Abstract
We have recently established a system for purifying minichromosome in a native state from S. cerevisiae. This system is extremely efficient, and a single-step purification yields samples with sufficient purity and quantity for mass spectrometry (MS) analysis of histones and non-histone proteins tightly associated with the minichromosome. The templates can also be used in various biochemical assays in vitro, such as transcription and recombination, and might be applicable to allow EM or other studies to be performed.
Keywords: chromatin, minichromosome, purification, histone modifications, episome, mass spectrometry, DNA replication, replication origin, TRP1-ARS1, FLAG tag
1. Introduction
The identification of chromatin-associated proteins and their modification status greatly facilitates the elucidation of the molecular mechanisms underlying DNA-dependent processes such as transcription, DNA replication and chromosome segregation. However, it has been difficult to obtain chromatin templates of specific genomic regions in sufficient purity and quantity for these analyses. We have recently established an improved TRP1-ARS1 system (1) using multimerized lac operators and an epitope-tagged LacI that allows purification of the mini-circles from S. cerevisiae in a native state (2) (see note 1). The purified samples are suitable for direct mass spectrometry (MS) analysis for the identification of chromatin-associated proteins and the detection of post-translational modifications on these proteins, as well as chromatin templates in biochemical assays in vitro.
2. Materials
2.1. Growing and harvesting cells
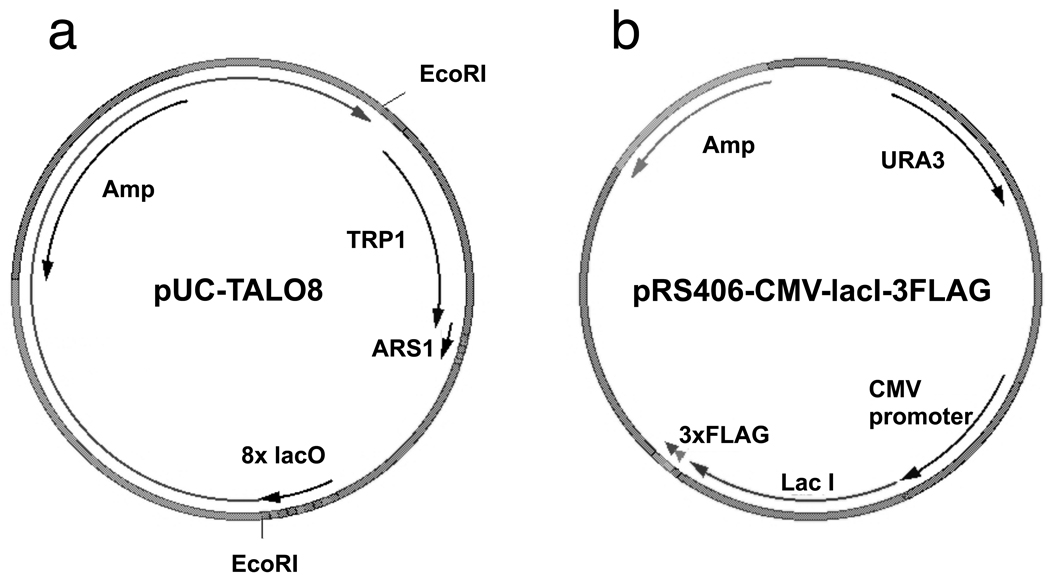
Yeast strain harboring pRS406-CMV-LacI-3FLAG and TALO8 (TRP1-ARS1-Lac Operator 8) (Figure 1).
TALO8 is propagated in E. coli as pUC-TALO8 as shown in Figure 1a. In order to preserve eight copies of lac operator (8xlacO) sequences, Stbl2 (Invitrogen) is used as a bacterial host for plasmid preparation and 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) is added to all bacterial culture media. After each preparation of pUC-TALO8, PCR is performed using primers surrounding 8xlacO (5'-CAGCTATGACCATGATTACG and 5'-AATGCGAGATCCGTTTAAC) to ensure that the 8xlacO are not lost by recombination (with intact lacO, the PCR product should be about 300 bp). The plasmid backbone is digested with EcoRI enzyme, ~1.7 kb fragment corresponding to TALO8 is gel purified, ligated in vitro under conditions that promote intramolecular ligation and then transformed into a yeast strain expressing epitope-tagged LacI. The sequence of the plasmid is available at http://labs.fhcrc.org/tsukiyama/protocols/TALO8_Protocol.pdf (a pdf file).
pRS406-CMV-LacI-3FLAG (LacI expression vector). As shown in Figure 1b, LacI protein with three copies of FLAG epitope at the C-terminal end is expressed from the plasmid pRS406-CMV-LacI-3FLAG. This plasmid is linearized within the URA3 gene by BstBI digestion and transformed into yeast. Integration of the plasmid is confirmed by detection of 3xFLAG-LacI by western blotting using FLAG M2 antibody (Sigma, F3165), which should recognize a band about 45 kDa. The sequence of the plasmid is available at http://labs.fhcrc.org/tsukiyama/protocols/TALO8_Protocol.pdf (a pdf file).
Yeast media: synthetic media without tryptophan (3).
Figure 1. Map of pUC-TALO8 and pRS406-CMV-LacI-3FLAG.
(a) TALO8 is inserted into the pUC18 vector to form pUC-TALO8 plasmid. To preserve eight copies of Lac operators, special care is taken during propagation of pUC-TALO8. TALO8 is released from pUC18 vector by EcoRI digestion. (b) LacI protein followed by three copies of FLAG epitope is expressed from the CMV promoter.
2.2. Preparation of whole cell extract
200 mM phenylmethanesulfonyl fluoride (PMSF) in 100% methanol
Buffer H 150: 25 mM HEPES KOH pH 7.6, 2 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% glycerol, 150 mM KCl, 0.02% NP40, freshly supplemented with 2 mM DTT. (see note 2)
100× Protease inhibitors: 100 mM PMSF, 200 µM pepstatin, 60 µM leupeptin, 200 mM benzamidine, 200 µg/ml chymostatin A in 100% methanol. Store at −20 °C.
100× Phosphatase inhibitors: 200 mM imidazole, 100 mM sodium fluoride, 115 mM sodium molybdate, 100 mM sodium orthovanadate, 400 mM sodium tartarate dihydrate in H2O. Store at −20 °C. 5.
1000× Phosphatase inhibitors: 2.5 mM (−)-p-bromotetramisole oxalate, 0.5 mM cantharidin, 500 nM microcystin in DMSO. Store at −20 °C.
1000× Histone deacetylase inhibitors: 500 µM Trichostatin A (Sigma), 25 mM Sirtinol (Calbiochem) in DMSO. Store at −20 °C.
Zirconia/silica beads (Research Products International Corp).
2ml screw cap tube.
2.3. Coupling anti-FLAG M2 antibody with magnetic beads
Dynabeads Protein G (Invitrogen).
Anti-FLAG M2 antibodies (Sigma, F3165).
0.1 M sodium phosphate pH 7.0.
0.1 M sodium phosphate pH 7.0, 0.01% Tween-20.
0.2 M triethanolamine pH 8.2 (Sigma).
20 mM Dimethyl pimelimidate (Sigma), 0.2M triethanolamine, pH 8.2. Freshly prepared.
50 mM Tris-HCl pH 7.5.
PBST: Phosphate buffered saline with 0.01% Tween-20.
Magnetic particle concentrator (MPC, Invitrogen).
2.4. Purification of TALO8 from cell extract
Buffer H 150: 25 mM HEPES KOH pH 7.6, 2 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% glycerol, 150 mM KCl, 0.02% NP40.
Buffer H 300: 25 mM HEPES KOH pH 7.6, 2 mM MgCl2, 0.5 mM EGTA, 0.1 mM EDTA, 10% glycerol, 300 mM KCl, 0.02% NP40.
Rinse Buffer: 25 mM HEPES KOH pH 7.6, 2 mM MgCl2, 10% glycerol, 150 mM KCl.
Elution Buffer: 50 mM Ammonium bicarbonate, 0.1% Rapigest (Waters Corporation).
3. Methods
3.1. Growing and harvesting cells
Grow yeast cells harboring TALO8 and pRS406-CMV-LacI-3FLAG to an appropriate cell density (OD660=0.7~1.2) in media lacking tryptophan.
Spin cells down at ~6,000 g for 5 minutes at 4 °C.
Suspend cells in ~20× packed cell volume of ice cold water supplemented with 2 mM phenylmethanesulfonyl fluoride (PMSF) and pellet them as above.
Suspend cells in ~10× packed cell volume of Buffer H 150 freshly supplemented with 1× protease inhibitors, phosphatase inhibitors and histone deacetylase inhibitors, and pellet them in 50 ml Falcon tubes at ~2,500 g for 5 minutes at 4 °C.
Whole cell extracts can be prepared immediately or the cell pellet can be frozen in liquid nitrogen and stored at −80 °C.
3.2. Preparation of whole cell extract
All the steps are done on ice or at 4 °C.
Thaw cells in room temperature water, then an equal volume of Buffer H 150 freshly supplemented with 1× protease inhibitors, phosphatase inhibitors and histone deacetylase inhibitors.
Aliquot equal volumes of cell suspension and zirconia/silica beads to fill up screw capped 2 ml tubes. Beat cells for 3–5 minutes using Mini-Beadbeater-96 (BioSpec Products) or equivalent until majority of the cells are broken as assessed under a light microscope.
Puncture holes at the bottom and top of the tubes, and place them on 12 × 75 mm tubes using microfuge tube locks. Recover the cell extract by spinning the tubes at ~285 g for 3 minutes.
Alternatively, frozen cell pellet in 3.1, step 5 can be ground in a blender or coffee grinder in the presence of dry ice for 20 minutes. Frozen ground cells are then thawed in Buffer H 150 freshly supplemented with 1× protease inhibitors, phosphatase inhibitors and histone deacetylase inhibitors.
Clarify the cell extract by centrifugation at ~125,000 g for 90 minutes in Beckman SW41 or equivalent at 4 °C.
Soluble cell extract is drawn out through a syringe. Insert needle just above the top of precipitates and slowly draw whole cell extract. Avoid taking up soft, fluffy precipitates on the top of firmly packed precipitates.
The cell extract can be used immediately in purification or be frozen in liquid nitrogen and stored at −80 °C.
3.3. Coupling anti-FLAG M2 antibody with magnetic beads
Typically, the antibody-conjugated beads are prepared immediately before use. Cross-linking of FLAG M2 antibody to beads is not essential for purification, but significantly reduces the amount of contaminating proteins in eluates. For each liter of cells from which extract was prepared, 25 µl of Dynabeads Protein G beads slurry and 11.5 µg of anti-FLAG M2 antibodies are used. Concentrate magnetic beads on a magnetic particle concentrator (MPC), then suspend and concentrate beads twice in 0.5 ml of 0.1 M sodium phosphate pH 7.0.
Mix antibody and magnetic beads in 0.1 M sodium phosphate pH 7.0 and gently shake them at room temperature for 30 minutes.
Suspend and concentrate beads twice in 0.5 ml of 0.1 M Sodium Phosphate pH 7.0, 0.01% Tween-20.
Suspend and concentrate the beads twice in 1ml 0.2 M triethanolamine pH 8.2.
Suspend the beads in 1 ml of 20 mM dimethyl pimelimidate in 0.2 M triethanolamine pH 8.2 (prepared fresh), and incubate them for 30 minutes at room temperature with constant rotational mixing.
Concentrate and suspend the beads in 1 ml 50 mM Tris-HCl pH 7.5 and incubate for 15 minutes at room temperature with constant rotational mixing.
Wash the beads three times in 1 ml PBST. The beads are ready for use in purification.
3.4. Purification of TALO8 from cell extract
Take small aliquots of the whole cell extract prior to mixing with beads for western blots and DNA analyses to monitor purification efficiency. Incubate antibody-conjugated beads and cell extract at 4 °C for 3 hours with constant rotational mixing.
Concentrate beads on an MPC and transfer the supernatant (unbound material) into a fresh tube. Ensure that the vast majority of beads are concentrated to the wall to minimize losses. At the same time, do not leave the beads on MPC for more than a few minutes as the beads will clump up, leading to increased background. Save small aliquots of unbound material for western blots and DNA analyses to monitor purification efficiency, freezing the rest in liquid nitrogen and saving them for troubleshooting purposes if required. Suspend the beads in 1 ml Buffer H 150 and transfer them into a siliconized 1.7 ml microfuge tube.
Suspend and concentrate the beads three times in 1ml Buffer H 150, freshly supplemented with protease inhibitors, phosphatase inhibitors, histone deacetylase inhibitors and 2mM dithiothreitol (DTT).
Suspend the beads in 1m Buffer H 300 freshly supplemented with protease inhibitors, phosphatase inhibitors, histone deacetylase inhibitors and 2 mM DTT and rotate them at 4 °C for 5 minutes. Concentrate the beads on MPC, and repeat this wash step three more times, for a total of 4 times.
Suspend and concentrate the beads three times in 1 ml of Rinse Buffer.
Mix beads with 50 µl Elution Buffer and agitate them vigorously for 30 minutes at room temperature. Concentrate the beads on an MPC, transfer the supernatant into a fresh microfuge tube. Take small aliquots from the eluted samples for western blots and DNA analyses to monitor purification efficiency, then immediately freeze the rest in liquid nitrogen and store them at −80 °C. Elution is performed a total of four times, with the first two done for 30 minutes each and the subsequent two for 15 minutes each (see note 1). Freeze the beads in liquid nitrogen and store them at −80 °C for troubleshooting purposes if required. The samples are ready for MS analyses.
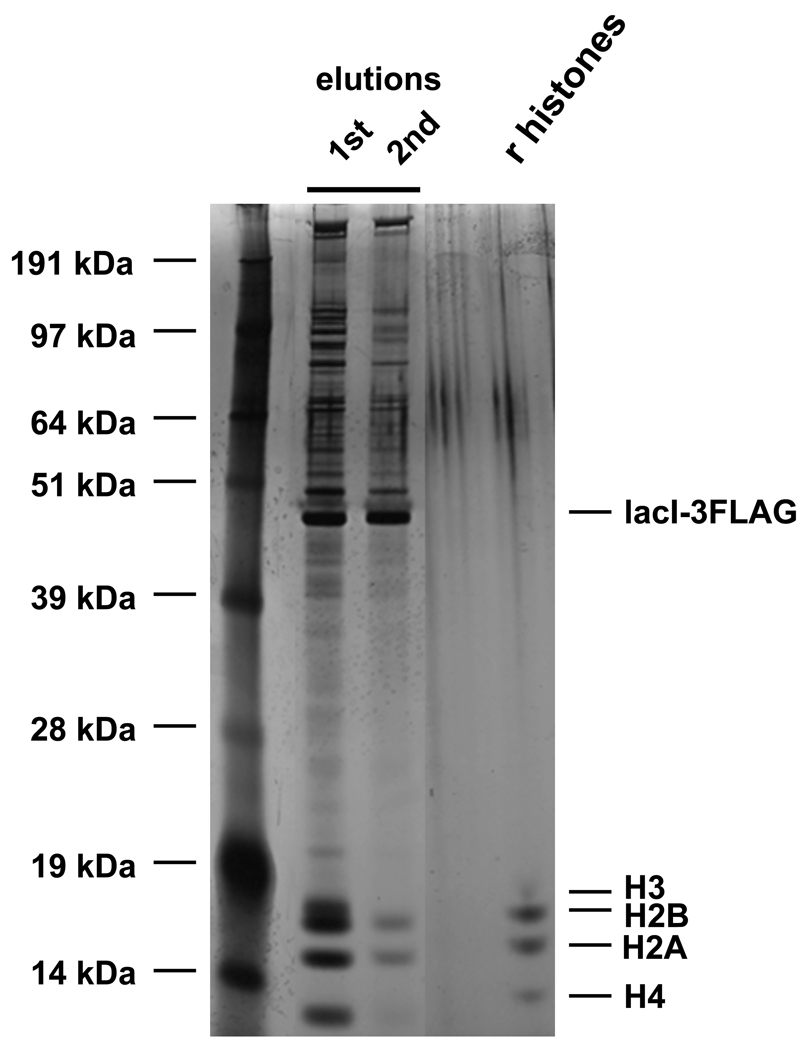
Determine the yield and purity of the sample by DNA preparation and SDS-PAGE gel electrophoresis followed by silver staining. Typical yield of TALO8 from 10 liter culture at OD660=0.7 is about 2–4 µg of chromatin (1–2 µg of core histones). See note 3 and 4 for troubleshooting.
If TALO8 is purified as a template for biochemical assays, such as in vitro transcription, it can be eluted by 3xFLAG peptide [(Met-Asp-Tyr-Lys-Asp-His-Asp-Gly-Asp-Tyr-Lys-Asp-His-Asp-Ile-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys), suspended at 5 mg / ml in Buffer H 0.1], which allows elution of the template in a native state. After the step 4 above, rinse beads three times with Buffer H with a desired salt concentration (usually 100–150 mM KCl), then incubate the beads with 0.5 mg/ml 3xFLAG peptide in the same buffer for 30 minutes with constant and gentle shaking at 4 °C. Repeat elution for total 3–4 times. For analyses of non-histone proteins associated with the templates, see note 5.
Figure 2. Typical SDS-PAGE gel of proteins eluted from purified TALO8.
Proteins eluted off TALO8 from ~700 ml culture were loaded onto NuPAGE 12% Bis-Tris gel (Invitrogen) and silver stained. “r histones” denotes 25 ng recombinant yeast histones.
ACKNOWLEDGMENTS
This work was supported in part by grants from NIGMS and Leukemia & Lymphoma Society to S.B. and T.T and a Beckman Young Investigator Award to S.B.
Footnotes
Affinity purification of TRP1-ARS1 minichromosome from S. cerevisiae using lacI was first reported by R. Simpson's lab to determine the stoichiometry of the Tup1 general transcriptional repressor to nucleosomes (1). In this system, point mutations were introduced within the ARS1 to create a single lac operator, and lacI was expressed and purified from E. coli as a recombinant protein to create an affinity resin in vitro. In our system, we have inserted eight copies of lac operators within a nucleosome-free region of the template (4), and expressed FLAG-epitope tagged lacI in vivo. These changes significantly increased the yield of the minichromosome. We have also tested Tet repressor and LexA as affinity modules to purify the TRP1-ARS1 mini-circle, but the lacI system was the most efficient in purification (data not shown).
Another strength of the TALO8 system is the purity of the samples. Southern blotting (data not shown) and SDS-PAGE gel followed by silver staining (Figure 2) showed that the level of contamination of non-specific DNA and proteins in eluted fractions is quite low. Using Rapigest in the elution buffer enables the direct use of eluted samples in the MS analyses.
All solutions are prepared in water that is highly purified (resistivity: ~18.2 ΩM-cm).
When the yield of TALO8 is too low, consider the following: low yield of TALO8 can be caused by inefficiency in either cell breakage, binding of TALO8 to antibody-coated magnetic beads, or elution of TALO8 from beads. To monitor cell breakage, examine cells under microscope before clarifying the cell extract by high-speed spin. The binding and elution efficiencies of TALO8 should be monitored in two ways: western blotting and DNA analysis using the starting materials, unbound extract, eluates and the beads after the final elution to determine how much lacI protein and TALO8, respectively, were bound and eluted during the process. If the efficiency of elution is low, increase the volume of Elution Buffer to 100 µl.
When the purity of the sample is too low (too much contaminant), consider the following. High levels of contamination can be caused by inefficient washing of beads before elution. Try washing beads more extensively or at a higher salt concentration. We have also found that excess levels of lacI lead to a large amount of proteins non-specifically co-purifying. If the level of free lacI in eluates is too high or the copy number of the template is too low, the expression construct for lacI (pRS406-CMV-LacI-3FLAG) needs to be modified to achieve optimal levels of expression. If the purification is successful, histone proteins should be the major bands when analyzed by silver stained gel (Figure 2).
Insertion of model genes or specific cis-elements into TALO8 will enable identification of proteins and their modification status that are specifically associated with these modules. For example, a modified TALO8 with a centromere sequence has been used to identify a previously unknown kinetochore protein (5). This purification required decreased salt concentrations throughout the process. To identify non-histone proteins associated with TALO8 or its derivative, optimal salt concentrations for binding and washing should be carefully determined. If the protein of interest survives 250 mM KCl wash, highly clean template can be obtained. If a lower salt concentration is required to retain the protein of interest, quantitative MS analysis, with mock purification as a negative control, maybe required for identification of proteins specifically associated with the template (5).
References
- 1.Ducker CE, Simpson RT. The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. Embo J. 2000;19:400–409. doi: 10.1093/emboj/19.3.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unnikrishnan A, Gafken PR, Tsukiyama T. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol. 2010;17:430–437. doi: 10.1038/nsmb.1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams A, Gottschling D, Stearns T. Methods in yeast genetics. New York: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 4.Thoma F, Bergman LW, Simpson RT. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984;177:715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- 5.Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]