Abstract
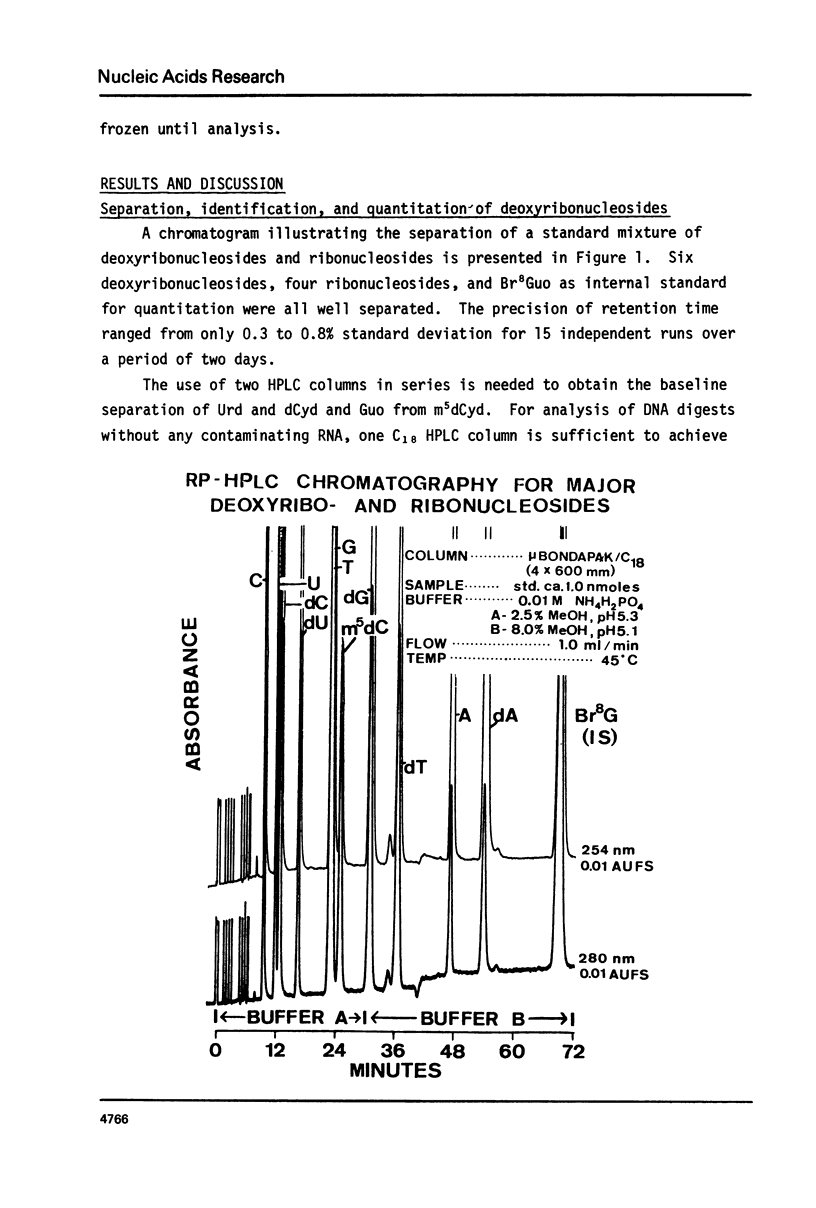
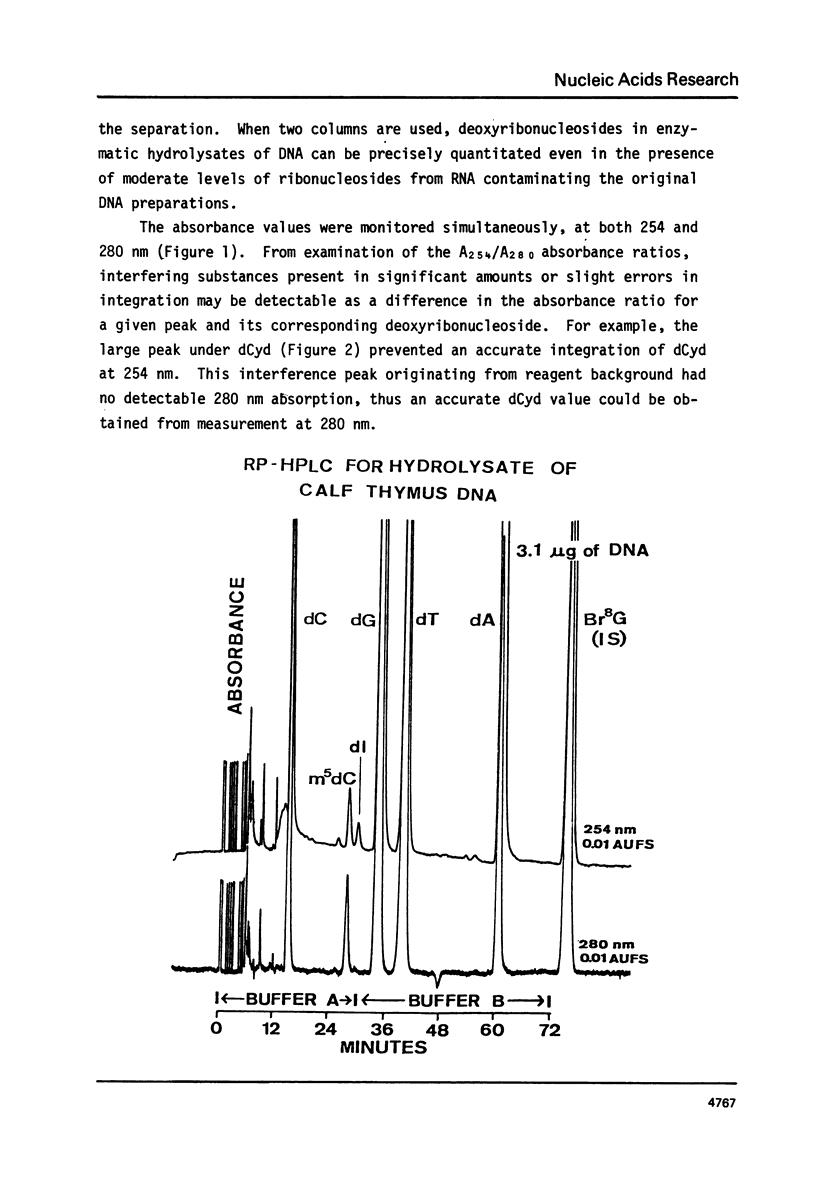
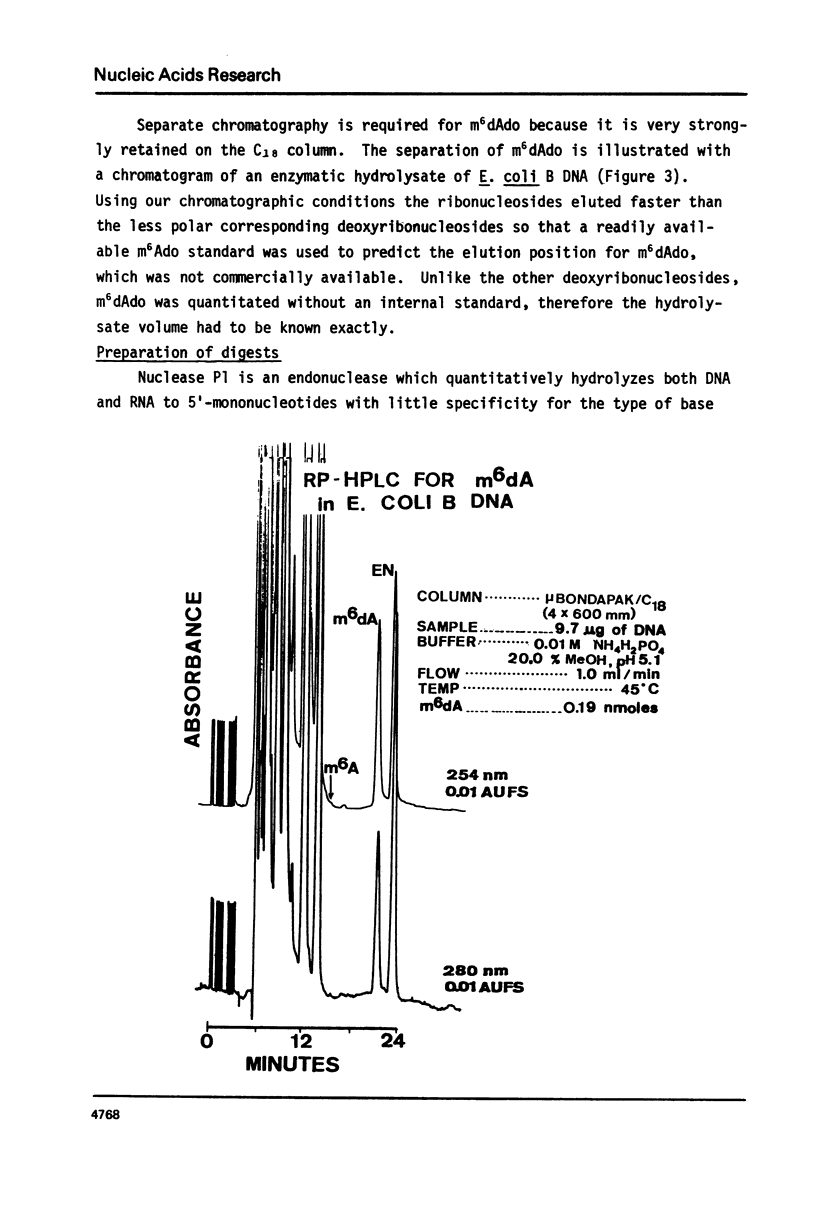
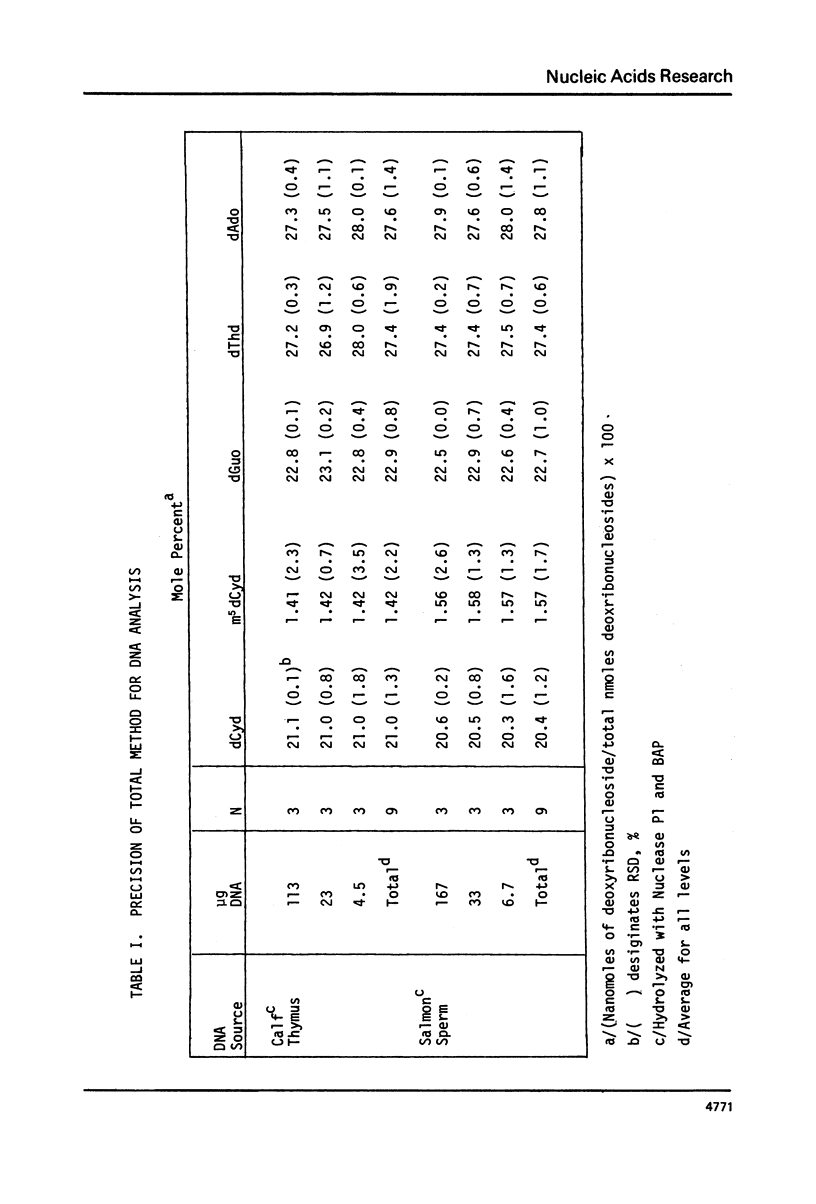
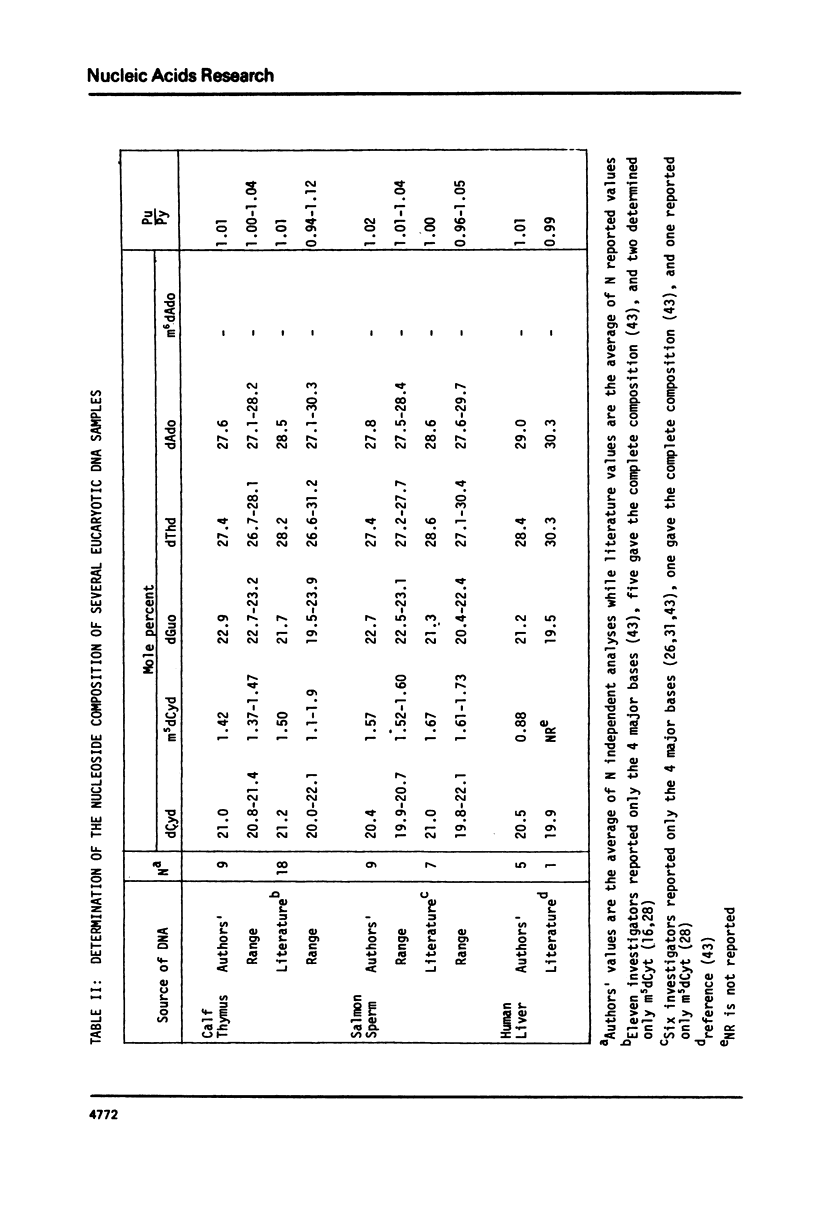
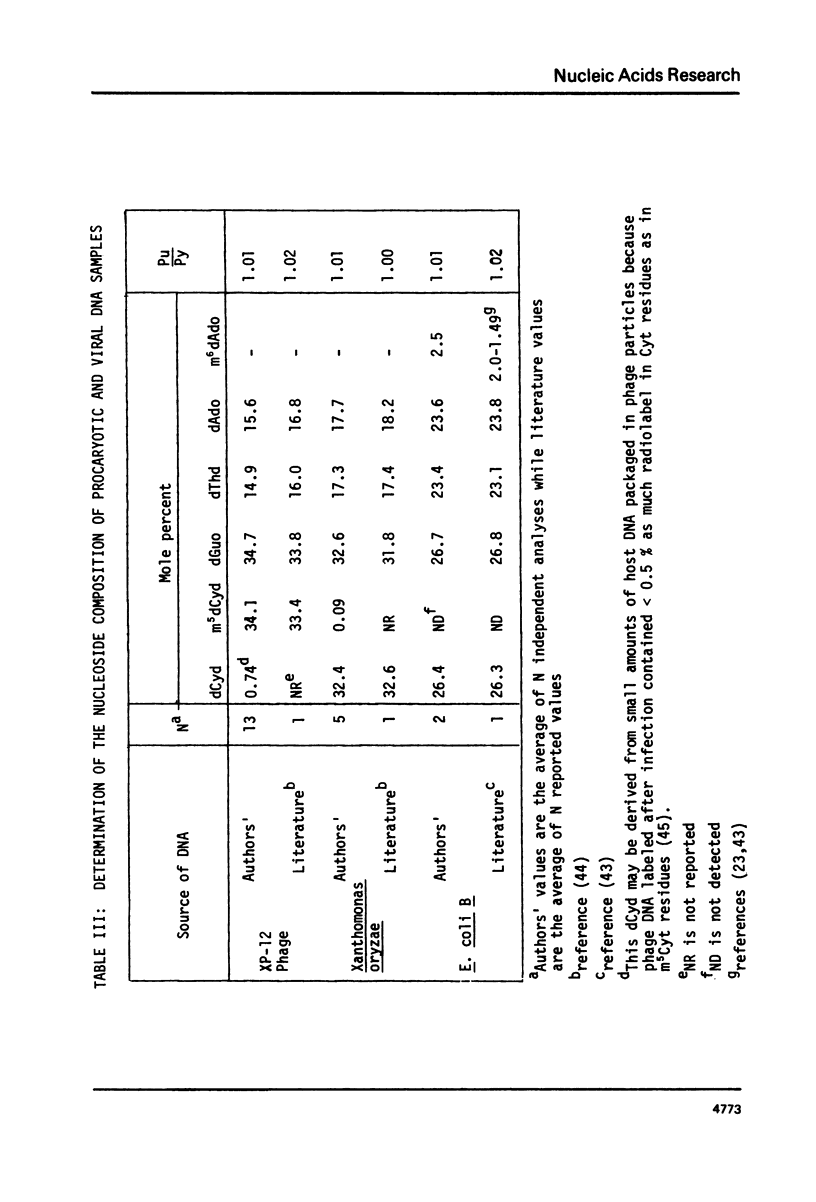
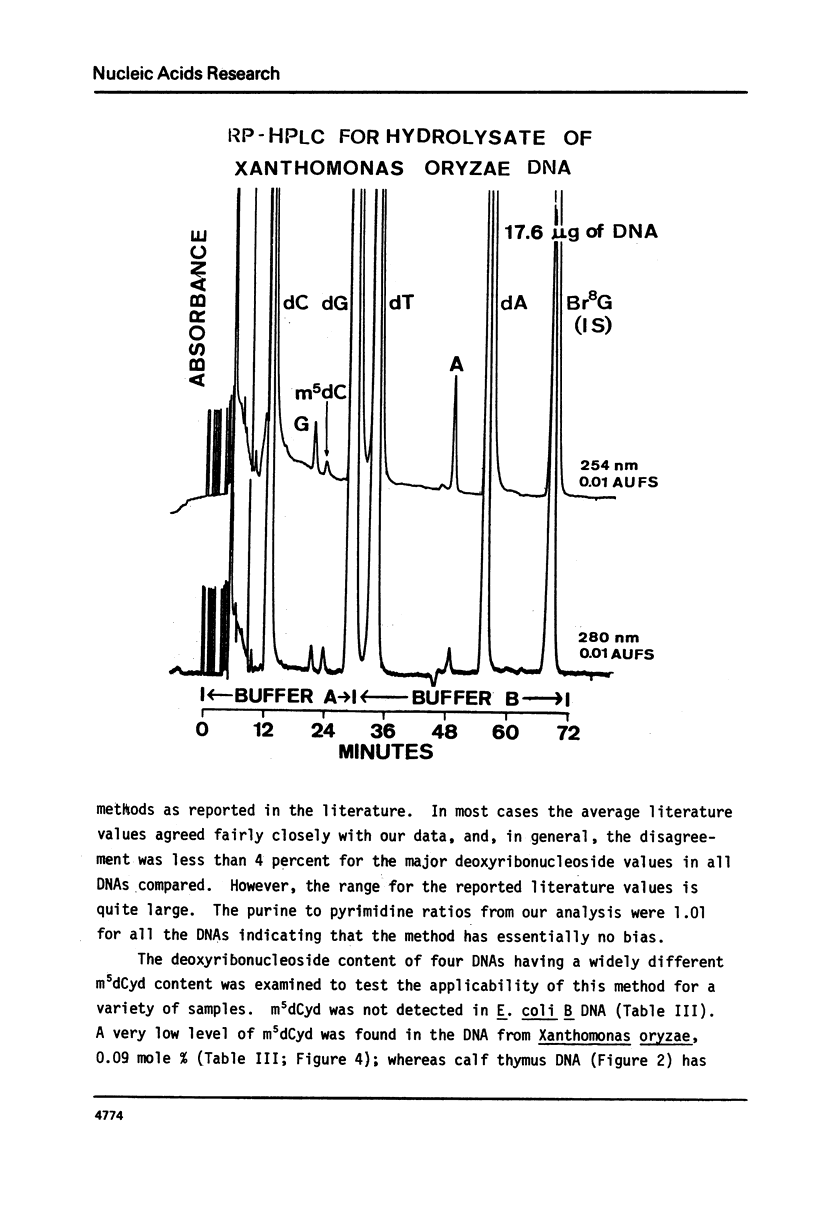
We have developed a method to accurately determine (< 3% RSD) the complete major and modified base composition of a few micrograms of unlabeled DNA. The DNA samples were quantitatively hydrolyzed with DNase 1, Nuclease P1, and bacterial alkaline phosphatase. The resulting deoxyribonucleosides were directly separated in 70 min by reversed-phase high performance liquid chromatography with detection by ultraviolet absorption at 254 nm and 280 nm (RP-HPLC). The highly sensitive and selective dual wavelength quantitation greatly enhances the precision and accuracy of the chromatographic analysis. Contamination of DNA preparations with RNA does not interfere with the DNA analysis due to the high resolution of the chromatography. We have used this method for the quantitation of m5dCyd in 5 microgram of calf thymus and salmon sperm DNA in which the m5dCyd comprises only 1 to 2% of the total bases. This method should be a useful research tool in studies on various DNAs and DNA subfractions and should help to elucidate the functions of methylation of DNA.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams R. L., McKay E. L., Craig L. M., Burdon R. H. Mouse DNA methylase: methylation of native DNA. Biochim Biophys Acta. 1979 Feb 27;561(2):345–357. doi: 10.1016/0005-2787(79)90143-6. [DOI] [PubMed] [Google Scholar]
- Breter H. J., Seibert G., Zahn R. K. Single-step separation of major and rare ribonucleosides and deoxyribonucleosides by high-performance liquid cation-exchange chromatography for the determination of the purity of nucleic acid preparations. J Chromatogr. 1977 Oct 21;140(3):251–256. doi: 10.1016/s0021-9673(00)93584-2. [DOI] [PubMed] [Google Scholar]
- Breter H. J., Seibert G., Zahn R. K. The use of high-pressure liquid cation-exchange chromatography for determination of the 5-methylcytosine content of DNA. J Chromatogr. 1976 Mar 17;118(2):242–249. doi: 10.1016/s0021-9673(00)81215-7. [DOI] [PubMed] [Google Scholar]
- Breter H. J., Zahn R. K. A rapid separation of the four major deoxynucleosides and deoxyinosine by high-pressure liquid cation-exchange chromatography. Anal Biochem. 1973 Aug;54(2):346–352. doi: 10.1016/0003-2697(73)90362-x. [DOI] [PubMed] [Google Scholar]
- Burtis C. A. The determination of the base composition of RNA by high-pressure cation-exchange chromatography. J Chromatogr. 1970 Sep 16;51(2):183–194. doi: 10.1016/s0021-9673(01)96853-0. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Tait A., Goddard J. M. Methylated bases in DNA from Paramecium aurelia. Biochim Biophys Acta. 1974 Nov 20;374(1):1–11. doi: 10.1016/0005-2787(74)90194-4. [DOI] [PubMed] [Google Scholar]
- Davis G. E., Gehrke C. W., Kuo K. C., Agris P. F. Major and modified nucleosides in tRNA hydrolysates by high-performance liquid chromatography. J Chromatogr. 1979 May 21;173(2):281–298. doi: 10.1016/s0021-9673(00)92297-0. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Brown D. D. The mitochondrial and ribosomal DNA components of oocytes of Urechis caupo. Dev Biol. 1970 May;22(1):1–14. doi: 10.1016/0012-1606(70)90002-3. [DOI] [PubMed] [Google Scholar]
- Deutsch J., Razin A., Sedat J. Analysis of 5-methylcytosine in DNA. I. Mass spectrometry. Anal Biochem. 1976 May 7;72:586–592. doi: 10.1016/0003-2697(76)90570-4. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Ehrlich K. Separation of six DNA bases by ion pair--reversed phase high pressure liquid chromatography. J Chromatogr Sci. 1979 Sep;17(9):531–534. doi: 10.1093/chromsci/17.9.531. [DOI] [PubMed] [Google Scholar]
- Farber M. B., Ehrlich M. Bacteriophage XP-12-induced exonuclease which preferentially hydrolyzes nicked DNA. J Virol. 1980 Feb;33(2):733–738. doi: 10.1128/jvi.33.2.733-738.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke C. W., Kuo K. C., Zumwalt R. W. Chromatography of nucleosides. J Chromatogr. 1980 Jan 25;188(1):129–147. doi: 10.1016/s0021-9673(00)88424-1. [DOI] [PubMed] [Google Scholar]
- Günthert U., Stutz J., Klotz G. Restriction and modification in B. subtilis. The biochemical basis of modification against endo R. Bsu R restriction. Mol Gen Genet. 1975 Dec 30;142(3):185–191. doi: 10.1007/BF00425644. [DOI] [PubMed] [Google Scholar]
- Harbers K., Harbers B., Spencer J. H. Nucleotide clusters in deoxyribonucleic acids. XII. The distribution of 5-methylcytosine in pyrimidine oligonucleotides of mouse L-cell satellite DNA and main band DNA. Biochem Biophys Res Commun. 1975 Sep 16;66(2):738–746. doi: 10.1016/0006-291x(75)90572-0. [DOI] [PubMed] [Google Scholar]
- Hashizume T., Sasaki Y. Base and phosphorus composition analysis of gas chromatography. Anal Biochem. 1968 Aug;24(2):232–242. doi: 10.1016/0003-2697(68)90176-0. [DOI] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Hawley D. M., Wiebers J. L. Quantitation of the common components of deoxyribonucleic acids by mass spectrometry: application to the analysis of DNAs of unusual composition. Nucleic Acids Res. 1978 Dec;5(12):4949–4956. doi: 10.1093/nar/5.12.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T. T., Huang T. C., Teng M. H. 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas oryzae. J Mol Biol. 1968 Jul 14;34(2):373–375. doi: 10.1016/0022-2836(68)90263-5. [DOI] [PubMed] [Google Scholar]
- Lapeyre J. N., Becker F. F. 5-Methylcytosine content of nuclear DNA during chemical hepatocarcinogenesis and in carcinomas which result. Biochem Biophys Res Commun. 1979 Apr 13;87(3):698–705. doi: 10.1016/0006-291x(79)92015-1. [DOI] [PubMed] [Google Scholar]
- Pollock J. M., Jr, Swihart M., Taylor J. H. Methylation of DNA in early development: 5-methyl cytosine content of DNA in sea urchin sperm and embryos. Nucleic Acids Res. 1978 Dec;5(12):4855–4861. doi: 10.1093/nar/5.12.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Steele R. E. Modified bases in the DNAs of unicellular eukaryotes: an examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems. 1978 Apr;10(1-2):37–53. doi: 10.1016/0303-2647(78)90027-8. [DOI] [PubMed] [Google Scholar]
- Rafalski A., Kohli J., Agris P., Söll D. The nucleotide sequence of a UGA suppressor serine tRNA from Schizosaccharomyces pombe. Nucleic Acids Res. 1979 Jun 25;6(8):2683–2695. doi: 10.1093/nar/6.8.2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2725–2728. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Sedat J. Analysis of 5-methylcytosine in DNA. II. Gas chromatography. Anal Biochem. 1977 Feb;77(2):370–377. doi: 10.1016/0003-2697(77)90250-0. [DOI] [PubMed] [Google Scholar]
- Rubery E. D., Newton A. A. DNA methylation in normal and tumour virus-transformed cells in tissue culture. I. The level of DNA methylation in BHK21 cells and in BHK21 cells transformed by polyoma virus (PyY cells). Biochim Biophys Acta. 1973 Sep 28;324(1):24–36. doi: 10.1016/0005-2787(73)90247-5. [DOI] [PubMed] [Google Scholar]
- Salomon R., Kaye A. M. Methylation of mouse DNA in vivo: di- and tripyrimidine sequences containing 5-methylcytosine. Biochim Biophys Acta. 1970 Apr 15;204(2):340–351. [PubMed] [Google Scholar]
- Singer J., Schnute W. C., Jr, Shively J. E., Todd C. W., Riggs A. D. Sensitive detection of 5-methylcytosine and quantitation of the 5-methylcytosine/cytosine ratio in DNA by gas chromatography--mass spectrometry using multiple specific ion monitoring. Anal Biochem. 1979 Apr 15;94(2):297–301. doi: 10.1016/0003-2697(79)90363-4. [DOI] [PubMed] [Google Scholar]
- Singer J., Stellwagen R. H., Roberts-Ems J., Riggs A. D. 5-Methylcytosine content of rat hepatoma DNA substituted with bromodeoxyuridine. J Biol Chem. 1977 Aug 10;252(15):5509–5513. [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. W., Potter V. R. Methylation of mammalian DNA: studies on Novikoff hepatoma cells in tissue culture. J Mol Biol. 1969 Jun 14;42(2):271–284. doi: 10.1016/0022-2836(69)90043-6. [DOI] [PubMed] [Google Scholar]
- Störl H. J., Simon H., Barthelmes H. Immunochemical detection of N6-methyladenine in DNA. Biochim Biophys Acta. 1979 Aug 29;564(1):23–30. doi: 10.1016/0005-2787(79)90184-9. [DOI] [PubMed] [Google Scholar]
- Vanyushin B. F., Mazin A. L., Vasilyev V. K., Belozersky A. N. The content of 5-methylcytosine in animal DNA: the species and tissue specificity. Biochim Biophys Acta. 1973 Mar 28;299(3):397–403. doi: 10.1016/0005-2787(73)90264-5. [DOI] [PubMed] [Google Scholar]
- Wakizaka A., Kurosaka K., Okuhara E. Rapid separation of DNA constituents, bases, nucleosides and nucleotides, under the same chromatographic conditions using high-performance liquid chromatography with a reversed-phase column. J Chromatogr. 1979 Mar 1;162(3):319–326. doi: 10.1016/s0378-4347(00)81518-2. [DOI] [PubMed] [Google Scholar]
- Yuki H., Kawasaki H., Imayuki A., Yajima T. Determination of 6-methyladenine in DNA by high-performance liquid chromatography. J Chromatogr. 1979 Jan 21;168(2):489–494. doi: 10.1016/0021-9673(79)80020-5. [DOI] [PubMed] [Google Scholar]



