Abstract
Human APOBEC3 (A3) proteins are cellular cytidine deaminases that potently restrict the replication of retroviruses by hypermutating viral cDNA and/or inhibiting reverse transcription. There are seven members of this family including A3A, B, C, DE, F, G, and H, all encoded in a tandem array on human chromosome 22. A3F and A3G are the most potent inhibitors of HIV-1, but only in the absence of the virus-encoded protein, Vif. HIV-1 utilizes Vif to abrogate A3 functions in the producer cells. More specifically, Vif, serving as a substrate receptor, facilitates ubiquitination of A3 proteins by forming a Cullin5 (Cul5)-based E3 ubiquitin ligase complex, which targets A3 proteins for rapid proteasomal degradation. The specificity of A3 degradation is determined by the ability of Vif to bind to the target. Several lines of evidence have suggested that three distinct regions of A3 proteins are involved in the interaction with Vif. Here, we review the biological functions of A3 family members with special focus on A3G and base our analysis on the available structural information.
Keywords: APOBEC3, Vif, APOBEC3G, HIV, retrovirus, ubiquitin, cytidine deaminase, structure
Introduction
During coevolution of mammalian organisms and retroelements, the hosts have developed defense systems to restrict replication of these elements. The restriction factors include the A3 family of DNA cytidine deaminases, which is characterized by the presence of one or two Zn clusters consisting of (H/C)XE(X)23–28CXXC motifs (reviewed in Wedekind et al., 2003). In humans, there are seven members of the A3 family (A3A, B, C, DE, F, G, and H), each encoded in a tandem array on chromosome 22 (Jarmuz et al., 2002; Figure 1A). Each protein has been found to have different inhibitory effects on various retroelements that are mediated by cytidine deamination and other mechanisms (reviewed in Goila-Gaur and Strebel, 2008; Albin and Harris, 2010). In order to overcome A3 antiviral activity, the viruses have acquired their own strategies (Yu, 2006). HIV-1 encodes the Vif protein to counteract the most potent inhibitors, human A3G (hA3G) and A3F (Sheehy et al., 2002; Wiegand et al., 2004; Zheng et al., 2004; Simon et al., 2005; Miyagi et al., 2010). Vif binds the A3 proteins and serves as a substrate receptor to recruit the Cul5-based E3 ubiquitin (Ub) ligase complex, which facilitates the polyubiquitination and subsequent proteasome-mediated degradation (Yu et al., 2003). The elimination of A3 during virus production prevents its encapsidation into progeny viruses. Thus, given Vif’s critical role in eliminating A3 function, it may be viewed as one of the most attractive pharmacologic targets for an anti-HIV drug, which would restore the activity of the intrinsic antiviral factor in the context of HIV infection. Here, we briefly review what is known about A3-Vif interactions and the subsequent ubiquitination, based on the available biological and structural information.
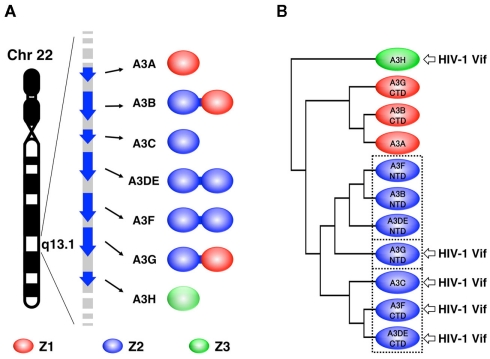
Figure 1.
Three types of Z domains in human A3 family. (A) Seven members of human A3 family, encoded by human chromosome 22, are illustrated. The protein products consist of one or two Z domains, each of which is phylogenetically grouped into three distinct types: Z1, Z2, and Z3. (B) Three subgroups of the Z2 domains are enclosed with dotted lines in the phylogenetic tree. The arrows point to the domains that are responsible for HIV-1 Vif interaction.
The Human A3 Family of Cytidine Deaminases
The seven members of the human A3 family have a defining feature: each protein has one or two conserved zinc (Z)-coordinating deaminase domains. Zinc coordination is mediated by a histidine and two cysteines, which form a catalytic center for a cytidine deaminase activity. Based on the phylogenetic analysis, the Z domains fall into three types: Z1 [A3A and the C-terminal half domains (CTD) of A3B and A3G], Z2 [A3C, both halves of A3DE and A3F, and the N-terminal domains (NTD) of A3B and A3G], and Z3 (A3H; LaRue et al., 2009; Figure 1A). Within the Z2 types, the domain can be further subdivided into three subgroups based on the identity of the amino acid sequence: (1) the A3F NTD; (2) the A3G NTD; (3) the A3F CTD (Figure 1B). Each subgroup has highly conserved amino acid sequences. For example, A3F CTD is 77 and 88% identical to A3C and A3DE CTD, respectively, whereas it is 42% identical to A3G NTD. As described in more detail below, only the Z3 type (A3H) and the Z2 type (more specifically, the A3G NTD and the A3F CTD subgroups) contain a critical interface for binding HIV-1 Vif (Figure 1B). It is thought that there is a common structural feature for the organization of cytidine deaminases: all Z domains are believed to have a conserved core structure composed of five β-strands (β1–β5) and six α-helices (α1–α6; Figure 2A). Although, to date, only the three-dimensional structure of the A3G CTD has been determined by NMR (Chen et al., 2008; Furukawa et al., 2009) or by X-ray crystallography (Holden et al., 2008; Shandilya et al., 2010), no structure of the Z domain having an HIV-1 Vif interface has been solved yet.
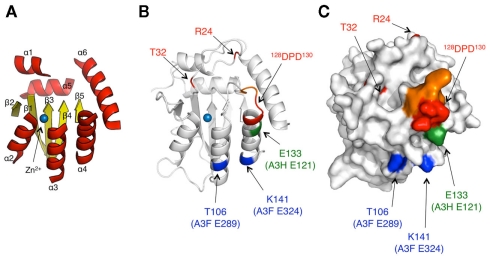
Figure 2.
Model structure of A3G NTD and location of responsible residues for Vif interaction. (A) Ribbon diagram of the A3 core structure for the model is depicted based on the crystal structure of the A3G CTD (PDB ID# 3IR2). The loop regions are removed. The α helices (α1–6) and β sheets (β1–5) are shown in red and yellow, respectively. (B,C) In the structure model, the amino acid determinants, T32, R24, and 128DPD130, for A3G are colored in red. The analogous residues T106 and K141 in A3F E289 and E324 (blue), and E133, corresponding to A3H E121 (green) are also shown. The critical residues, 125YFW127, for nucleic acid binding are colored in orange. (C) Predicted surface of the A3G NTD model is shown. All the critical residues are predicted to be surface exposed.
Three Distinct Interfaces of A3 Z Domains Interact with HIV-1 Vif
The region in A3G responsible for binding to HIV-1 Vif was initially identified by comparative studies of the species specificity of A3G degradation by Vif. A single amino acid difference in hA3G, aspartic acid at position 128 (D128) versus lysine in the A3G of African green monkeys, determines species specificity by influencing Vif–A3G binding (Bogerd et al., 2004; Mangeat et al., 2004; Schrofelbauer et al., 2004; Xu et al., 2004). In addition, extensive site-direct mutagenesis revealed that the 128DPD130 motif of A3G, located at the loop between β4 and α4 shown in red (Figures 2B,C), is crucial for direct binding to HIV-1 Vif (Huthoff and Malim, 2007). Furthermore, there is a report that residue T32, which may be potentially phosphorylated by protein kinase A, is also involved in the Vif–A3G interaction by collaborating with R24 (Shirakawa et al., 2008). All of these critical residues are mapped on the variable loop structure in proximity to the nucleic acid binding surface (Figure 2B).
In contrast, two independent studies have shown that two C-terminal A3F residues, E289 and E324, located in helices α3 and α4, respectively, are critical for the interaction with HIV-1 Vif (Albin et al., 2010; Smith and Pathak, 2010). Interestingly, in the structural model of the A3F CTD, these two residues are close to some negatively charged surface residues although the location of these residues is separate from 128DPD130. In accord with phylogenetic similarities, A3F E289 and E324 are highly conserved in hA3C (E106 and E141) and hA3DE CTD (E302 and E337). These findings suggest that structural features of the Vif-binding interfaces might be conserved among the A3F CTD, A3C, and A3DE CTD, but different from the A3G NTD. In the case of A3H, the interface for Vif is likely to have a surface area close to that of 128DPD130 in the A3G NTD. The A3H gene is polymorphic, with four major haplotypes in humans. The four proteins have different levels of antiviral activity and sensitivity to HIV-1 Vif, in which haplotype II has the highest antiviral activity (Dang et al., 2008a; OhAinle et al., 2008; Harari et al., 2009; Tan et al., 2009; Li et al., 2010). By comparing the A3H variants, Zhen et al. (2010) identified a critical residue, E121, in A3H haplotype II for binding to HIV-1 (Figures 2B,C). Because the identity of amino acid sequences between Z2 and Z3 types, particularly the β4–α4 loop and the α4, is quite low, it is assumed that the putative Vif interface structure of A3H might be different from those of the A3G NTD or A3F CTD.
Residues of HIV-1 Vif that are Critical for Binding A3 Proteins
Extensive mutational analysis of HIV-1 Vif has led to the characterization of several distinct motifs in HIV-1 Vif that are required for formation of the Cul5-based E3 Ub ligase complex and recruitment of human A3 proteins (reviewed in Albin and Harris, 2010; Figure 3). The C-terminal half of Vif contains three conserved motifs: (1) the HCCH domain, which chelates zinc mediates the interaction with Cul5; (2) the SLQ motif, which binds elongin C (Elo C); and (3) the PPLP motif, which is important for Vif dimerization and recruitment of A3G, albeit by an unknown mechanism. Meanwhile, the N-terminal half of Vif is involved in the interaction with A3 proteins. Figure 3 shows the compiled map of the critical residues that have been identified by several groups (Schrofelbauer et al., 2006; Tian et al., 2006; Russell and Pathak, 2007; He et al., 2008; Zhang et al., 2008; Chen et al., 2009; Pery et al., 2009; Zhen et al., 2010; Binka et al., 2011). Overall, it appears that the critical residues for binding to each Z domain type are discontinuous. This suggests that the interfaces in Vif might be determined by conformational constraints.
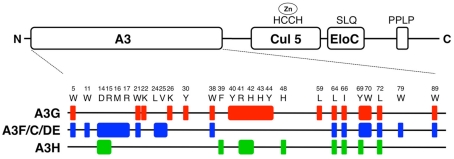
Figure 3.
Map of critical residues in HIV-1 Vif. Important domains in Vif (HIV-1 HXB2) are illustrated in the schematic diagram. The domains labeled “A3,” “Cul5,” and “Elo C” are responsible for Vif’s functional interactions. The “A3” domain is responsible for binding to A3 family proteins. The amino acid sequence motifs, “HCCH,” “SLQ,” and “PPLP” are important for binding to Cul5, and Elo C, and Vif dimerization, respectively. The positions and the names of amino acid residues that were identified by several groups are shown. The residues that were identified as important for forming an interface with human A3 proteins are mapped in Vif, i.e., A3G (red), A3F, A3C, and A3DE (blue), and A3H (green).
Interestingly, some residues are unique among the three types of Z domains, suggesting that such residues might determine binding specificity. For example, K22, K26, Y30, Y40, H43, Y44, and L59 are important for interaction only with A3G, whereas the residues W11, M16, R17, L24, V25, and W79 are required for specific interactions with A3F. These residues tend to have two characteristics: they are positively charged or hydrophobic. Interestingly, the electrostatic charge of the Vif residues is mostly positive whereas the key residues that comprise the A3 interfaces are negatively charged. Thus, such electrostatic interactions might be one of the common features for binding between human A3 proteins and HIV-1 Vif. This could also determine the species-specific interactions between Vif and A3 proteins, as demonstrated in the case of Vif–A3G (Bogerd et al., 2004; Mangeat et al., 2004; Schrofelbauer et al., 2004; Xu et al., 2004). In contrast to the aforementioned regions, there are some aromatic or hydrophobic residues, W5, W21, W38, W70, and W89, which are commonly responsible for binding to two (the A3G NTD and the A3F CTD), or L64, I66, Y69, and L72 to all types of Z domains. These regions could play fundamental roles in maintaining the overall conformation of A3-binding interfaces or could form a partly shared structure of the interface for either two or all Z domains.
Vif-Mediated Ubiquitination/Degradation
Information concerning the specific interaction between Vif and A3G in the E3 Ub ligase complex suggests that there may be a specific mechanism for Ub conjugation of the A3G protein. Previously, our studies using structure-guided mutagenesis demonstrated that four lysine residues (K297, 301, 303, and 334) near the C-terminal end of A3G are critical sites for HIV-1 Vif-mediated A3G ubiquitination and degradation (Iwatani et al., 2009). The data suggested that these sites, which are located on the opposite end of A3G relative to the Vif-binding interface, might be specifically dictated by the rigid structure of the Cul5-based scaffold. In addition, the study clearly demonstrated that the additional residues, particularly lysines near the C-terminal tag of A3G could be potential targets for Vif-induced ubiquitination. However, recently, Wang et al. (2011) showed that a detectable level of Vif-mediated ubiquitination occurs at the N-terminus in addition to certain lysine residues within 20 lysine residues of A3G. In contrast, another group could detect no N-terminal ubiquitination (Dang et al., 2008b). Although these apparent contradictions might be due to differences in the detection level of ubiquitination, it is important to consider the possibility that different tags attached at the C-terminal end of an A3G protein could create potential ubiquitination sites and/or mask the area where the four lysine residues are located (Iwatani et al., 2009). Moreover, while ubiquitination is associated primarily with the lysines in the CTD, the observation that there is also N-terminal ubiquitination could be of interest. The evidences of two distal ubiquitination sites in A3G may provide important structural insight, which implies two distinct types of structural configuration for Vif–A3G interaction. Further studies are needed to clarify Vif-mediated ubiquitination in the context of the E3 Ub ligase complex and to allow us to answer the following two questions: (1) How can we rationalize the relationship between the configuration of the N-terminal end and the proximal Vif-binding interface? (2) In particular, how can the Ub molecules be conjugated at the N-terminus, which is predicted to be a structurally flexible end?
Conclusion
Phylogenetic analyses and genetic studies of A3 and Vif have provided important evidence for three distinct types of interactions between human A3 and HIV-1 Vif proteins, which are determined by the characteristic Z domain types of A3G, A3F/C/DE, and A3H. Further understanding of Vif–A3 interactions could advance efforts to develop novel anti-HIV drugs, which would function as anti-Vif inhibitors. Although presenting a tremendous challenge, complementary studies focusing on the structure of the Vif-interactive A3 domain and Vif are also critical to accelerate future progress in this field.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Judith G. Levin (NIH, NICHD) for helpful discussions. This work was supported in part by a grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a grant for HIV/AIDS research from the Ministry of Health, Labor, and Welfare of Japan.
References
- Albin J. S., Harris R. S. (2010). Interactions of host APOBEC3 restriction factors with HIV-1 in vivo: implications for therapeutics. Expert Rev. Mol. Med. 12, e4. 10.1017/S1462399409001343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin J. S., LaRue R. S., Weaver J. A., Brown W. L., Shindo K., Harjes E., Matsuo H., Harris R. S. (2010). A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J. Biol. Chem. 285, 40785–40792 10.1074/jbc.M110.173161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binka M., Ooms M., Steward M., Simon V. (2011). The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F and APOBEC3H. J. Virol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H. P., Doehle B. P., Wiegand H. L., Cullen B. R. (2004). A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. U.S.A. 101, 3770–3774 10.1073/pnas.0307713101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., He Z., Wang T., Xu R., Yu X. F. (2009). A patch of positively charged amino acids surrounding the human immunodeficiency virus type 1 Vif SLVx4Yx9Y motif influences its interaction with APOBEC3G. J. Virol. 83, 8674–8682 10.1128/JVI.01775-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., Matsuo H. (2008). Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452, 116–119 10.1038/nature06757 [DOI] [PubMed] [Google Scholar]
- Dang Y., Siew L. M., Wang X., Han Y., Lampen R., Zheng Y. H. (2008a). Human cytidine deaminase APOBEC3H restricts HIV-1 replication. J. Biol. Chem. 283, 11606–11614 10.1074/jbc.M708728200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Siew L. M., Zheng Y. H. (2008b) APOBEC3G is degraded by the proteasomal pathway in a Vif-dependent manner without being polyubiquitylated. J. Biol. Chem. 283, 13124–13131 10.1074/jbc.M708728200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa A., Nagata T., Matsugami A., Habu Y., Sugiyama R., Hayashi F., Kobayashi N., Yokoyama S., Takaku H., Katahira M. (2009). Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 28, 440–451 10.1038/emboj.2008.290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goila-Gaur R., Strebel K. (2008). HIV-1 Vif, APOBEC, and intrinsic immunity. Retrovirology 5, 51. 10.1186/1742-4690-5-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari A., Ooms M., Mulder L. C., Simon V. (2009). Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83, 295–303 10.1128/JVI.01665-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Zhang W., Chen G., Xu R., Yu X. F. (2008). Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 381, 1000–1011 10.1016/j.jmb.2008.06.061 [DOI] [PubMed] [Google Scholar]
- Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., Chen X. S. (2008). Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456, 121–124 10.1038/nature07357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H., Malim M. H. (2007). Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 81, 3807–3815 10.1128/JVI.02795-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y., Chan D. S., Liu L., Yoshii H., Shibata J., Yamamoto N., Levin J. G., Gronenborn A. M., Sugiura W. (2009). HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc. Natl. Acad. Sci. U.S.A. 106, 19539–19544 10.1073/pnas.0906652106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmuz A., Chester A., Bayliss J., Gisbourne J., Dunham I., Scott J., Navaratnam N. (2002). An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79, 285–296 10.1006/geno.2002.6718 [DOI] [PubMed] [Google Scholar]
- LaRue R. S., Andresdottir V., Blanchard Y., Conticello S. G., Derse D., Emerman M., Greene W. C., Jonsson S. R., Landau N. R., Lochelt M., Malik H. S., Malim M. H., Munk C., O’Brien S. J., Pathak V. K., Strebel K., Wain-Hobson S., Yu X. F., Yuhki N., Harris R. S. (2009). Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83, 494–497 10.1128/JVI.01976-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. M., Wu L. I., Emerman M. (2010). The range of human APOBEC3H sensitivity to lentiviral Vif proteins. J. Virol. 84, 88–95 10.1128/JVI.01561-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B., Turelli P., Liao S., Trono D. (2004). A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279, 14481–14483 10.1074/jbc.C400060200 [DOI] [PubMed] [Google Scholar]
- Miyagi E., Brown C. R., Opi S., Khan M., Goila-Gaur R., Kao S., Walker R. C., Jr., Hirsch V., Strebel K. (2010). Stably expressed APOBEC3F has negligible antiviral activity. J. Virol. 84, 11067–11075 10.1128/JVI.01249-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- OhAinle M., Kerns J. A., Li M. M., Malik H. S., Emerman M. (2008). Antiretroelement activity of APOBEC3H was lost twice in recent human evolution. Cell Host Microbe 4, 249–259 10.1016/j.chom.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pery E., Rajendran K. S., Brazier A. J., Gabuzda D. (2009). Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J. Virol. 83, 2374–2381 10.1128/JVI.01898-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. A., Pathak V. K. (2007). Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81, 8201–8210 10.1128/JVI.00395-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B., Chen D., Landau N. R. (2004). A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. U.S.A. 101, 3927–3932 10.1073/pnas.0307132101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B., Senger T., Manning G., Landau N. R. (2006). Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80, 5984–5991 10.1128/JVI.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya S. M., Nalam M. N., Nalivaika E. A., Gross P. J., Valesano J. C., Shindo K., Li M., Munson M., Royer W. E., Harjes E., Kono T., Matsuo H., Harris R. S., Somasundaran M., Schiffer C. A. (2010). Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure 18, 28–38 10.1016/j.str.2009.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Shirakawa K., Takaori-Kondo A., Yokoyama M., Izumi T., Matsui M., Io K., Sato T., Sato H., Uchiyama T. (2008). Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nat. Struct. Mol. Biol. 15, 1184–1191 10.1038/nsmb.1497 [DOI] [PubMed] [Google Scholar]
- Simon V., Zennou V., Murray D., Huang Y., Ho D. D., Bieniasz P. D. (2005). Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1, e6. 10.1371/journal.ppat.0010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. L., Pathak V. K. (2010). Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J. Virol. 84, 12599–12608 10.1128/JVI.01108-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Sarkis P. T., Wang T., Tian C., Yu X. F. (2009). Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. FASEB J. 23, 279–287 10.1096/fj.07-088781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Yu X., Zhang W., Wang T., Xu R., Yu X. F. (2006). Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 80, 3112–3115 10.1128/JVI.80.6.3112-3115.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Shao Q., Yu X., Kong W., Hildreth J. E., Liu B. (2011). N-terminal hemagglutinin tag renders lysine-deficient APOBEC3G resistant to HIV-1 Vif-induced degradation by reduced polyubiquitination. J. Virol. 85, 4510–4519 10.1128/JVI.02031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind J. E., Dance G. S., Sowden M. P., Smith H. C. (2003). Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19, 207–216 10.1016/S0168-9525(03)00054-4 [DOI] [PubMed] [Google Scholar]
- Wiegand H. L., Doehle B. P., Bogerd H. P., Cullen B. R. (2004). A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23, 2451–2458 10.1038/sj.emboj.7600246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Svarovskaia E. S., Barr R., Zhang Y., Khan M. A., Strebel K., Pathak V. K. (2004). A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. U.S.A. 101, 5652–5657 10.1073/pnas.0308745101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003). Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 10.1126/science.1092951 [DOI] [PubMed] [Google Scholar]
- Yu X. F. (2006). Innate cellular defenses of APOBEC3 cytidine deaminases and viral counter-defenses. Curr. Opin. HIV AIDS 1, 187–193 10.1097/01.COH.0000221590.03670.32 [DOI] [PubMed] [Google Scholar]
- Zhang W., Chen G., Niewiadomska A. M., Xu R., Yu X. F. (2008). Distinct determinants in HIV-1 Vif and human APOBEC3 proteins are required for the suppression of diverse host anti-viral proteins. PLoS ONE 3, e3963. 10.1371/journal.pone.0003963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen A., Wang T., Zhao K., Xiong Y., Yu X. F. (2010). A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J. Virol. 84, 1902–1911 10.1128/JVI.01509-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B. M. (2004). Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78, 6073–6076 10.1128/JVI.78.13.6908-6914.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]