Abstract
The Ikzf1 gene encodes Ikaros – a DNA-binding zinc finger protein. Ikaros functions as a regulator of gene expression and chromatin remodeling. The biological roles of Ikaros include regulating the development and function of the immune system and acting as a master regulator of hematopoietic differentiation. Genomic profiling studies identified Ikzf1 as an important tumor suppressor in acute lymphoblastic leukemia (ALL), particularly in ALL that is associated with poor prognosis. This review summarizes currently available data regarding the structure and function of Ikaros, the clinical relevance of genetic inactivation of Ikzf1, and signal transduction pathways that regulate Ikaros function.
Keywords: Ikaros, leukemia, ALL, CK2, microarray, high-risk, deletion, phosphorylation, casein kinase, tumor suppression
I. INTRODUCTION
The Ikzf1 gene encodes Ikaros protein. Since its discovery, independently, by K. Georgopoulos et al. and S. Smale's group, Ikzf1 has attracted tremendous attention from the scientific community. This interest is due to the biological roles of Ikaros in hematopoiesis, immune function, and tumor suppression, as well as its complex role in the regulation of transcription and chromatin remodeling.1-3 During the past several years, Ikzf1 has been established as one of the most clinically relevant tumor suppressors in high-risk acute lymphoblastic leukemia (ALL). This review will summarize our current understanding of the structure and function of Ikaros protein and the clinical relevance of its inactivation, as well as insights into the signal transduction pathways that regulate Ikaros activity.
II. IKAROS MOLECULAR STRUCTURE
The Ikaros proteins contain several functional domains:
A. DNA-Binding Domain
The N-terminal end of Ikaros contains a DNA-binding domain that consists of three zinc finger motifs with a typical C2H2 structure, and one CCHC-type zinc finger. A point mutation in the fourth zinc finger has been associated with primary immunodeficiency and pancytopenia in man.4
B. Dimerization Domain
The C-terminal end of Ikaros contains two zinc finger motifs that are essential for protein-protein interaction with other Ikaros isoforms or Ikaros family members.5 This allows for the formation of very diverse protein complexes among different Ikaros family members and/or isoforms.
C. Bipartite Activation Domain
This bipartite activation domain lies adjacent to C-terminal zinc finger region. The presence of this domain stimulates basal levels of transcriptional activation of Ikaros target genes.5,6
D. Human Ikaros Activation Domain
This 20-amino acid domain adjacent to the N-terminal zinc fingers regulates DNA-binding specificity and function in transcriptional activation and chromatin remodeling of Ikaros target genes in humans.7,8
III. IKAROS IN GENE REGULATION AND CHROMATIN REMODELING
Ikaros has been shown to bind DNA and directly regulate expression of its target genes.2,3,9,10 Subsequent experiments established that Ikaros regulates transcription of its target genes primarily via chromatin remodeling. Ikaros is abundantly localized in pericentromeric heterochromatin in the nucleus.11 Experiments by several groups have shown that Ikaros regulates expression of its target genes by recruiting them to pericentromeric heterochromatin, resulting in their activation or repression.8,11,12
Ikaros associates with histone deacetylase (HDAC)-containing complexes by direct interaction with the NuRD complex ATPase, Mi-2β, and with Sin3A and Sin3B.13,14 It has been suggested that Ikaros recruits histone deacetylase complex to the upstream regulatory elements of its target genes, which results in chromatin remodeling and repression of the Ikaros target gene.11,15
It has been demonstrated that Ikaros can function as transcriptional repressor in a HDAC-independent way. Ikaros interacts with the corepressor, CtBP 16 and the Ikaros-CtBP complex acts to repress transcription without HDAC involvement, thus Ikaros can function as a transcriptional repressor of its target genes through both HDAC-dependent and HDAC-independent mechanisms.16
Ikaros interacts with Brg-1, a catalytic subunit of the SWI/SNF chromatin remodeling complex that functions as an activator of gene expression.14,17 It has been suggested that Ikaros functions as transcriptional activator by recruiting the SWI/SNF nucleosome remodeling complex to the upstream regions of its target genes resulting in chromatin remodeling and activation of the gene Thus, Ikaros can both activate or repress transcription of its target genes via chromatin remodeling, depending on whether it associates with the NuRD, the CtBP or the SWI/SNF complex.
IV. IKAROS IN B-CELL ALL
Since the discovery that Ikaros functions as a master regulator of lymphocyte differentiation and a tumor suppressor in the mouse, human studies have been focused on determining whether Ikaros acts as a tumor suppressor in human leukemia. Initial studies focused on the expression of small dominant negative Ikzf1 isoforms. These studies found that expression of DN isoforms was associated with adult B cell ALL,18 as well as with myelodysplastic syndrome,19 AML,20 and adult and juvenile CML.21 However, due to an absence of functional data and the small numbers of patients as well as the lack of genetic evidence for alteration of Ikzf1, these studies did not have a profound effect on clinical practice.
During the last several years multiple microarray-based analyses of genetic changes and alterations in gene expression have been conducted by several groups. These genomic profiling studies have produced strong evidence that that Ikaros plays a key role in tumor suppression in pediatric B-cell ALL and in particular in high-risk B-cell ALL. These findings can be summarized as follows:
Deletion of a single Ikzf1 allele or mutation of a single copy of Ikzf1 were detected in 15% of all cases of pediatric B-cell ALL.22 It should be noted that all of the described mutations were either nonsense, or frameshift mutations, or mutations that functionally inactivated a particular Ikzf1 allele. Thus, each of these defects resulted in haploinsufficiency of the Ikzf1 gene, along with expression of a functionally inactive form of Ikaros which could potentially act as a dominant-negative form.
Deletion or mutation of a single copy of the Ikzf1 allele was detected in over 80% of BCR-ABL1 ALL, a subtype of ALL that are associated with a poor outcome. Deletion or mutation of an Ikzf1 allele was also identified in 66% of chronic myeloid leukemia (CML) patients during lymphoid blast crisis.23-25
Deletion or mutation of Ikzf1 was identified in one-third of cases of BCR-ABL1 negative ALL. Haploinsufficiency of Ikzf1 was associated with a three-fold increase in relapse of ALL following treatment.26-28
Expression profiles of BCR-ABLL1 negative cases with haploinsufficiency of Ikzf1 and poor prognosis were noted to have similar expression profiles to BCR-ABL1 positive ALL.26 This led to the definition of the BCR-ABL1-like subtype of B-cell ALL with haploinsufficiency of Ikzf1 or other transcriptional regulators.28
Inherited genetic variations of Ikzf1 are associated with the risk of childhood ALL and poor outcome of the disease.29,30 Genetic variations have been shown to affect the expression level of Ikaros, suggesting a potential mechanism for leukemogenesis.30
In 14% of pediatric high-risk leukemia with a poor outcome,31 the CRLF2 gene is overexpessed due to rearrangement. This CRLF2 defect is significantly associated with JAK mutations and with deletions or mutations of Ikzf1.31,32
The functional, leukemogenic significance of Ikzf1 haploinsufficiency and/or expression of dominant-negative Ikaros isoforms has been confirmed by several animal models. These models demonstrated that the expression of the dominant negative Ikzf1 allele in CD34+ cells results in impaired lymphoid differentiation.33 These models also demonstrate that the haploinsufficiency of Ikzf1 accelerates the development of leukemia in both retrovirally transduced bone marrow transplants and in a transgenic model of BCR-ABL1 ALL.34,35
Overall, the above data established that:
Ikaros acts as a highly clinically-relevant tumor suppressor in B-cell ALL and particularly in high-risk B-cell ALL
The modest decrease in Ikaros activity (e.g. haploinsufficiency) is sufficient to contribute to leukemogenesis
Genetic alterations of Ikzf1 might serve as a prognostic marker for B-cell ALL outcome.
Based on these results, testing for genetic alteration of Ikzf1 is currently performed in prospective clinical trials.
V. IKAROS IN T-CELL ALL
Initial studies of Ikaros in T-cell ALL produced somewhat conflicting data: All 18 T-ALL patients in the first study were reported to express dominant negative Ikaros isoforms (assessed by Western blot and RT-PCR),36 suggesting a strong correlation of loss of Ikaros function with the development of T-cell ALL. However, in subsequent studies on a total of 14 T-ALL patients (both adult and pediatric) dominant-negative isoforms were not detected by Western blot and RT-PCR.18,37 However, the expression of a dominant-negative isoform of the Ikaros-family member – Helios – was associated with T-cell ALL in one study.38
Deletion of one copy of Ikaros was detected in 5% of T-cell ALL patients in more comprehensive studies that utilized high-resolution CGH-arrays on a total of 81 patients.23,39,40 The most recent study combined Western blot, CGH-array analysis, and sequencing of Ikaros cDNA following RT-PCR to provide a more complete view of the relation of Ikaros and T-cell ALL evaluate. That study of 25 cases of human T-cell ALL detected one patient (4%) in which one Ikaros allele had been deleted. The Ikaros protein that was produced by the other intact allele exhibited association with an abnormal cytoplasmic structure and a loss of nuclear localization.41 This study provided the first definitive functional evidence to link the complete loss of Ikaros function with human T-cell ALL.42
In summary, these studies of human T-cell ALL demonstrate that inactivation of the Ikzf1 gene by deletion occurs in human T-cell ALL in at least 5% of cases. Although Ikaros deletion is less frequent in T-ALL, when compared to B-cell ALL (15%) or BCR-ABL1 ALL (80%), its occurrence in T-Cell All is a notable cause of T-cell ALL, and testing for genetic alteration of Ikzf1 in newly diagnosed patients with this disease is warranted. It remains to be determined whether Ikaros deletion will have prognostic significance in T-cell ALL.
VI. MECHANISMS OF IKAROS TUMOR SUPPRESSOR ACTIVITY
The mechanism by which Ikaros suppresses malignant transformation and the development of ALL is largely unknown. The discovery of several Ikaros target genes provided potential mechanisms of the tumor suppressor action of Ikaros in ALL. These are summarized below.
A. Positive Regulation of B cell Differentiation
Expression of several genes that are essential for normal B cell differentiation are directly regulated by Ikaros
Ikaros has been shown to bind the Igll1 promoter and to regulate expression of this gene in early B lineage cells.43,44 The Ikaros binding site at the Igll1 promoter overlaps the binding site of the EBF transcriptional activator. Thus, Ikaros regulates Igll1 transcription by competing with EBF for binding to the Igll1 promoter and subsequently regulating Igll1 expression. The Igll1 gene encodes Lambda5, a component of the pre-B cell receptor (pre-BCR). Pre-BCR expression is essential for progression beyond the pre-B cell stage of differentiation. Thus, Ikaros controls this critical step in early stages of B cell differentiation.
Ikaros binds to the promoter region of the recombinase activating genes (rag) and positively regulates transcription of both rag1 and rag2.45 Upregulation of expression of RAG1 and RAG2, along with Ikaros-mediated control of the compaction of the immunoglobulin heavy-chain locus, as well as accessibility of the variable gene segments, promotes immunoglobulin heavy-chain gene rearrangement.45 Thus Ikaros controls another essential step in normal B cell differentiation.
B. Positive Regulation of T cell Differentiation
Ikaros has been shown to regulate expression of multiple genes that are essential for T cell differentiation.
The regulation of dntt (terminal deoxynucleotide transferase -TdT) gene expression during thymocyte differentiation by Ikaros has been studied by several groups.9,46-48 Ikaros binds to the D’ upstream regulatory element of the dntt gene promoter. This region contains contains a consensus binding site that is bound, in vivo, by the Elf-1 activator – a member of the Ets family of transcription factors. Ikaros and Elf-1 have been shown to compete for the occupancy of the D’ upstream regulatory element of the dntt gene during thymocyte development. Ikaros binding to the dntt upstream regulatory element results in repression of TdT transcription, which is associated with repositioning of the dntt gene to pericentromeric heterochromatin.46 During induction of thymocyte differentiation, Ikaros displaces Elf-1 from the D’ upstream regulatory element of dntt, which results in downregulation of TdT expression. Phosphorylation of Ikaros has been shown to regulate Ikaros’ affinity toward the dntt upstream regulatory region.49
During thymocyte development, Ikaros binds to the regulatory element of the CD8α gene. It has been suggested that Ikaros positively regulates transcription of the CD8α gene during T cell development,50 and thus, plays an important role in CD4 versus CD8 lineage commitment. This hypothesis has been supported by decreased numbers of CD8+ T cells in Ikaros-deficient mice.50
Studies by Georgopoulos’ group demonstrated that Ikaros binds to the upstream regulatory region of the CD4 gene. Ikaros binding at this site, in complex with the Mi-2β chromatin remodeler, results in expression of CD4, suggesting that Ikaros positively regulates transcription of CD4 via chromatin remodeling.51
C. Downregulation of the Notch Pathway
The Notch pathway is essential for T cell development. Activation of the Notch-1 gene has been found in over 50% of T-cell ALL.52 In addition, T-cell ALL cells have high expression of the Notch target genes Hes-1 and pT.53 In T-cell leukemia derived from Ikaros-deficient mice, the Notch pathway is activated.54
The synergism between Notch activation and the loss of Ikaros function in T cell leukemogenesis has been demonstrated by Beverly and Capobianco.55 Since the consensus binding sequences for the Notch-associated transcriptional activator, CSL, and Ikaros were highly similar, Ikaros was hypothesized to interfere with CSL binding and Notch signaling.55 Ikaros directly binds to the upstream regulatory element of a Notch target gene Hes-1, and downregulates its expression. Ikaros competes with the transcriptional activator CSL for binding to the upstream regulator element of Hes-1 in a manner similar to that demonstrated for EBF1 and Elf1 (described above).56 It has been suggested that transcriptional repression of Hes-1 by Ikaros involves chromatin remodeling, since Ikaros binding to the upstream regulatory region of Hes-1 leads to decreased histone H3 acetylation at the Hes-1 locus.57
Ikaros competes with CSL for the binding to the upstream regulatory region of Deltex1, another target gene for the Notch signaling pathway.57 Ikaros represses transcription of Deltex1 by chromatin remodeling as evidenced by decreased histone H3 acetylation at the Deltex1 locus following Ikaros binding to the upstream region of Deltex1.57
D. Negative Regulation of Cellular Proliferation
The negative regulation of pre-B cell proliferation by Ikaros has been demonstrated by Ma et al. The mechanism of inhibition of cellular proliferation involves direct binding of Ikaros to the promoter of the c-Myc gene which results in direct suppression of c-Myc expression in pre-B cells.58 Repression of c-Myc by Ikaros in pre-B cells also leads to induction of expression of p27, as well as downregulation of cyclin D3.58 These data provided a potential mechanism by which Ikaros can suppress proliferation of pre-B cells in vivo.
It has also been shown that Ikaros can negatively regulate cell cycle progression at the G1/S transition,59 suggesting that Ikaros has a role in the regulation of the G1/S check point of the cell cycle.
E. Regulation of apoptosis
The loss of Ikaros function is associated with increased Bcl-xL expression, which suggests that Ikaros downregulates Bcl-xL expression.20,60,61 These data led to the hypothesis that Ikaros regulates apoptosis, and that decreased Ikaros activity in leukemia cells would increase resistance to chemotherapy. This hypothesis remains speculative due to a lack of mechanistic data to back up this assertion.
F. Post-Translational Modifications Regulate Ikaros Tumor Suppressor Function
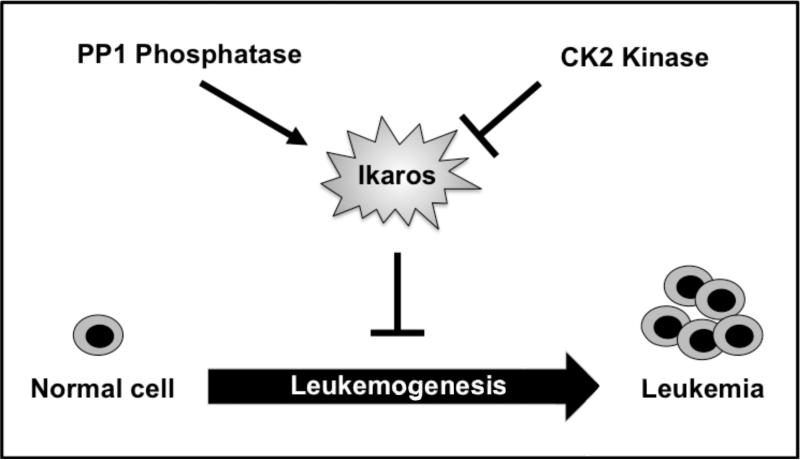
Post-translational modifications have been shown to regulate Ikaros’ activity. Sumoylation regulates Ikaros repressor function.62 The cell cycle-specific phosphorylation of Ikaros regulates its DNA-binding ability and nuclear localization during mitosis.63 In cycling cells Ikaros is a direct substrate for pro-oncogenic kinase CK2. Phosphorylation of Ikaros by CK2 regulates the subcellular localization of Ikaros to pericentromeric heterochromatin, and its DNA-binding affinity toward the upstream regulatory element of the Ikaros’ target gene, TdT,49 as well as its ability to control G1/S cell cycle progression.59 More recent data showed that Ikaros is a substrate for PP1 phosphatase, and that CK2 and PP1 exert opposite effects on Ikaros function in DNA binding, pericentromeric localization, and chromatin remodeling.64 Overexpression of CK2 has been shown to increase degradation of Ikaros protein via the ubiquitin pathway, while PP1 counteracts this process (Fig.1).64 These data led to the development of a model whereby the loss of Ikaros activity in leukemia can result from genetic defects (deletion, mutation) or functional inactivation of Ikaros due to hyperphosphorylation by CK2.65 More studies are needed to test this model.
FIGURE 1. Phosphorylation regulates the tumor suppressor function of Ikaros.
CK2 kinase directly phosphorylates and functionally inactivates Ikaros, while PP1 phosphatase counteracts this process. Functional inactivation of Ikaros by CK2 kinase promotes leukemogenesis.
VII. CONCLUSION
Genomic profiling of ALL identified Ikaros as a major tumor suppressor in ALL. Functional studies revealed possible mechanisms of tumor suppression by Ikaros, as well as the regulatory pathways that control the tumor suppressor function of Ikaros. Future studies will be directed toward evaluating genetic changes in Ikaros as a prognostic marker for ALL, as well as a factor in the decision-making process to design appropriate therapy. Regulatory pathways that control the tumor suppressor function of Ikaros are a potential target for a novel chemotherapy for ALL.
ACKNOWLEDGEMENTS
The work has been supported in whole or in part by an R01 HL095120 grant, a St. Baldrick's Foundation Career Development Award, the Four Diamonds Fund of the Pennsylvania State University, College of Medicine, and the John Wawrynovic Leukemia Research Scholar Endowment (to S.D.) This work was also supported by the Department of Pathology and Human Anatomy and the Center for Health Disparities and Molecular Medicine at Loma Linda University School of Medicine (to K.J.P). This work was also supported by a Grants for Research and School Partnerships from Loma Linda University (to K.J.P).
ABBREVIATIONS
- ALL
acute lymphoblastic leukemia
- HDAC
histone deacetylase
- CML
chronic myeloid leukemia
- pre-BCR
pre-B cell receptor
- rag
recombinase activating genes
- TdT
terminal deoxynucleotide transferase
Footnotes
THIS ARTICLE HAS NOT BEEN PUBLISHED ELSEWHERE AND HAS NOT BEEN SUBMITTED ELSEWHERE.
REFERENCES
- 1.Lo K, Landau NR, Smale ST. LyF-1, a transcriptional regulator that interactswith a novel class of promoters for lymphocyte-specific genes. Mollecular Cellular Biology. 1991;11:5229–5243. doi: 10.1128/mcb.11.10.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258(5083):808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 3.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14(11):7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman FD, Gurel Z, Al-Zubeidi D, Fried AJ, Icardi M, Song C, Dovat S. Congenital pancytopenia and absence of B lymphocytes in a neonate with a mutation in the ikaros gene. Pediatr Blood Cancer. 2011 doi: 10.1002/pbc.23160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. Embo J. 1996;15(19):5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 6.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 7.Ronni T, Payne KJ, Ho S, Bradley MN, Dorsam G, Dovat S. Human Ikaros function in activated T cells is regulated by coordinated expression of its largest isoforms. J Biol Chem. 2007;282(4):2538–2547. doi: 10.1074/jbc.M605627200. [DOI] [PubMed] [Google Scholar]
- 8.Kim JH, Ebersole T, Kouprina N, Noskov VN, Ohzeki J, Masumoto H, Mravinac B, Sullivan BA, Pavlicek A, Dovat S, Pack SD, Kwon YW, Flanagan PT, Loukinov D, Lobanenkov V, Larionov V. Human gamma-satellite DNA maintains open chromatin structure and protects a transgene from epigenetic silencing. Genome Res. 2009;19(4):533–544. doi: 10.1101/gr.086496.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst P, Hahm K, Smale ST. Both LyF-1 and an Ets protein interact with a critical promoter element in the murine terminal transferase gene. Mol Cell Biol. 1993;13(5):2982–2992. doi: 10.1128/mcb.13.5.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molnár A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14(12):8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91(6):845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 12.Koipally J, Heller EJ, Seavitt JR, Georgopoulos K. Unconventional potentiation of gene expression by ikaros. J Biol Chem. 2002;277(15):13007–13015. doi: 10.1074/jbc.M111371200. [DOI] [PubMed] [Google Scholar]
- 13.Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. Embo J. 1999;18(11):3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10(3):345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 15.Liberg D, Smale ST, Merkenschlager M. Upstream of Ikaros. Trends Immunol. 2003;24(11):567–570. doi: 10.1016/j.it.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Koipally J, Georgopoulos K. Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J Biol Chem. 2000;275(26):19594–19602. doi: 10.1074/jbc.M000254200. [DOI] [PubMed] [Google Scholar]
- 17.O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, Han S, Seong RH, Park SD, Agalioti T, Munshi N, Thanos D, Erdjument-Bromage H, Tempst P, Bank A. An ikaros-containing chromatin-remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20(20):7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakase K, Ishimaru F, Avitahl N, Dansako H, Matsuo K, Fujii K, Sezaki N, Nakayama H, Yano T, Fukuda S, Imajoh K, Takeuchi M, Miyata A, Hara M, Yasukawa M, Takahashi I, Taguchi H, Matsue K, Nakao S, Niho Y, Takenaka K, Shinagawa K, Ikeda K, Niiya K, Harada M. Dominant negative isoform of the Ikaros gene in patients with adult B- cell acute lymphoblastic leukemia. Cancer Res. 2000;60(15):4062–4065. [PubMed] [Google Scholar]
- 19.Crescenzi B, La Starza R, Romoli S, Beacci D, Matteucci C, Barba G, Aventin A, Marynen P, Ciolli S, Nozzoli C, Martelli MF, Mecucci C. Submicroscopic deletions in 5q- associated malignancies. Haematologica. 2004;89(3):281–285. [PubMed] [Google Scholar]
- 20.Yagi T, Hibi S, Takanashi M, Kano G, Tabata Y, Imamura T, Inaba T, Morimoto A, Todo S, Imashuku S. High frequency of Ikaros isoform 6 expression in acute myelomonocytic and monocytic leukemias: implications for up-regulation of the antiapoptotic protein Bcl-XL in leukemogenesis. Blood. 2002;99(4):1350–1355. doi: 10.1182/blood.v99.4.1350. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama H, Ishimaru F, Avitahl N, Sezaki N, Fujii N, Nakase K, Ninomiya Y, Harashima A, Minowada J, Tsuchiyama J, Imajoh K, Tsubota T, Fukuda S, Sezaki T, Kojima K, Hara M, Takimoto H, Yorimitsu S, Takahashi I, Miyata A, Taniguchi S, Tokunaga Y, Gondo H, Niho Y, Nakao S, Kyo T, Dohy H, Kamada N, Harada M. Decreases in Ikaros activity correlate with blast crisis in patients with chronic myelogenous leukemia. Cancer Res. 1999;59(16):3931–3934. [PubMed] [Google Scholar]
- 22.Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui CH, Relling MV, Evans WE, Shurtleff SA, Downing JR. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 23.Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J, White D, Hughes TP, Le Beau MM, Pui CH, Relling MV, Shurtleff SA, Downing JR. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 24.Martinelli G, Iacobucci I, Storlazzi CT, Vignetti M, Paoloni F, Cilloni D, Soverini S, Vitale A, Chiaretti S, Cimino G, Papayannidis C, Paolini S, Elia L, Fazi P, Meloni G, Amadori S, Saglio G, Pane F, Baccarani M, Foa R. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: a GIMEMA AL WP report. J Clin Oncol. 2009;27(31):5202–5207. doi: 10.1200/JCO.2008.21.6408. [DOI] [PubMed] [Google Scholar]
- 25.Iacobucci I, Storlazzi CT, Cilloni D, Lonetti A, Ottaviani E, Soverini S, Astolfi A, Chiaretti S, Vitale A, Messa F, Impera L, Baldazzi C, D'Addabbo P, Papayannidis C, Lonoce A, Colarossi S, Vignetti M, Piccaluga PP, Paolini S, Russo D, Pane F, Saglio G, Baccarani M, Foa R, Martinelli G. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: on behalf of Gruppo Italiano Malattie Ematologiche dell'Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood. 2009;114(10):2159–2167. doi: 10.1182/blood-2008-08-173963. [DOI] [PubMed] [Google Scholar]
- 26.Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, Harvey RC, Chen IM, Clifford RJ, Carroll WL, Reaman G, Bowman WP, Devidas M, Gerhard DS, Yang W, Relling MV, Shurtleff SA, Campana D, Borowitz MJ, Pui CH, Smith M, Hunger SP, Willman CL, Downing JR. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuiper RP, Waanders E, van der Velden VH, van Reijmersdal SV, Venkatachalam R, Scheijen B, Sonneveld E, van Dongen JJ, Veerman AJ, van Leeuwen FN, van Kessel AG, Hoogerbrugge PM. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia. 2010;24(7):1258–1264. doi: 10.1038/leu.2010.87. [DOI] [PubMed] [Google Scholar]
- 28.Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, Horstmann MA, Janka-Schaub GE, Kamps WA, Evans WE, Pieters R. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10(2):125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, Willman C, Neale G, Downing J, Raimondi SC, Pui CH, Evans WE, Relling MV. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1001–1005. doi: 10.1038/ng.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, Kinsey SE, Lightfoot T, Roman E, Irving JA, Allan JM, Tomlinson IP, Taylor M, Greaves M, Houlston RS. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41(9):1006–1010. doi: 10.1038/ng.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harvey RC, Mullighan CG, Chen IM, Wharton W, Mikhail FM, Carroll AJ, Kang H, Liu W, Dobbin KK, Smith MA, Carroll WL, Devidas M, Bowman WP, Camitta BM, Reaman GH, Hunger SP, Downing JR, Willman CL. Rearrangement of CRLF2 is associated with mutation of JAK kinases, alteration of IKZF1, Hispanic/Latino ethnicity, and a poor outcome in pediatric B-progenitor acute lymphoblastic leukemia. Blood. 2010;115(26):5312–5321. doi: 10.1182/blood-2009-09-245944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cario G, Zimmermann M, Romey R, Gesk S, Vater I, Harbott J, Schrauder A, Moericke A, Izraeli S, Akasaka T, Dyer MJ, Siebert R, Schrappe M, Stanulla M. Presence of the P2RY8-CRLF2 rearrangement is associated with a poor prognosis in non-high-risk precursor B-cell acute lymphoblastic leukemia in children treated according to the ALL-BFM 2000 protocol. Blood. 2010;115(26):5393–5397. doi: 10.1182/blood-2009-11-256131. [DOI] [PubMed] [Google Scholar]
- 33.Tonnelle C, Bardin F, Maroc C, Imbert AM, Campa F, Dalloul A, Schmitt C, Chabannon C. Forced expression of the Ikaros 6 isoform in human placental blood CD34(+) cells impairs their ability to differentiate toward the B- lymphoid lineage. Blood. 2001;98(9):2673–2680. doi: 10.1182/blood.v98.9.2673. [DOI] [PubMed] [Google Scholar]
- 34.Collins-Underwood JR, Miller CB, Downing JR, Mullighan CG. Ikzf1 Haploinsufficiency Contributes to the Pathogenesis of BCR-ABL1 Positive Acute Lymphoblastic Leukemia. Blood. 2009:114. (abstract 678) [Google Scholar]
- 35.Virely C, Moulin S, Cobaleda C, Lasgi C, Alberdi A, Soulier J, Sigaux F, Chan S, Kastner P, Ghysdael J. Haploinsufficiency of the IKZF1 (IKAROS) tumor suppressor gene cooperates with BCR-ABL in a transgenic model of acute lymphoblastic leukemia. Leukemia. 2010;24(6):1200–1204. doi: 10.1038/leu.2010.63. [DOI] [PubMed] [Google Scholar]
- 36.Sun L, Crotty ML, Sensel M, Sather H, Navara C, Nachman J, Steinherz PG, Gaynon PS, Seibel N, Mao C, Vassilev A, Reaman GH, Uckun FM. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin Cancer Res. 1999;5(8):2112–2120. [PubMed] [Google Scholar]
- 37.Ruiz A, Jiang J, Kempski H, Brady HJ. Overexpression of the Ikaros 6 isoform is restricted to t(4;11) acute lymphoblastic leukaemia in children and infants and has a role in B-cell survival. Br J Haematol. 2004;125(1):31–37. doi: 10.1111/j.1365-2141.2004.04854.x. [DOI] [PubMed] [Google Scholar]
- 38.Nakase K, Ishimaru F, Fujii K, Tabayashi T, Kozuka T, Sezaki N, Matsuo Y, Harada M. Overexpression of novel short isoforms of Helios in a patient with T- cell acute lymphoblastic leukemia. Exp Hematol. 2002;30(4):313–317. doi: 10.1016/s0301-472x(01)00796-2. [DOI] [PubMed] [Google Scholar]
- 39.Maser RS, Choudhury B, Campbell PJ, Feng B, Wong KK, Protopopov A, O'Neil J, Gutierrez A, Ivanova E, Perna I, Lin E, Mani V, Jiang S, McNamara K, Zaghlul S, Edkins S, Stevens C, Brennan C, Martin ES, Wiedemeyer R, Kabbarah O, Nogueira C, Histen G, Aster J, Mansour M, Duke V, Foroni L, Fielding AK, Goldstone AH, Rowe JM, Wang YA, Look AT, Stratton MR, Chin L, Futreal PA, DePinho RA. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447(7147):966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuiper RP, Schoenmakers EF, van Reijmersdal SV, Hehir-Kwa JY, van Kessel AG, van Leeuwen FN, Hoogerbrugge PM. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 2007;21(6):1258–1266. doi: 10.1038/sj.leu.2404691. [DOI] [PubMed] [Google Scholar]
- 41.Marcais A, Jeannet R, Hernandez L, Soulier J, Sigaux F, Chan S, Kastner P. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk Res. 2010;34(4):426–429. doi: 10.1016/j.leukres.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 42.Dovat S, Payne KJ. Tumor suppression in T cell leukemia--the role of Ikaros. Leuk Res. 2010;34(4):416–417. doi: 10.1016/j.leukres.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, Smale ST, Fisher AG, Merkenschlager M. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26(3):335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Sabbattini P, Lundgren M, Georgiou A, Chow C, Warnes G, Dillon N. Binding of Ikaros to the lambda5 promoter silences transcription through a mechanism that does not require heterochromatin formation. Embo J. 2001;20(11):2812–2822. doi: 10.1093/emboj/20.11.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reynaud D, I AD, K LR, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9(8):927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trinh LA, Ferrini R, Cobb BS, Weinmann AS, Hahm K, Ernst P, Garraway IP, Merkenschlager M, Smale ST. Down-regulation of TdT transcription in CD4+CD8+ thymocytes by Ikaros proteins in direct competition with an Ets activator. Genes and Development. 2001;15:1817–1832. doi: 10.1101/gad.905601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst P, Hahm K, Trinh L, Davis JN, Roussel MF, Turck CW, Smale ST. A potential role for Elf-1 in terminal transferase gene regulation. Mol Cell Biol. 1996;16(11):6121–6131. doi: 10.1128/mcb.16.11.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su RC, Sridharan R, Smale ST. Assembly of silent chromatin during thymocyte development. Semin Immunol. 2005;17(2):129–140. doi: 10.1016/j.smim.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 49.Gurel Z, Ronni T, Ho S, Kuchar J, Payne KJ, Turk CW, Dovat S. Recruitment of ikaros to pericentromeric heterochromatin is regulated by phosphorylation. J Biol Chem. 2008;283(13):8291–8300. doi: 10.1074/jbc.M707906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harker N, Naito T, Cortes M, Hostert A, Hirschberg S, Tolaini M, Roderick K, Georgopoulos K, Kioussis D. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10(6):1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 51.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27(5):723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 53.Chiaramonte R, Basile A, Tassi E, Calzavara E, Cecchinato V, Rossi V, Biondi A, Comi P. A wide role for NOTCH1 signaling in acute leukemia. Cancer Lett. 2005;219(1):113–120. doi: 10.1016/j.canlet.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 54.Dumortier A, Jeannet R, Kirstetter P, Kleinmann E, Sellars M, dos Santos NR, Thibault C, Barths J, Ghysdael J, Punt JA, Kastner P, Chan S. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol. 2006;26(1):209–220. doi: 10.1128/MCB.26.1.209-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3(6):551–564. doi: 10.1016/s1535-6108(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 56.Kleinmann E, Geimer Le Lay AS, Sellars M, Kastner P, Chan S. Ikaros represses the transcriptional response to Notch signaling in T-cell development. Mol Cell Biol. 2008;28(24):7465–7475. doi: 10.1128/MCB.00715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kathrein KL, Chari S, Winandy S. Ikaros directly represses the notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem. 2008;283(16):10476–10484. doi: 10.1074/jbc.M709643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 30(17):4149–4158. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomez-del Arco P, Maki K, Georgopoulos K. Phosphorylation controls Ikaros's ability to negatively regulate the G(1)-S transition. Mol Cell Biol. 2004;24(7):2797–2807. doi: 10.1128/MCB.24.7.2797-2807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ezzat S, Zhu X, Loeper S, Fischer S, Asa SL. Tumor-derived Ikaros 6 acetylates the Bcl-XL promoter to up-regulate a survival signal in pituitary cells. Mol Endocrinol. 2006;20(11):2976–2986. doi: 10.1210/me.2006-0265. [DOI] [PubMed] [Google Scholar]
- 61.Kano G, Morimoto A, Takanashi M, Hibi S, Sugimoto T, Inaba T, Yagi T, Imashuku S. Ikaros dominant negative isoform (Ik6) induces IL-3-independent survival of murine pro-B lymphocytes by activating JAK-STAT and up-regulating Bcl-xl levels. Leuk Lymphoma. 2008;49(5):965–973. doi: 10.1080/10428190801993462. [DOI] [PubMed] [Google Scholar]
- 62.Gomez-del Arco P, Koipally J, Georgopoulos K. Ikaros SUMOylation: switching out of repression. Mol Cell Biol. 2005;25(7):2688–2697. doi: 10.1128/MCB.25.7.2688-2697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dovat S, Ronni T, Russell D, Ferrini R, Cobb BS, Smale ST. A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes Dev. 2002;16(23):2985–2990. doi: 10.1101/gad.1040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Popescu M, Gurel Z, Ronni T, Song C, Hung KY, Payne KJ, Dovat S. Ikaros stability and pericentromeric localization are regulated by protein phosphatase 1. J Biol Chem. 2009;284(20):13869–13880. doi: 10.1074/jbc.M900209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Perez-Casellas LA, Savic A, Song C, Dovat S. Ikaros isoforms: The saga continues. World J Biol Chem. 2011;2(6):140–145. doi: 10.4331/wjbc.v2.i6.140. [DOI] [PMC free article] [PubMed] [Google Scholar]