Abstract
Hantaviruses, of the family Bunyaviridae, are present throughout the world and cause a variety of infections ranging from the asymptomatic to mild and severe hemorrhagic fevers. Hantaviruses are enveloped anti-sense RNA viruses that contain three genomic segments that encode for a nucleocapsid protein, two membrane glycoproteins (Gn and Gc), and an RNA polymerase. Recently, the pathogenicity of hantaviruses has been mapped to the carboxyl end of the 150 residue Gn cytoplasmic tail. The Gn tail has also been shown to play a role in binding the ribonucleoprotein (RNP), a step critical for virus assembly. In this study, we use NMR spectroscopy to compare the structure of a Gn tail zinc finger domain of both a pathogenic (Andes) and a non-pathogenic (Prospect Hill) hantavirus. We demonstrate that despite a stark difference in the virulence of both of these viruses, the structure of the Gn core zinc finger domain is largely conserved in both strains. We also use NMR backbone relaxation studies to demonstrate that the regions of the Andes virus Gn tail immediately outside the zinc finger domain, sites known to bind the RNP, are disordered and flexible, thus intimating that the zinc finger domain is the only structured region of the Gn tail. These structural observations provide further insight into the role of the Gn tail during viral assembly as well as its role in pathogenesis.
Keywords: hantavirus, andes virus, Prospect Hill virus, zinc finger, glycoprotein
Introduction
Infection by a hantavirus can occur by inhalation of aerosolized urine or excrement from an infected rodent, thus leading a wide range of hemorrhagic fevers. European and Asian strains, for example, cause hemorrhagic fever with renal syndrome (HFRS), which is fatal in 1–15% of patients (Khaiboullina et al., 2005). The American strains, by contrast, cause the more severe hantavirus cardiopulmonary syndrome (HCPS), which is more aggressive and carries up to 40% mortality rate (Khaiboullina et al., 2005). Perhaps even more interesting are the two well known hantaviruses non-pathogenic to humans: Tula virus (present in European common voles) and the Prospect Hill virus (PHV; present in the American meadow vole; Mackow and Gavrilovskaya, 2009). In general, pathogenic and non-pathogenic hantaviruses appear to have a similar organization. They are enveloped and contain an anti-sense segmented RNA genome composed of a small (S) segment, medium (M) segment, and large (L) segment (Elliott et al., 2000). These in turn encode for the viral nucleocapsid protein, a glycoprotein polyprotein (subsequently processed into Gn and Gc proteins), and the RNA polymerase, respectively. RNA segments are bound to repeating trimers of the nucleocapsid protein into an assembled ribonucleocapsid (RNP) particle (Elliott et al., 2000).
As do most of the hantavirus structural proteins, the Gn glycoprotein has multiple functions in the viral replication cycle. As part of the virus envelope, it forms the structural core of a spike complex that consists of a multimeric arrangement of Gn and Gc glycoproteins, with a Gn tetramer at the center of the complex (Hepojoki et al., 2010a). On the cytoplasmic side of this complex, the inward facing 150 residue Gn cytoplasmic tail associates with the RNP during viral assembly (Hepojoki et al., 2010b,c; Wang et al., 2010; Battisti et al., 2011). Previously, we reported that a conserved array of cysteine and histidine residues (the dual CCHC motifs) within the Andes virus Gn tail folds into a compact dual ββα-type zinc fingers (Estrada et al., 2009a; Estrada and De Guzman, 2011). While ββα-type zinc fingers are primarily known for binding nucleic acids, the core zinc finger domain formed by 57-residues of the Andes virus Gn tail (residues 543–599) did not bind RNA in vitro (Estrada et al., 2009a), thus suggesting a role in protein–protein binding during the Gn–RNP interaction. Recently, Hepojoki et al. (2010c) showed that the Gn tail does bind the nucleocapsid (N) protein, the principal component of the RNP. Specifically, they showed that residues flanking the core zinc finger domain contain three binding sites (Binding sites 1, 2, and 3, Figure 1) for the N-protein (Hepojoki et al., 2010c). Others showed that the proper folding of the zinc finger domain was required for the ability of the Gn cytoplasmic tail to interact with the N-protein (Wang et al., 2010). These data strongly suggest a role for Gn tail in mediating an interaction with the N-protein.
Figure 1.
Sequence alignment of representative hantaviruses with the Gn tail–RNP binding sites indicated by brackets. The constructs used in this study are represented by the shaded gray boxes.
Additionally, the Gn tails of hantaviruses also participate in determination of virulence, specifically by helping to modulate the host cell immune response to infection (Geimonen et al., 2003b; Alff et al., 2006, 2008). The non-pathogenic PHV tail fails to co-precipitate tumor necrosis factor receptor-associated factor 3 (TRAF3), as is the case for the New York hantavirus (Alff et al., 2008). TRAF3 is a key component of the host cell’s interferon response to viral infection. In addition, the Gn tail of PHV was found not to be degraded, as is the case for typical pathogenic hantaviruses (Sen et al., 2007). However, the mutations of four residues at the carboxyl terminus of the Gn tail effectively targeted the PHV tail for proteasomal degradation (Sen et al., 2007). The observation of a virulence contribution by the Gn tail raises the possibility of potentially important differences between the predicted dual CCHC-type zinc finger domain of PHV and the structure determined for the pathogenic Andes virus (Estrada et al., 2009a). The two Gn tails overall are highly conserved between both viruses (75% identity, 84% similarity for the entire tail; 70% identity, 77% similarity for the zinc finger domain alone; Figure 1).
In this study, we used 2D and 3D NMR spectroscopy to compare the structures of two constructs representing segments of the cytoplasmic Gn tail for both a pathogenic (Andes virus) and a non-pathogenic hantavirus (PHV). Our NMR data suggests that, similar to the Andes virus, the dual CCHC motif of PHV forms an independently folded zinc binding domain. The Cα secondary chemical shifts of the PHV zinc finger domain are remarkably similar to those of the Andes structure, suggesting there is no appreciable difference in the two structures. These findings further support reports that the virulence determinants are located further toward the C-terminal end of the Gn tails. Furthermore, we also report the backbone assignment of an extended form of the Andes virus Gn tail (76 residues, from residues 534–610) that includes all of RNP Binding site 2 and part of RNP Binding site 1 (Figure 1). We demonstrate that Binding site 2 includes a short helix, while Binding site 1 appears to be largely disordered. Our results of NMR backbone dynamics indicate that both binding sites are flexible and undergo motion on a faster timescale than that of the core zinc finger domain. This enhanced motion may confer some degree of modularity in binding a crowded RNP complex. Taken together, these results provide novel structural insight into both the structural and immunogenic functions of the hantavirus Gn cytoplasmic tail.
Materials and Methods
Protein expression and purification
For NMR data collection, the soluble Gn construct spanning residues 534–610 of the Andes virus Gn cytoplasmic tail (GenBank #AF291703) and residues 548–602 of the PHV Gn tails (GenBank #X55129) were expressed as fusion proteins with the Streptococcal GB1 domain linked with a tobacco etch virus (TEV) protease cleavage site (Estrada et al., 2009b). The fusion proteins were expressed and purified under native conditions following closely the method reported previously for the Andes virus zinc finger domain (Estrada et al., 2009b). Briefly, 15N- and 15N/13C-labeled proteins were expressed in E. coli BL21(DE3) grown in 1 L M9 minimal media supplemented with 0.1 mM ZnSO4 before and after induction. Cells were grown at 37°C, induced with 1 mM isopropyl-β-d-thiogalactopyranoside at A600 ∼ 0.8, and cell growth was continued in a 15°C shaker overnight (to a final A600 ∼ 2.0). Cells were centrifuged, resuspended in nickel column binding buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 5 mM imidazole, 0.1 mM ZnSO4), and lysed by sonication. Cellular debris was removed by centrifugation, and to the supernatant was added 1/10 volume of 1% polyethyleneimine (pH 8) to precipitate the nucleic acids. Following centrifugation, the supernatant was bound to a charged 10 mL nickel-affinity column and eluted with buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 250 mM imidazole, 0.1 mM ZnSO4). For TEV protease digestion, fractions containing the fusion protein were pooled and dialyzed at 25°C overnight in buffer (50 mM Tris–HCl pH 8.0, 20 mM NaCl, 1 mM DTT, 1 mM ZnSO4) with 0.16 mg recombinant TEV protease (Geisbrecht et al., 2006) per 10 mL of fusion protein. The TEV digestion mixture was dialyzed back into the nickel column binding buffer and passed again through a charged 10 mL nickel resin. The His-tagged GB1 protein was retained on the column while hantavirus Gn tail constructs were present in the flow-through. The flow-through fractions were analyzed by SDS-PAGE and key fractions pooled and concentrated using Ultra-15 centrifugal filters (Amicon) and dialyzed into NMR buffer (20 mM NaPO4 pH 7.0, 20 mM NaCl, 1 mM DTT, 0.1 mM ZnSO4). Both proteins retained two residues (Gly–His) cloning artifacts at the N-terminus.
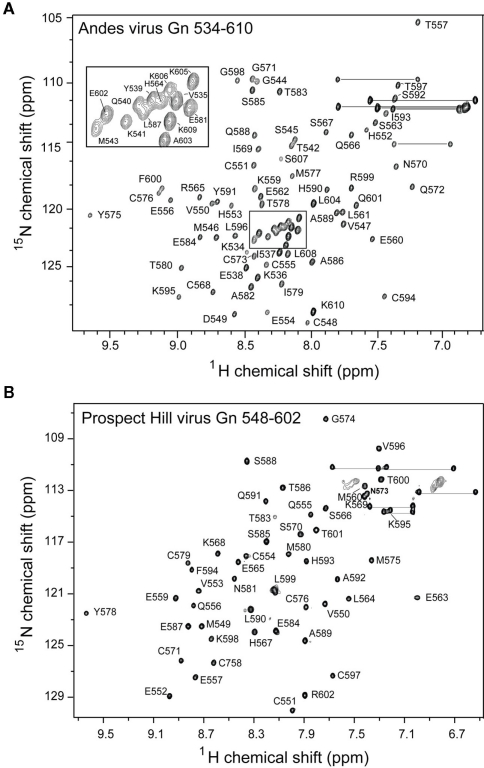
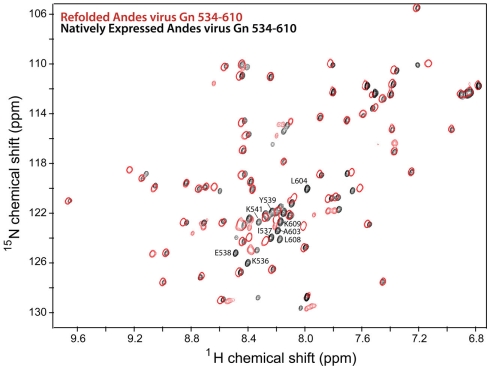
Notably, approximately half of the expressed Andes virus Gn 534–610 remained in the insoluble portion upon lysis. This fraction was solubilized in 6 M urea and 10 mM DTT, then refolded by step-wise dialysis to remove the denaturant and digested with TEV protease as described above. While the 1H–15N HSQC spectrum of the refolded protein showed a similar peak pattern for the core zinc finger domain (residues 543–599), several of the additional peaks corresponding to the binding sites had different chemical shifts compared to the natively expressed protein (Figure 2). Therefore, the 3D NMR data used for analysis was collected exclusively on the natively expressed Andes virus Gn 534–610.
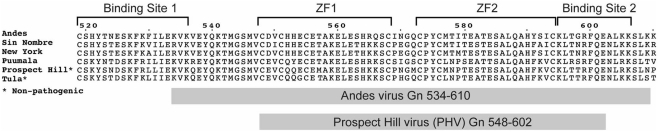
Figure 2.
Assigned 2D 1H–15N HSQC spectra for (A) the Andes Gn 534–610 and (B) the Prospect Hill virus Gn 548–602. Well-dispersed and resolved spectra for both constructs allowed complete assignment of the protein backbone.
NMR spectroscopy
NMR data were acquired at 25°C using a Bruker Avance 800 MHz spectrometer equipped with a cryoprobe, processed with NMRPipe (Delaglio et al., 1995), and analyzed with NMRView (Johnson, 2004). Backbone assignments for PHV Gn 548–602 were obtained from 2D 1H–15N HSQC (Grzesiek and Bax, 1993), 3D HNCA (Grzesiek et al., 1992), and HNCACB (Wittekind and Mueller, 1993) while assignments for Andes virus Gn 534–610 were obtained from the 2D 1H–15N HSQC and 3D HNCA alone. Secondary structures were identified from the Cα chemical shifts (Wishart and Nip, 1998). The heteronuclear {1H}–15N NOE for Andes virus Gn 534–610 was acquired as described (Stone et al., 1992) with 2048 (1H) × 128 (15N) complex points, 32 scans per point, and a 5-s recycle delay. Error bars were estimated using the SD of the background signal of each spectrum.
CD spectroscopy
CD spectra were collected in triplicate at 25° on a JASCO J-815 Spectropolarimeter using a scanning speed of 50 nm/min. Protein concentrations were kept at 5 μM in buffer (10 μM NaPO4, 10 μM NaCl, 0.1 mM ZnSO4). EDTA and ZnSO4 titrations were applied to the same sample.
PHV ZF structural modeling
Modeling of the structure of PHV Gn 548–602 was achieved by using the Andes dual zinc finger domain (PDB ID: 2K9H) as a template for the I-TASSER structure prediction software (Zhang, 2008). The model backbone was aligned with the backbone of the Andes virus Gn zinc finger structure using TM-Align (Zhang and Skolnick, 2005). Surface electrostatics were calculated using APBS (Baker et al., 2001) and visualized using Pymol (DeLano, 2002).
Results
Protein expression
The PHV Gn tail construct was designed similarly to the previously published structure of the Andes virus core zinc finger domain (Estrada et al., 2009a), yielding a construct of residues 548–602. The Andes virus Gn tail construct used in this study include as much of the reported binding sites as possible, yielding a protein containing residues 534–610. Both proteins expressed well in E. coli as N-terminal GB1 fusion recombinant proteins and were purified under native conditions via ion exchange chromatography. Our attempts to express longer full-length Gn tail in Andes virus and other hantaviruses have been unsuccessful. However, the Andes virus Gn tail construct used for this study (76 residues) does represent the longest piece of the hantavirus Gn tail that is structurally characterized at atomic resolution. Incidentally, expression of the Andes virus Gn 548–602 construct yielded approximately half of the protein in inclusion bodies. Upon removal of the GB1 tag via cleavage by treatment with TEV protease, the Andes virus Gn 548–602 protein that was purified from the inclusion body and the natively purified fraction gave well resolved and well-dispersed 1H–15N HSQC spectra (Figure 3).
Figure 3.
Overlay of 2D 1H–15N HSQC spectra of the Andes virus Gn (534–610) that has been refolded from insoluble inclusion bodies (red) and that was natively expressed in the soluble fraction (black). Notably, the chemical shift values for the flanking sequences are different for each spectra, indication two conformations for these sequences.
PHV Gn tail relies on Zn2+ for proper refolding
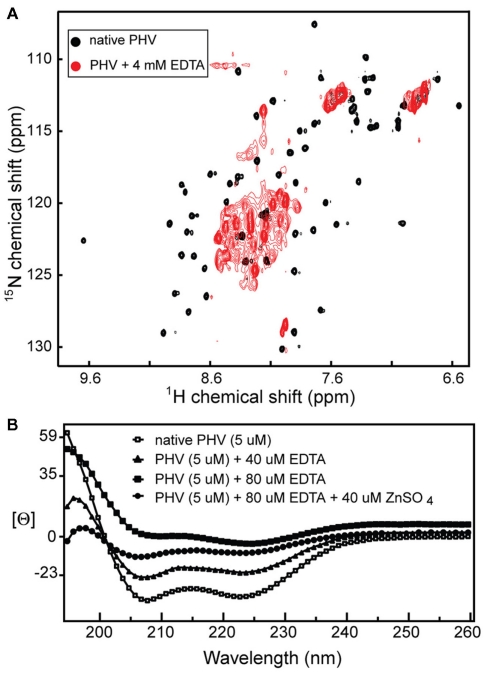
As expected, treatment of the 15N labeled PHV Gn 548–602 NMR sample with excess EDTA resulted in a near complete collapse of the 2D 1H–15N HSQC spectrum (Figure 4A), suggesting a loss of tertiary structure. Similarly, the CD trace of the PHV Gn 548–602 was also affected by the presence of EDTA (Figure 4B). The addition of Zn2+ back into the sample was only partially able to recover the original helical trace. This behavior of the PHV domain resembled that of the Andes protein (Estrada et al., 2009a), suggesting the confirmed cysteines and histidines in Gn tails were required for metal coordination.
Figure 4.
(A) EDTA titration of the 2D 1H–15N HSQC spectrum of PHV Gn 548–602 causes a dramatic narrowing of the spectrum, suggesting a loss of tertiary structure and (B) the same effect on the protein CD trace [Θ is mean molar residue ellipticity (×107, degree·cm2/dmol·residue)].
NMR data and backbone assignments
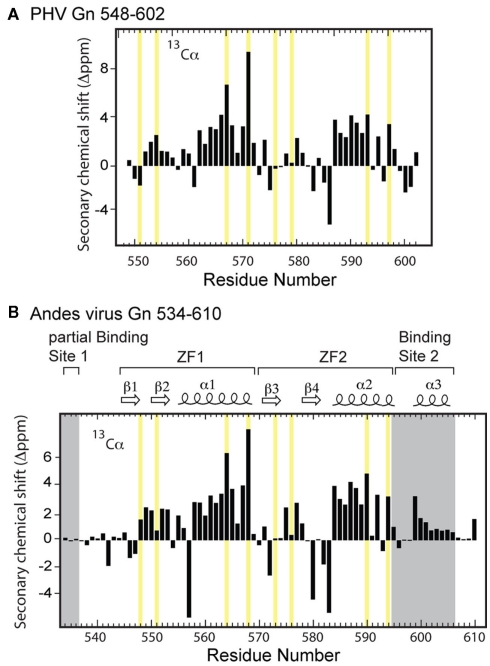
Prospect Hill virus Gn 548–602 and Andes virus Gn 534–610 both show a well-dispersed two-dimensional (2D) 1H–15N HSQC (Figure 2). However, an interesting feature of the natively folded Andes virus Gn 534–610 spectrum is that it differed from the spectrum of protein refolded from inclusion bodies. An overlay of the 1H–15N HSQC spectra of the refolded and natively expressed Andes virus Gn 534–610 confirmed that the majority of peaks are in identical positions and that the core zinc finger domain (residues 543–599; Estrada et al., 2009a) was properly folded (Figure 3). However, all subsequent data and assignments were conducted on the natively Andes virus Gn 534–610 expressed protein. Nearly complete backbone assignments were obtained from 2D and 3D NMR datasets for PHV Gn 548–620 and Andes virus Gn 534–610. Analysis of the Cα secondary chemical shifts for the PHV protein indicated the presence of two short α-helices and two random coil regions flanking the central domain (Figure 5A; Wishart and Nip, 1998). This pattern closely resembles that of the Andes virus Gn zinc finger domain (Figure 5A). Given the high degree of secondary structural similarity between the zinc binding domains in both structures (77% similarity) and the remarkable similarity of the Cα secondary chemical shift profile, full side chain assignments and structure determination of PHV Gn 548–620 was not continued.
Figure 5.
Secondary Cα chemical shift profile for (A) PHV Gn 548–602 and (B) Andes virus Gn 534–610 zinc finger structures. The zinc coordinating residues are highlighted in yellow.
Secondary chemical shift profile of the RNP binding sites
Based on the HNCA-derived assignments of the Andes virus Gn 534–610 backbone, we constructed the secondary Cα chemical shift profile of both the core zinc finger domain (residues 543–599) along with the flanking sequences (Figure 5B). Notably, analysis of the profile suggested the N-terminal sequence (Lys534–Met543) was disordered. However, the C-terminus contained an intact helix between residues Arg599 and Lys606. Another interesting feature was that the intervening sequence corresponding to the core zinc finger domain appeared largely unchanged from that of the protein without the flanking sequences, indicating that the presence of the binding sites reported by Hepojoki et al. (2010c) did not affect the local environment and had minimal contact with the core zinc finger domain.
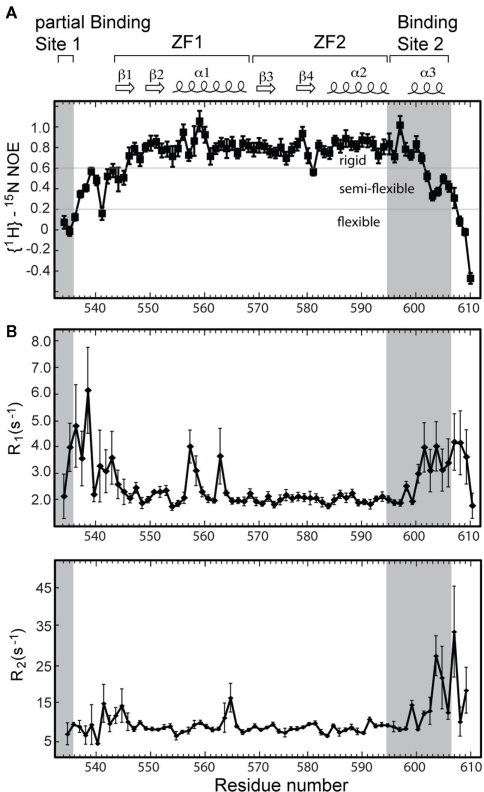
Backbone dynamics of the RNP binding sites
Heteronuclear {1H}–15N NOE analysis of Andes virus Gn 534–610, which provides a probe on protein backbone dynamics, suggests that the core zinc finger domain is largely a rigid structure (Figure 6A). However, the flanking sequences are considerably more flexible. The disordered N-terminus is almost entirely flexible relative to the core structure as evident by the heteronuclear {1H}–15N NOE values below 0.2 in this region (Figure 6A). Notably, gradually increasing flexibility at the C-terminus corresponds to the tapering of helix α3, with greatest flexibility at the disordered five terminal residues. Backbone R1 and R2 relaxation results also indicate that the flanking sequences undergo motion on a faster time scale than the core zinc finger, with ZF1 and ZF2 both reflecting R1 values of approximately 2.0, while the flanking sequences having R1 values of approximately 4.0 (Figure 6B).
Figure 6.
(A) Heteronuclear {1H}–15N NOE values for the Andes virus zinc finger protein indicate enhanced flexibility corresponding to both binding sites. (B) The relative motions of each binding site are quantified with R1 and R2 relaxation rates. The spin–lattice relaxation rates R1 suggest each binding site undergoes motions at least twice as fast as the rigid core zinc finger domain.
Modeling the PHV Gn zinc finger structure
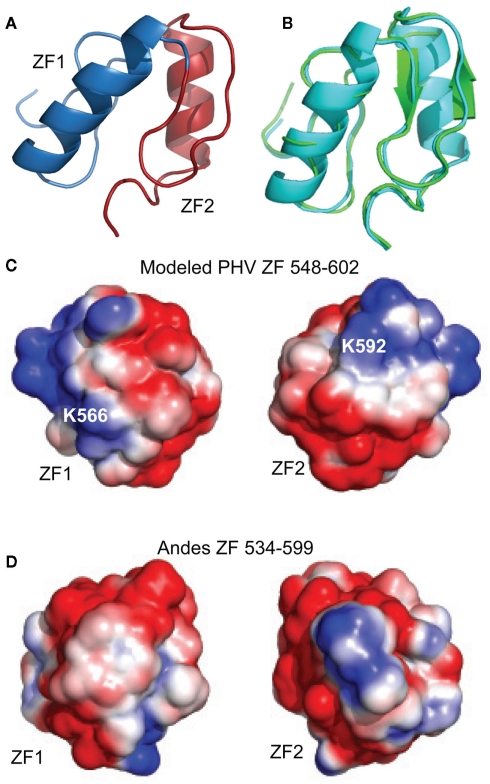
The similarity between the secondary chemical shift profiles of the PHV and the Andes virus zinc binding domains suggest the two proteins have similar folds. Therefore, the structure of the Andes virus core zinc finger domain (residues 543–599, PDB ID 2K9H; Estrada et al., 2009a) was used as a template for the structural modeling of the PHV structure. The program I-TASSER (Zhang, 2008) was used to thread the PHV sequence onto the structure. The resulting structure is shown in Figure 7A. It closely resembles that of the Andes structure, with a backbone rmsd of 0.68 between the two (Figure 7B). Notably, the surface electrostatics of the PHV model closely resemble those of the Andes virus structure (Figures 7C,D) in that neither exhibit extensive clustering of charges.
Figure 7.
Model of (A) PHV Gn 548–602 superimposed on the (B) Andes virus zinc finger structure. The PHV model (cyan) matched the Andes structure (green) with a backbone rmsd of 0.68 for all Cα’s. The PHV model was constructed using I-TASSER (Zhang, 2008) using the Andes structure as a template. Structural alignment was carried out using I-TASSER TM-align (Zhang and Skolnick, 2005). Surface electrostatic comparison between the (C) PHV model (residues 548–602) and (D) the Andes virus zinc finger domain (residues 534–599). Despite the presence of two charges (K566 and K592), the net distribution of charges in the PHV model is not appreciably different from that of the pathogenic.
Discussion
Studies into the virulence of hantaviruses reveal several notable differences in cells infected by pathogenic and non-pathogenic hantaviruses. For example, while the non-pathogenic PHV still manages to infect endothelial cells, the virus apparently does not successfully replicate in humans (Mackow and Gavrilovskaya, 2009). This suggests PHV does not manage to evade the host immunogenic response. This notion is supported by evidence that early infection by PHV causes the induction of up to 24 early interferon genes, as opposed to only three by the pathogenic New York and Hantaan viruses (Geimonen et al., 2002; Spiropoulou et al., 2007).
Recent studies have begun to map virulence in hantaviruses to the Gn cytoplasmic tail (Geimonen et al., 2003a,b; Alff et al., 2006, 2008). In light of these findings, our structural studies were conducted on the predicted Gn tail zinc finger domain of the non-pathogenic PHV for the purpose of comparing the structure to that of the pathogenic Andes virus (Figure 1). Additionally, due to the recent report of the presence of Gn tail–RNP binding sites located proximal and on either side of the zinc finger domain of the Andes virus, the Andes protein used in this study represents a larger construct design to contain as much of the reported binding sites as possible. Due to the toxicity of the full-length Gn tail in bacteria, this latter construct (76 residues) represents the largest soluble segment of the Gn tail that we have been able to produce.
With respect to the PHV zinc finger domain, the CD data and the spectrum of the protein in the presence of chelator (Figure 4A) confirmed the presence of a metal binding domain. Specifically, the fact that titration of zinc sulfate back into the CD sample (Figure 4B) partially recovers the original spectrum indicates the domain binds zinc. These results compare favorably with the behavior of the core zinc finger domain in the Andes virus (Estrada et al., 2009a).
The 2D 1H–15N HSQC of the PHV Gn tail consisting of residues 548–602 showed a well-dispersed spectrum (Figure 4A), suggesting a monomeric and independently folded protein. Likewise, the 1H–15N HSQC spectrum of the extended Andes virus Gn 534–610 also showed well-dispersed and resolvable peaks (Figure 2). The high quality of the NMR data facilitated assignment of the backbone resonances, yielding the Cα chemical shift profile for both proteins. The profile of the PHV zinc finger domain is compared to that of the Andes virus in Figure 5. Given the high degree of sequence conservation, as well as conservation in the spacing of the dual CCHC motif (Figure 1), it is not surprising to find that the profiles of the core domain are remarkably similar. The only notable differences occur in the first and final coordinating cysteines (Cys551 and Cys597, respectively). These differences may be due to the presence of two non-conserved residues, Glu552 and Lys598, immediately following each cysteine.
One interesting feature of the PHV Gn amino acid sequence is the presence of a proline (Pro572) in the linker. The proline is somewhat conserved; it is present in Tula and Hantaan viruses but absent in the Andes virus, which has an isoleucine in the same position. Interestingly, a residue with a rigid peptide bond in this position has little effect on the Cα secondary chemical shift of Cys571. The secondary chemical shift profile for this part of the ZF1 array is virtually identical to that of the ZF1 array in the Andes virus structure (Figure 5), suggesting the tight turn between the β hairpin and helix of ZF1 is maintained despite the difference in residues in this region. This observation was an early indicator that the dual zinc finger fold is preserved despite large differences in chemical composition (Figure 1).
Due to the apparent similarity of the structures, we did not solve the three-dimensional (3D) structure of the PHV zinc finger domain. However, a model using I-TASSER (Zhang, 2008) and the Andes structure as a template was constructed to check the surface electrostatics of the protein which could then influence the molecular interactions of this protein. The modeled PHV zinc finger domain contains two similar short α-helices in a relative orientation as seen in the Andes structure (Figure 7). Previously, we have reported that the core zinc finger domain of a related Bunyavirus, the Crimean Congo Hemorrhagic Fever virus, contains a vastly different arrangement of basic charged residues on its surface when compared to the Andes virus structure (Estrada and De Guzman, 2011). With respect to the PHV model, we find that the surface electrostatics resemblance those of the Andes virus structure, with dispersed charges covering the zinc fingers. Only two non-conserved basic charges, K566 and K592, are present which are not on the Andes virus structure. Despite these, the PHV structure is also acidic, with a theoretical isoelectric point of approximately 6.0, thus suggesting that differences in electrostatics of the core zinc finger domain are not likely to play a role in pathogenicity.
Overall, the PHV Gn zinc finger domain appears to adopt the same dual zinc finger fold to that of the pathogenic Andes virus. There were no obvious differences in either the secondary chemical shift profile or the predicted surface electrostatics. Based on this data we conclude that, given the widespread nature of the dual zinc finger fold in hantaviruses, its role is likely general in nature and apparently does not play a role in the determination of virulence. These conclusions are consistent with studies mapping virulence to the C-terminus of the Gn tails (Sen et al., 2007; Spiropoulou et al., 2007).
With respect to the role of the Gn tail in virus assembly, the amino acid sequences flanking the hantavirus zinc finger domain have been shown to be important for Gn–N protein interaction (Hepojoki et al., 2010b). Specifically, short peptides representing these regions of the Gn tail disrupted the Gn–N protein interaction as detected using a SPOT peptide array (Hepojoki et al., 2010b). Generation of the extended zinc finger domain (534–610) in the Andes virus allowed us to characterize one full RNP binding site (Lys595–Lys610) and a portion of a second (Lys534 and Lys536).
The reported Binding site 2 (Hepojoki et al., 2010c) contains the short helix α3 located near the carboxyl terminus. Helix α3 contains three conserved basic residues. Arg599 is located at the beginning turn of the helix, while the lysine pair located at positions 605 and 606 approximate the end of the helix. A helical wheel projection suggests that Arg599 and Lys606 are likely oriented in the same direction and may form a conserved basic patch. Notably, the surface opposite these charges contains the two conserved hydrophobic residues Phe600 and Leu604, suggesting the helix is amphipathic. Despite this, the helix does not appear to form part of the independently folded core zinc finger domain.
Significantly, half of the protein expressed for this larger segment was insoluble. The solubilized and refolded fraction appeared to have a normal core zinc finger fold, as measured by an overlay of the previously published spectrum and that of the refolded Andes virus Gn (534–610). However, comparison of the spectrum of the refolded protein with that of the native protein revealed two distinct set of chemical shift values for the flanking sequences, thus indicating two separate conformations (Figure 3). While this was likely an artifact of refolding, it suggested the arms can be arranged independent of the core zinc finger structure. This notion is supported by the heteronuclear {1H}–15N NOE plot (Figure 6A), which suggests that much of the flanking sequence is either somewhat or fully flexible.
The presence of a Gn tetramer in the hantavirus spike complex suggests that four Gn cytoplasmic tails may participate in binding the RNP (Hepojoki et al., 2010a). Meanwhile, the RNP itself consists primarily of an N-protein trimer complexed with RNA (Alfadhli et al., 2001; Kaukinen et al., 2004; Mir and Panganiban, 2004) thus making the Gn–N protein binding site a crowded space where the important intra-molecular contacts may be complex and multivalent. Our NMR results indicate that the Gn tail consists of a structured core zinc finger domain that is flanked by flexible regions where the reported N-protein binding sites are located (Hepojoki et al., 2010c). The flexibility in Binding site 2 and part of Binding site 1 as described here may contribute to modularity of the Gn tail in binding at multiple sites. Thus, our NMR results, together with the results of others (Hepojoki et al., 2010c; Wang et al., 2010) suggest that backbone flexibility may be important in the molecular recognition of the Gn tail.
In summary, the work presented here strongly indicates that the conserved dual CCHC motif of hantavirus Gn cytoplasmic tails correlates to a structurally conserved dual zinc finger domain that likely represents the only structured region of the tail. The widespread nature of this domain within Bunyaviridae and the high degree of structural conservation both suggest an important role in viral assembly, possibly by helping to mediate inter-molecular contacts with the RNP. The RNP binding sites characterized here were shown to be flexible and move independently of the core Andes domain, thus suggesting some degree of modularity in the function of the Gn tail.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge funding from the American Heart Association 0755724Z (Roberto N. De Guzman), K-INBRE NIH P20 RR016475 (Roberto N. De Guzman), NIH AI065359 (Stephen C. St. Jeor), and the Madison and Lila Self Graduate Fellowship (D. Fernando Estrada). We are grateful to Gerard Kroon (Scripps Research Institute) and Asokan Anbanandam (University of Kansas) for helpful discussions.
References
- Alfadhli A., Love Z., Arvidson B., Seeds J., Willey J., Barklis E. (2001). Hantavirus nucleocapsid protein oligomerization. J. Virol. 75, 2019–2023 10.1128/JVI.75.4.2019-2023.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alff P. J., Gavrilovskaya I. N., Gorbunova E., Endriss K., Chong Y., Geimonen E., Sen N., Reich N. C., Mackow E. R. (2006). The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J. Virol. 80, 9676–9686 10.1128/JVI.02162-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alff P. J., Sen N., Gorbunova E., Gavrilovskaya I. N., Mackow E. R. (2008). The NY-1 hantavirus Gn cytoplasmic tail coprecipitates TRAF3 and inhibits cellular interferon responses by disrupting TBK1-TRAF3 complex formation. J. Virol. 82, 9115–9122 10.1128/JVI.00290-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. A., Sept D., Joseph S., Holst M. J., Mccammon J. A. (2001). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 10.1073/pnas.031419998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battisti A. J., Chu Y. K., Chipman P. R., Kaufmann B., Jonsson C. B., Rossmann M. G. (2011). Structural studies of Hantaan virus. J. Virol. 85, 835–841 10.1128/JVI.01847-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- DeLano W. L. (2002). The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific [Google Scholar]
- Elliott R. M., Bouloy M., Calisher C. H., Goldbach R., Moyer J. T., Nichol S. T., Pettersson R., Plyusnin A., Schmaljohn C. (2000). “Bunyaviridae,” in Virus Taxonomy: The classification and Nomenclature of Viruses. The Seventh Report of the International Committee on Taxonomy of Viruses, eds Van Regenmortel M. H. V., Fauquet C. M., Bishop D. H. L., Carsten E. B., Estes M. K., Lemon S. M., Maniloff J., Mayo M. A., Mcgeoch D. J., Pringle C. R., Wickner R. B. (San Diego: Academic Press; ), 599–621 [Google Scholar]
- Estrada D. F., Boudreaux D. M., Zhong D., St Jeor S. C., De Guzman R. N. (2009a). The hantavirus glycoprotein G1 tail contains dual CCHC-type classical zinc fingers. J. Biol. Chem. 284, 8654–8660 10.1074/jbc.M808081200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada D. F., Boudreaux D. M., Zhong D., St. Jeor S. C., De Guzman R. N. (2009b). The hantavirus glycoprotein G1 tail contains dual CCHC-type classical zinc fingers. J. Biol. Chem. 284, 8654–8660 10.1074/jbc.M808081200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada D. F., De Guzman R. N. (2011). Structural characterization of the Crimean-Congo hemorrhagic fever virus Gn tail provides insight into virus assembly. J. Biol. Chem. 286, 21678–21686 10.1074/jbc.M110.216515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimonen E., Fernandez I., Gavrilovskaya I. N., Mackow E. R. (2003a). Tyrosine residues direct the ubiquitination and degradation of the NY-1 hantavirus G1 cytoplasmic tail. J. Virol. 77, 10760–10868 10.1128/JVI.77.2.1638-1643.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimonen E., Lamonica R., Springer K., Farooqui Y., Gavrilovskaya I. N., Mackow E. R. (2003b). Hantavirus pulmonary syndrome-associated hantaviruses contain conserved and functional ITAM signaling elements. J. Virol. 77, 1638–1643 10.1128/JVI.77.2.1638-1643.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimonen E., Neff S., Raymond T., Kocer S. S., Gavrilovskaya I. N., Mackow E. R. (2002). Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc. Natl. Acad. Sci. U.S.A. 99, 13837–13842 10.1073/pnas.192298899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht B. V., Bouyain S., Pop M. (2006). An optimized system for expression and purification of secreted bacterial proteins. Protein Expr. Purif. 46, 23–32 10.1016/j.pep.2005.08.011 [DOI] [PubMed] [Google Scholar]
- Grzesiek S., Bax A. (1993). The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115, 12593–12594 10.1021/ja00063a068 [DOI] [Google Scholar]
- Grzesiek S., Dobeli H., Gentz R., Garotta G., Labhardt A. M., Bax A. (1992). 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-gamma. Biochemistry 31, 8180–8190 10.1021/bi00150a009 [DOI] [PubMed] [Google Scholar]
- Hepojoki J., Strandin T., Vaheri A., Lankinen H. (2010a). Interactions and oligomerization of hantavirus glycoproteins. J. Virol. 84, 227–242 10.1128/JVI.00481-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepojoki J., Strandin T., Wang H., Vapalahti O., Vaheri A., Lankinen H. (2010b). Cytoplasmic tails of hantavirus glycoproteins interact with the nucleocapsid protein. J. Gen. Virol. 91, 2341–2350 10.1099/vir.0.021006-0 [DOI] [PubMed] [Google Scholar]
- Hepojoki J. M., Strandin T., Wang H., Vapalahti O., Vaheri A., Lankinen H. (2010c). The cytoplasmic tails of hantavirus glycoproteins interact with the nucleocapsid protein. J. Gen. Virol. 91, 2341–2350 10.1099/vir.0.021006-0 [DOI] [PubMed] [Google Scholar]
- Johnson B. A. (2004). Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- Kaukinen P., Kumar V., Tulimaki K., Engelhardt P., Vaheri A., Plyusnin A. (2004). Oligomerization of hantavirus N protein: C-terminal alpha-helices interact to form a shared hydrophobic space. J. Virol. 78, 13669–13677 10.1128/JVI.78.24.13669-13677.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaiboullina S. F., Morzunov S. P., St Jeor S. C. (2005). Hantaviruses: molecular biology, evolution and pathogenesis. Curr. Mol. Med. 5, 773–790 10.2174/156652405774962317 [DOI] [PubMed] [Google Scholar]
- Mackow E. R., Gavrilovskaya I. N. (2009). Hantavirus regulation of endothelial cell functions. Thromb. Haemost. 102, 1030–1041 [DOI] [PubMed] [Google Scholar]
- Mir M. A., Panganiban A. T. (2004). Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. J. Virol. 78, 8281–8288 10.1128/JVI.78.15.8281-8288.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen N., Sen A., Mackow E. R. (2007). Degrons at the C terminus of the pathogenic but not the nonpathogenic hantavirus G1 tail direct proteasomal degradation. J. Virol. 81, 4323–4330 10.1128/JVI.01124-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiropoulou C. F., Albarino C. G., Ksiazek T. G., Rollin P. E. (2007). Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J. Virol. 81, 2769–2776 10.1128/JVI.02402-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M. J., Fairbrother W. J., Palmer A. G., Iii, Reizer J., Saier M. H., Jr., Wright P. E. (1992). Backbone dynamics of the Bacillus subtilis glucose permease IIA domain determined from 15N NMR relaxation measurements. Biochemistry 31, 4394–4406 10.1021/bi00133a003 [DOI] [PubMed] [Google Scholar]
- Wang H., Alminaite A., Vaheri A., Plyusnin A. (2010). Interaction between hantaviral nucleocapsid protein and the cytoplasmic tail of surface glycoprotein Gn. Virus Res. 151, 205–212 10.1016/j.virusres.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Wishart D. S., Nip A. M. (1998). Protein chemical shift analysis: a practical guide. Biochem. Cell Biol. 76, 153–163 10.1139/o98-038 [DOI] [PubMed] [Google Scholar]
- Wittekind M., Mueller L. (1993). HNCACB, a high sensitivity 3D NMR experiment to correlate amide proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J. Magn. Reson. 101B, 201–205 [Google Scholar]
- Zhang Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9, 40. 10.1186/1471-2105-9-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. (2005). TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 10.1093/nar/gki479 [DOI] [PMC free article] [PubMed] [Google Scholar]