Abstract
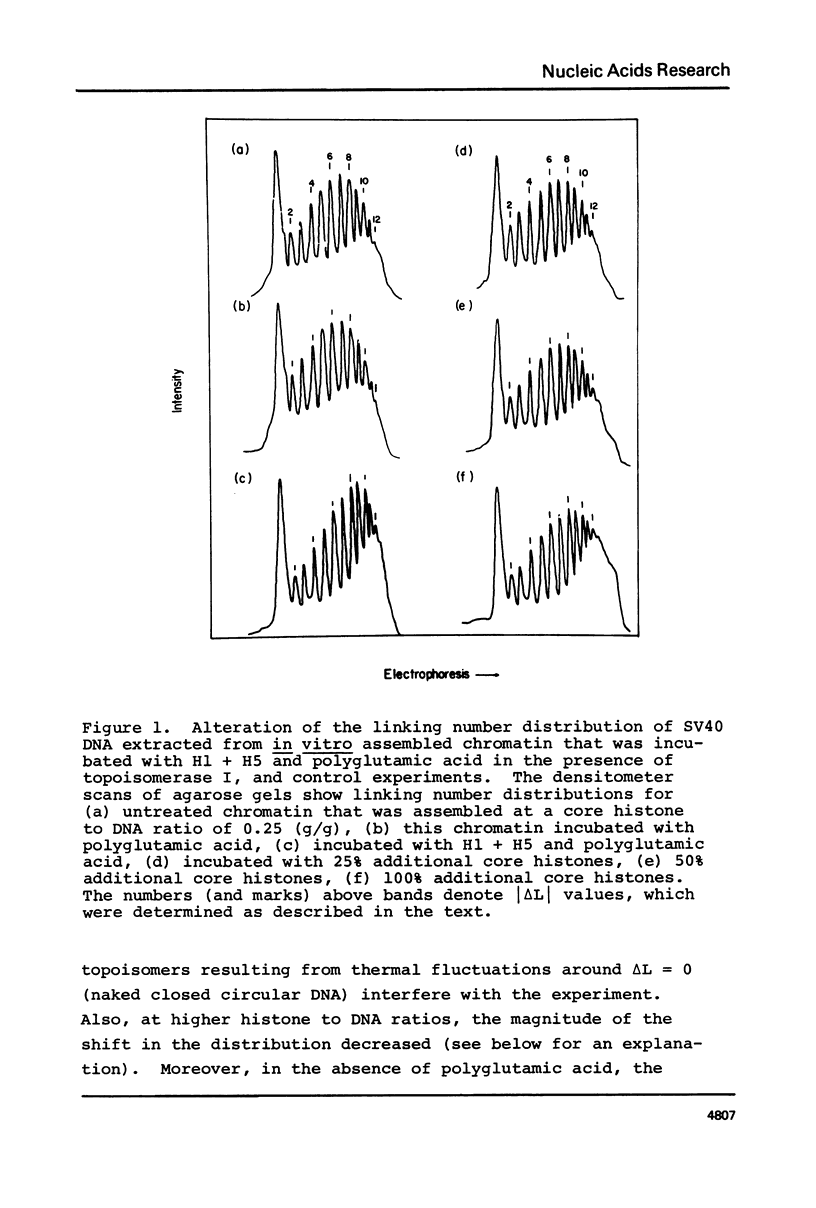
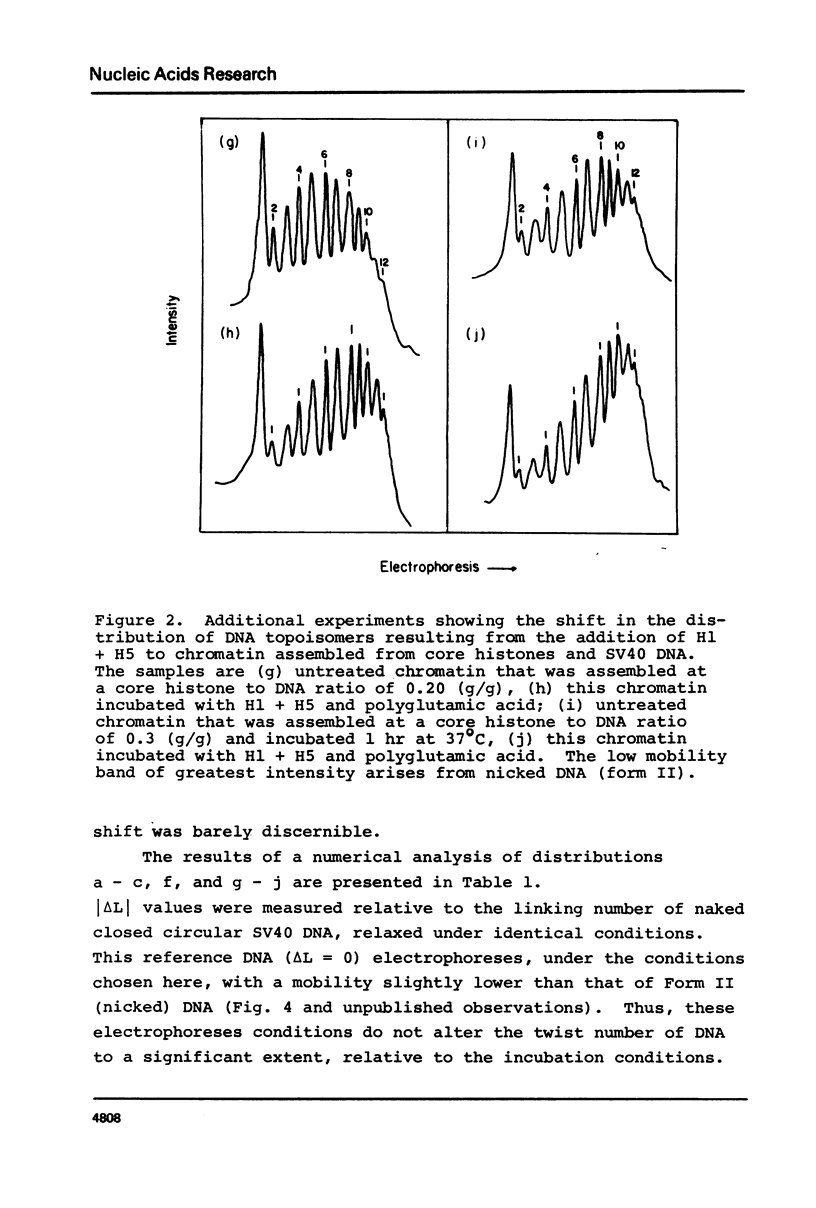
Chromatin was assembled in vitro from relaxed closed circular DNA (SV40) and core histones at histone to DNA ratios of 0.2 to 0.3 (g/g) and incubated with topoisomerase I to relax supercoils in DNA regions not constrained by protein. Addition of histones H1 + H5 to the chromatin at an ionic strength of 0.1 M, in the presence of the solubilizing agent, polyglutamic acid, and topoisomerase I, increased the magnitude of the DNA linking number change, relative to protein-free DNA. No change in the linking number distribution occurred for relaxed protein-free DNA under these conditions. Control experiments indicated that the increase in the absolute value of the DNA linking number change in the chromatin could not be attributed to an increase in the number of nucleosomes per DNA molecule. These data suggest a solution to the linking number problem associated with models of chromatin structure.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allan J., Staynov D. Z., Gould H. Reversible dissociation of linker histone from chromatin with preservation of internucleosomal repeat. Proc Natl Acad Sci U S A. 1980 Feb;77(2):885–889. doi: 10.1073/pnas.77.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenburger W., Hörz W., Zachau H. G. Nuclease cleavage of chromatin at 100-nucleotide pair intervals. Nature. 1976 Dec 9;264(5586):517–522. doi: 10.1038/264517a0. [DOI] [PubMed] [Google Scholar]
- Bellard M., Oudet P., Germond J. E., Chambon P. Subunit structure of simian-virus-40 minichromosome. Eur J Biochem. 1976 Nov 15;70(2):543–553. doi: 10.1111/j.1432-1033.1976.tb11046.x. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M., Singer M. F. The effect of H1 histone on the action of DNA-relaxing enzyme. Nucleic Acids Res. 1977 Jan;4(1):117–127. doi: 10.1093/nar/4.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Griffith J. Salt and divalent cations affect the flexible nature of the natural beaded chromatin structure. Nucleic Acids Res. 1977 Jun;4(6):1837–1851. doi: 10.1093/nar/4.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman H. A., Shelton E. R., Wassarman P. M., DePamphilis M. L. Structure of simian virus 40 chromosomes. Appendix. J Mol Biol. 1978 Nov 15;125(4):511–514. doi: 10.1016/0022-2836(78)90313-3. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Germond J. E., Bellard M., Oudet P., Chambon P. Stability of nucleosomes in native and reconstituted chromatins. Nucleic Acids Res. 1976 Nov;3(11):3173–3192. doi: 10.1093/nar/3.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyin Y. V., Varshavsky A. Y., Mickelsaar U. N., Georgiev G. P. Studies on deoxyribonucleoprotein structure. Redistribution of proteins in mixtures of deoxyribonucleoproteins, DNA and RNA. Eur J Biochem. 1971 Sep 24;22(2):235–245. doi: 10.1111/j.1432-1033.1971.tb01537.x. [DOI] [PubMed] [Google Scholar]
- Keller W. Characterization of purified DNA-relaxing enzyme from human tissue culture cells. Proc Natl Acad Sci U S A. 1975 Jul;72(7):2550–2554. doi: 10.1073/pnas.72.7.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Müller U., Eicken I., Wendel I., Zentgraf H. Biochemical and ultrastructural analysis of SV40 chromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):227–244. doi: 10.1101/sqb.1978.042.01.025. [DOI] [PubMed] [Google Scholar]
- Keller W., Wendel I. Stepwise relaxation of supercoiled SV40 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):199–208. doi: 10.1101/sqb.1974.039.01.026. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D., Morris N. R. Assembly of SV40 chromatin in a cell-free system from Xenopus eggs. Cell. 1977 Feb;10(2):237–243. doi: 10.1016/0092-8674(77)90217-3. [DOI] [PubMed] [Google Scholar]
- Lohr D., Van Holde K. E. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6326–6330. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Nelson P. P., Albright S. C., Wiseman J. M., Garrard W. T. Reassociation of histone H1 with nucleosomes. J Biol Chem. 1979 Nov 25;254(22):11751–11760. [PubMed] [Google Scholar]
- Noll M. DNA folding in the nucleosome. J Mol Biol. 1977 Oct 15;116(1):49–71. doi: 10.1016/0022-2836(77)90118-8. [DOI] [PubMed] [Google Scholar]
- Noll M. Internal structure of the chromatin subunit. Nucleic Acids Res. 1974 Nov;1(11):1573–1578. doi: 10.1093/nar/1.11.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noll M., Zimmer S., Engel A., Dubochet J. Self-assembly of single and closely spaced nucleosome core particles. Nucleic Acids Res. 1980 Jan 11;8(1):21–42. doi: 10.1093/nar/8.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudet P., Germond J. E., Sures M., Gallwitz D., Bellard M., Chambon P. Nucleosome structure I: all four histones, H2A, H2B, H3, and H4, are required to form a nucleosome, but an H3-H4 subnucleosomal particle is formed with H3-H4 alone. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):287–300. doi: 10.1101/sqb.1978.042.01.031. [DOI] [PubMed] [Google Scholar]
- Pardon J. F., Worcester D. L., Wooley J. C., Cotter R. I., Lilley D. M., Richards R. M. The structure of the chromatin core particle in solution. Nucleic Acids Res. 1977 Sep;4(9):3199–3214. doi: 10.1093/nar/4.9.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D., Lutter L., Klug A., Levitt M., Crick F. H. Periodicity of deoxyribonuclease I digestion of chromatin. Science. 1979 May 25;204(4395):855–858. doi: 10.1126/science.441739. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Whitlock J. P. Mapping DNAase l-susceptible sites in nucleosomes labeled at the 5' ends. Cell. 1976 Oct;9(2):347–353. doi: 10.1016/0092-8674(76)90124-0. [DOI] [PubMed] [Google Scholar]
- Stein A., Page D. Core histone associations in solutions of high salt. An osmotic pressure study. J Biol Chem. 1980 Apr 25;255(8):3629–3637. [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz M., Streeck R. E., Zachau H. G. Closely spaced nucleosome cores in reconstituted histone.DNA complexes and histone-H1-depleted chromatin. Eur J Biochem. 1978 Feb;83(2):615–628. doi: 10.1111/j.1432-1033.1978.tb12131.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. The nucleosome core protein. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):119–125. doi: 10.1101/sqb.1978.042.01.013. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Chumackov P. M., Georgiev G. P. Minichromosome of simian virus 40: presence of histone HI. Nucleic Acids Res. 1976 Aug;3(8):2101–2113. doi: 10.1093/nar/3.8.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaneva M., Tasheva B., Dessev G. Nuclease digestion of reconstituted chromatin. FEBS Lett. 1976 Nov;70(1):67–70. doi: 10.1016/0014-5793(76)80727-2. [DOI] [PubMed] [Google Scholar]