Abstract
Acute myeloid leukemia (AML) is one of the most common leukemias with a 20% 5-year event-free survival in adults and 50% overall survival in children, despite aggressive chemotherapy treatment and bone marrow transplantation. The incidence and mortality rates for acute leukemia have only slightly decreased over the last 20 years and therefore greater understanding of the molecular mechanisms associated with leukemic progression is needed. To this end, a number of transcription factors which appear to play a central role in leukemogenesis are being investigated; among them is the cAMP response element binding protein (CREB). CREB is a transcription factor that can regulate downstream targets involving in various cellular functions including cell proliferation, survival, and differentiation. In several studies, the majority of bone marrow samples from patients with acute lymphoid and myeloid leukemia demonstrate CREB overexpression. Moreover, CREB overexpression is associated with a poor outcome in AML patients. This review summarizes the role of CREB in leukemogenesis.
Keywords: CREB, Leukemia, Oncogenesis, Transcription Factors
I. ACUTE LEUKEMIA
Leukemia develops when a malfunction in the normal regulatory mechanisms of mitosis occurs and allows bone marrow progenitor blood cells to expand in an uncontrolled fashion. The immature blasts proliferate more than normal cells and fail to differentiate normally. Although leukemia affects approximately 10 times more adults than children, it is the most common cancer among children. The most common type of leukemia in adults is Acute Myeloid Leukemia (AML), while Acute Lymphoblastic Leukemia (ALL) accounts for nearly 70% of childhood leukemia.1, 2
Leukemia is the sixth most common cause of cancer deaths in men and seventh most common cause of death among women in the U.S. The treatment for ALL and AML has improved with the use of chemotherapy based on stratified risk, molecular markers for prognosis prediction, and supportive care. Generally, the event-free survival rate is lower and the relapse rate is higher in adults than children. The response to treatment for leukemia is variable and associated with the age of the patient, as well as a number of other factors on presentation. The 5-year event-free survival is 70%–85% in some of the successful clinical trials in children with ALL and 30%–60% in children diagnosed with AML.1–3 According to the SEER Cancer Statistic Review, for ALL patients, the five-year relative survival rate for adults under 45 years old is about 75%, and for patients over 45 years old it is less than 20%; for AML patients, the five-year relative survival rate is over 50% for adults under 45 years old and it is less than 40% for patients over 45 years old.
II. MOLECULAR MECHANISMS OF LEUKEMIA
Acute leukemia had previously been classified by morphology, cytogenetics, and cell surface markers; more recently, it has become clear that molecular characterization of genetic mutations in ALL and AML may relate more strongly to clinical prognosis, and can provide information for potential targeted therapies. The advantage of characterizing the phenotype of human leukemia stems from the observation that it is a heterogeneous disease consisting of a variety of accumulated DNA alterations in progenitor blood cells. Primary genetic defects have been detected by molecular analyses, and these somatic mutations often alter crucial functions of the progenitor cells such as self-renewal and differentiation.
Similar to other cancers, it appears that leukemia arises from the accumulation and synergy of more than one genetic alteration. Many genetic alterations in AML are loss of function mutations in transcription factors critical for normal hematopoiesis.1,4 Data suggest that mutations which alter proliferation and survival functions of the progenitor cells cooperate with the mutations of the transcription factors and result in acute leukemia.4,5 Examples of genes that are found mutated in AML include biallelic mutations in CCAAT/enhancer binding protein alpha gene (CEBPA), inactivating mutations in Wilms’ tumor gene (WT1), activating mutations in fms-like tyrosine kinase 3 gene (FLT3), and mutation of nucleophosmin (NPM1), which produces mislocalized protein product in cells.5–9 In recent studies, apart from alterations of genes, differential microRNA expression is also involved in leukemia progression. 10
Acute leukemias are hypothesized to be the consequence of cooperation between mutations that alter proliferation and survival functions of hematopoietic cells and mutations that result from defective differentiation and loss of apoptosis in cells. These mutations may be found at many levels of cellular processes, such as growth factor receptors, kinase phosphorylation cascades, or cellular transcription programs. Central to all processes, however, are transcription factors, which integrate extranuclear signals and directly influence DNA transcription. Thus, understanding the function of transcription factors in blood malignancies can provide a wealth of information for treatment modalities. CREB, the cAMP-Response Element Binding Protein, has become of particular interest in leukemias, as it is known to play a broad range of roles in many critical cellular processes, and the majority of tissue samples from patients with ALL and AML overexpress the CREB in the bone marrow. CREB overexpression is associated with poor outcome in AML patients and increased survival and growth of myeloid cells.11,12 Transgenic mice expressing CREB in myeloid cells develop aberrant monocytosis and, after a prolonged latency, myeloproliferative disease. Thus, there is both clinical and laboratory data which implicates CREB as a potential critical regulator of leukemogenesis.
III. CREB
CREB is a 43-kDa leucine zipper transcription factor that belongs to the CREB/ATF family and regulates proliferation, differentiation, and survival in a variety of cell types, including neuronal and hematopoietic cells.13,14
CREB is a modular protein that contains a kinase-inducible domain (KID), two Glutamine-rich domains, and a basic Leucine zipper (bZIP) domain. The KID and Glutamine-rich domains are critical for transactivation and phosphorylation of CREB. 13,14 A serine 133 (Ser-133) residue within the KID domain is phosphorylated by various kinases and this phosphorylation promotes the interaction of CREB with a number of transcription coactivators, especially the histone acetyltransferases CREB-binding protein (CBP) or p300.15,16 CREB can be phosphorylated and thus activated in response to various stimuli such as growth factors, neurotransmitters, stress signals that increase intracellular cAMP or calcium levels. CREB is also activated by phosphorylation at Ser-133 through nuclear translocation of transducer of regulated CREB activity (TORC) coactivators, which occurs through a Ser-133 phosphorylation independent mechanism.17,18
CREB family member proteins, when activated, bind to the cAMP response elements, and promote the recruitment of coactivators such as CBP/p300, thereby initiating the transcriptional machinery and inducing CREB target genes.19
IV. PHOSPHORYLATION AND ACTIVATION OF CREB
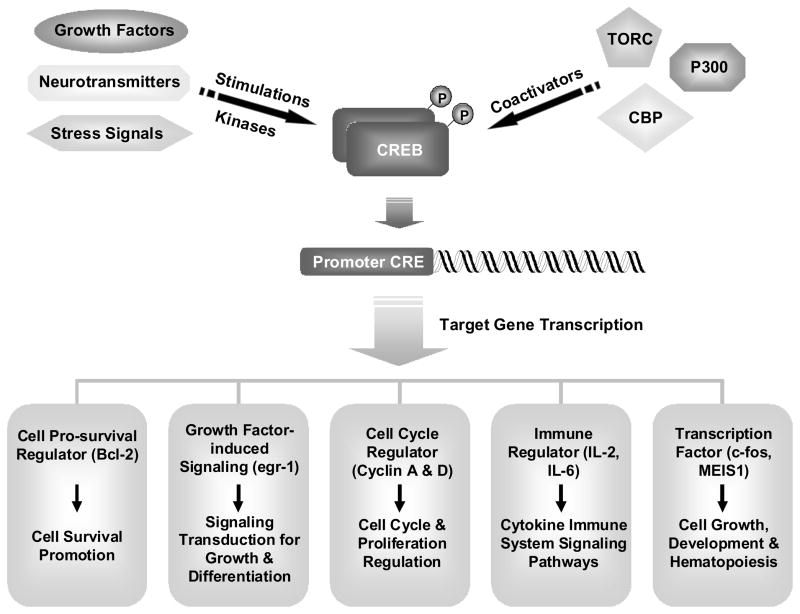
Phosphorylation is one of the most important post-translational modifications that can modulate the charge, activity, stability, cellular localization, and even downstream signal transduction of its target proteins, or have impact on proteins through crosstalk with other post-translational modifications.20 CREB was one of the first transcription factors shown to be regulated by phosphorylation and act as an intracellular signaling second messenger in cells.21–23 In late 1980s, CREB was found to be phosphorylated by the cAMP-dependent protein kinase (PKA) in vitro, and then phosphorylated by forskolin in cells.23 CREB is phosphorylated at Ser-133, and various kinases including ribosomal protein S6 kinase (pp90RSK), protein kinase C (PKC), protein kinase B/AKT, and mitogen- and stress-activated protein kinase (MSK-1) can all phosphorylate CREB at Ser-133.13,24 Numerous stimuli, including stress signals that increase intracellular cAMP or calcium levels, such as neurotransmitters and growth factors, were found to activate CREB in cells. Different growth factors such as mast/stem cell growth factor, basic fibroblast growth factor, and Granulocyte-macrophage colony-stimulating factor (GM-CSF), can all induce phosphorylation of CREB.25,26 CREB, when activated, dimerizes and binds to the promoter regions of its target gene that contains cAMP response element (CRE site), TGACGTCA, or CRE half sites CGTCA/TGACG, and promotes the recruitment of its transcriptional coactivators, CBP/p300, for CREB-mediated transcription. Therefore, CREB can regulate various cellular mechanisms through modulating its target genes (Figure 1).
Figure 1. The regulation of CREB activation.
A variety of extracellular stimuli can promote CREB phosphorylation and activation through different kinases. CREB can then interact with coactivators to promote the transcription of CREB responsive genes. CREB target genes have been shown to mediate effects including cellular proliferation, survival, differentiation, immune response, and hematopoiesis.
V. CREB TARGET GENES
Genome-wide analysis revealed that CREB can occupy approximately 4,000 promoter sites in vivo, emphasizing the broad array of functions CREB may exert; it is important in controlling well-known cell cycle regulators such as Ras, 14-3-3, cyclins and heat-shock proteins.27,28 Consistent with that, CREB is involved in a variety of cellular functions, including cell proliferation, survival, apoptosis, differentiation, metabolism, glucose homeostasis, hematopoiesis, immune response, and neuronal activities such as memory and learning.29,30
A. Transcription Factors, Metabolic, and Immune Response Regulators
Phosphorylation of CREB at Ser-133 is linked to regulation of transcription factors including c-fos and MEIS1, which contain CREB binding motif on their promoters and can be modulated by CREB.31–33 CRE binding sites are critical for c-fos transcription and it was suggested that CREB is a general mediator of stimulus-dependent transcription of c-fos.31 MEIS1 was upregulated in a microarray analysis in CREB overexpressing cells, and CREB can induce MEIS1 expression in normal and malignant hematopoietic cells.32 The importance of CREB in metabolism was also suggested as numerous CRE-containing genes were found to function in metabolic regulation. 34 Moreover, genes regulating immune response including IL-2 and IL-6, also possess consensus sites for CREB binding and can be modulated by CREB.13,35,36
B. Cell Cycle and Proliferation Regulators
CREB is capable of binding to and regulating the promoter regions of cell cycle genes such as cyclin A, D1, and D2, and thus impacts cell proliferation.37–39 For example, both PI3K and CREB can regulate cyclin D2 promoter activity. 38 Phosphorylation of CREB at Ser-133 is critical for IL-2 induced cyclin D2 transactivation, and the CREB-binding site on cyclin D2 is also important for cyclin D2 promoter activity.38 PKA inhibitors reduce lymphocyte proliferation and CREB phosphorylation, and thereby CREB and PKA regulate lymphocyte proliferation.38 Cyclin A1 is also upregulated in leukemia cells that overexpress CREB, while mRNA levels of both cyclin A and D were decreased in CREB shRNA transduced leukemia cells, suggesting that CREB can promote proliferation of leukemic cells through its downstream targets.11,12
C. Growth Factors and Signaling Modulators
Both GM-CSF and interleukin 3 (IL-3) stimulate the proliferation and maturation of myeloid progenitor cells, and each of them can activate signaling pathways involving a CREB-binding site of the early growth response-1 gene (egr-1) promoter.40 Also, CREB is phosphorylated on Ser-133 in response to GM-CSF or IL-3 stimulation, and that phosphorylation of CREB on Ser-133 substantially contributes to egr-1 transcriptional activation in response to GM-CSF. In addition, GM-CSF induces pp90RSK activation and phosphorylation of CREB in the human myeloid cell line, TF-1.11 In TF-1 cells, GM-CSF induces CREB phosphorylation and egr-1 transcription by activating pp90RSK through an MEK-dependent signaling pathway.11 These studies suggest that phosphorylation of CREB impacts on signal transduction in myeloid cells.
D. Cell Survival Regulation
The role of CREB in cell survival has also been described in a number of tissues. Neurotrophins such as nerve growth factor (NGF) induces phosphorylation of CREB at Ser-133, and it was proposed that Ser-133 phosphorylated CREB induces genes that confer specificity to neurotrophin signals and promote the survival and differentiation of neurons.41 In addition, CREB-mediated gene expression is necessary for NGF-dependent survival and crucial to promote survival of sympathetic neurons.42 Moreover, Bcl-2 is activated by NGF and other neurotrophins in a CREB-dependent fashion, and overexpression of Bcl-2 reduces the death-promoting effects of CREB inhibition.42 Therefore, it appears that activation of CREB promotes survival of neuron cells through activating downstream transcriptional target genes that encode pro-survival factors.
VI. CREB IN HEMATOPOIESIS AND LEUKEMOGENESIS
A. GM-CSF Signaling and CREB Activation
Genetic alterations are involved in leukemogenesis, and it can lead to dysregulated cytokine/growth factor dependent signal-transduction pathways in leukemic cells.43–45 Growth factors are produced by myeloid leukemic cells as well as stromal cells and bind their own receptors in an autocrine fashion to activate signaling pathways that promote cell growth and survival. Both GM-CSF and IL-3 stimulation result in the proliferation and maturation of early bone marrow progenitor cells. CREB is phosphorylated at Ser-133 in response to GM-CSF or IL-3 stimulation although with different kinetics, and this phosphorylation substantially contributes to transcriptional activation of egr-1 in response to GM-CSF but not IL-3.40,46 Moreover, egr-1 induced expression by GM-CSF is a PKA-independent event.47 In TF-1 cells, GM-CSF can induce CREB phosphorylation and egr-1 transcription by activating pp90RSK through an MEK-dependent mechanism.26 Furthermore, CREB-binding sites have been identified in the promoter of genes regulating proliferation and survival such as Bcl-2 and egr-1, which suggests multiple layers of CREB regulation in leukemic cells. Overall, the role of CREB activation in regulating hematopoietic growth factor signaling in myeloid cells is clearly demonstrated.
B. CREB is A Proto-Oncogene in Hematopoiesis and AML
Our laboratory showed that the majority of bone marrow samples from patients with acute lymphoid and myeloid leukemia overexpress CREB protein and mRNA.48 In addition, CREB overexpression is associated with poor outcome of clinical disease in AML patients.11,48 To understand the role of CREB in leukemogenesis and the biological consequences of CREB overexpression in primary human leukemia cells, leukemia cell lines and transgenic mice were investigated.11 Overexpression of CREB promotes growth and survival in leukemia cells, while its downregulation leads to suppression of myeloid cell proliferation and survival. Furthermore, CREB transgenic mice developed myeloproliferative disease after one year, but not leukemia, suggesting that CREB contributes to leukemic phenotype, but is not sufficient for complete transformation to leukemia.11 CREB promotes abnormal proliferation and survival of myeloid cells in vitro and in vivo through upregulation of specific downstream target genes such as cyclin A1.11,49 It appears that CREB acts as a proto-oncogene to regulate hematopoiesis and contributes to the leukemia phenotype, and therefore the results also suggest that CREB-dependent pathways may be targets for directed therapies for leukemia in the future.
C. CREB as a Critical Regulator of Normal Hematopoiesis and Leukemogenesis
CREB appears to be most highly expressed in lineage negative hematopoietic stem cells (HSCs). CREB RNA interference (RNAi) and shRNA techniques were used to knockdown CREB to elucidate its role in hematopoietic progenitors and leukemia cells. Transduction of primary HSCs or myeloid leukemia cells with lentiviral CREB shRNAs resulted in decreased proliferation of stem cells, cell cycle abnormalities, and inhibition of CREB transcription.12 Transplantation of bone marrow transduced with CREB shRNA in irradiated mice had decreased committed progenitors compared to scrambled control shRNA. However, there was no effect on long-term engraftment, suggesting that CREB insufficiency is not required for HSC activity. Therefore CREB is critical for normal myelopoiesis and leukemia cell proliferation, but not essential for normal function of HSCs.12
Compared to patients with leukemia remission or without leukemia, CREB was expressed more highly in bone marrow cells from patients with acute lymphoid or myeloid leukemia.48 Therefore CREB expression is a potential marker of malignant disease. In an effort to define the target genes of CREB in leukemias, genome-wide analyses were performed and CREB target genes were described; numerous candidate genes have been identified such as transcription regulators and histones, though these await in vivo validation.27,28,50 To identify potential downstream target genes, a microarray analysis with RNA from leukemia K562 cells overexpressing CREB was performed.51 Approximately 896 genes were differentially expressed in the CREB overexpressing cells compared to control parental cells. Among these, 702 genes were upregulated and they included members from the MEIS1 and the PBX1 family, which have both been reported to be critical for hematopoietic stem cell self-renewal and leukemogenesis.51–53
VII. MICRORNAS AND ONCOGENESIS
Although CREB is overexpressed in leukemia cells, the underlying mechanisms of how CREB regulates leukemogenesis remain largely unknown. Small regulatory non-coding RNA molecules, known as microRNAs, are single-stranded 20–24 nucleotide length RNA molecules that can regulate gene expression in many cellular mechanisms. These microRNAs can modulate gene function at the post-transcriptional level, as they typically reduce the stability of mRNAs that mediate various cellular processes including cell cycle regulation, proliferation, differentiation, and apoptosis and thus have an impact on oncogenesis.54,55
Specifically, differential expression of microRNAs in AML appears to have functional relevance in leukemogenesis.10 MiR-193a, which binds to c-kit proto-oncogene mRNA, was repressed by promoter hypermethylation in AML cell lines and primary AML blasts, but not in normal bone marrow cells.56 MiR-193a levels were inversely correlated with c-kit levels. Moreover, restoring miR-193a expression in AML cells containing mutated or overexpressed c-kit resulted in reduction in c-kit expression as well as inhibition of cell growth. The growth inhibition activity of miR-193a was suggested to be associated with apoptosis and granulocytic differentiation.56 CREB pathways are regulated by microRNAs in different cellular backgrounds.57–59 In myeloid cells that have higher CREB expression levels, miR-34b was expressed less, while overexpression of miR-34b resulted in a reduction of the CREB protein levels.57 Moreover, miR-34b expression caused abnormal cell cycle progression, reduced cell growth, and altered expression of CREB targets such as Bcl-2, cyclins, protein kinases, and cell survival signaling pathways.57 The miR-34b promoter is also methylated, which then regulates miR-34b expression level in the leukemia cell lines. The study therefore provides a possible mechanism for CREB overexpression. In another study, miR-301 was found to indirectly regulate ERK/CREB pathway, thereby controlling the transcription and function of its host gene, ska2, a CREB target, in lung cancer cells.59 Furthermore, inhibition of miR-301 or ska2 leads to an increase of the mitotic index and a decrease in colony formation, which could contribute to lung cancer transformation.59
VIII. CREB AS A POTENTIAL TARGET FOR THERAPY
As described, several lines of evidence support the notion that elevated CREB expression is associated with pathologic growth and survival of hematopoietic cells in primary human leukemic cells, human leukemia cell lines and transgenic mice and that CREB and pathways downstream of CREB may represent novel therapeutic targets. CREB levels were found to be elevated at diagnosis, and intriguingly, were also high in patients with relapsed AML. Patients in remission have similar CREB levels to unaffected controls. Recent evidence also implicates CBP as another important determinant in ALL disease relapse and prognosis. In pediatric patients with relapsed ALL, some 18% demonstrated a focal deletion or gene sequence alteration in the CBP gene.60
These alterations were rare in children with ALL who did not relapse, suggesting that the presence of CBP mutations may influence treatment responsiveness. These data demonstrate that CREB and its binding partners influence treatment responsiveness, and suggest that CREB signaling pathways may represent a novel therapeutic target. To this end, small molecules that inhibit binding of CREB and CBP have already been identified; since this interaction is critical in CREB signaling, and interruption at this step is postulated to reduce CREB activity. Studies on the compound 2-napthol-AS-E-phosphate (KG-501) showed that this molecule specifically inhibits the interaction between the KID of CREB and the helical ‘KIX’ domain of CBP in a dose-dependent and reversible manner.61 This molecule does not inhibit forskolin-stimulated phosphorylation of CREB at Ser-133. Furthermore, cAMP-dependent CREB target gene expression was inhibited in the presence of micromolar amounts of this drug, without off-target inhibition of transcriptional machinery. Thus, CREB appears to be a druggable target, and small molecules that inhibit CREB signaling may useful in the clinical setting.30
IX. CONCLUSION
In summary, CREB is an important target of growth factor signaling in myeloid cells and promotes the proliferation and differentiation of myeloid progenitor cells. CREB overexpression is observed in the majority of AML and ALL bone marrow cells from patients with leukemia. Ectopic expression of CREB in mice results in myeloproliferative disease but not leukemia, suggesting that additional cooperating oncogenes are required for full transformation. Knockdown of CREB appears to affect myeloid differentiation and myeloid leukemia cell proliferation but does not interfere with long-term engraftment. These results support the possibility of CREB being a potential target for drug development to treat AML. Future directions will focus on understanding how CREB specifically regulates leukemogenesis and targeting this critical protein to treat acute leukemia.
Acknowledgments
K.M.S. is supported by NIH R01 HL83077, R01 HL75826, R13 HL103078, R13 CA 159800, ASH Alternative Training Pathway Award, and UCLA Stein-Oppenheimer Award. B. M. is a recipient of an ASH Trainee Research award. This manuscript has not been published elsewhere and that it has not been simultaneously submitted for publication elsewhere.
Abbreviation
- AML
acute myeloid leukemia
- CREB
cAMP response element binding protein
- ALL
acute lymphoblastic leukemia
- CEBPA
CCAAT/enhancer binding protein alpha
- WT
Wilms’ tumor
- FLT3
fms-like tyrosine kinase 3
- NPM
nucleophosmin
- ATF
activating transcription factor
- KID
kinase-inducible domain
- bZIP
basic Leucine zipper
- Ser-133
serine 133 amino acid
- CBP
CREB-binding protein
- p300
adenovirus E1A-associated cellular p300 transcriptional co-activator protein
- TORC
transducer of regulated CREB activity coactivator
- cAMP
cyclic adenosine monophosphate
- PKA
cAMP-dependent protein kinase
- pp90RSK
ribosomal protein S6 kinase
- PKC
protein kinase C
- AKT
a serine-threonine protein kinase and is called protein kinase B
- MSK
mitogen- and stress-activated protein kinase
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- CRE
cAMP response element
- Ras
RAt Sarcoma, an oncogene
- 14-3-3
This name was assigned based on fractionation on DEAE cellulose and electrophoretic mobility upon starch gel electrophoresis when purifying brain proteins
- c-fos
proto-oncogene whose viral homologue, v-fos, was identified from FBJ-murine osteosarcoma virus
- MEIS1
a homeobox gene found to be activated in myeloid leukemia by retroviral insertion
- Cyclin
cell cycle regulator
- IL
interleukin
- PI3K
phosphoinositide 3-kinase
- shRNA
small hairpin RNA
- egr-1
early growth response-1 gene
- TF-1
a human myeloid cell line
- MEK
mitogen-activated protein kinase
- NGF
nerve growth factor
- Bcl-2
B-cell lymphocytic-leukaemia proto-oncogene
- HSCs
hematopoietic stem cells
- RNAi
RNA interference
- K562
a human leukemia cell line
- PBX1
Pre-B-cell leukemia transcription factor 1
- mRNA
messenger RNA
- MiR
microRNA
- c-kit
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
- ERK
extracellular signal-regulated kinase
- ska
Spindle and KT Associated
- KG-501
2-napthol-AS-E-phosphate compound
- KIX
helical CREB-binding domain of CBP
Footnotes
The authors of this manuscript declare no financial conflicts of interest.
Contributor Information
Er-Chieh Cho, Email: echo@mednet.ucla.edu.
Bryan Mitton, Email: bmitton@mednet.ucla.edu.
References
- 1.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011 Feb 10;29(5):551–65. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011 Feb 10;29(5):487–94. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 3.Ofran Y, Rowe JM. Induction and postremission strategies in acute myeloid leukemia: what is new? Curr Opin Hematol. 2011 Mar;18(2):83–8. doi: 10.1097/MOH.0b013e32834399d9. [DOI] [PubMed] [Google Scholar]
- 4.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J Clin Oncol. 2011 Feb 10;29(5):475–86. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 5.Scholl S, Fricke HJ, Sayer HG, Hoffken K. Clinical implications of molecular genetic aberrations in acute myeloid leukemia. J Cancer Res Clin Oncol. 2009 Apr;135(4):491–505. doi: 10.1007/s00432-008-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barjesteh van Waalwijk van Doorn-Khosrovani S, Erpelinck C, Meijer J, van Oosterhoud S, van Putten WL, Valk PJ, Berna Beverloo H, Tenen DG, Löwenberg B, Delwel R. Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J. 2003;4(1):31–40. doi: 10.1038/sj.thj.6200216. [DOI] [PubMed] [Google Scholar]
- 7.Chillon MC, Fernandez C, Garcia-Sanz R, Balanzategui A, Ramos F, Fernandez-Calvo J, González M, Miguel JF. FLT3-activating mutations are associated with poor prognostic features in AML at diagnosis but they are not an independent prognostic factor. Hematol J. 2004;5(3):239–46. doi: 10.1038/sj.thj.6200382. [DOI] [PubMed] [Google Scholar]
- 8.Frohling S, Schlenk RF, Breitruck J, Benner A, Kreitmeier S, Tobis K, Döhner H, Döhner K AML Study Group Ulm. Acute myeloid leukemia. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: a study of the AML Study Group Ulm. Blood. 2002 Dec 15;100(13):4372–80. doi: 10.1182/blood-2002-05-1440. [DOI] [PubMed] [Google Scholar]
- 9.Falini B, Mecucci C, Tiacci E, Alcalay M, Rosati R, Pasqualucci L, La Starza R, Diverio D, Colombo E, Santucci A, Bigerna B, Pacini R, Pucciarini A, Liso A, Vignetti M, Fazi P, Meani N, Pettirossi V, Saglio G, Mandelli F, Lo-Coco F, Pelicci PG, Martelli MF GIMEMA Acute Leukemia Working Party. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005 Jan 20;352(3):254–66. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 10.Marcucci G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011 Jan 27;117(4):1121–9. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shankar DB, Cheng JC, Kinjo K, Federman N, Moore TB, Gill A, Rao NP, Landaw EM, Sakamoto KM. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005 Apr;7(4):351–62. doi: 10.1016/j.ccr.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Cheng JC, Kinjo K, Judelson DR, Chang J, Wu WS, Schmid I, Shankar DB, Kasahara N, Stripecke R, Bhatia R, Landaw EM, Sakamoto KM. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood. 2008 Feb 1;111(3):1182–92. doi: 10.1182/blood-2007-04-083600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayr B, Montminy M. Transcriptional regulation by the phosphorylationdependent factor CREB. Nat Rev Mol Cell Biol [Review] 2001 Aug;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 14.Sandoval S, Pigazzi M, Sakamoto KM. CREB: A Key Regulator of Normal and Neoplastic Hematopoiesis. Adv Hematol. 2009;2009:634292. doi: 10.1155/2009/634292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993 Oct 28;365(6449):855–9. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 16.Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol Cell. 2000 Sep;6(3):551–61. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 17.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–61. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 18.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003 Aug;12(2):413–23. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004 Nov;16(11):1211–27. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007 Dec 14;28(5):730–8. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001 Aug;2(8):599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto KK, Gonzalez GA, Biggs WH, 3rd, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988 Aug 11;334(6182):494–8. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 23.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987 Jul 9–15;328(6126):175–8. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 24.Bohm M, Moellmann G, Cheng E, Alvarez-Franco M, Wagner S, Sassone-Corsi P, Halaban R. Identification of p90RSK as the probable CREB-Ser133 kinase in human melanocytes. Cell Growth Differ. 1995 Mar;6(3):291–302. [PubMed] [Google Scholar]
- 25.Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science. 1996 Aug 16;273(5277):959–63. doi: 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 26.Kwon EM, Raines MA, Blenis J, Sakamoto KM. Granulocyte-macrophage colony-stimulating factor stimulation results in phosphorylation of cAMP response element-binding protein through activation of pp90RSK. Blood. 2000 Apr 15;95(8):2552–8. [PubMed] [Google Scholar]
- 27.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004 Dec 29;119(7):1041–54. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005 Mar 22;102(12):4459–64. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siu YT, Jin DY. CREB--a real culprit in oncogenesis. FEBS J. 2007 Jul;274(13):3224–32. doi: 10.1111/j.1742-4658.2007.05884.x. [DOI] [PubMed] [Google Scholar]
- 30.Xiao X, Li BX, Mitton B, Ikeda A, Sakamoto KM. Targeting CREB for cancer therapy: friend or foe. Curr Cancer Drug Targets. 2010 Jun;10(4):384–91. doi: 10.2174/156800910791208535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998 Feb;18(2):967–77. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esparza SD, Chang J, Shankar DB, Zhang B, Nelson SF, Sakamoto KM. CREB regulates Meis1 expression in normal and malignant hematopoietic cells. Leukemia. 2008 Mar;22(3):665–7. doi: 10.1038/sj.leu.2404933. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Iwasaki M, Ficara F, Lin C, Matheny C, Wong SH, Smith KS, Cleary ML. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell. 2010 Jun 15;17(6):597–608. doi: 10.1016/j.ccr.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011 Mar;12(3):141–51. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powell JD, Lerner CG, Ewoldt GR, Schwartz RH. The -180 site of the IL-2 promoter is the target of CREB/CREM binding in T cell anergy. J Immunol. 1999 Dec 15;163(12):6631–9. [PubMed] [Google Scholar]
- 36.Wen AY, Sakamoto KM, Miller LS. The role of the transcription factor CREB in immune function. J Immunol. 2010 Dec 1;185(11):6413–9. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desdouets C, Matesic G, Molina CA, Foulkes NS, Sassone-Corsi P, Brechot C, Sobczak-Thepot J. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol Cell Biol. 1995 Jun;15(6):3301–9. doi: 10.1128/mcb.15.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White PC, Shore AM, Clement M, McLaren J, Soeiro I, Lam EW, Brennan P. Regulation of cyclin D2 and the cyclin D2 promoter by protein kinase A and CREB in lymphocytes. Oncogene. 2006 Apr 6;25(15):2170–80. doi: 10.1038/sj.onc.1209255. [DOI] [PubMed] [Google Scholar]
- 39.Fox KE, Colton LA, Erickson PF, Friedman JE, Cha HC, Keller P, MacDougald OA, Klemm DJ. Regulation of cyclin D1 and Wnt10b gene expression by cAMP-responsive element-binding protein during early adipogenesis involves differential promoter methylation. J Biol Chem. 2008 Dec 12;283(50):35096–105. doi: 10.1074/jbc.M806423200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto KM, Fraser JK, Lee HJ, Lehman E, Gasson JC. Granulocyte-macrophage colony-stimulating factor and interleukin-3 signaling pathways converge on the CREB-binding site in the human egr-1 promoter. Mol Cell Biol. 1994 Sep;14(9):5975–85. doi: 10.1128/mcb.14.9.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonni A, Ginty DD, Dudek H, Greenberg ME. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol Cell Neurosci. 1995 Apr;6(2):168–83. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- 42.Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD. Mediation by a CREB family transcription factor of NGF-dependent survival of sympathetic neurons. Science. 1999 Dec 17;286(5448):2358–61. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 43.Dai CH, Krantz SB, Dessypris EN, Means RT, Jr, Horn ST, Gilbert HS. Polycythemia vera. II. Hypersensitivity of bone marrow erythroid, granulocyte-macrophage, and megakaryocyte progenitor cells to interleukin-3 and granulocyte-macrophage colony-stimulating factor. Blood. 1992 Aug 15;80(4):891–9. [PubMed] [Google Scholar]
- 44.Marvin J, Swaminathan S, Kraker G, Chadburn A, Jacobberger J, Goolsby C. Normal bone marrow signal-transduction profiles: a requisite for enhanced detection of signaling dysregulations in AML. Blood. 2011 Apr 14;117(15):e120–30. doi: 10.1182/blood-2010-10-316026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inukai T, Sugita K, Mitsui K, Iijima K, Goi K, Tezuka T, Kojika S, Kagami K, Mori T, Kinoshita A, Suzuki T, Okazaki-Koyama T, Nakazawa S. Participation of granulocyte colony-stimulating factor in the growth regulation of leukemia cells from Philadelphia chromosome-positive acute leukemia and blast crisis of chronic myeloid leukemia. Leukemia. 2000 Aug;14(8):1386–95. doi: 10.1038/sj.leu.2401837. [DOI] [PubMed] [Google Scholar]
- 46.Lee HJ, Mignacca RC, Sakamoto KM. Transcriptional activation of egr-1 by granulocyte-macrophage colony-stimulating factor but not interleukin 3 requires phosphorylation of cAMP response element-binding protein (CREB) on serine 133. J Biol Chem. 1995 Jul 7;270(27):15979–83. doi: 10.1074/jbc.270.27.15979. [DOI] [PubMed] [Google Scholar]
- 47.Wong A, Sakamoto KM. Granulocyte-macrophage colony-stimulating factor induces the transcriptional activation of egr-1 through a protein kinase A-independent signaling pathway. J Biol Chem. 1995 Dec 22;270(51):30271–3. doi: 10.1074/jbc.270.51.30271. [DOI] [PubMed] [Google Scholar]
- 48.Crans-Vargas HN, Landaw EM, Bhatia S, Sandusky G, Moore TB, Sakamoto KM. Expression of cyclic adenosine monophosphate response-element binding protein in acute leukemia. Blood. 2002 Apr 1;99(7):2617–9. doi: 10.1182/blood.v99.7.2617. [DOI] [PubMed] [Google Scholar]
- 49.Kinjo K, Sandoval S, Sakamoto KM, Shankar DB. The role of CREB as a proto-oncogene in hematopoiesis. Cell Cycle [Review] 2005 Sep;4(9):1134–5. doi: 10.4161/cc.4.9.1991. [DOI] [PubMed] [Google Scholar]
- 50.Pellegrini M, Cheng JC, Voutila J, Judelson D, Taylor J, Nelson SF, Sakamoto KM. Expression profile of CREB knockdown in myeloid leukemia cells. BMC Cancer. 2008;8:264. doi: 10.1186/1471-2407-8-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Esparza SD, Chang J, Shankar DB, Zhang B, Nelson SF, Sakamoto KM. CREB regulates Meis1 expression in normal and malignant hematopoietic cells. Leukemia. 2008 Mar;22(3):665–7. doi: 10.1038/sj.leu.2404933. [DOI] [PubMed] [Google Scholar]
- 52.Wong P, Iwasaki M, Somervaille TC, So CW, Cleary ML. Meis1 is an essential and rate-limiting regulator of MLL leukemia stem cell potential. Genes Dev. 2007 Nov 1;21(21):2762–74. doi: 10.1101/gad.1602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008 May 8;2(5):484–96. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011 Jan;223(2):102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011 May;1812(5):592–601. doi: 10.1016/j.bbadis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Gao XN, Lin J, Li YH, Gao L, Wang XR, Wang W, Kang HY, Yan GT, Wang LL, Yu L. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011 Aug 4;30(31):3416–28. doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- 57.Pigazzi M, Manara E, Baron E, Basso G. miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 2009 Mar 15;69(6):2471–8. doi: 10.1158/0008-5472.CAN-08-3404. [DOI] [PubMed] [Google Scholar]
- 58.Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010 Aug 26;466(7310):1105–9. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia W, Li J, Li S, Chen L, Ding H, Zhao Q, Fan M, Shen B, Shao N. Intronic miR-301 feedback regulates its host gene, ska2, in A549 cells by targeting MEOX2 to affect ERK/CREB pathways. Biochem Biophys Res Commun. 2010 Jun 11;396(4):978–82. doi: 10.1016/j.bbrc.2010.05.037. [DOI] [PubMed] [Google Scholar]
- 60.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, Buetow KH, Pui CH, Baker SD, Brindle PK, Downing JR. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011 Mar 10;471(7337):235–9. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Best JL, Amezcua CA, Mayr B, Flechner L, Murawsky CM, Emerson B, Zor T, Gardner KH, Montminy M. Identification of small-molecule antagonists that inhibit an activator: coactivator interaction. Proc Natl Acad Sci U S A. 2004 Dec 21;101(51):17622–7. doi: 10.1073/pnas.0406374101. [DOI] [PMC free article] [PubMed] [Google Scholar]