Abstract
MicroRNAs (miRNAs) are short, non-coding RNAs that post-transcriptionally silence gene expression by binding to target mRNAs. Previous studies have identified the miRNA miR-8 as a pleiotropic regulator of Drosophila development, controlling body size and neuronal survival by targeting multiple mRNAs. Here we demonstrate that miR-8 is required for proper spatial patterning of pigment on the adult abdominal cuticle. Female adult flies lacking miR-8 exhibit decreased pigmentation of the dorsal abdomen, with a pattern of pigmentation similar to wild type flies grown at higher temperatures. This pigmentation defect in miR-8 mutants is independent of the previously reported body size defect, and miR-8 acts directly in the developing cuticle to regulate pigmentation patterning. Loss of miR-8 increases thermal sensitivity of Drosophila for both pigmentation patterning and the ability to eclose. Together, these data suggest miR-8 acts as a buffer to stabilize gene expression patterns in the midst of environmental variation.
Keywords: microRNA, miRNA, pigmentation, patterning, phenotypic plasticity, eclosion
Introduction
The study of adult insect cuticle pigmentation has led to key insights into the genetic differences underlying morphological diversity (Wittkopp and Beldade, 2009). Insects display a wide range of pigmentation patterns, and variation in pigmentation is seen both between species and also within species groups. Body pigments or pigment precursors are produced in the epidermis, and these pigments are then deposited in the hard exoskeleton. The genetic regulation of the pattern and timing of pigment production in adult abdominal segments has been studied most extensively in Drosophila melanogaster (Wittkopp et al., 2003). Sexually dimorphic, D. melanogaster adult males have fully pigmented posterior abdominal segments A5 and A6, whereas adult females have variable degrees of pigmentation in these segments. In addition to genetic differences, temperature is another factor that affects the degree of pigmentation in the posterior segments of female abdomens (David et al., 1990; Gibert et al., 2000). Growth at lower temperatures causes increased pigmentation, whereas higher temperatures result in less pigmentation.
MicroRNAs (miRNAs) are one class of genes that have been implicated in buffering developmental processes against the effects of environmental fluctuations such as temperature changes (Hornstein and Shomron, 2006; Wu et al., 2009). MiRNAs are short, noncoding RNAs that regulate gene expression by binding target mRNAs and preventing translation or destabilizing the mRNA (Du and Zamore, 2005). After processing, mature miRNAs, in concert with the RISC complex, generally bind to their target mRNAs by base pairing with complementary regions in the 3’untranslated region (3’UTR). A single miRNA can bind multiple mRNA targets and thus regulate the expression of multiple genes at one time (Bartel, 2009; Smibert and Lai, 2010). MiRNAs have been implicated in many developmental processes, but prior to this study, no miRNA has been implicated in regulating the complex process of cuticle pigmentation in insects.
Here we report that the miRNA miR-8 is a positive regulator of pigmentation in D. melanogaster, in particular, regulating the spatial patterning of the posterior abdominal segments of adult females. This effect on pigmentation is direct and is independent of the previously reported small body size phenotype of miR-8 mutants. Overall, loss of miR-8 affects the thermosensitivity of Drosophila for both pigmentation and the ability to eclose, suggesting miR-8 acts to buffer these complex processes against environmental variation.
Results
MiR-8 is required for proper spatial patterning of pigment on adult female abdomens
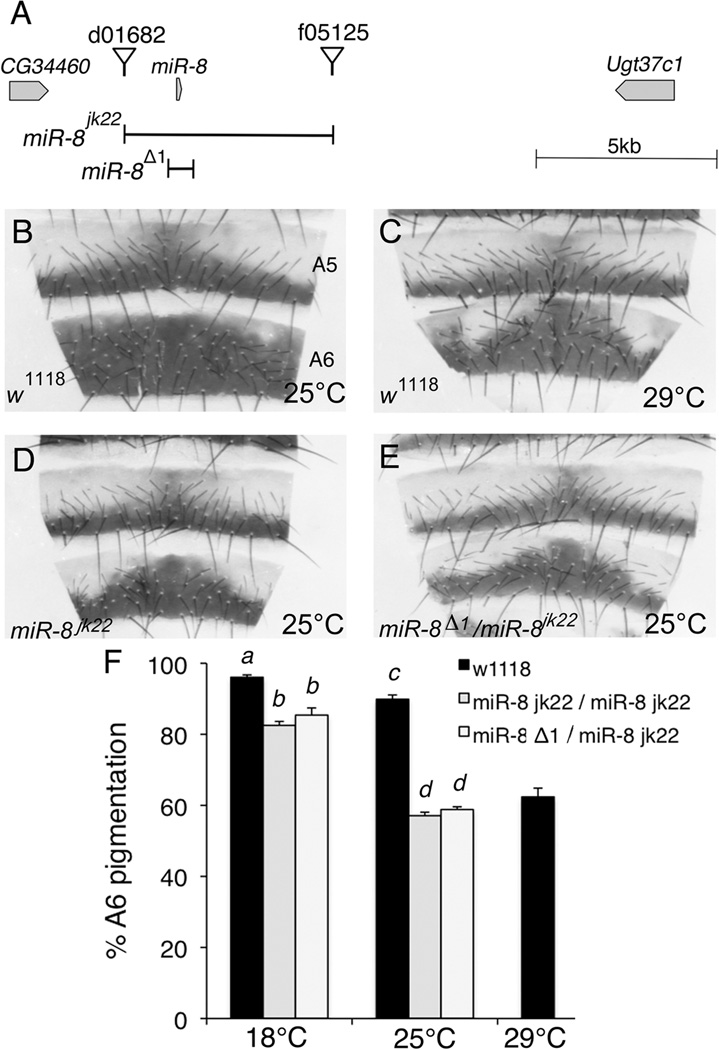
We previously identified miR-8 as a negative regulator of Wingless signaling in a misexpression screen (Kennell et al., 2008). To determine the function of miR-8 in flies, we generated a deletion of the entire predicted miR-8 locus using the FLP/FRT deletion method (Parks et al., 2004; Thibault et al., 2004). The resulting miR-8jk22 allele is a deletion that removed 5.6 kb of genomic DNA surrounding the miR-8 hairpin (Figure 1A). Consistent with previous reports of miR-8 mutants, flies homozygous for the miR-8jk22 allele were proportionately smaller in size, had defective 3rd legs and demonstrated decreased survival and ability to eclose (data not shown; (Karres et al., 2007; Hyun et al., 2009)). In addition to these previously reported phenotypes, we found that female flies lacking miR-8 showed alterations in the spatial pigmentation pattern of the dorsal abdomen, most evident in the A6 segment (Figure 1). At 25°C, the A6 segment of control flies is pigmented throughout most of the segment whereas the A6 segment of miR-8 mutants is widest at the dorsal midline but tapers off laterally (Figure 1B vs. D). This decreased pigmentation lateral to the dorsal midline was completely penetrant in miR-8 mutant females; however, no alteration in pigmentation was evident in mutant adult male fly abdomens (data not shown).
Figure 1. MiR-8 is required for proper spatial patterning of pigment on adult female abdomens.
(A) Schematic representation of the genomic region surrounding the miR-8 locus on Chromosome 2R. The null allele miR-8jk22 was generated with FLP/FRT mediated deletion using the FRT containing elements P{XP}d01682 and PBac{WH}f05125. MiR-8jk22 lacks the indicated 5.6kb region containing the miR-8 hairpin between the two elements. The previously reported miR-8Δ1 allele is missing the 400bp fragment indicated in the schematic (Karres et al., 2007). (B–E) Dorsal abdominal cuticle segments 5 and 6 (A5 and A6) from 4- to 6- day old adult Drosophila melanogaster females. The dorsal midline is in the center of each panel. Wild type (w1118) flies were reared at 25°C (B) or 29°C (C). Flies homozygous for the miR-8jk22 allele (D) or transheterozygous for the miR-8Δ1 and miR-8jk22 (E) were reared at 25°C. To control for genetic background, both miR-8jk22 and miR-8Δ1 lines were backcrossed to w1118 flies for over 20 generations. (F) Percent pigmentation of A6 segments was determined by analyzing 15–20 cuticles for each of the indicated genotype/temperature combinations. Mean percent pigmentation is displayed, with error bars representing SEM. Multivariate analysis revealed a miR-8xtemperature interaction (ANOVA, F2,98=15.24, p<0.001). Shared letters indicate no significant difference (Tukey post-hoc, α=0.05).
To verify the phenotype was not due to genetic background differences, we also analyzed flies that were transheterozygous for two independent deletions of miR-8 and found a similar loss of pigmentation (Figure 1E). Consistent with this finding, flies transheterozygous for the miR-8jk22 or miR-8D1 allele and a deficiency on Chromosome 2R encompassing miR-8 (Df(2R)ED2747) had a similar phenotype, with 59% and 52% A6 pigmentation at 25°C, respectively (data not shown).
Interestingly, loss of miR-8 caused a similar pigmentation pattern to rearing control flies at a higher temperature (29°C, Figure 1C), suggesting that miR-8 may act as a buffer against the effects of temperature on expression of genes involved in pigmentation. We quantified the percent pigmentation of the A6 segment in flies grown at 18°C and 25°C (Figure 1F). Consistent with the hypothesis that miR-8 buffers against the effects of temperature on pigmentation, multivariate analysis revealed a miR-8xtemperature interaction (ANOVA, F2,98=15.24, p<0.0001). Loss of miR-8 resulted in a statistically significant decrease in A6 pigmentation at both 18°C and 25°C (Tukey post-hoc, α=0.01). However, the decrease in pigmentation in miR-8 mutants compared to controls was more pronounced at 25°C than at 18°C, suggesting that loss of miR-8 sensitizes the flies to the effects of growth at higher temperatures.
Pigmentation defect in flies lacking miR-8 is independent of the small body size defect
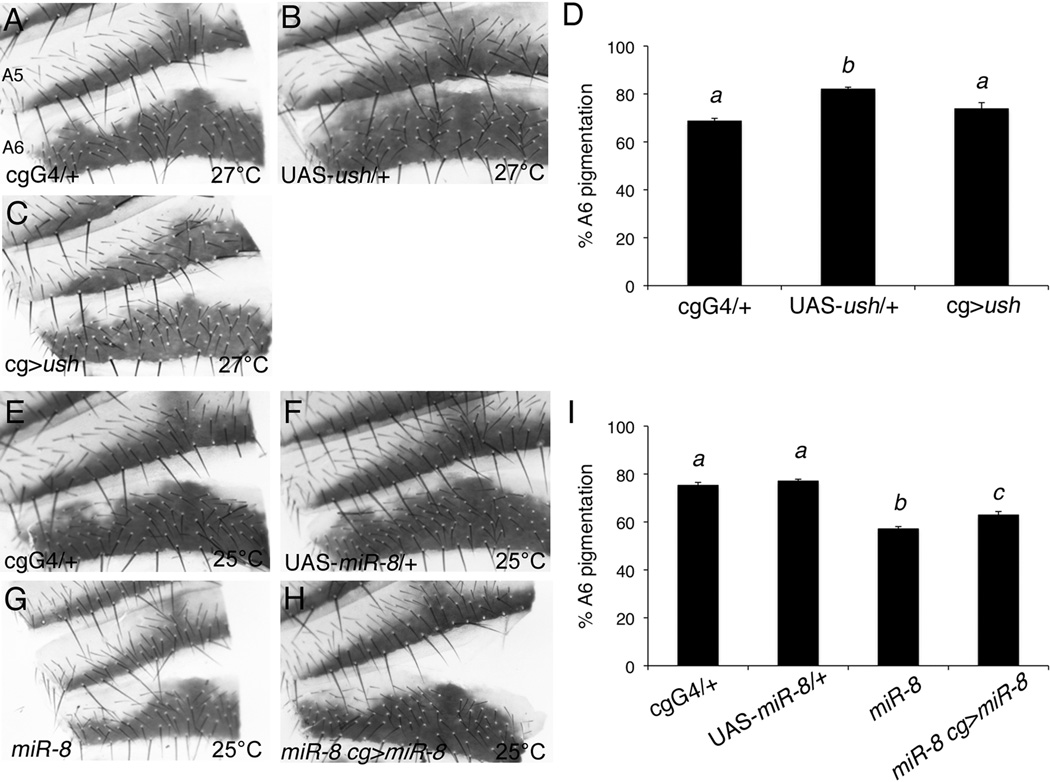
A previous study suggested that loss of miR-8 results in decreased body size due to loss of insulin signaling in the fat body, caused by increased expression of the miR-8 target, u-shaped (ush, (Hyun et al., 2009)). To determine whether the pigmentation defect in miR-8 mutants is related to the proportionately smaller size of the mutants, we analyzed the pigmentation pattern of females overexpressing ush in their fat bodies during development (Figure 2A to D). As predicted, overexpression of ush in the larval fat body using the fat body driver cg-Gal4 at 27°C resulted in flies that were proportionally smaller than control flies (data not shown). Despite the decrease in body size, the pigmentation pattern of female flies overexpressing ush in the fat body was similar to cg-Gal4/+ controls, with no significant difference in pigmentation of the A6 segment (Figure 2C, 2D and data not shown). The parental lines used in these experiments were not introgressed into the w1118 stock to control for genetic background. Consequently, the statistically significant difference in pigmentation between UAS-ush/+ and the other genotypes is most likely due to effects of unknown genetic modifiers on pigmentation patterning in those strains.
Figure 2. Spatial pigmentation patterning in miR-8 mutants is independent of body size.
(A–C and E–H) Dorsal abdominal cuticle segments A5 and A6 from 4- to 6- day old adult females. The dorsal midline is toward the right of each panel for better visualization of the region lateral to the midline. (A) cg-Gal4/+, (B) UAS-ush/+ and (C) cg-Gal4/UAS-ush flies were reared at 27°C. Expression of ush in the fat body by the cg-Gal4 driver resulted in decreased body size (data not shown). (D) Quantification of A6 segment pigmentation from 10–12 cuticles per genotype reared at 27°C. Mean percent pigmentation is displayed, with error bars representing SEM. Though there was a statistically significant difference between all three groups (ANOVA, F2,31=12.8, p<0.0001), shared letters indicate no significant difference (Tukey post-hoc, α=0.05). (E) cg-Gal4/+, (F) UAS-miR-8/+, (G) miR-8jk22/miR-8jk22, and (H) miR-8jk22, cg-Gal4/ miR-8jk22; UAS-miR-8/ + flies were reared at 25°C. Overexpression of miR-8 in the fat body of miR-8 mutant flies rescued the body size defect (data not shown). (I) Quantification of A6 pigmentation from 13–19 cuticles per genotype reared at 25°C. Mean percent pigmentation is displayed, with error bars representing SEM. Though there was a statistically significant difference between all groups (ANOVA, F3,62=75.78, p<0.0001), shared letters indicate no significant difference (Tukey post-hoc, α=0.05).
To verify that the pigmentation defect in miR-8 mutants is not caused by the decrease in body size, we overexpressed miR-8 in the fat bodies of miR-8 mutants (Figure 2E to I). As previously reported, expression of miR-8 in the larval fat body using the cg-Gal4 driver rescued the small body size phenotype caused by loss of miR-8 (data not shown, (Hyun et al., 2009)). However, the pigmentation defect was still present despite the rescue to normal body size (Figure 2H), and there was still a statistically significant decrease in A6 segment pigmentation in fat body rescued flies versus either control (Figure 2I; Tukey post hoc, α=0.01). The slight increase in pigmentation of fat body rescued flies compared to miR-8 mutants may indicate that some, but not all, of the pigmentation defect is due to non-autonomous effects in the fat body. However, the failure to completely rescue the pigmentation defect suggests that miR-8 functions independently of its role in the fat body to regulate the spatial pattern of pigmentation in the female abdomen. We were unable to determine whether the pigmentation defect could be rescued by expressing miR-8 in the developing cuticle. Flies misexpressing miR-8 in the cuticle with the epidermal drivers pnr-Gal4 or en-Gal4 died prior to eclosion or pupation, respectively, even in the absence of endogenous miR-8 expression (data not shown).
Loss of miR-8 in the developing cuticle results in cell-autonomous loss of pigmentation
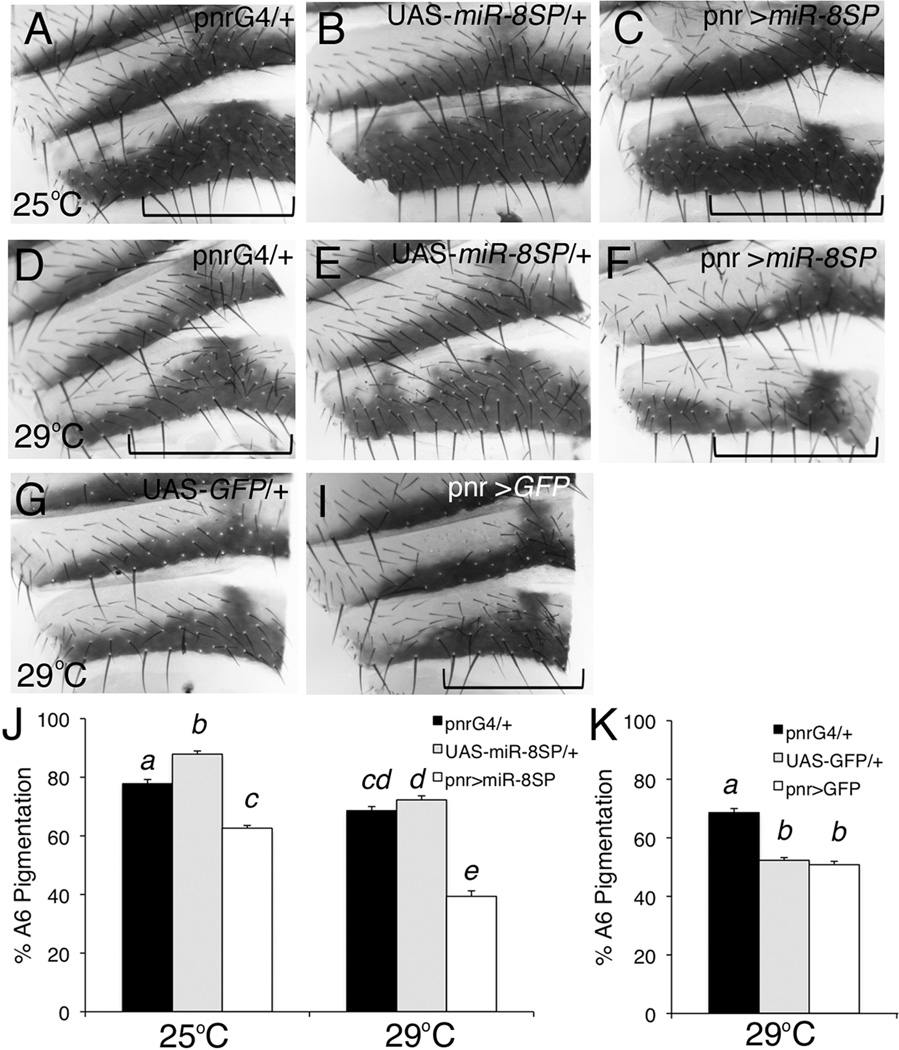
To determine whether loss of miR-8 in the developing cuticle was sufficient for the decreased pigmentation seen in mutants, we expressed a miR-8 sponge to inhibit miR-8 function directly in the developing cuticle (Figure 3; (Loya et al., 2009)). MicroRNA sponges act as competitive inhibitors to prevent the microRNA from targeting endogenous mRNAs (Ebert and Sharp, 2010). The miR-8 sponge transgene was engineered to express, under the control of the UAS promoter, EGFP with 10 target sites complementary to miR-8 in the EGFP 3’UTR (Loya et al., 2009). Expression of the miR-8 sponge in a broad stripe along the dorsal midline by pnr-Gal4 resulted in a decrease in A6 segment pigmentation, similar to the phenotype caused by loss of miR-8 throughout the entire fly (Figure 3C, F). Regions proximal to the dorsal midline, in the region of pnr-Gal4 expression, were affected whereas the central region and lateral edges remained pigmented. Loss of pigmentation is visible in miR-8 sponge expressing flies reared at either 25°C or 29°C. We quantified this effect by measuring pigmentation only in the presumed pnr expression domain of the A6 segment (Figure 3J). Expression of the miR-8 sponge caused a greater decrease in pigmentation at 29°C than at 25°C compared to parental line controls (ANOVA, F2,93=23.85, p<0.0001). This difference is consistent with an interaction between miR-8 and temperature, though this could be explained by greater expression of the miR-8 sponge at 29°C due to UAS/Gal4 system temperature sensitivity. Misexpression of GFP by pnr-Gal4 did not cause decreased A6 segment pigmentation in the region proximal to the dorsal midline (Figure 3I and K), suggesting the effect is specific to the presence of the binding sites for miR-8 in the sponge transgene and not due to overexpression of GFP alone. In addition, loss of miR-8 specifically in the developing cuticle by expression of the miR-8 sponge had no obvious effect on pigmentation patterning in males (data not shown), consistent with the lack of pigmentation defect in male miR-8 mutants.
Figure 3. MiR-8 acts as a cell-autonomous regulator of pigmentation patterning.
Dorsal abdominal cuticle segments A5 and A6 from 4- to 6- day old adult females. The dorsal midline is toward the right of each panel. The pnr-Gal4 transgene drives expression along the dorsal midline of the developing animal, and the UAS-miR-8SP transgene expressed EGFP with 10 miR-8 binding sites in its 3’UTR to soak up endogenous miR-8. Approximate regions of pnr-Gal4 expression are indicated with brackets. (A, D) pnr-Gal4/+, (B, E) UAS-miR-8SP, and (C, F) UAS-miR-8SP/+; pnr-Gal4/UAS-miR-8SP flies were reared at 25°C (A–C) or 29°C (D–F). (G) UAS-GFP/+ and (H) pnr-Gal4/UAS-GFP flies were reared at 29°C. (J–K) Percent pigmentation of the A6 segment in the presumed pnr expression domain was measured using 15–18 cuticles per genotype/temperature combination. Mean percent pigmentation is displayed, with error bars representing SEM. (J) Multivariate analysis indicates there is a significant difference between genotypes (ANOVA, F2,93=197.92, p<0.0001). Shared letters indicate no significant difference (Tukey post-hoc, α=0.05). (K) Though there was a statistically significant difference between genotypes (ANOVA, F=68.99, p<0.0001), shared letters indicate no significant difference (Tukey post-hoc α=0.05).
MiR-8 is expressed in the epidermis underlying the dorsal abdominal cuticle
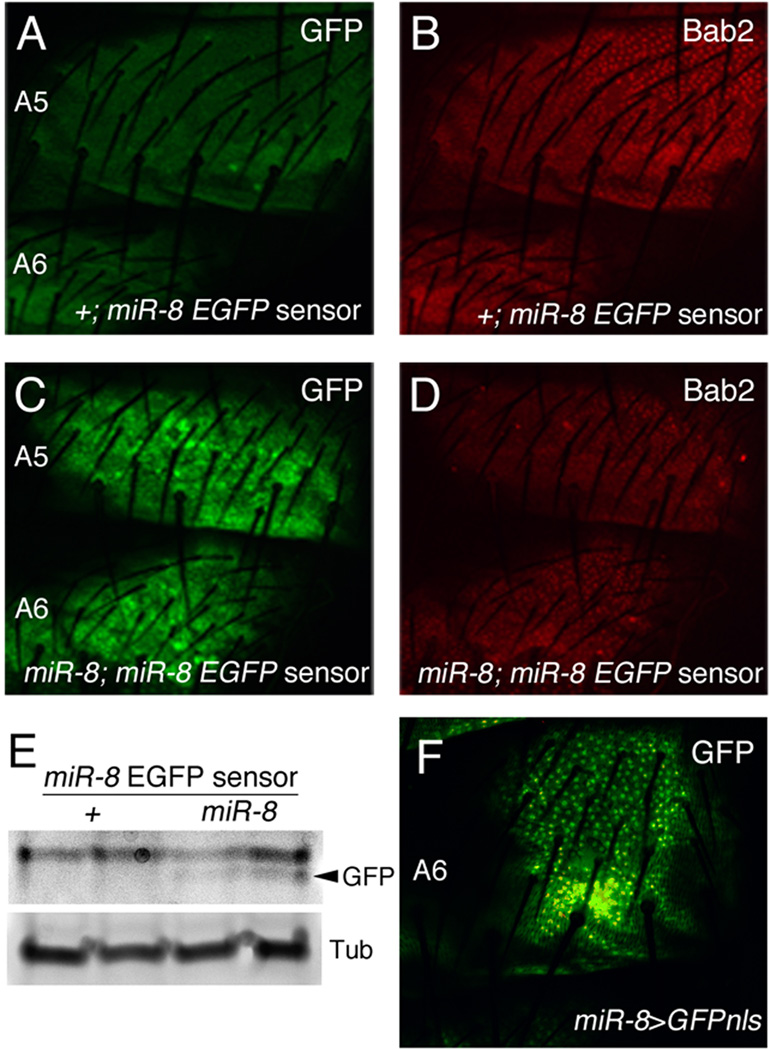
Loss of miR-8 could cause altered spatial patterning of pigmentation either directly in the developing cuticle or indirectly by altering a target in another tissue that has non-autonomous effects. The result of inhibiting miR-8 function specifically in the developing cuticle with expression of a miR-8 sponge is consistent with the hypothesis that miR-8 is functioning directly in the epidermis underlying the developing cuticle. Pigmentation of the dorsal abdomen is spatially patterned during the late stages of pupal development (Wittkopp et al., 2003). To determine the location of miR-8 expression and activity in the developing cuticle of late pupae, we generated an in vivo sensor in which EGFP, containing two miR-8 binding sites in its 3’UTR, is expressed ubiquitously under the control of the α-tubulin promoter (miR-8 EGFP sensor). We expect decreased expression of the EGFP sensor anywhere miR-8 is normally expressed during development, due to targeting by miR-8 (Brennecke et al., 2003; Li and Carthew, 2005). MiR-8 sensor expression was very low in control flies throughout the abdominal dorsal epidermis but was increased in miR-8 mutant epidermis (Figure 4A and C), indicating that miR-8 is expressed and active when cuticle pigment patterning is occurring. The transcription factor Bab2, a known regulator of the pigmentation pathway (Couderc et al., 2002), was visualized as a control to confirm that miR-8 was expressed in the epidermal layer. No consistent change in Bab2 expression was observed with the loss of miR-8 (data not shown), suggesting Bab2 is not a target of miR-8 in the developing cuticle. Increased expression of the miR-8 EGFP sensor in miR-8 mutants was confirmed by immunoblot (Figure 4E). We also used a previously reported miR-8-Gal4 enhancer trap line to verify that miR-8 is expressed in the epidermis of the dorsal abdomen in late pupae (Figure 4F, (Karres et al., 2007)). The expression of miR-8 in the developing cuticle is not sex-specific as miR-8 is expressed throughout the dorsal abdomen of both males and females (data not shown). A previous study has also reported that miR-8 is expressed in the cuticle of third instar larvae (Hyun et al., 2009).
Figure 4. MiR-8 is expressed in the epidermis underlying the dorsal abdominal cuticle.
(A–D, F) Confocal images of the epidermis underlying the dorsal abdominal cuticle in female late pupae (88–96 h APF). All images were taken with the same confocal settings at 40X magnification. (A–E) MiR-8 activity was detected using a miR-8 EGFP sensor transgene. The sensor gene contains EGFP with two binding sites for miR-8 in its 3’UTR, driven by the α-tubulin promoter. Expression of the miR-8 sensor in a wild type background (+/+; miR-8 EGFP sensor/+) (A) was compared with miR-8 sensor expression in miR-8 mutants (miR-8jk22/miR-8jk22; miR-8 EGFP sensor/+) (C). Bab2 expression was used to visualize the epidermis underneath the cuticle (B and D). (E) Western blot analysis of GFP and α-Tubulin from isolated dorsal abdominal cuticles of wild type or miR-8 mutant flies containing the miR-8 sensor. The GFP band is indicated by an arrowhead. A strong non-specific band is located above the GFP band in both wild type and miR-8 mutant backgrounds. (F) GFPnls visualized in dorsal abdominal epidermis of a miR-8-Gal4/ UAS-GFPnls pupa. The miR-8-Gal4 enhancer trap line has an element containing the Gal4 gene upstream of the miR-8 locus and drives expression of Gal4, and hence GFPnls, in a pattern similar to miR-8 expression (Karres et al., 2007).
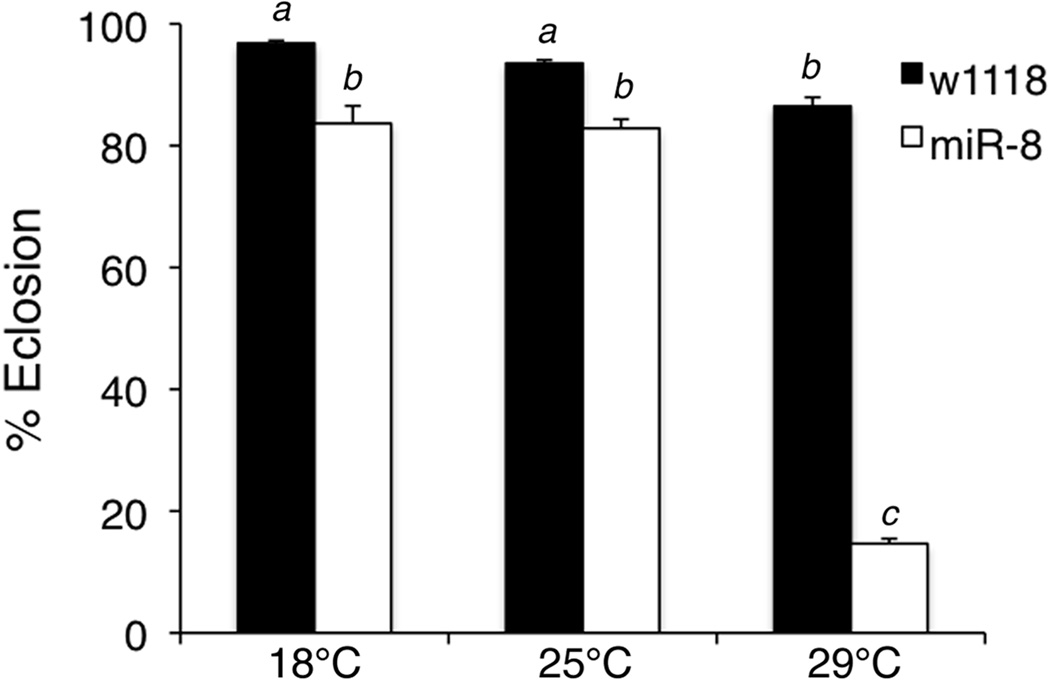
Loss of miR-8 sensitizes flies to the effects of high temperatures on eclosion success
A previous study reported that 14% of miR-8 mutants failed to eclose or died during eclosion, possibly due to elevated apoptosis in the nervous system and impaired motor coordination (Karres et al., 2007). When investigating the effect of miR-8 on spatial pigmentation patterns at different rearing temperatures, we were unable to collect miR-8 mutants grown at 29°C for analysis of cuticle pigmentation. We noticed very few miR-8 mutants successfully emerged from the pupal cases and those that did eclose died within a few hours. Most of the flies that failed to eclose died as late stage pupae (data not shown). To study more closely the interaction between miR-8 and temperature in regulating eclosion rates, we quantified the percent eclosion in control and miR-8 mutant flies reared at different temperatures (Figure 5). Across the temperatures tested, fewer miR-8 mutants successfully eclosed than controls. However, the effect was most pronounced when flies were grown at 29°C, as only 15% of miR-8 mutants successfully eclosed (Figure 5). Multivariate analysis revealed a strong miR-8xtemperature interaction for eclosion (ANOVA, F2,30=138.75, p<0.001). These results suggest that miR-8 mutants are more sensitive to high temperatures, and the underlying defects in leg development and motor activity caused by loss of miR-8 are more severe when flies are grown at 29°C.
Figure 5. Loss of miR-8 sensitizes flies to the effects of high temperatures on eclosion success.
The percentage of eclosed flies from wild type (w1118) and miR-8 mutants (miR-8jk22/miR-8jk22) was determined after rearing flies at 18°C, 25°C or 29°C. Percent eclosion was determined by dividing the number of empty pupal cases by the total number of cases. The experiment was performed with 6 batches of flies per genotype/temperature combination and the mean percent eclosion was calculated. Error bars indicate SEM. Results are representative of three separate trials. Statistical analysis revealed a strong interaction between miR-8 and temperature (ANOVA, F2,30=138.75, p<0.001). Shared letters indicate no significant difference (Tukey post-hoc, α=0.05).
Discussion
Prior studies have identified miR-8 as a regulator of growth, apoptosis and neuronal survival by targeting multiple mRNAs (Karres et al., 2007; Kennell et al., 2008; Hyun et al., 2009; Loya et al., 2009; Vallejo et al., 2011). Our study reveals the previously unidentified role of miR-8 in regulating the spatial patterning of pigmentation. Indeed, miR-8 is the first microRNA to be identified as a regulator of pigmentation in insects. The role of miR-8 in pigmentation patterning appears to be independent of the previously reported growth defect in miR-8 mutants and highlights the pleiotropic functions of the miR-8 gene during Drosophila development. We found that miR-8 is expressed in the pupal epidermis when genes involved in pigmentation and patterning are expressed. In addition, tissue-specific inhibition of miR-8 function by expression of a miR-8 sponge caused a decrease in pigmentation similar to full body miR-8 mutants. Together these data suggest that miR-8 directly regulates the pigmentation pathway in the developing cuticle and does not exert its effects on pigmentation indirectly through effects on another tissue.
Pigmentation is a complex trait and is impacted by multiple factors (Wittkopp et al., 2003). In particular, the female dorsal abdomen of D. melanogaster demonstrates a high degree of phenotypic plasticity in response to changes in temperature (David et al., 1990; Gibert et al., 2000). Interestingly, the spatial pigmentation pattern in female miR-8 mutant abdomens is similar to the pattern of females grown at 29°C. Because miR-8 mutants fail to eclose/survive at 29°C, we were unable to assess the extent of pigmentation in mutants at this high temperature. However, inhibition of miR-8 by overexpression of a miR-8 sponge at 29°C caused cell-autonomous loss of much of the pigment proximal to the dorsal midline. Yet we did not see an obvious difference in the pigmentation patterning of males lacking miR-8, which is consistent with temperature having less impact on male pigmentation patterning. Our analysis of the effects of loss of miR-8 at 18°C versus 25°C suggests that miR-8 interacts with temperature to regulate spatial patterning of pigmentation, and this possible interaction may be indicative of an unexpected role for miR-8 in the plasticity and possible evolution of pigmentation patterning in Drosophila.
In addition to our finding that miR-8 may interact with temperature to regulate pigmentation patterning, we found that the previously reported eclosion defect in miR-8 mutants (Karres et al., 2007) is much more severe when the mutants are grown at 29°C than at 18°C or 25°C. Together these data suggest that miR-8 may buffer against the effects of temperature on gene expression, and, in the absence of miR-8, temperature has a greater impact on the complex genetic pathways regulating pigmentation and eclosure. Other studies of microRNAs have shown a similar role for microRNAs in fine-tuning gene-environment interactions. For example, the microRNA miR-7 appears to buffer gene expression against the effects of temperature fluctuations in Drosophila (Li et al., 2009). Loss of miR-7 affected sensory organ development in the presence of an environmental stressor (temperature fluctuations) but had very little effect under uniform laboratory conditions. Our results are consistent with a model that some microRNAs, such as miR-8 and miR-7, serve to integrate multiple environmental factors that impact developmental processes and buffer against the destabilizing effects of those factors on genetic networks.
The mechanism for miR-8 regulation of pigmentation is not yet known. Individual microRNAs have the potential to target multiple mRNAs. In fact, each of the previously identified phenotypes of miR-8 mutants is caused by upregulation of a different target mRNA (e.g. ush for small body size, atrophin for neurodegeneration; Karres et al., 2007; Hyun et al., 2009; Loya et al., 2009; Vallejo et al., 2011). We have found no evidence that upregulation of the previously identified targets of miR-8 is causing the pigmentation defect in miR-8 mutants (data not shown), suggesting that a novel target of miR-8 is responsible. The increased thermosensitivity of miR-8 mutants may be indicative of miR-8 playing a more general role in temperature response, possibly in regulating chaperones and other genes involved more broadly in thermosensitive developmental processes (Gibert et al., 2007). Loss of the chromatin regulators cramped or corto, or the chaperone Hsp83, causes a similar decrease in pigmentation as seen with loss of miR-8 (Gibert et al., 2007; Gibert et al., 2011), suggesting miR-8 may act in the same pathway as these genes to regulate spatial patterning of pigmentation.
Experimental Procedures
Drosophila genetics
All flies were maintained at 25°C unless otherwise indicated and all flies contained a mutation in the white locus (w1118). pnr-GAL4, cg-Gal4, UAS-GFPnls and Df(2R)ED2747 lines were obtained from the Bloomington Stock Center, miR-8-GAL4 from the Kyoto Stock Center (stock# NP5247) and P{XP}d01682 and PBac{WH}f05125 from the Exelixis Collection at Harvard. The following lines were kindly provided by various researchers: UAS-GFPmiR-8SP#9; UAS-GFPmiR-8SP#10 (Loya et al., 2009), miR-8Δ1 (Karres et al., 2007) and UAS-ush (Tokusumi et al., 2007).
UAS-miR-8 contains a 500bp genomic fragment encompassing the miR-8 locus (Kennell et al., 2008). miR-8jk22 is a null allele that contains a 5.6 kb deletion of the genome that includes the miR-8 locus. The deletion was generated by FLP/FRT based deletion using P{XP}d01682 and PBac{WH}f05125 lines (Parks et al., 2004). The resulting allele contains a transposable element that is a hybrid of the original XP and WH elements and contains two copies of the mini-white gene. Deletion was verified by PCR. The miR-8jk22 and miR-8Δ1 stocks were backcrossed to w1118 flies for over twenty generations to isogenize the stocks with the control w1118 line. The miR-8 EGFP sensor line contains two target sites complementary to miR-8 inserted downstream of the EGFP coding region of tub-EGFP in pCaSpeR4 (Brennecke et al., 2003). Transgenic Drosophila lines were generated by standard P-element mediated genomic integration (Bestgene Inc.), and multiple lines were analyzed for each construct.
Abdominal cuticle preparation and immunofluorescence
Adult female flies were fixed in 10% glycerol in ethanol 4- to 6- days after eclosion. Abdominal cuticles were dissected, mounted in PVA mounting medium (BioQuip) and imaged with a Leica S8APO dissecting scope and Leica DFC295 camera. All settings were kept constant between images. Quantification of dorsal A6 segment pigmentation was performed using NIH ImageJ software. Images of ten to twenty cuticles were analyzed for each genotype/temperature combination to determine percent pigmentation. Quantification of pigmentation in Figures 1 and 2 was conducted by measuring the percent pigmentation of the A6 segment for half of the tergite, from the dorsal midline to one lateral edge. Percent pigmentation was determined in Figure 3 for the region from the dorsal midline to halfway to one lateral edge of the tergite to approximate the expression domain of pnr-Gal4.
To visualize gene expression, late pupal cuticles (APF 88–96 h; determined by morphological markers) were dissected in 1X PBS, fixed in 4% formaldehyde and blocked with normal donkey serum. Fixed cuticles were incubated with rat anti-Bab2 (1:1000, Frank Laski) followed by incubation with Alexa Fluor 568 anti-rat (1:500, Invitrogen). Pupal cuticles were mounted in vectashield (Vector Labs) and imaged using a Nikon PCM2000 confocal microscope.
Immunoblotting
Dorsal abdominal cuticles from female late pupae (APF 88–96 h) were homogenized in lysis buffer (125 mM Tris, pH 6.8; 6% SDS). Following centrifugation the supernatant was combined with equal amounts of loading buffer (125 mM Tris, pH 6.8; 6% SDS; 20% glycerol; 0.04% Bromophenol Blue; 10% β-mercapatoethanol), and heated to 95°C. Samples were loaded onto Precise Protein gels (Pierce) and transferred to Low fluorescence PVDF membrane (Pierce). Primary antibodies used were rabbit anti-GFP (1:500; Santa Cruz) and mouse anti-α-tubulin (1:500; Sigma) followed by Dylight 488 conjugated anti-mouse or anti-rabbit secondary antibodies (1:2500; Immunoresearch Labs). Dried blots were imaged with a VersaDoc 4000MP (Biorad).
Eclosion assay
Ten each of males and females were added to new vials and incubated at 25°C for 3 days. Adults were discarded and vials were transferred to the indicated temperature. Pupal cases were counted 15, 16 and 28 days after vials were transferred to 29°C, 25°C, and 18°C, respectively. Empty pupal cases were counted as successful eclosion events, whereas full or partially filled cases were considered unsuccessful. Six vials were prepared for each genotype/temperature combination and the reported results presented are representative of three separate trials.
Statistical analysis
The calculated values for A6 pigmentation were arcsine transformed prior to statistical analysis. Multivariate analysis was performed using the statistical tools on the VassarStats website (http://faculty.vassar.edu/lowry/VassarStats.html).
Acknowledgments
We thank D. VanVactor for miR-8 sponge lines; S. Cohen for the miR-8Δ1 line; R. Schulz for the UAS-ush line; F. Laski for the Bab2 antibody; the Bloomington Stock Center, Kyoto Stock Center and Exelixis Collection at Harvard for various lines; the Cadigan lab, T. Wittkopp and J. Davis for helpful discussions. This work was supported by an NIH NRSA individual postdoctoral fellowship (J.A.K.) and start-up funds from Vassar College.
Grant sponsor: NIH-NIGMS; Grant number: F32GM074465
References
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Couderc JL, Godt D, Zollman S, Chen J, Li M, Tiong S, Cramton SE, Sahut-Barnola I, Laski FA. The bric a brac locus consists of two paralogous genes encoding BTB/POZ domain proteins and acts as a homeotic and morphogenetic regulator of imaginal development in Drosophila. Development. 2002;129:2419–2433. doi: 10.1242/dev.129.10.2419. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P, Gauthier JP. Abdominal Pigmentation and Growth Temperature in Drosophila-Melanogaster - Similarities and Differences in the Norms of Reaction of Successive Segments. Journal of Evolutionary Biology. 1990;3:429–445. [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert JM, Karch F, Schlotterer C. Segregating variation in the polycomb group gene cramped alters the effect of temperature on multiple traits. PLoS genetics. 2011;7 doi: 10.1371/journal.pgen.1001280. e1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert JM, Peronnet F, Schlotterer C. Phenotypic plasticity in Drosophila pigmentation caused by temperature sensitivity of a chromatin regulator network. PLoS genetics. 2007;3:e30. doi: 10.1371/journal.pgen.0030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert P, Moreteau B, David JR. Developmental constraints on an adaptive plasticity: reaction norms of pigmentation in adult segments of Drosophila melanogaster. Evolution & development. 2000;2:249–260. doi: 10.1046/j.1525-142x.2000.00064.x. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. Canalization of development by microRNAs. Nature genetics. 2006;38 Suppl:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee JH, Jin H, Nam J, Namkoong B, Lee G, Chung J, Kim VN. Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell. 2009;139:1096–1108. doi: 10.1016/j.cell.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15417–15422. doi: 10.1073/pnas.0807763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loya CM, Lu CS, Van Vactor D, Fulga TA. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nature methods. 2009;6:897–903. doi: 10.1038/nmeth.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Smibert P, Lai EC. A view from Drosophila: multiple biological functions for individual microRNAs. Seminars in cell & developmental biology. 2010;21:745–753. doi: 10.1016/j.semcdb.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Tokusumi T, Russell M, Gajewski K, Fossett N, Schulz RA. U-shaped protein domains required for repression of cardiac gene expression in Drosophila. Differentiation; research in biological diversity. 2007;75:166–174. doi: 10.1111/j.1432-0436.2006.00120.x. [DOI] [PubMed] [Google Scholar]
- Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. The EMBO journal. 2011;30:756–769. doi: 10.1038/emboj.2010.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Seminars in cell & developmental biology. 2009;20:65–71. doi: 10.1016/j.semcdb.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Carroll SB, Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends in genetics : TIG. 2003;19:495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- Wu CI, Shen Y, Tang T. Evolution under canalization and the dual roles of microRNAs: a hypothesis. Genome research. 2009;19:734–743. doi: 10.1101/gr.084640.108. [DOI] [PMC free article] [PubMed] [Google Scholar]