) were maintained under a standard 12-hour dark and 12-hour light cycle with water and chow provided ad libitum. Handling was in accordance with study protocols approved by the University of Kansas Medical Center Institutional Animal Care and Use Committee. WT and Cyp3a-null mice were treated with RTV (25 mg/kg, p.o.), and housed separately in metabolic cages for 18 hours. 2-Isopropylthiazole-4-carbaldehyde, thiazole-5-carbaldehyde, and 5-(hydroxymethyl)-thiazole are proposed as RTV metabolites. WT mice were also treated with these chemicals (10 mg/kg, i.p.) to verify the metabolic pathways of RTV. Urine and feces were collected for metabolite analysis. The methods for analysis of metabolites from urine and feces have been described in previous reports (18, 19). Briefly, urinary samples were prepared by mixing 40 µl of urine with 160 µl of 50% acetonitrile and were centrifuged at 18,000 relative centrifugal forces (rcf) for 10 min. Feces were homogenized in water (10 mg feces in 100 µl of H2O). Subsequently, 200 µl of acetonitrile was added to 200 µl of the resulting mixture, followed by centrifugation at 18,000 rcf for 10 min. The supernatant was transferred to a new Eppendoff vial for a second centrifugation (18,000 rcf for 10 min). Each supernatant was transferred to an auto sampler vial and 5.0 µl was injected into an analytical system combining ultra performance liquid chromatography (UPLC) and time of flight mass spectrometry TOFMS) for metabolite analysis.
RTV or RTV-D6 Metabolism in HLM
Incubations were conducted in 1X phosphate-buffered saline (pH 7.4), containing 10 µM RTV or 10 µM RTV-D6, 0.1 mg HLM in a final volume of 190 µl. After 5 min of pre-incubation at 37 °C, the reaction was initiated by the addition of 10 µl of 20 mM NADPH (final concentration 1.0 mM) and continued for 30 min with gentle shaking. Methoxylamine or semicarbazide (final concentration 2.5 mM) was used to trap aldehydes. Incubations were terminated by adding 200 µl of acetonitrile and vortexing for 30 seconds and centrifuging at 18,000 rcf for 10 min. Each supernatant was transferred to an auto sampler vial, and 5.0 µl was injected onto the UPLC-TOFMS system for metabolite analysis. All the incubations were performed in duplicate.
UPLC-TOFMS Analysis
RTV and its metabolites were separated using a 100 mm × 2.1 mm (Acquity 1.7 µm) UPLC BEH C-18 column (Waters, Milford, MA). The flow rate of the mobile phase was 0.3 ml/min with a gradient ranging from 2% to 98% aqueous acetonitrile containing 0.1% formic acid in a 10-min run. SYNAPT G1 TOFMS (Waters, Milford, MA) was operated in a positive mode with electrospray ionization. The source temperature and desolvation temperature were set at 120 °C and 350 °C, respectively. Nitrogen was applied as the cone gas (10 L/hour), and desolvation gas (700 L/hour). Argon was applied as the collision gas. TOFMS was calibrated with sodium formate and monitored by the intermittent injection of lock mass leucine enkephalin in real time. The capillary voltage and the cone voltage were set at 3.5 kV and 35 V in positive ion mode. Screening of metabolites was performed by using MarkerLynx software (Waters, Milford, MA) based on accurate mass measurement (mass errors less than 10 ppm). The structures of RTV and its metabolites were elucidated by tandem mass spectrometry fragmentation with collision energy ramp ranging from 10 to 40 eV.
Data Analysis
Mass chromatograms and mass spectra were acquired by MassLynx software in centroid format from m/z 50 to 1000. Centroid and integrated mass chromatographic data were processed by MarkerLynx software to generate a multivariate data matrix. The corresponding data matrices were then exported into SIMCA-P+12 (Umetrics, Kinnelon, NJ) for multivariate data analysis. Orthogonal projection to latent structures-discriminant analysis (OPLS-DA) was conducted on Pareto-scaled data. For the quantification of RTV metabolites, peak areas of RTV and its metabolites were added up and set as 100% for each sample. The percentage of each metabolite in WT was compared with that of the corresponding one in Cyp3a-null mice.
RESULTS AND DISCUSSION
Profiling RTV metabolism in mice
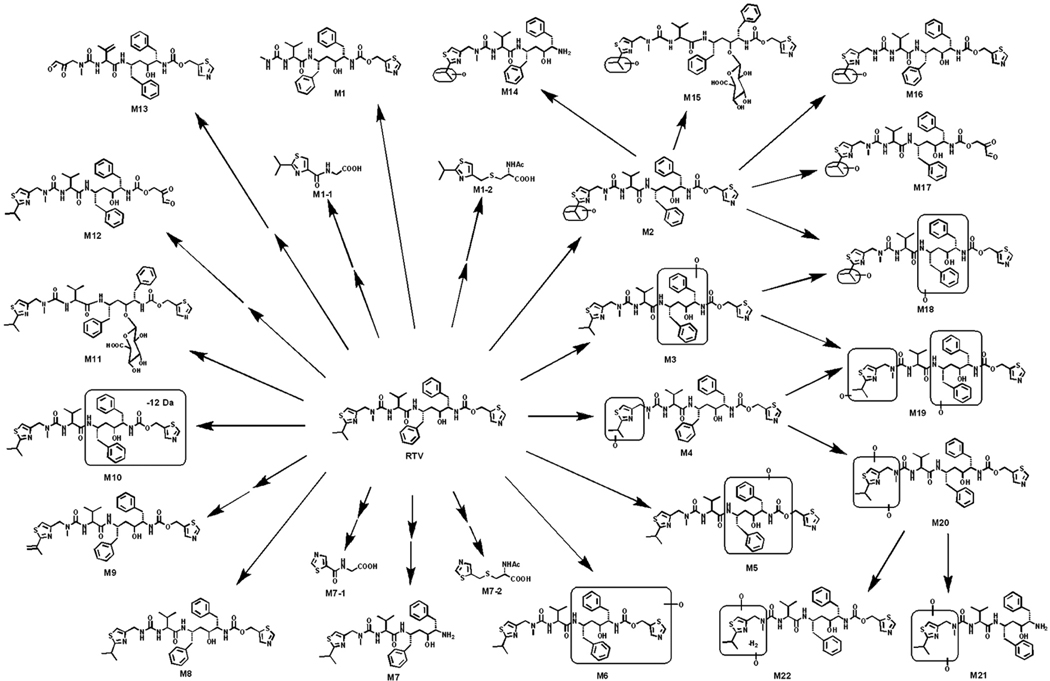
RTV metabolites were found in mouse feces and urine, but mainly in the feces. The results of chemometric analysis on the ions produced by UPLC-TOFMS assay of control and RTV-treated mouse feces are shown in Supplemental Figure 1. The OPLS-DA analysis revealed two clusters corresponding to the control and RTV-treated groups (Supplemental Figure 1A). The corresponding Splots (Supplemental Figure 1B) display the ion contribution to the group separation in the feces. The top ranking ions were identified as RTV and its metabolites, which are marked in S-plots (Supplemental Figure 1B). The urine samples were analyzed also using chemometric methods, which unraveled the small molecular metabolites (M1-1, M1-2, M7-1, and M7-2). Overall, 26 RTV metabolites were identified (Figure 1 and Supplemental Table 1), including 13 previously reported metabolites (M1, M2, M4-M11, M14, M16, and M20) (10–14) and 13 novel metabolites (M1-1, M1-2, M3, M7-1, M7-2, M12, M13, M15, M17-M19, M21, and M22). These data reflect the power of metabolomic strategies in studying drug metabolism (20). Among these novel RTV metabolites, two glycine conjugated (M1-1 and M7-1), two N-acetylcysteine conjugated (M1-2 and M7-2), and three ring-open (M12, M13, and M17) metabolites were of interest because they may be associated with RTV bioactivation.
Figure 1. The metabolic map of RTV in mice.
All metabolites were determined by accurate mass and mass fragmentations. Twenty-six RTV metabolites were identified, including 13 previously reported metabolites (M1, M2, M4-M11, M14, M16, and M20) and 13 novel metabolites (M1-1, M1-2, M3, M7-1, M7-2, M12, M13, M15, M17-M19, M21, and M22). M1-1, M1-2, M7-1, and M7-2 were detected in the urine. The rest of the metabolites were mainly found in the feces.
Mechanism of M1-1 and M1-2 formation
2-(2-Isopropylthiazole-4-carboxamido)acetic acid (M1-1) and (R)-2-acetamido-3-((2-isopropylthiazol-4-yl)methylthio)propanoic acid (M1-2) were identified in the urine of mice treated with RTV. Metabolite M1-1 had a protonated molecule [M+H]+ at m/z = 229 Da. MS/MS of M1-1 produced major ions at m/z 182, 154, 127, and 78 (Supplemental Figure 2A). The ion at m/z 211 was formed by loss of H2O. Metabolite M1-2 had a protonated molecule [M+H]+ at m/z = 303 Da. MS/MS of M1-2 produced major ions at m/z 261, 174, 140, and 70 (Supplemental Figure 2B).
M1-1 is a glycine conjugated metabolite and M1-2 is an N-acetylcysteine conjugated product. We hypothesized that both M1-1 and M1-2 are further metabolites of 2-isopropylthiazole-4-carbaldehyde that is generated from RTV dealkylation. A dealkylated RTV metabolite (M1, Figure 1) has been reported in previous reports and it is a major metabolic pathway of RTV in humans (10, 11). Theoretically, an aldehyde should be accompanied with the dealkylated RTV metabolite M1. We confirmed the existence of 2-isopropylthiazole-4-carbaldehyde in the incubation of RTV in HLM using methoxylamine as a trapping agent. The formed oxime had a mass of [M+H]+ = 185 m/z, with major MS/MS fragments at 153 (loss of CH3OH) and 127 (loss of C2H4NO) (Supplemental Figure 2C). In addition, in the urine of mice treated with 2-isopropylthiazole-4-carbaldehyde, both M1-1 and M1-2 were detected.
The mechanism of M1-1 and M1-2 formation was proposed as follows. 2-Isopropylthiazole-4-carbaldehyde is reduced to an alcohol that can be further sulfated. Sulfate anion is a good leaving group. Decomposition of sulfates forms reactive cationic intermediates that can bind to nucleophiles (21) including protein, DNA, and GSH. The GSH adduct is further degraded to M1-2 that can be detected in the urine. For M1-1, 2-isopropylthiazole-4-carbaldehyde is further oxidized to an acid, which then conjugates with glycine to form M1-1 (Supplemental Scheme 2). The chromatographic and MS/MS data of the acid and alcohol intermediates of M1-1 and M1-2 are presented in Supplemental Figure 3.
Mechanism of M7-1 and M7-2 formation
2-(Thiazole-5-carboxamido)acetic acid (M7-1) and 2-acetamido-3-(thiazol-5-ylmethylthio)propanoic acid (M7-2) were detected in the urine of mice treated with RTV. M7-1, eluted at 2.56 min (Supplemental Figure 4A), had a protonated molecule [M+H]+ at m/z = 187 Da. The major MS/MS fragments of M7-1 are interpreted in Supplemental Figure 4B. Metabolite M7-2, eluted at 3.30 min (Supplemental Figure 4A), had a protonated molecule [M+H]+ at m/z = 261 Da. MS/MS of M7-2 produced major ions at m/z 219, 184, 131, and 98 (Supplemental Figure 4C).
M7-1 is a glycine conjugated metabolite and M7-2 is an N-acetylcysteine conjugated product. The generation of M7-1 and M7-2 is associated with the metabolic pathway of M7 (Figure 1). Previous literature proposed that M7 was produced by amide hydrolysis, concurrently releasing CO2 and thiazole-5-carbaldehyde (10). However, this aldehyde was not detected in the HLM incubation system containing methoxylamine or semicarbazide as trapping agents, suggesting that the product accompanying M7 may not be thiazole-5-carbaldehyde. We hypothesized that the accompanying product is 5-hydroxymethyl)-thiazole. In order to test this hypothesis, WT mice were treated with thiazole-5-carbaldehyde and 5-(hydroxymethyl)-thiazole, respectively. In the urine of mice treated with thiazole-5-carbaldehyde, only glycine conjugated adduct (M7-1) was observed. However, in the urine of 5-(hydroxymethyl)-thiazole treated mice, both glycine conjugated (M7-1) and N-acetylcysteine conjugated (M7-2) adducts were detected. These data indicate that the released thiazole unit is more likely to be an alcohol. The mechanism of M7-1 and M7-2 formation may be similar to that of M1-1 and M1-2 (Supplemental Scheme 2).
Mechanism of M12, M13 and M17 formation
Three ring-open metabolites (M12, M13, and M17) were identified in RTV metabolism for the first time (Figure 1).
2,3-Dioxopropyl (2S,3S,5S)-3-hydroxy-5-((S)-2-(3-((2-isopropylthiazol-4-yl)methyl)-3-methylureido)-3-methylbutanamido)-1,6-diphenylhexan-2-ylcarbamate (M12)
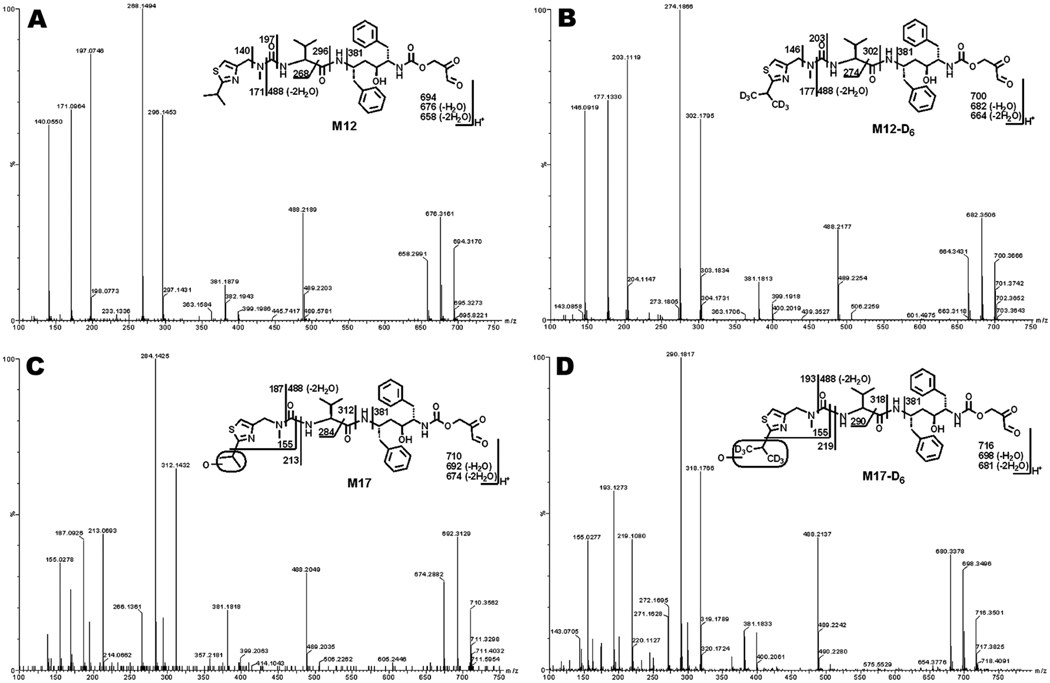
Metabolite M12 had a protonated molecule [M+H]+ at m/z = 694 Da. MS/MS of M12 produced major ions at m/z 676 (loss of H2O), 488, 381, 296, 268, 197, 171, and 140 Figure 2A). In the incubation of RTV-D6 in HLM, M12-D6 was detected and displayed a similar fragmental pattern to that of M12 (Figure 2B). The ions at m/z 302, 274, 203, 177, and 146 were 6 mass units higher than those fragments generated from M12. These ions indicate that the left hand of the structure is intact. The ion at m/z 381 suggests the formation of a thiazole ring-open metabolite (Figures 2A and 2B).
Figure 2. Identification of thiazole ring-open metabolites M12 and M17.
Metabolites M12 and M17 were recapitulated in the incubations of RTV or RTV-D6 in HLM. Structural elucidations were based on accurate mass measurement and MS/MS fragmentations. (A) MS/MS of M12. (B) MS/MS of M12-D6. (C) MS/MS of M17. (D) MS/MS of M17-D6.
Mono-hydroxylated thiazole ring-open metabolite (M17)
M17 had a protonated molecule [M+H]+ at m/z = 710 Da, 16 Da higher than that of M12. MS/MS of M17 produced major ions at m/z 692 (loss of H2O), 488, 381, 296, 268, 197, 171, and 140 Figure 2C). Compared with the MS/MS of M12, the ions at m/z 187 and 155 indicate that the hydroxylation occurred in the isopropyl unit (Figure 2C). In the incubation of RTV-D6 in HLM, M17-D6 displayed a similar fragment pattern to that of M17, except for mass units differences (Figure 2D). The ion at m/z 155 in M17-D6 further confirmed that hydroxylation took place in the isopropyl unit (Figure 2D). The ion at m/z 381 was also observed in the MS/MS fragments of M17 and M17-D6 (Figures 2C and 2D), suggesting that M17 and M17-D6 are also thiazole ring-open metabolites.
Based on the finding of thiazole ring-open metabolites (Figure 2), epoxidations at the thiazole ring of RTV are proposed (Supplemental Scheme 3). Epoxides are electrophilic species that react with nucleophiles, including water and GSH (22, 23). The epoxides of RTV are subject to hydrolysis to give the unstable diol. The resulting diol is then decomposed to form the corresponding α-keto aldehydes (M12 and M17). The generation of ring-open metabolites M12 and M17 is expected to be accompanied with the formation of methanethioamides (Supplemental Scheme 3).
Thiazol-5-ylmethyl (2S,3S,5S)-5-((S)-2-(3-(2,3-dioxopropyl)-3-methylureido)-3-methylbut-3-enamido)-3-hydroxy-1,6-diphenylhexan-2-ylcarbamate (M13)
M13 was eluted at the retention time of 5.64 min (Supplemental Figure 5A), having a protonated molecule [M+H]+ at m/z 650. MS/MS of M13 produced major ions at m/z 606, 535, 250, 197, and 143. The ion at m/z 143 suggests the formation of an isopropylthiazole ring-open metabolite (Supplemental Figure 5B). The ions at m/z 197 and 143 indicate that dehydrogenation happened in the isopropyl moiety in the middle. In the incubation of RTV-D6 in HLM, the metabolite M13-D6 was detected (Supplemental Figure 5A), which had the exact same fragments as M13, indicating that the isopropylthiazole-D6 moiety lost 2-methylpropanethioamide-D6. A similar mechanism of M13 formation is proposed as that of M12 and M17 (Supplemental Scheme 2) with the production of an epoxide. The generation of ring-open metabolite M13 is expected to be accompanied with the formation of 2-methylpropanethioamide.
Ring-open of thiazole derivatives has been proposed as an important bioactivation pathway, which produces thioamide analogs that can be further oxidized and cause liver and kidney injury (24–27). We predict the formation of methanethioamide and 2-methylpropanethioamide to be accompanied with the generation of ring-open metabolites M12, M13 and M17. These results may offer new avenues for investigating RTV-induced liver and kidney injury (5–7).
Role of CYP3A in RTV bioactivation pathways
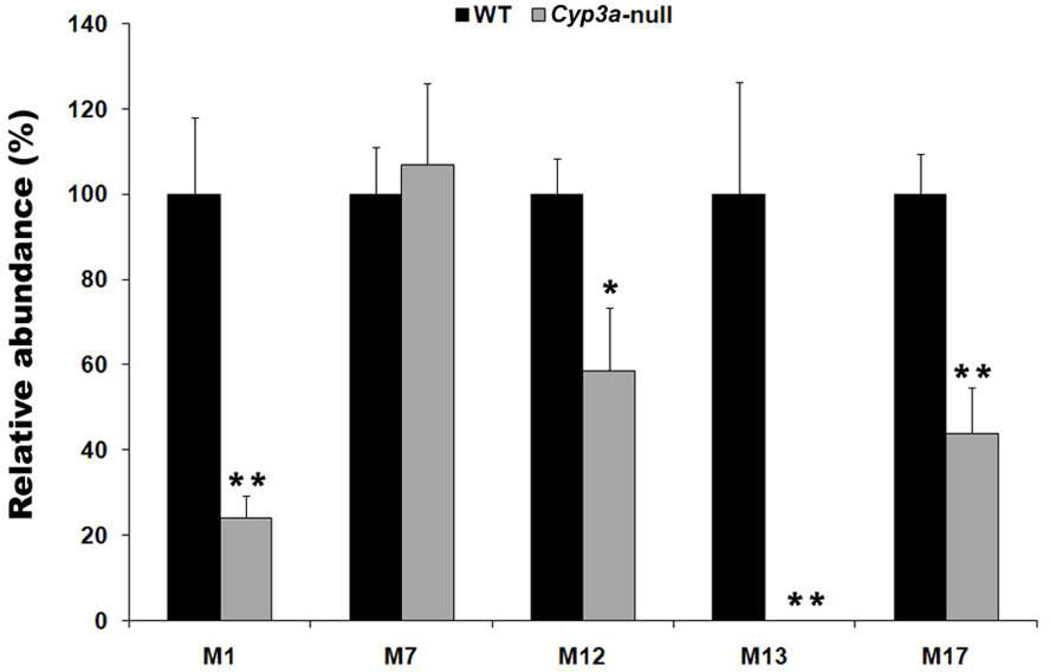
Because RTV is a potent CYP3A inhibitor (3), the contribution of CYP3A in RTV metabolism may not be fully reflected by in vitro experiments. Cyp3a-null mice were used in the current study to determine the contribution of CYP3A in the formation of M1-2, M7-2, M12, M13 and M17. The formation of M1-2 and M7-2 is dependent on the formation of M1 and M7, respectively. Therefore, M1 and M7 were used to evaluate the contribution of CYP3A in the formation of M1-2 and M7-2. The relative abundances of M1, M12, M13, and M17 in the feces of Cyp3a-null mice were 24%, 59%, 1%, and 44%, respectively, of WT mice (Figure 3). There is no significant difference in the production of M7 between the two genotypes. These data suggest that CYP3A played an important role in RTV bioactivation, especially for the pathways of M1-2 and M13. These data also suggest that although RTV is a potent CYP3A inhibitor, the bioactivation of RTV is still dependent on CYP3A.
Figure 3. The role of CYP3A in RTV bioactivation.
WT mice and Cyp3a-null mice were treated with 25 mg/kg RTV (p.o.). The feces were collected 18 hours after RTV treatment. All samples were analyzed by UPLC-TOFMS. The relative abundance of M1, M7, M12, M13, and M17 was set as 100% respectively in WT mice. *P<0.05; **P<0.01 vs WT mice, n = 4.
CYP3A is one of the major CYP450 isoenzymes in mammals and it is highly expressed in the liver and small intestine (28, 29). Due to the contribution of CYP3A in RTV bioactivation, the expression pattern of CYP3A may be associated with the gastrointestinal adverse effects of RTV, such as diarrhea, nausea, vomiting, and abdominal pain (30, 31). CYP3A expression can be up-regulated by multiple nuclear receptors, including pregnane X receptor, constitutive androstane receptor, vitamin D receptor and glucocorticoid receptor (32, 33). It is expected that pre- or co-treatment with a CYP3A inducer will increase RTV bioactivation and potentiate RTV toxicity. In a recent series of clinical trials, hepatotoxicity of RTV-boosted PI regimens was observed in all subjects who were pre-treated with rifampicin or efavirenz (34–37). Both rifampicin and efavirenz are activators of pregnane X receptor (38). It is possible that pre-treatment of rifampicin or efavirenz up-regulates CYP3A expression, which subsequently accelerates RTV bioactivation and potentiates RTV hepatotoxicity.
CONCLUSIONS
Thirteen known and thirteen novel RTV metabolites were identified in mice using metabolomic strategies. Five bioactivation pathways are proposed and all of them were recapitulated in the incubation of RTV in HLM. CYP3A is involved in four pathways of RTV bioactivation. Further studies are suggested to determine the role of CYP3A and RTV bioactivation in RTV toxicity.
Supplementary Material
Acknowledgements
We thank the National Institutes of Health AIDS Research and Reference Reagent Program for providing ritonavir. We thank Dr. Martha Montello for editing the manuscript.
Funding support. This work was supported by the National Institutes of Health National Center for Research Resources [COBRE 5P20-RR021940].
ABBREVIATIONS
- RTV
ritonavir
- PI
protease inhibitor
- WT
wild-type
- GSH
glutathione
- HLM
human liver microsomes
- OPLS-DA
orthogonal projection to latent structures-discriminant analysis
- TOFMS
time-of-flight mass spectrometry
- UPLC
ultraperformance liquid chromatography
Footnotes
Supporting information available. Supplemental Table 1, Figures 1–5, and Schemes 1–3. This information is available free of charge via the internet at http://pubs.acs.org/.
REFERENCES
- 1.Markowitz M, Mo H, Kempf DJ, Norbeck DW, Bhat TN, Erickson JW, Ho DD. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J Virol. 1995;69:701–706. doi: 10.1128/jvi.69.2.701-706.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierman WF, van Agtmael MA, Nijhuis M, Danner SA, Boucher CA. HIV monotherapy with ritonavir-boosted protease inhibitors: a systematic review. AIDS. 2009;23:279–291. doi: 10.1097/QAD.0b013e32831c54e5. [DOI] [PubMed] [Google Scholar]
- 3.Merry C, Barry MG, Mulcahy F, Ryan M, Heavey J, Tjia JF, Gibbons SE, Breckenridge AM, Back DJ. Saquinavir pharmacokinetics alone and in combination with ritonavir in HIV-infected patients. AIDS. 1997;11:F29–F33. doi: 10.1097/00002030-199704000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Bruno R, Sacchi P, Maiocchi L, Patruno S, Filice G. Hepatotoxicity and antiretroviral therapy with protease inhibitors: A review. Dig Liver Dis. 2006;38:363–373. doi: 10.1016/j.dld.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 5.Sulkowski MS. Hepatotoxicity associated with antiretroviral therapy containing HIV-1 protease inhibitors. Semin Liver Dis. 2003;23:183–194. doi: 10.1055/s-2003-39949. [DOI] [PubMed] [Google Scholar]
- 6.Duong M, Sgro C, Grappin M, Biron F, Boibieux A. Renal failure after treatment with ritonavir. Lancet. 1996;348:693. doi: 10.1016/S0140-6736(05)65125-2. [DOI] [PubMed] [Google Scholar]
- 7.Shafi T, Choi MJ, Racusen LC, Spacek LA, Berry C, Atta M, Fine DM. Ritonavir-induced acute kidney injury: kidney biopsy findings and review of literature. Clin Nephrol. 2011;75 Suppl 1:60–64. doi: 10.5414/cn106469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guengerich FP, MacDonald JS. Applying mechanisms of chemical toxicity to predict drug safety. Chem Res Toxicol. 2007;20:344–369. doi: 10.1021/tx600260a. [DOI] [PubMed] [Google Scholar]
- 9.Zhou S, Chan E, Duan W, Huang M, Chen YZ. Drug bioactivation, covalent binding to target proteins and toxicity relevance. Drug Metab Rev. 2005;37:41–213. doi: 10.1081/dmr-200028812. [DOI] [PubMed] [Google Scholar]
- 10.Kumar GN, Rodrigues AD, Buko AM, Denissen JF. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J Pharmacol Exp Ther. 1996;277:423–431. [PubMed] [Google Scholar]
- 11.Denissen JF, Grabowski BA, Johnson MK, Buko AM, Kempf DJ, Thomas SB, Surber BW. Metabolism and disposition of the HIV-1 protease inhibitor ritonavir (ABT-538) in rats, dogs, and humans. Drug Metab Dispos. 1997;25:489–501. [PubMed] [Google Scholar]
- 12.Koudriakova T, Iatsimirskaia E, Utkin I, Gangl E, Vouros P, Storozhuk E, Orza D, Marinina J, Gerber N. Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug Metab Dispos. 1998;26:552–561. [PubMed] [Google Scholar]
- 13.Gangl E, Utkin I, Gerber N, Vouros P. Structural elucidation of metabolites of ritonavir and indinavir by liquid chromatography-mass spectrometry. J Chromatogr A. 2002;974:91–101. doi: 10.1016/s0021-9673(02)01243-8. [DOI] [PubMed] [Google Scholar]
- 14.Yao M, Ma L, Humphreys WG, Zhu M. Rapid screening and characterization of drug metabolites using a multiple ion monitoring-dependent MS/MS acquisition method on a hybrid triple quadrupole-linear ion trap mass spectrometer. J Mass Spectrom. 2008;43:1364–1375. doi: 10.1002/jms.1412. [DOI] [PubMed] [Google Scholar]
- 15.von Moltke LL, Durol AL, Duan SX, Greenblatt DJ. Potent mechanism-based inhibition of human CYP3A in vitro by amprenavir and ritonavir: comparison with ketoconazole. Eur J Clin Pharmacol. 2000;56:259–261. doi: 10.1007/s002280000125. [DOI] [PubMed] [Google Scholar]
- 16.Kalgutkar AS, Obach RS, Maurer TS. Mechanism-based inactivation of cytochrome P450 enzymes: chemical mechanisms, structure-activity relationships and relationship to clinical drug-drug interactions and idiosyncratic adverse drug reactions. Curr Drug Metab. 2007;8:407–447. doi: 10.2174/138920007780866807. [DOI] [PubMed] [Google Scholar]
- 17.van Herwaarden AE, Wagenaar E, van der Kruijssen CM, van Waterschoot RA, Smit JW, Song JY, van der Valk MA, van Tellingen O, van der Hoorn JW, Rosing H, Beijnen JH, Schinkel AH. Knockout of cytochrome P450 3A yields new mouse models for understanding xenobiotic metabolism. J Clin Invest. 2007;117:3583–3592. doi: 10.1172/JCI33435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Lu J, Wang L, Ma X. CYP3A-mediated generation of aldehyde and hydrazine in atazanavir metabolism. Drug Metab Dispos. 2011;39:394–401. doi: 10.1124/dmd.110.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Wang L, Guo GL, Ma X. Metabolism-mediated drug interactions associated with ritonavir-boosted tipranavir in mice. Drug Metab Dispos. 2010;38:871–878. doi: 10.1124/dmd.109.030817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39:581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glatt H. Sulfotransferases in the bioactivation of xenobiotics. Chem Biol Interact. 2000;129:141–170. doi: 10.1016/s0009-2797(00)00202-7. [DOI] [PubMed] [Google Scholar]
- 22.Williams DP, Naisbitt DJ. Toxicophores: groups and metabolic routes associated with increased safety risk. Curr Opin Drug Discov Devel. 2002;5:104–115. [PubMed] [Google Scholar]
- 23.Stepan AF, Walker DP, Bauman J, Price DA, Baillie TA, Kalgutkar AS, Aleo MD. Structural Alert/Reactive Metabolite Concept as Applied in Medicinal Chemistry to Mitigate the Risk of Idiosyncratic Drug Toxicity: A Perspective Based on the Critical Examination of Trends in the Top 200 Drugs Marketed in the United States. Chem Res Toxicol. 2011 doi: 10.1021/tx200168d. [DOI] [PubMed] [Google Scholar]
- 24.Mizutani T, Yoshida K, Kawazoe S. Possible role of thioformamide as a proximate toxicant in the nephrotoxicity of thiabendazole and related thiazoles in glutathione-depleted mice: structure-toxicity and metabolic studies. Chem Res Toxicol. 1993;6:174–179. doi: 10.1021/tx00032a006. [DOI] [PubMed] [Google Scholar]
- 25.Ji T, Ikehata K, Koen YM, Esch SW, Williams TD, Hanzlik RP. Covalent modification of microsomal lipids by thiobenzamide metabolites in vivo. Chem Res Toxicol. 2007;20:701–708. doi: 10.1021/tx600362h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizutani T, Suzuki K. Relative hepatotoxicity of 2-(substituted phenyl)thiazoles and substituted thiobenzamides in mice: evidence for the involvement of thiobenzamides as ring cleavage metabolites in the hepatotoxicity of 2-phenylthiazoles. Toxicol Lett. 1996;85:101–105. doi: 10.1016/0378-4274(96)03646-6. [DOI] [PubMed] [Google Scholar]
- 27.Mizutani T, Yoshida K, Ito K. Nephrotoxicity of thiazoles structurally related to thiabendazole in mice depleted of glutathione by treatment with buthionine sulfoximine. Res Commun Chem Pathol Pharmacol. 1992;75:29–38. [PubMed] [Google Scholar]
- 28.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu Rev Pharmacol Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 29.de Wildt SN, Kearns GL, Leeder JS, van den Anker JN. Cytochrome P450 3A: ontogeny and drug disposition. Clin Pharmacokinet. 1999;37:485–505. doi: 10.2165/00003088-199937060-00004. [DOI] [PubMed] [Google Scholar]
- 30.Struble KA, Pratt RD, Gitterman SR. Toxicity of antiretroviral agents. Am J Med. 1997;102:65–67. doi: 10.1016/s0002-9343(97)00065-x. discussion 68–69. [DOI] [PubMed] [Google Scholar]
- 31.Baker R. Highlights from the FDA antiviral drug advisory committee meetings, February 27-March 1. BETA. 1996:7–13. [PubMed] [Google Scholar]
- 32.Tompkins LM, Wallace AD. Mechanisms of cytochrome P450 induction. J Biochem Mol Toxicol. 2007;21:176–181. doi: 10.1002/jbt.20180. [DOI] [PubMed] [Google Scholar]
- 33.Urquhart BL, Tirona RG, Kim RB. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol. 2007;47:566–578. doi: 10.1177/0091270007299930. [DOI] [PubMed] [Google Scholar]
- 34.Nijland HM, L'Homme RF, Rongen GA, van Uden P, van Crevel R, Boeree MJ, Aarnoutse RE, Koopmans PP, Burger DM. High incidence of adverse events in healthy volunteers receiving rifampicin and adjusted doses of lopinavir/ritonavir tablets. AIDS. 2008;22:931–935. doi: 10.1097/QAD.0b013e3282faa71e. [DOI] [PubMed] [Google Scholar]
- 35.Haas DW, Koletar SL, Laughlin L, Kendall MA, Suckow C, Gerber JG, Zolopa AR, Bertz R, Child MJ, Hosey L, Alston-Smith B, Acosta EP. Hepatotoxicity and gastrointestinal intolerance when healthy volunteers taking rifampin add twice-daily atazanavir and ritonavir. J Acquir Immune Defic Syndr. 2009;50:290–293. doi: 10.1097/QAI.0b013e318189a7df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jamois C, Riek M, Schmitt C. Potential Hepatotoxicity of Efavirenz and Saquinavir/Ritonavir Coadministration in Healthy Volunteers. Arch Drug Inf. 2009;2:1–7. doi: 10.1111/j.1753-5174.2009.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt C, Riek M, Winters K, Schutz M, Grange S. Unexpected Hepatotoxicity of Rifampin and Saquinavir/Ritonavir in Healthy Male Volunteers. Arch Drug Inf. 2009;2:8–16. doi: 10.1111/j.1753-5174.2009.00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hariparsad N, Nallani SC, Sane RS, Buckley DJ, Buckley AR, Desai PB. Induction of CYP3A4 by efavirenz in primary human hepatocytes: comparison with rifampin and phenobarbital. J Clin Pharmacol. 2004;44:1273–1281. doi: 10.1177/0091270004269142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.