Abstract
Acute myeloid leukemia (AML) is a heterogeneous disease with highly variable prognosis. Identification of recurring chromosomal translocations provides some prognostic information for individual AML subjects. Population based gene expression profiling studies also identified abnormalities relevant to prognosis. Such studies associate increased expression of a set of homeodomain transcription factors with poor prognosis in AML. This set includes HoxB3, B4, A7–11 and Meis1, which are dysregulated as a group in the bone marrow in poor prognosis AML. Aberrant expression of these homeodomain transcription factors is found in AML with chromosomal translocations involving the MLL, MYST3 and CREBBP genes, and in a poor prognosis subset with normal cytogenetics. Studies in murine models suggest that Hox protein overexpression is functionally significant for myeloid malignancies. Overexpression of individual Hox proteins expanded various bone marrow populations in vitro, leading to myeloproliferation and in some cases differentiation block and AML in vivo. Therefore, dysregulated expression of key Hox target genes may contribute to adverse prognosis in AML. Identification of these genes will provide insights into the pathobiology of prognosis in AML. Studies are beginning to identify Hox target genes which may be rational targets for therapeutic approaches to this poor prognosis leukemia subset.
Keywords: leukemogenesis, gene regulation, transcription factor, Hox, myelopoiesis
I. HOX PROTEINS ARE HOMEODOMAIN TRANSCRIPTION FACTORS
A. Characteristics of Hox Proteins
Gene expression profiling studies in human AML correlated increased expression of a subset of Hox proteins with poor prognosis.1–5 These studies led to increased interest in understanding this family of homeodomain transcription factors and their roles in normal and leukemic myelopoiesis.
1. Highly Conserved Homeodomain Transcription Factors
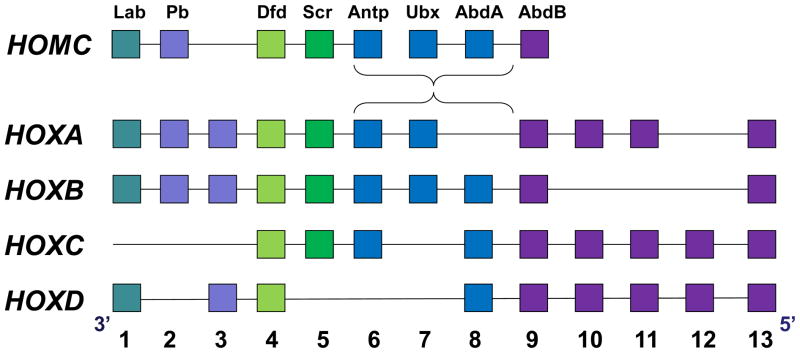
HOX genes encode homeodomain transcription factors which are highly conserved from Drosophila to man. HOX genes were initially identified due to homology to Drosophila HOMC genes, and naming of mammalian HOX genes follows this homology (Figure 1) (reviewed 6,7). Human and murine HOX genes are arranged in four groups (referred to as paralog groups A–D) which are found on four different chromosomes. Numbering of individual HOX genes follows homology between groups, with greatest similarity between Hox proteins of different groups with the same number. During embryogenesis, Hox1–4 are most highly expressed in the head, Hox5–7 in the thorax and Hox8–11 in the abdomen and pelvis. 6,7 This tight spacial regulation is hypothesized to result in regulation of organ specific genes by groups of Hox proteins, although few such genes have been identified.
Figure 1. Homology of mammalian HOX genes to Drosophila HOMC genes.
Mammalian HOX genes are found on four different chromosomes which are arranged similarly to homologous Drosophila HOMC genes.
Similarly, HOX gene transcription during definitive hematopoiesis is tightly regulated, but in a temporal manner. Maximal expression of Hox1–4 occurs in hematopoietic stem cells (CD34+CD38− in humans), with down regulation of these genes during CD38+ differentiation. Hox7–11 expression is maximal in lineage committee progenitors (CD34+/−CD38+) with down regulation as differentiation proceeds.2,8 In AML, increased expression of HoxB3, B4, A7–11 is found in the most primitive progenitors with expression of A7–11 aberrantly sustained in differentiating progenitors.1,2 These observations suggest that identification of Hox target genes would provide useful insights into stem cell biology, myelopoiesis, and myeloid leukemogenesis. Recent studies have identified Hox target genes which begin to explain the crucial role these proteins play in hematopoiesis.
2. Mechanisms of Gene Regulation by Hox Proteins
a. Conserved domains in Hox proteins
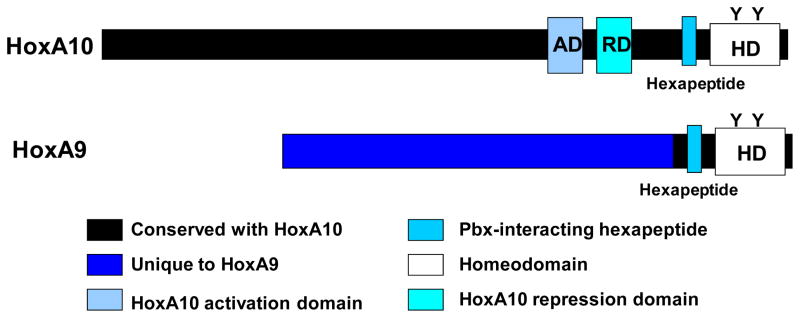
Hox proteins with the same number but from different groups are well conserved in comparison to adjacent Hox proteins in the same group (Figure 2). Hox proteins bind to DNA through a homeodomain (HD) which is found in the C-terminus of the protein. The HD is highly conserved between Hox proteins, and Hox-HD are highly conserved between species. The HD includes conserved tyrosine residues which may regulate Hox activity.9–11 For example, HD-tyrosine phosphorylation of HoxA10 decreases the binding affinity for cis elements in phagocyte effector genes, but increases the binding affinity of HoxA9 to the same genes.9,10 Tyrosine phosphorylation of HoxA9 and HoxA10 occurs in a Jak2 dependent manner in myeloid progenitor cells in response to differentiating cytokines.12 HD-tyrosine phosphorylation alters both protein-DNA and protein-protein interactions which may change the profile of interacting target genes.13
Figure 2. Hox proteins share conserved domains.
HoxA9 and HoxA10 share conserved domains, including the DNA-binding homeodomain and the hexapeptide domain which mediates interaction with Pbx proteins. The remainder of the HoxA9 and HoxA10 proteins are divergent.
Hox proteins also include a conserved hexapeptide domain which is N-terminal to the HD (Figure 2). This domain interacts with proteins of the Pbx family of transcription factors. Pbx proteins are frequent DNA-binding partners for Hox proteins, and participate in binding site selection, as is discussed below.14 Meis proteins are also frequent Hox partners, but the domains involved in Hox/Meis interactions have not been well defined.
Hox proteins can either activate or repress transcription, depending on the sequence of the binding site, the partner proteins, and cellular context. An activation domain was identified for HoxA10 which is conserved with other Hox10 proteins. This domain facilitates interaction with the Creb-binding protein (CBP) and is homologous to PQ domains in E1a interacting proteins.15 This domain does not exist in HoxA9, suggesting that another mechanism is used for transcriptional activation by this protein. A HoxA10 repression domain was described between the activation domain and the hexapeptide (Figure 2). This domain facilitates interaction between a HoxA10/Pbx1 heterodimer and histone deacetylase 2.16
b. Binding site consensus sequences
Even before identification of the first genuine Hox target gene, studies were performed to identify the Hox-DNA binding site consensus sequences. These studies used iterative binding site selection techniques to identify DNA sequences preferentially selected by various Hox proteins. A relatively loose consensus (5′-TNATNN-3′) was identified by these studies.17 These studies also determined a binding site preference across each Hox locus from 5′-TTAT-3′ on the Hox1 end of each locus to 5′-TGAT-3′ on the Hox13 end of the locus.17 Given the extensive homology between Hox-HDs, this suggests that other, less conserved domains are also involved in binding site selection. The short length and relative degeneracy of the Hox-DNA binding consensus make genome wide sequence analysis of limited use for identifying candidate Hox target genes. Other approaches, including gene expression studies and chromatin immuno-precipitation based assays have been employed for this purpose, as discussed below.
c. DNA-binding partners
Hox proteins often bind DNA as heterodimers with Pbx proteins.14 DNA-binding consensus sequences for Hox-Pbx heterodimers were also identified by binding site selection studies.17 In these studies, 5′-ATGATTNATNN-3′ was identified as the composite consensus sequence, with Pbx proteins recognizing the 5′-ATGAT-3′ end of the sequence.17 Identified target genes for Hox-Pbx dimers include the ITGB3 and DUSP4 (activated by HoxA10+Pbx2), and CYBB and NCF2 (repressed by HoxA10+Pbx1).15,16,18,19 Pbx proteins have not been found to directly influence transcriptional repression or activation of any target gene, but are hypothesized to assist the Hox protein in selecting DNA-binding sites and/or increasing affinity of Hox-DNA binding.
AbdB-Hox proteins (i.e. A9–13) may also bind DNA as a heterodimer with Meis proteins, or as a trimer with both Pbx and Meis. Binding site selection studies determined that the consensus for the Meis half of the DNA-binding site is 5′-TGACAG-3′; quite different than the Pbx recognition sequence.20 However, activation of the CYBB gene by HoxA9 involves interaction with Meis1 at a binding site which overlaps the HoxA10-Pbx1 repressor element on this gene.11
B. Mechanisms of HOX Gene Regulation
Differentiation stage specific transcription of various HOX genes is an important mechanism which regulates Hox protein activity. Although expression proceeds 5′ to 3′ through each HOX locus during hematopoiesis, mechanisms which regulate this process have not been completely defined. No control regions for HOX loci have been identified, but several transcription factors have been identified which regulate the promoter regions of a number of HOX genes.
1. Regulation by the Mixed Lineage Leukemia (Mll) Protein
Increased and sustained Hox expression is found in a subset of leukemia with chromosomal translocations involving the MLL gene.1–3 This observation suggested that the MLL gene product (Mixed Lineage Leukemia or Mll protein) might be involved in HOX regulation. Consistent with this hypothesis, Mll binds to the promoter region of multiple HOX genes of the AbdA and AbdB groups.21 Mll is a complex proteins with AT hooks (involved in DNA binding), DNA methyltransferase domain (DNMT), a PHD domain, and a SET domain (reviewed 22). Functional studies suggest that Mll binding to HOX promoters maintains differentiation stage specific transcription, but does not initiate transcription.21 Leukemia associated Mll-fusion proteins are hypothesized to induce aberrant HOX transcription because the Mll domains involved in maintaining transcription are present in the fusion protein, but domains essential for stage specific down regulation are not. The SET domain has been implicated in this latter function, since it is reproducibly deleted in leukemia associated Mll-fusion proteins.
Since “anterior” HOX genes (Hox1–5) also exhibit aberrant transcription in AML with MLL translocations, Mll-fusion proteins must influence events which indirectly dysregulate transcription of these genes. Conditional knock out of the MLL gene is an embryonic lethal at day 16.5 in mice. These mice have a reduced number of hematopoietic stem cells (HSC) which are unable to compete in repopulation assays.23 These results also suggest that Mll influences 5′HOX genes.
2. Regulation by Cdx Proteins
Another family of transcription factors which influence HOX gene transcription and/or Hox protein expression are Cdx proteins. Cdx are HD transcription factors and increased expression of Cdx1, 2 and 4 have been variously documented in human AML. Gene disruption studies identified roles for Cdx proteins in HOX gene regulation and hematopoiesis. Knock out of individual CDX genes impairs hematopoiesis during embryogenesis in murine models, and impairs transcription of multiple HOX genes, including HoxB2–5 and HoxA7–13.24,25 Conversely, overexpression of Cdx4 in murine bone marrow induces a myeloproliferative disorder and AML in murine transplantation experiments.26 Interestingly, overexpression of Cdx4 rescues the hematopoietic defect in murine Mll−/− bone marrow.27
Mechanisms for cross regulation by Cdx and Hox proteins have been recently defined. HOXA10 has been identified as a direct Cdx4 target gene in myeloid progenitor cells.28 In these studies, Cdx4 overexpression activated the HOXA10 promoter and increased HoxA10 expression. Additionally, these studies identified CDX4 as a HoxA10 activation target gene, establishing a positive feedback relationship between Cdx and Hox proteins.28 This has implications for understanding myeloid leukemogenesis with Hox overexpression.
4. Regulation by Polycomb Proteins
Studies in Drosophila determined that HOMC gene transcription is regulated by Polycomb Group (PcG) proteins (reviewed 29). Subsequent studies in mammalian models determined that PcG proteins bind to HOX promoter regions where they are thought to generally repress transcription. This has been described for YY1 binding to the HOXB4 promoter and Bmi1 binding to the HOXA9 promoter.30,31 PcG knockout in murine models impairs HSC function in various assays, which is hypothesized to be due to dysregulated HOX gene transcription.29 Molecular mechanisms for these effects remain to be clarified.
C. Function of Hox Proteins During Hematopoiesis
Investigation of specific activities for various Hox proteins is complicated by functional redundancy between groups and between adjacent members of the same group. This makes gene disruption studies difficult to interpret unless multiple members of a group and knocked out, which produces additional complications for expression of other members of the locus. More information has been obtained from overexpression studies, as is discussed below.
1. HoxB3 and B4
a. Studies in knockout mice
Homologous recombination was used to generate murine models with disruption of HOXB4 or double knockout of HOXB3/B4.32 These mice have a minor hemato-phenotype and mild abnormalities in hematopoiesis. These abnormalities include impairment of HSC function as indicted by decreased competitive repopulating activity. The results of studies with either HOXB4 or HOXB3/B4 knockout are similar, suggesting functional redundancy between these two Hox proteins. The mild nature of the phenotype suggests there is redundancy with other HoxB proteins, or anterior Hox proteins of other groups.
b. Overexpression studies
More information was obtained with gain of function studies for HoxB3 and HoxB4. These studies determined that overexpression of either HoxB3 or HoxB4 in murine or human bone marrow expanded the most immature cell populations in vitro.33–35 Consistent with these results, mice that were transplanted with HoxB3 or HoxB4 overexpressing bone marrow developed a myeloproliferative neoplasm (MPN) with expansion of the long term repopulating HSC population in the bone marrow.33–35 However, the MPN did not progress to AML in these mice.
2. HoxA9 and A10
a. Studies in knockout mice
Constitutive knockout of the HOXA9 or HOXA10 gene in mice results in abnormal development of the genitourinary system, impaired fertility in heterozygous knockout animals, and a very mild hemato-phenotype. Mice with knockout of either gene are reported to have either mild pancytopenia, or unremarkable blood counts, depending upon the report. Studies of bone marrow function are similarly unremarkable. There is some decrease in competitive repopulating ability in HOXA9−/− bone marrow compared to normal, but the difference is not profound.36 Similar studies have not been performed with HOXA10−/− bone marrow, but serial plating assays are only slightly less efficient in comparison to Wt bone marrow. Double knockout of HOXA9 and HOXA10 has not been studied nor has conditional knockout in bone marrow cells only.
b. Overexpression studies
Investigations of HoxA9 and HoxA10 by in vitro and in vivo overexpression have been quite informative. In vitro, overexpression of either protein in murine or human bone marrow preferentially expands the granulocyte/monocyte progenitor (GMP) population and immortalizes the cells.37–40 Mice transplanted with HoxA10-overexpressing bone marrow develop a MPN with mature neutrophils which progresses to differentiation block and AML over 6–8 mos.40 Mice transplanted with HoxA9-overexpressing bone marrow also develop a MPN which only progresses to AML after an extremely long lag time, or if the bone marrow is co-overexpressing Meis1.36 These results suggest some redundancy and some unique functions for HoxA9 and HoxA10. Additional studies of murine models suggest the hypothesis that HoxA10 induces differentiation block while HoxA9 is involved in selection of myeloid versus lymphoid lineage commitment.
II. HOX PROTEINS ARE ABERRANTLY EXPRESSED IN AML
A. Association Between Hox Expression and MLL-Translocation
The MLL gene is involved in leukemia associated translocations with over 70 different partner genes (the most common of which are listed in Table 1) (reviewed 22). These leukemias are characterized by expression of a fusion protein which include the same set of domains from the Mll N-terminus, and C-terminal domains from the fusion partner. Gene expression profiles and clinical characteristics fairly similar for all leukemias with translocations involving the MLL gene, suggesting that Mll domains are the dominant factor driving pathogenesis. Since MLL is located on the q23 region of chromosome 11, these are referred to as 11q23 leukemias. In adults, 11q23 leukemia is often associated with prior exposure to topoisomerase II inhibitors and has extremely poor prognosis, even among therapy related leukemias. In pediatric patients, MLL translocations are found predominantly in a poor prognosis subset of infant leukemias with both lymphoid and myeloid blasts (hence the term mixed lineage leukemia).3 As discussed above, 11q23 leukemias are associated with increased expression of a set of HD transcription factors in CD34+ bone marrow cells and CD34+CD38+ circulating blasts; therefore expression of these genes is both increased and aberrantly sustained in differentiating myeloid cells. This set includes HoxB3, B4, A7–11 and Meis1. A similar gene expression profile is found in AML with normal cytogenetics, but tandem duplication of the MLL gene.41
Table 1.
| Mll Fusion Proteins | |
|---|---|
| MLL-ELL | MLL-SEPTIN6 |
| MLL-GAS | MLL-AF4 |
| MLL-CBP | MLL-AF6 |
| MLL-EEN | MLL-AF9 |
| MLL-LTG9 | MLL-AF17 |
B. Other Forms of Leukemia with Increased Hox Expression
Studies of the pathogenesis of 11q23 leukemias suggest the hypothesis that aberrant HD-transcription factor expression is prognosis driving in AML. Additional studies in forms of leukemia without MLL translocations confirmed this hypothesis. Increased expression of similar sets of Hox proteins and Meis1 were also described in AML with translocations involving MYST3 and CREBBP genes.5 Statistical analysis also revealed a poor prognosis subset of AML subjects with normal cytogenetics (and without tandem duplications of MLL) with increased Hox and Meis1 expression.4 These studies suggest that Hox proteins may regulate a network of genes which influence molecular events especially associated with adverse outcomes in AML.
C. Murine Models to of Leukemogenesis
The functional contribution of various Hox proteins to myeloid leukemogenesis has been studied in murine models. These murine models have been crucial to develop the causal links between increased Hox expression and the pathogenesis of myeloid malignancy.
1. HoxA10 and Shp2-PTP
As discussed above, overexpression of HoxA10 or HoxA9 + Meis1in murine bone marrow expands the GMP population in vitro and results in a myeloproliferative neoplasm in murine transplantation experiments. However, AML does not occur immediately in mice transplanted with HoxA10 or HoxA9 overexpressing bone marrow, but develops over the course of months.38,40 These results suggest that Hox protein overexpression is sufficient to induce myeloproliferation, and also predisposes to acquisition of additional mutations which are necessary for differentiation block and AML. Post translational modification of HoxA10 is involved in modulation of both transcriptional repression and activation functions.9–13 In vitro studies suggest that post translational modification of HoxA10 prevents the overexpressed protein from participating in gene regulatory activities which might lead to differentiation block. This suggests the hypothesis that mutations which impair cytokine induced tyrosine phosphorylation of HoxA10 might lead to AML in HoxA10 overexpressing bone marrow.
HoxA10 is a substrate for Shp2 protein tyrosine phosphatase.10 Down regulation of Shp2-PTP activity during myelopoiesis is one factor which contributes to HoxA10 tyrosine phosphorylation in response to differentiating cytokines. Leukemia associated mutations in the gene encoding Shp2 have been described which result in constitutive activation of the PTP42. These mutations are found in 10% of AML and perhaps a higher percent of 11q23-AML. Consistent with this hypothesis, mice transplanted with bone marrow overexpressing HoxA10 plus a constitutively active form of Shp2 develop leukemia immediately.43
2. Mll-Fusion Proteins
Various leukemia associated Mll-fusion proteins have been overexpressed in murine bone marrow cells and analyzed in vitro and in vivo. Similar to overexpression studies with individual Hox proteins, Mll-fusion protein expressing murine bone marrow has increased serial plating capacity and is immortalized in vitro. Mice transplanted with such bone marrow develop a MPN with predominance of mature neutrophils. This MPN progresses to differentiation block and AML over a 6–8 mos.44,45 This in vivo process is delayed in mice transplanted with Mll-fusion protein expressing, Cdx4−/− bone marrow. 27 Various MLL-fusion partners have been tested in such experiments, and aberrant Hox expression has been identified which follows a very similar pattern to that observed in human 11q23-AML.46 Results from studies of Mll-fusion protein expression in murine bone marrow with knock out of various Hox proteins has been inconsistent, but the majority indicate development of MPN and AML in mice transplanted with HoxA7−/− or HoxA9−/− bone marrow expressing various Mll-fusion proteins.47, 48
III. HOX TARGET GENES AND DOWNSTREAM REGULATORY EVENTS
A. HoxB3 and B4
Despite the documented importance of HoxB3 and HoxB4 in HSC maintenance and function, relatively few relevant target genes have been identified which explain these activities (Table 2). HoxB3 interacts with the promoter region of OTX2 which encodes a homeodomain protein involved in regulating development of eye structures, but without a documented role in hematopoiesis.49 The gene encoding DNA methyltransferase 3B is activated by HoxB3 and may be involved in epigenetic regulation of HSC relevant genes.50 A number of HoxB4 target genes have been identified which impact cell proliferation and survival (Figure 2), including the genes encoding myc, IGF binding protein 1, FLASH and Rap1.51 –54 Interestingly, expression of myc and Rap1 are relatively decreased in 11q23-AML. HoxB4 also activates the HEMGN gene encoding a nuclear protein of unknown significance which is expressed in bone marrow progenitor cells.55
Table 2.
increased in 11q23
decreased in 11q23
no difference
B. HoxA9
A number of target genes for HoxA9 have been identified, generally using expression microarray screening for altered gene expression in HoxA9 overexpressing cells. HoxA9 activates several genes of potential significance for the pathogenesis of AML, including genes encoding Flt3, Pim1 and miR155. 56– 60 Flt3 is the receptor for Flt3L and is expressed on HSC and myeloid progenitor cells. Aberrant activation of this receptor due to FLT3 gene mutation is associated with poor prognosis in AML. Gene expression studies indicate a statistically significant association between the presence of FLT3 mutation and HoxA9 and HoxA10 overexpression in AML.61 Pim1 is a kinase which is frequently activated by the Moloney murine leukemia virus and participates in leukemic transformation of such cells. Expression of miR155 has been described in AML and a number of putative target mRNAs have been identified which may mediate a functional contribution to leukemogenesis. HoxA9 also activates several genes which confer the mature phagocyte phenotype, including genes encoding the NADPH oxidase component gp91phox, and E selectin.11,62 This is consistent with the hypothesis that HoxA9 is involved in selection of myeloid lineage commitment.
C. HoxA10
A slightly larger number of HoxA10 target genes have been identified (Table 4). Similar to HoxA9, HoxA10 activates a number of genes involved in cell proliferation and survival.63 HoxA10 activates transcription of several genes involved in proliferation of HSC and myeloid progenitors, including the genes encoding Tgfβ2 and one a cognate receptor, and the gene encoding β3 integrin.11,64 HoxA10 also activates the CDX4 gene in myeloid progenitor cells, which suggests an important role for HoxA10 in regulation of the Hox-code.63 HoxA10 also activates the gene encoding Mkp2 (the DUSP4 gene) in myeloid progenitors.65 Mkp2 inactivates Jnk and p36 Map-kinases, resulting in HoxA10-dependent apoptosis resistance of progenitor cells. Hoxa10 also regulates genes which confer the mature phagocyte phenotype (Table 4). However, in contrast to HoxA9, HoxA10 represses transcription of these genes in myeloid progenitor cells. This is consistent with descriptive observations suggesting that HoxA10 induces differentiation block and identifies potentially antagonistic functions for HoxA9 and HoxA10 for regulating this aspect of myelopoiesis.
Table 4.
| HoxA10 Target Genes
| ||
|---|---|---|
| Cytokines/receptors | Proliferation | Inflammation |
| ITGB3* | MAPK6PS1* | ATPGV1H |
| SELL* | NUDT6^ | CYBB* |
| TGFB2* | PLCB1# | IL11^ |
| TGFBR3* | PRKAR2A^ | NCF2* |
| TMEM8^ | TBXAS1* | |
|
|
|
|
| Ubiquitination | DNA-binding proteins | Apoptosis |
| ARIH2^ | CDX4^ | DUSP4* |
| UBE2S^ | CUX1* | BRSK1# |
| MEIS1^ | PDCD5^ | |
| PBX2^ | ||
increased in 11q23,
decreased in 11q23,
no difference
IV SUMMARY
Dysregulated Hox expression is found in a subset of poor prognosis AML, including leukemias with MLL gene translocations. Studies in murine models and human primary cells suggest that increased and sustained expression of Hox proteins plays a functional role in leukemogenesis through dysregulation of HSC and GMP populations. Studies have identified some Hox target genes which may be relevant to this process. The products of such genes, and cognate pathways, may be rational therapeutic targets for personalized therapeutic approaches to AML with dysregulated Hox expression.
Table 3.
| HoxA9 Target Genes
| ||
|---|---|---|
| Cytokines/receptors | Proliferation | Inflammation |
| EPHB4# | miR155 | CYBB* |
| FLT3^ | PIM1^ | NCF2* |
| OPN | SELE* | |
increased in 11q23,
decreased in 11q23,
no difference
ABBREVIATIONS
- AML
acute myeloid leukemia
- MLL
mixed lineage leukemia
- HD
homeodomain
- Jak2
Janus kinase 2
- CBP
CREB-binding protein
- PCG
polycomb group proteins
- MPN
myeloproliferative neoplasm
- GMP
granulocyte/monocyte progenitors
- DNMT
DNA-methyl transferase
LITERATURE CITED
- 1.Drabkin HA, Parsy C, Ferguson K, Guilhot F, Lacotte L, Roy L, Zeng C, Baron A, Hunger SP, Varella-Garcia M, Gemmill R, Brizard F, Brizard A, Roche J. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia. 2002;16:186–195. doi: 10.1038/sj.leu.2402354. [DOI] [PubMed] [Google Scholar]
- 2.Kawagoe H, Humphries RK, Blair A, Sutherland HJ, Hogge DE. Expression of HOX genes, HOX cofactors and MLL in phenotypically and functionally defined subpopulations of leukemic and normal human hematopoietic cells. Leukemia. 1999;13:687–698. doi: 10.1038/sj.leu.2401410. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 4.Riche J, Zeng C, Baron A, Gadgil S, Gemmill RM, Tigaud I, Thomas X, Drabkin HA. Hox expression in AML identified a distinct subset of patients with intermediate cytogenetics. Leukemia. 2004;18:1059–63. doi: 10.1038/sj.leu.2403366. [DOI] [PubMed] [Google Scholar]
- 5.Camos M, Esteve J, Jares P, Colomer D, Rozman M, Villamor N, Costa D, Carrio A, Nomdedue J, Montserrat E, Campo E. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;q13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–54. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- 6.Eklund EA. The role of Hox proteins in myeloid leukemia. Curr Opin Hematol. 2006;13:67–73. doi: 10.1097/01.moh.0000208467.63861.d6. [DOI] [PubMed] [Google Scholar]
- 7.Eklund EA. The role of HOX genes in malignant myeloid disease. Curr Opin Hematol. 2007;14:85–9. doi: 10.1097/MOH.0b013e32801684b6. [DOI] [PubMed] [Google Scholar]
- 8.Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Largman C. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. PNAS USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eklund EA, Jalava A, Kakar R. Tyrosine phosphorylation decreases HoxA10 DNA-binding and transcriptional repression during IFNγ induced differentiation in myeloid cell lines. J Biol Chem. 2000;275:20117–26. doi: 10.1074/jbc.M907915199. [DOI] [PubMed] [Google Scholar]
- 10.Lindsay S, Huang W, Wang H, Horvath E, Zhu C, Eklund EA. Activation of SHP2 Protein-tyrosine Phosphatase Increases HoxA10-induced Repression of the Genes Encoding gp91PHOX and p67PHOX. J Biol Chem. 2007;282:2237–49. doi: 10.1074/jbc.M608642200. [DOI] [PubMed] [Google Scholar]
- 11.Bei L, Lu YF, Eklund EA. HoxA9 activates transcription of the gene encoding gp91PHOX during myeloid differentiation. J Biol Chem. 2005;280:12359–70. doi: 10.1074/jbc.M408138200. [DOI] [PubMed] [Google Scholar]
- 12.Kakar R, Kautz B, Eklund EA. JAK2 is necessary and sufficient for interferon gamma-induced transcription of the gene encoding gp91PHOX. J Leukocyte Biol. 2005;77:120–127. doi: 10.1189/jlb.0704429. [DOI] [PubMed] [Google Scholar]
- 13.Eklund EA, Goldenberg I, Lu Y, Andrejic J, Kakar R. SHP1 protein tyrosine phosphatase regulates HoxA10 DNA-binding and transcriptional repression activity in undifferentiated myeloid cells. J Biol Chem. 2002;39:36878–36888. doi: 10.1074/jbc.M203917200. [DOI] [PubMed] [Google Scholar]
- 14.Chang CP, Shen WF, Rozenfeld S, Lawrence HJ, Largman C, Cleary ML. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox Proteins. Genes Dev. 1995;9:663–674. doi: 10.1101/gad.9.6.663. [DOI] [PubMed] [Google Scholar]
- 15.Bei L, Lu YF, Bellis SL, Zhou W, Horvath E, Eklund EA. Identification of a HoxA10 activation domain necessary for transcription of the gene encoding Beta 3 integrin during myeloid differentiation. J Biol Chem. 2007;282:16846–59. doi: 10.1074/jbc.M609744200. [DOI] [PubMed] [Google Scholar]
- 16.Lu Y, Goldenberg I, Bei L, Andrejic J, Eklund EA. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with Histone deacetylase 2. J Biol Chem. 2003;278:47792–802. doi: 10.1074/jbc.M305885200. [DOI] [PubMed] [Google Scholar]
- 17.Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox Locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Lu YF, Huang W, Papoutsakis ET, Fuhrken P, Eklund EA. HoxA10 activates transcription of the gene encoding Mkp2 in myeloid cells. J Biol Chem. 2007;282:16164–76. doi: 10.1074/jbc.M610556200. [DOI] [PubMed] [Google Scholar]
- 19.Lindsey S, Zhu C, Lu YF, Eklund EA. HoxA10 represses transcription of the gene encoding p67phox in phagocytic cells. J Immunol. 2005;175:5269–5279. doi: 10.4049/jimmunol.175.8.5269. [DOI] [PubMed] [Google Scholar]
- 20.Shen WF, Montgomery JC, Rozenfeld S, Moscow JJ, Lawrence HJ, Buchberg AM, Largman C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6458. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Molecular Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 22.Marschalek R. Mechanisms of leukemogenesis by MLL fusion proteins. Br J Haem. 2010;152:141–154. doi: 10.1111/j.1365-2141.2010.08459.x. [DOI] [PubMed] [Google Scholar]
- 23.McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes G, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJM. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Davidson AJ, Ernst P, Wang Y, Dekens MPS, Kingsley PD, Palis J, Korsmeyer SJ, Daley GQ, Zon LI. Cdx4 mutants fail to specify blood progenitors and can be rescued by multiple Hox genes. Nature. 2003;425:300–306. doi: 10.1038/nature01973. [DOI] [PubMed] [Google Scholar]
- 25.Bansal D, Scholl C, Frohling S, McDowell E, Lee BH, Dohner K, Ernst P, Davidson AJ, Daley GQ, Zon LI, Gilliland DG, Huntly BJP. Cdx4 dysregulates Hox gene expression and generates acute myeloid leukemia alone and in cooperation with Meis1a in a murine model. PNAS. 2006;103:16924–16929. doi: 10.1073/pnas.0604579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Yabuuchi A, McKinney-Freeman S, Ducharme DMK, Ray MK, Chawengsaksophak K, Archer TK, Daley GQ. Cdx gene deficiency compromises embryonic hematopoiesis in the mouse. PNAS. 2008;105:7756–7761. doi: 10.1073/pnas.0708951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo S, Huntly BJ, Wang Y, Chen J, Brumme K, Ball B, McKinney-Freeman SL, Yabuuchi A, Scholl C, Bansal D, Zon LI, Fröhling S, Daley GQ, Gilliland DG, Mercher T. Cdx4 is dispensable for murine adult hematopoietic stem cells but promotes MLL-AF9-mediated leukemogenesis. Haematologica. 2010;95:1642–1650. doi: 10.3324/haematol.2010.023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bei L, Huang W, Wang H, Shah C, Horvath E, Eklund EA. HoxA10 activates CDX4 transcription and Cdx4 activates HOXA10 transcription in myeloid cells. J Biol Chem. 2011;286:19047–64. doi: 10.1074/jbc.M110.213983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takihara Y. Role of Polycomb-group genes in sustaining activities of normal and malignant stem cells. Int J Hematol. 2008;87:25–34. doi: 10.1007/s12185-007-0006-y. [DOI] [PubMed] [Google Scholar]
- 30.Gilthorpe J, Vandromme M, Brend T, Gutman A, Summerbell1 D, Totty N, Rigby PWJ. Spatially specific expression of Hoxb4 is dependent on the ubiquitous transcription factor NFY. Development. 2002;129:3887–3899. doi: 10.1242/dev.129.16.3887. [DOI] [PubMed] [Google Scholar]
- 31.Smith LL, Yeung J, Zeisig BB, Popov N, Huijbers I, Barnes J, Wilson AJ, Taskesen E, Delwel R, Gil J, Van Lohuizen M, So CWE. Functional Crosstalk between Bmi1and MLL/Hoxa9 Axis in Establishment of Normal Hematopoietic and Leukemic Stem Cells. Cell Stem Cell. 2011;8:649–662. doi: 10.1016/j.stem.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Bjornsson JM, Larsson N, Brun ACM, Magnusson M, Andersson E, Lundstrom P, Larsson J, Repetowska E, Ehinger M, Humphries RK, Karlsson S. Reduced proliferative capacity of hematopoietic stem cells deficient in Hoxb3 and Hoxb4. Mol Cell Biol. 2003;23:3872–3883. doi: 10.1128/MCB.23.11.3872-3883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvageau G, Thorsteinsdottir U, Hough MR, Hugo P, Lawrence HJ, Largman C, Humphries RK. Overexpression of HOXB3 in hematopoietic cells causes defective lymphoid development and progressive myeloproliferation. Immunity. 1997;6:13–22. doi: 10.1016/s1074-7613(00)80238-1. [DOI] [PubMed] [Google Scholar]
- 34.Antonchuka J, Sauvageau G, Humphries RK. HOXB4 overexpression mediates very rapid stem cell regeneration and competitive hematopoietic repopulation. Expl Hem. 2001;29:125–1134. doi: 10.1016/s0301-472x(01)00681-6. [DOI] [PubMed] [Google Scholar]
- 35.Schmittwolf C, Porsch M, Greiner A, Avots A, Muller AM. HOXB4 confers a constant rate of in vitro proliferation to transduced bone marrow cells. Oncogene. 2005;24:561–572. doi: 10.1038/sj.onc.1208202. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89:1922–30. [PubMed] [Google Scholar]
- 37.Thorsteinsdottir U, Mamo A, Kroon E, Jerome L, Bijl J, Lawrence HJ, Humphries K, Sauvageau G. Overexpression of the myeloid leukemia-associated HoxA9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 38.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. HoxA9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–25. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bjornsson JM, Andersson E, Lundstrom P, Larsson N, Xu X, Repetowska E, Humpthries KR, Karlsson S. Proliferation of primitive myeloid progenitors can be reversibly induced by HOXA10. Blood. 2001;98:3301–3308. doi: 10.1182/blood.v98.12.3301. [DOI] [PubMed] [Google Scholar]
- 40.Thorsteinsdottir U, Sauvageau G, Hough MR, Dragowska W, Lansdorp PM, Lawrence HJ, Largman C, Humphries RK. Overexpression of HoxA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol Cell Biol. 1995;17:495–505. doi: 10.1128/mcb.17.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dorrance AM, Liu S, Yuan W, Becknell B, Arnoczky KJ, Guimond M, Strout MP, Feng L, Nakamura T, Yu L, Rush LJ, Weinstein M, Leone G, Wu L, Ferketich A, Whitman SP, Marcucci G, Caligiuri MA. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006;116:2707–2716. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tartaglia M, Niemeyer CM, Fragale A, Song X, Beuchner J, Jung A, Hahlen K, Hasle H, Licht JD, Gleb BD. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nature Genet. 2003;34:148–150. doi: 10.1038/ng1156. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Lindsey S, Konieczna I, Bei L, Horvath E, Huang W, Saberwal G, Eklund EA. Constitutively Active SHP2 Cooperates with HoxA10-overexpression to Induce Acute Myeloid Leukemia. J Biol Chem. 2009;284:2549–67. doi: 10.1074/jbc.M804704200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horton SJ, Grier DG, McGonigle GJ, Thompson A, Morrow M, De Silva I, Moulding DA, Kioussis D, Lappin TR, Brady HJ, Williams O. Continuous MLL-ENL expression is necessary to establish a Hox code and maintain immortalization of hematopoietic progenitor cells. Cancer Res. 2005;65:9245–9252. doi: 10.1158/0008-5472.CAN-05-1691. [DOI] [PubMed] [Google Scholar]
- 45.Lavau C, Luo RT, Du C, Thirman MJ. Retrovirus mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemia in mice. Proc Natl Acad Sci USA. 2000;60:10984–9. doi: 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Luo RT, Mi S, Sun M, Chen P, Bao J, Neilly MB, Jayathilaka N, Johnson DS, Wang L, Lavau C, Zhang Y, Tseng C, Zhang X, Wang J, Yu J, Yang H, Wang SM, Rowley JD, Chen J, Thirman MJ. Consistent deregulation of gene expression between human and murine MLL rearrangement leukemias. Cancer Res. 2009;69:1109–1116. doi: 10.1158/0008-5472.CAN-08-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.So CH, Karsunky H, Wong P, Weissman IL, Cleary ML. Leukemic transformation of hematopoietic progenitors by MLL-GAS7 in the absence of HoxA7 or HoxA9. Blood. 2004;103:3192–3199. doi: 10.1182/blood-2003-10-3722. [DOI] [PubMed] [Google Scholar]
- 48.Kumar AR, Hudson WA, Chen W, Nishiuchi R, Yao Q, Kersey JH. HoxA9 influences the phenotype but not the incidence of MLL-AF9 fusion gene leukemia. Blood. 2004;103:1823–1828. doi: 10.1182/blood-2003-07-2582. [DOI] [PubMed] [Google Scholar]
- 49.Guazzi S, Pintonello ML, Vigano A, Boncinelli E. Regulatory interactions between the human HOXB1, HOXB2, and HOXB3 proteins and the upstream sequence of the Otx2 gene in embryonal carcinoma cells. J Biol Chem. 1998;273:11092–9. doi: 10.1074/jbc.273.18.11092. [DOI] [PubMed] [Google Scholar]
- 50.Palakurthy RK. Epigenetic silencing of the RASSF1A tumor suppressor gene through HOXB3-mediated induction of DNMT3B expression. Molecular Cell. 2009;36:219–30. doi: 10.1016/j.molcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan Q, Simpson RU. c-myc intron element-binding proteins are required for 1, 25-dihydroxyvitamin D3 regulation of c-myc during HL-60 cell differentiation and the involvement of HOXB4. J Biol Chem. 1999;274:8437–44. doi: 10.1074/jbc.274.13.8437. [DOI] [PubMed] [Google Scholar]
- 52.Gao J, Mazella J, Tseng L. Hox proteins activate the IGFBP-1 promoter and suppress the function of hPR in human endometrial cells. DNA Cell Biol. 2002;21:819–25. doi: 10.1089/104454902320908469. [DOI] [PubMed] [Google Scholar]
- 53.Morgan R, Nalliah A, Morsi El-Kadi AS. FLASH, a component of the FAS-CAPSASE8 apoptotic pathway, is directly regulated by Hoxb4 in the notochord. Dev Biol. 2004;265:105–12. doi: 10.1016/j.ydbio.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 54.Morsi El-Kadi AS, in der Reiden P, Durston A, Morgan R. The small GTPase Rap1 is an immediate downstream target for Hoxb4 transcriptional regulation. Mech Dev. 2002;113:131–9. doi: 10.1016/s0925-4773(02)00047-3. [DOI] [PubMed] [Google Scholar]
- 55.Jiang J, Yu H, Shou Y, Neale G, Zhou S, Lu T, Sorrentino BP. Hemgn is a direct transcriptional target of HOXB4 and induces expansion of murine myeloid progenitor cells. Blood. 2010;116:711–9. doi: 10.1182/blood-2009-07-235341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruhl T. Homeobox A9 transcriptionally regulates EphB4 to modulate endothelial cell migration and tube formation. Circ Res. 2004;94:743–51. doi: 10.1161/01.RES.0000120861.27064.09. [DOI] [PubMed] [Google Scholar]
- 57.Gwin K, Frank E, Bossou A, Medina KL. Hoxa9 regulates Flt3 in lymphohematopoietic progenitors. J Immunol. 2010;185:6572–83. doi: 10.4049/jimmunol.0904203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi X, Bai S, Li L, Cao X. Hoxa-9 represses transforming growth factor-beta-induced osteopontin gene transcription. J Biol Chem. 2001;276:850–5. doi: 10.1074/jbc.M005955200. [DOI] [PubMed] [Google Scholar]
- 59.Hu YL, Fong S, Largman C, Shen WF. HOXA9 regulates miR-155 in hematopoietic cells. Nucleic Acids Res. 2010;38:5472–8. doi: 10.1093/nar/gkq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu YL, Passegué E, Fong S, Largman C, Lawrence HJ. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood. 2007:1094732–8. doi: 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oncomine 4.4 Research Edition. Compendia Biosciences. www.oncomine.org/resource/
- 62.Bandyopadhyay S, Ashraf MZ, Daher P, Howe PH, DiCorleto PE. HOXA9 participates in the transcriptional activation of E-selectin in endothelial cells. Mol Cell Biol. 2007;27:4207–16. doi: 10.1128/MCB.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bei L, Huang W, Wang H, Shah C, Horvath E, Eklund EA. HoxA10 activates CDX4 transcription and Cdx4 activates HOXA10 transcription in myeloid cells. J Biol Chem. 2011;286:19047–64. doi: 10.1074/jbc.M110.213983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shah CA, Wang H, Bei L, Platanias LC, Eklund EA. HoxA10 regulates transcription of the gene encoding transforming growth factor beta 2 (TGFB2) in myeloid cells. J Biol Chem. 2011;286:3161–76. doi: 10.1074/jbc.M110.183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Lu YF, Huang W, Papoutsakis ET, Fuhrken P, Eklund EA. HoxA10 activates transcription of the gene encoding Mkp2 in myeloid cells. J Biol Chem. 2007;282:16164–76. doi: 10.1074/jbc.M610556200. [DOI] [PubMed] [Google Scholar]