Abstract
E2F family of transcription factors are best known for regulating genes involved in cell cycle control, cell proliferation, tumorigenesis, and apoptosis. Recent evidences have revealed their critical involvement in modulating cellular response to hypoxia and ischemia in a variety of physiological and pathological processes. Of particular interest are findings that E2Fs act as both regulators and targets of microRNAs that govern hypoxic/ischemic angiogenesis. This review focuses on the crosstalk between E2Fs and microRNAs that have been shown to participate in the regulation of angiogenesis, hypoxia response and ischemic disease.
Keywords: E2F, microRNA, angiogenesis, ischemia, hypoxia
Introduction
Exposure of a cell or an organism to hypoxia, or decrease in the level of oxygen, leads to a massive well studied response characterized by a change of expression of certain genes [1]. Hypoxia may be a necessary part of a physiological process, as in development, or may be a characteristic of some pathological situation such as solid tumor growth or ischemic disease. One of the well characterized organism's responses to hypoxia is an increase of blood supply to tissues through angiogenesis, the process of developing new blood vessels from pre-existing ones.
Recently it became clear that many processes triggered by hypoxia are regulated by microRNAs (miRNAs, or miRs). MiRNAs are short non-coding RNAs ∼22 nt long that modulate the stability and/or translation potential of their targets [2]. At present there are more than 1400 miRNAs discovered in humans (miRBase release 17, April 2011) that are predicted to target more than 60% of all mRNAs [3], and this number is constantly growing.
Most mammalian miRNAs are transcribed by RNA polymerase II into primary miRNA transcripts (pri-miRNA) that consists of one or more hairpin structures [4-5]. Many pri-miRNAs are 5'-capped and polyadenylated and often produce more than one functional miRNA. pri-miRNAs are cleaved by the nuclear microprocessor complex containing RNase III enzyme Drosha and DGCR8 protein into ∼ 70 nt long hairpin precursor pre-miRNAs which are carried into cytoplasm by Exportin-5. In the cytoplasm pre-miRNAs get further processed and loaded into RISC (RNA-induced silencing complex) by the RISC loading complex (RLC). RLC is a multi-protein complex consisting of RNase III enzyme Dicer and proteins TRBP/PACT and core component Argonaut-2. The pre-miRNA is cleaved into ∼ 22 nt miRNA duplex which is separated into guide and passenger strands. The passenger strand typically gets degraded, and the guide strand directs RISC to the complimentary sites within target mRNAs. Usually miRNAs basepair with their target imperfectly and induce translation inhibition. The process of target recognition by miRNAs is not clearly understood yet, but it is generally accepted that Watson-Crick pairing between the “seed region” of miRNA (between nucleotides 2-7) and 3'-UTR of target mRNA is necessary for the inhibition.
According to E2F pathway dogma, E2F transcription factors are tightly linked to cell cycle regulation and cell proliferation, controlling the expression of genes essential for transition from G1 to S and initiation of DNA replication [6-7]. E2F transcription factors belong to a large family consisting of eight members (E2F1-8). All family members possess signature winged-helix DNA-binding domain. E2F1-5 factors can interact with retinoblastoma family of proteins, also called pocket proteins. When E2F is bound to a pocket protein, it functions as a transcription repressor. Upon phosphorylation of pocket protein by cyclin-dependent kinases, E2Fs are released and can act as transactivators. Most E2F family members (E2F1-6) bind DNA as heterodimers with one of three dimerization partner proteins (i.e., DP1-3). Traditionally, the E2F family has been divided into activator (E2F1-3) and repressor (E2F4-8) subclasses. Alternative promoters at the E2F3 locus drive the expression of two highly related isoforms, E2F3a and E2F3b. Paradoxically, overexpression of E2F1 can promote both cell proliferation and apoptosis. The apoptotic response is p53-dependent in most cases, through transcriptional activation of p19Arf that inhibits p53 degradation and therefore indirectly increases its protein levels. Consistently, mice deficient for E2F1 were shown to have tendency to develop tumors.
We had previously shown that E2F1 can regulate ischemic angiogenesis in-vivo [8]. Mice deficient for E2F1 demonstrated enhanced angiogenesis in hind-limb ischemia and tumor graft models, and we showed that the E2F1-dependent regulation of VEGF expression was responsible for this angiogenic response. Another E2F family member, E2F2 regulates endothelial function, arterial contractility, and blood pressure [9].
In recent years E2F transcription factors were identified as both targets of miRNAs and regulators of their biogenesis. In this review we describe some of well-known hypoxia-, angiogenesis-, and ischemia-related miRNAs that can regulate or are regulated by E2Fs (Figure 1).
Figure 1.
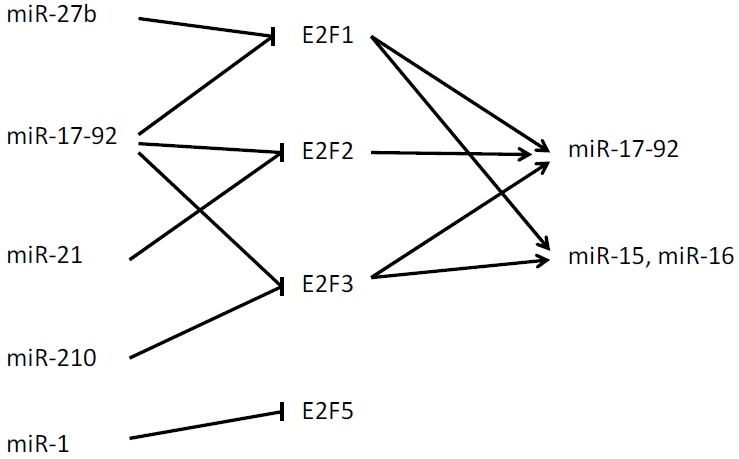
Schematic representation of E2F family members and microRNA regulatory network.
Hypoxia-induced miRNA
miR-210
Hypoxia inducible factor (HIF) is a key mediator of gene expression following hypoxia [1]. HIF is a heterodimeric complex consisting of an α and a β subunits. HIF-β is constitutively expressed in all cell types, but α-subunit levels are tightly controlled by oxygen. Under normoxia, prolyl hydroxylases (PHDs) catalyze hydroxylation of HIF-α. The hydroxylated HIF-α subunits are then polyubiquitilated and targeted for proteasome mediated degradation. If the level of oxygen drops, the α subunit does not undergo prolyl hydroxylation and, as a result, forms a stable complex with the β subunit. This complex binds as a heterodimer to hypoxia response elements (HREs) in the promoter regions of hypoxia-sensitive genes to induce gene transcription. Significant number of HIF target genes have been identified and validated in humans. HIF-dependent transcriptional changes regulate a broad spectrum of cellular functions, including metabolism, proliferation, apoptosis, and angiogenesis.
A number of microRNAs have been shown to be induced by hypoxia, and the term “hypoxamiRs” have been coined to describe them [10]. One of the most consistently and robustly induced miRNA is miR-210 [11]. Its regulation has been shown in a variety of aspects of hypoxia biology: angiogenesis, apoptosis, DNA damage repair, tumor biology, cell cycle regulation and stem cell biology (reviewed in [12]). The hypoxic regulation of miR-210 was first identified in cancer cell lines and HIF-1α was shown to directly bind the HRE on the proximal miR-210 promoter [13]. miR-210 has been shown to play an important role in endothelial cell response to hypoxia. It is strongly upregulated by hypoxia in HUVECs and shown to regulate VEGF-induced chemotaxis and ability of endothelial cells to form capillary-like structures [14]. The importance of miR -210 regulation was confirmed in in-vivo studies. miR-210 had been shown to be upregulated in mouse hind limb ischemia and rat brain transient focal ischemia [15-16]. Given the universal regulation of miR-210 by hypoxia, not surprisingly, a variety of miR-210 targets have been identified. Among them are Ephrin A3 [14, 16], RAD52 [17], ACVR1B [18], MNT [19] and FGFRL2[20].
Involvement of E2F family in the miR-210 signaling pathway was demonstrated by Giannakakis et al. [21]. In luciferase assays miR-210 directly targets transcription factor E2F3. This regulation was confirmed at the protein level in Western blot experiments where authors showed the suppression of E2F3 expression by miR-210. Using antibody against the C–terminal region of the E2F3 protein that recognizes both E2F3a and E2F3b, the authors showed that overexpression of miR-210 in HeLa cells caused significant decrease of the protein levels of both isoforms of E2F3.
Angiogenesis-related miRNAs
The crucial role played by microRNAs in the regulation of angiogenesis became obvious after several studies demonstrated that deletion of Dicer, enzyme required for miRNA biogenesis, resulted in severe in-vivo and in-vitro angiogenic defects [22-25]. Since then miRNAs were shown to play important roles in regulation of angiogenesis during development and normal physiological processes, as well as pathological angiogenesis (for review, see [26-28]).
miR-17∼92 cluster
The miR-17∼92 cluster is a polycistronic miRNA gene. In the human genome, the miR-17∼92 cluster encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1). In vertebrates the sequences of these mature miRNAs are highly conserved. The human miR-17∼92 cluster is located in the third intron of an approximately 7 kb primary transcript known as C13orf25 [29]. The miR-17∼92 cluster first attracted attention following a series of observations linking these miRNAs to cancer pathogenesis [30]. Mice deficient for miR-17∼92 die after birth with lung hypoplasia and a ventricular septal defect [31].
The roles of the cluster members appear to be different in physiological angiogenesis and tumor angiogenesis. Dews et al. [32] showed that overexpression of miR-17∼92 in murine carcinoma cells resulted in enhanced tumor angiogenesis. The mechanism involved downregulation of the potent endogenous inhibitor of angiogenesis thrombospondin-1 together with several proteins containing thrombospondin type 1 repeats. miR-17∼92 downregulate thrombospondin type 1 repeat protein clusterin indirectly through transforming growth factor-β (TGFβ) pathway. The expression of type II TGFβ receptor was suppressed by miR-17-5p and miR -20a, and miR-18a reduced Smad4 level [33]. Similarly, injection of miR-17-5p in combination with let-7b into the ovaries of Dicer-deficient mice partially normalized corpus luteum angiogenesis [34]. However, Bonauer et al [35] demonstrated that overexpression of miR-92a in endothelial cells blocked angiogenesis in vitro and in vivo. In mouse models of limb ischemia and myocardial infarction, systemic administration of miR-92a antagomiR led to enhanced blood vessel growth and functional recovery of the injured tissues. Besides, same group [36] showed that overexpression of miR-17, -18a, -19a, and -20a significantly inhibited HUVEC sprouting in a 3-dimensional spheroid model and inhibition of miR-17, -18a, and -20a increased sprouting. In-vivo inhibition of miR-17 and miR-20a increased the number of blood vessels in Matrigel plugs, but antagomiRs that target miR-18a and miR-19a were less effective. Comparing the angiogenic properties of conditioned media obtained from tumor cells overexpressing miR-17, miR-19a, and miR-20a with that of likewise transfected ECs the authors found that conditioned media of LLC1 tumor cells slightly enhanced angiogenic sprouting of ECs, but media derived from transfected endothelial cells showed a trend toward reduction of angiogenic activity of ECs. In Lewis lung carcinoma tumor model inhibition of miR-17 and miR -20 slightly increased tumor size but did not increase angiogenesis.
Several E2F family members were shown to be both targets and regulators of miR-17∼92 cluster. O'Donnell et al. [37] demonstrated that members of miR-17∼92 cluster directly regulate expression of E2F1. Inhibition of miR-17-5p and miR-20a led to approximately 4-fold increase in E2F1 protein level, and overexpression of miR-17 cluster resulted in decrease of E2F1 protein. Another group [38] also showed that miR-20a can bind to 3'-UTR region of E2F1, E2F2 and E2F3 factors and inhibition of miR-17 and miR-20a led to increase in protein levels of E2F1 and E2F2. Interestingly, the authors demonstrated that E2F1-3 themselves directly regulate the expression of miR-17∼92 cluster and overexpression of E2F1-3 in HeLa cells led to increase of miR-20a. These results suggest a self-regulatory mechanism, where miR-20a controls the translation of E2F1-3 which, in turn, regulate miR-20a transcription. The binding of E2F3 to miR-17∼92 cluster promoter region was reported by another group [39]. Interestingly, Aurora kinase A upregulates miR-17∼92 cluster by increasing E2F1 binding to its promoter region [40]. Pickering et al [41] demonstrated that in normal human fibroblasts inhibition of miR-17 and miR-20a led to earlier peak accumulation of E2F1 after serum starvation (9-12 hrs vs. 15-18 hrs in control cells), and this premature accumulation resulted in a DNA damage induced cell cycle arrest. Interestingly, according to their data, it was the miRNA-regulated timing of E2F1 accumulation but not the gross level of protein, required for initiation of DNA damage response.
miR-27b
E2F1 levels has also been shown to be regulated by another angiogenesis-related microRNA, miR-27b.
miR-27b belongs to the intronic miR-23b∼miR-27b∼miR-24-1 cluster and have been shown as an angiogenesis-related microRNA in experiments using Dicer depletion to identify endothelial miRNAs [24]. Its inhibition was shown to significantly reduce endothelial sprouting. Recent data show that inhibition of miR-27b impairs angiogenesis in vitro and postnatal vascular retinal development in-vivo [42]. In developing mouse heart miR-27b was highly expressed in myocardium at day E10.5 and its expression remained high at later developmental stages [43]. Cheng et al [44] also observed high levels of miR-27b expression in normal mouse heart. miR-27b was upregulated in a rat model of early heart hypertrophy [45].
In addition, miR-27b expression was regulated by shear stress [46]. In this paper the authors found that the upregulation of miR-23b and miR -27b was correlated with the shear flow-induced growth arrest of endothelial cells. Pulsatile shear flow led to a reduction of E2F1 protein level and Rb hypophosphorylation. The Rb hypo-phosphorylation was reversed by inhibition of miR-23b, but not miR-27b, and inhibition of both microRNAs could reverse shear flow-induced decrease of E2F1 levels.
miR-15, miR-16
Though miR-15 and miR-16 are better known for their involvement in cancer [47-48], several studies profiling miRNA expression in endothelial cells demonstrated that these microRNAs are expressed at high levels, indicating their potential importance in angiogenesis regulation [23, 27, 49]. miR-15 and miR-16 were first reported to inhibit VEGF expression in human carcinoma cell line [50]. Hypoxia-induced VEGF expression was significantly decreased by transfection of miR-15,16 and inhibition of these microRNAs under normoxia led to increase of VEGF level. Later it was demonstrated that miR-16 negatively regulates VEGF translation by targeting VEGF 3'-UTR [51]. miR-15, miR-16 were shown to be involved in various cancers. In chronic lymphocytic leukemia both miR-15a and miR-16-1 negatively regulate antiapoptotic B cell lymphoma 2 (Bcl2) protein at a posttranscriptional level [52], and Bcl2 repression by these microRNAs induces apoptosis in a leuke-mic cell line model. Growing experimental evidence suggest that miR-15a/miR-16 constitute key tumor suppressors whose deletion contributes to cancer. mRNAs targeted by miR-15 and miR-16 include CCND3, CCNE1, CDK6, CA-PRIN1, and HMGA1 (reviewed in [53]). During Xenopus embryonic development, miR-15, miR-16 were shown to interfere with TGFβ signaling by targeting Acvr2a, receptor for Nodal, ligand belonging to TGFβ superfamily. At the same time, miR-15 and miR-16 are negatively regulated by Wnt/beta-catenin pathway, linking these two crucial developmental pathways [54].
MiR-15 and miR-16 were shown to participate in the regulation of cell-cycle. Thus, Linsley et al. [55] demonstrated that miR-16 family negatively regulates cell cycle progression. Recently Ofir et al. [56] demonstrated that miR-15a, miR-15b and miR-16 are significantly induced by E2F1 in human lung carcinoma cells. In chromatin immunoprecipitation experiments E2F1 was found binding to human Dleu2 and SMC4 gene promoters, therefore regulating miR-15b, miR-16-2 cluster and miR-15a, miR-16-1 cluster. Interestingly, ectopic expression of miR-15 resulted in reduced levels of cyclin E, a key direct transcriptional target of E2F pivotal for the G1/S transition [57]. This observation suggests that E2F1, miR-15, and cyclin E form a self-regulating loop that modulates E2F activity. Bueno et al. [58], analyzing the transcription profiles of miRNAs in response to mitogenic stimulation in primary fibroblasts, demonstrated that miR-15b and miR-16-2 miRNAs are direct targets of E2F1 and E2F3. These miRNAs were specifically induced by E2F1 or E2F3 during the G1/S transition and repressed in E2F1-, and E2F3-knockout cells.
Ischemia-related miRNAs
Another microRNAs involved in E2F signaling are miR-1 and miR-21, known for their important role in cardiac ischemia.
miR-1
miR-1 involvement in cardiac disorders is well documented [59]. Downregulation of miR-1 after pressure overload in mouse heart was reported by Sayed et al [60]. Ikeda et al [61] showed miR-1 decrease in cardiac hypertrophy, where it targets calmodulin and Mef2a. Reduction of miR-1 was shown to be required for an increase of cell mass [62-63]. miR-1 was demonstrated to lead to pro-apoptotic signaling in cardiomyocytes [64], and in cardiac ischemia miR-1 was shown to regulate apoptotic pathways through regulation of Bcl2 [65]. In the failing human heart the reported levels of miR-1 have been inconsistent [59], but overall miR-1 regulates the genes involved in cardiac hypertrophy. Targeted deletion of miR-1-2 was demonstrated to result in disrupted cardiac morphogenesis and, surprisingly, cell cycle abnormalities [66]. Adult miR-1-2 mutants had significant increase in heart/body weight ratio due to hyperplasia. Analysis showed 20% increase in the myocytes number in the mutants, and many adult cardiomyocytes were undergoing division. Authors observed significant increase in the number of mitotic marker-positive myocytes at the postnatal day P10 compared to wild type animals, and even at the age of 2-3 months there were mitotic myocytes in the hearts of miR -1-2-/- mutants.
It turned out that E2F factors are among miR-1 targets. Thus, Zhang et al [67] have shown that in hepatoma cells overexpression of miR-1 led to the cell cycle arrest at the G1 phase. E2F5 was identified as a direct target of miR-1, and overexpression of miR-1 resulted in marked dose-dependent decrease of E2F5 protein. pRb levels were also decreased by miR-1 overexpression.
miR-21
miR-21 is among most significantly upregulated microRNAs in hypertrophic mouse heart [44, 60]. Following aortic banding, its expression increased more than 4-fold at day 7. These results were confirmed in in-vitro experiments with rat neonatal cardiac myocytes with stimulated hypertrophy [44, 68]. Upregulation of miR-21 was shown in the stimulated cells, and inhibition of miR-21 led to a decrease in hypertrophy. miR-21 was shown to target pro-apoptotic PDCD4 [69], tumor suppressor PTEN and anti-angiogenic Spry-2 [70-71].
Comparing microRNAs expression of left ventricular tissue in failing, healthy and fetal human heart Thum et al found several fold increase in miR-21 expression levels in fetal and failing sample versus healthy ones [72]. In human in-farcted hearts there was a striking increase of miR-21 expression in border zone [73]. Following experimental data showing marked increase of miR-21 in mouse heart failure models as well as in human heart, Thum et al [74] showed that this increase happens mostly in cardiac fibroblasts, and not in myocytes. miR-21 was shown to have an anti-apoptotic role that was dependent on ERK activity. Spry1, an inhibitor of RAS/ MEK/ERK pathway was identified as a direct target of miR-1 that was responsible for ERK activation. Treatment of mice with chemically modified antisense oligonucleotide (antagomiR) specific for miR-21 following pressure overload of left ventricle by transverse aortic constriction had reversed the negative effects of procedure such as myocyte size, heart weight and interstitial fibrosis. Moreover, the authors reported that treatment with miR-21 antagomiR had curative effect in pressure overload model. Surprisingly, these results were not confirmed by genetic deletion of miR-21 [75]. miR-21-null mice had no abnormalities in heart size, structure or cardiac contractility, and demonstrated no significant differences in pathologic remodeling from wild type control after being subjected to 4 different cardiac stresses: acute pressure overload, chronic calcineurin activation, infusion of Ang II and myocardial infarction. Similarly, inhibition of miR-21 using LNA-antimiR resulted in comparable cardiac hypertrophy between control and treated groups. These discrepancies might be explained in part by the possible compensatory mechanisms activated in mice due to the deletion of miR-21 and difference between cholesterol-conjugated antagomiR used by Thum et al. and short 8-nt LNA-modified antimiR used by Patrick et al. Another explanation might be a redundancy of microRNAs, resulting in inhibition of multiple microRNAs sharing similar seed sequence [76].
Involvement of miR-21 in E2F regulation was demonstrated by Bhat-Nakshatri et al [77]. In experiments with MCF-7 breast cancer cells authors showed that inhibition of miR-21 increased E2F2 basal level, but had no effect on E2F1 protein. Estradiol-stimulated increase of E2F1 and E2F2 proteins was not significantly affected by miR-21 inhibition.
Outlook
E2F family of transcription factors have long been a focus of intensive research, and its role in the cell cycle regulation and cell survival is well established. It is known that members of the family are involved in the signaling cascades induced by hypoxia. Rapidly accumulating data show that miRNAs actively participate in the modulation of E2F activity as both regulators of E2F transcription and their targets, forming intricate signaling network. Studying these complex interactions gives a new dimension to our understanding of the role of E2F transcription factors in hypoxic conditions and their mode of action, and can potentially lead to the development of new therapeutic targets in cardiovascular and cancer medicine.
Acknowledgments
Dr. Gangjian Qin is supported by NIH grant HL093439 and American Heart Association Grant 0430135N”.
References
- 1.Rocha S. Gene regulation under low oxygen: holding your breath for transcription. Trends Biochem Sci. 2007;32:389–397. doi: 10.1016/j.tibs.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 6.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–657. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin G, Kishore R, Dolan CM, Silver M, Wecker A, Luedemann CN, Thorne T, Hanley A, Curry C, Heyd L, Dinesh D, Kearney M, Martelli F, Murayama T, Goukassian DA, Zhu Y, Losordo DW. Cell cycle regulator E2F1 modulates angiogenesis via p53-dependent transcriptional control of VEGF. Proc Natl Acad Sci USA. 2006;103:11015–11020. doi: 10.1073/pnas.0509533103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou J, Zhu Y, Cheng M, Dinesh D, Thorne T, Poh KK, Liu D, Botros C, Tang YL, Reisdorph N, Kishore R, Losordo DW, Qin G. Regulation of vascular contractility and blood pressure by the E2F2 transcription factor. Circulation. 2009;120:1213–1221. doi: 10.1161/CIRCULATIONAHA.109.859207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loscalzo J. The cellular response to hypoxia: tuning the system with microRNAs. J Clin Invest. 2010;120:3815–3817. doi: 10.1172/JCI45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan SY, Loscalzo J. MicroRNA-210: A unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:6. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Le QT, Giaccia AJ. MiR-210-micromanager of the hypoxia pathway. Trends Mol Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 16.Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008;582:2397–2401. doi: 10.1016/j.febslet.2008.05.048. [DOI] [PubMed] [Google Scholar]
- 17.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y, Suda T, Fukuda T, Katagiri T, Kondoh Y, Amemiya T, Tashiro H, Okazaki Y. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583:2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, Burchard J, Dai X, Chang AN, Diaz RL, Marszalek JR, Bartz SR, Carleton M, Cleary MA, Linsley PS, Grandori C. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 20.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O'Brien-Jenkins A, Katsaros D, Weber BL, Simon C, Coukos G, Zhang L. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280:9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 23.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 24.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 25.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicerdependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105:14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonauer A, Boon RA, Dimmeler S. Vascular microRNAs. Curr Drug Targets. 2010;11:943–949. doi: 10.2174/138945010791591313. [DOI] [PubMed] [Google Scholar]
- 27.Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. microRNA: emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther. 2010;125:92–104. doi: 10.1016/j.pharmthera.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 2004;64:3087–3095. doi: 10.1158/0008-5472.can-03-3773. [DOI] [PubMed] [Google Scholar]
- 30.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, Cleary MA, Thomas-Tikhonenko A. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010;70:8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otsuka M, Zheng M, Hayashi M, Lee JD, Yoshino O, Lin S, Han J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Invest. 2008;118:1944–1954. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 36.Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, Hofmann WK, Zeiher AM, Dimmeler S. Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115:4944–4950. doi: 10.1182/blood-2010-01-264812. [DOI] [PubMed] [Google Scholar]
- 37.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 38.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 39.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 40.He S, Yang S, Deng G, Liu M, Zhu H, Zhang W, Yan S, Quan L, Bai J, Xu N. Aurora kinase A induces miR-17-92 cluster through regulation of E2F1 transcription factor. Cell Mol Life Sci. 2010;67:2069–2076. doi: 10.1007/s00018-010-0340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickering MT, Stadler BM, Kowalik TF. miR-17 and miR-20a temper an E2F1-induced G1 checkpoint to regulate cell cycle progression. Oncogene. 2009;28:140–145. doi: 10.1038/onc.2008.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci U S A. 2011;2:2. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chinchilla A, Lozano E, Daimi H, Esteban FJ, Crist C, Aranega AE, Franco D. MicroRNA profiling during mouse ventricular maturation: a role for miR-27 modulating Mef2c expression. Cardiovasc Res. 2011;89:98–108. doi: 10.1093/cvr/cvq264. [DOI] [PubMed] [Google Scholar]
- 44.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart: do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busk PK, Cirera S. MicroRNA profiling in early hypertrophic growth of the left ventricle in rats. Biochem Biophys Res Commun. 2010;396:989–993. doi: 10.1016/j.bbrc.2010.05.039. [DOI] [PubMed] [Google Scholar]
- 46.Wang KC, Garmire LX, Young A, Nguyen P, Trinh A, Subramaniam S, Wang N, Shyy JY, Li YS, Chien S. Role of microRNA-23b in flowregulation of Rb phosphorylation and endothelial cell growth. Proc Natl Acad Sci U S A. 2010;107:3234–3239. doi: 10.1073/pnas.0914825107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down -regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 49.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 50.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNAdirected regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15:249–254. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR -15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martello G, Zacchigna L, Inui M, Montagner M, Adorno M, Mamidi A, Morsut L, Soligo S, Tran U, Dupont S, Cordenonsi M, Wessely O, Piccolo S. MicroRNA control of Nodal signalling. Nature. 2007;449:183–188. doi: 10.1038/nature06100. [DOI] [PubMed] [Google Scholar]
- 55.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ofir M, Hacohen D, Ginsberg D. miR-15 and miR-16 Are Direct Transcriptional Targets of E2F1 that Limit E2F-Induced Proliferation by Targeting Cyclin E. Mol Cancer Res. 2011;9:440–447. doi: 10.1158/1541-7786.MCR-10-0344. [DOI] [PubMed] [Google Scholar]
- 57.Ohtani K, DeGregori J, Nevins JR. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci U S A. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bueno MJ, Gomez de Cedron M, Laresgoiti U, Fernandez-Piqueras J, Zubiaga AM, Malumbres M. Multiple E2F-induced microRNAs prevent replicative stress in response to mitogenic signaling. Mol Cell Biol. 2010;30:2983–2995. doi: 10.1128/MCB.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han M, Toli J, Abdellatif M. MicroRNAs in the cardiovascular system. Curr Opin Cardiol. 2011;26:181–189. doi: 10.1097/HCO.0b013e328345983d. [DOI] [PubMed] [Google Scholar]
- 60.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 61.Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR, Pu WT. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MV, Hoydal M, Autore C, Russo MA, Dorn GW, 2nd, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 63.Elia L, Contu R, Quintavalle M, Varrone F, Chimenti C, Russo MA, Cimino V, De Marinis L, Frustaci A, Catalucci D, Condorelli G. Reciprocal regulation of microRNA-1 and insulinlike growth factor-1 signal transduction cascade in cardiac and skeletal muscle in physiological and pathological conditions. Circulation. 2009;120:2377–2385. doi: 10.1161/CIRCULATIONAHA.109.879429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu C, Lu Y, Pan Z, Chu W, Luo X, Lin H, Xiao J, Shan H, Wang Z, Yang B. The musclespecific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–3052. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 65.Tang Y, Zheng J, Sun Y, Wu Z, Liu Z, Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int Heart J. 2009;50:377–387. doi: 10.1536/ihj.50.377. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 67.Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476–1485. doi: 10.1002/hep.24195. [DOI] [PubMed] [Google Scholar]
- 68.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen JF, Newman M, Rojas M, Hammond SM, Wang DZ. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J Mol Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 70.Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3:251–255. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 targets Sprouty2 and promotes cellular outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 73.van Rooij E, Sutherland LB, Thatcher JE, Di-Maio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 75.Patrick DM, Montgomery RL, Qi X, Obad S, Kauppinen S, Hill JA, van Rooij E, Olson EN. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morrisey EE. The magic and mystery of miR-21. J Clin Invest. 2010;120:3817–3819. doi: 10.1172/JCI44596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, Brown M, Hammond S, Srour EF, Liu Y, Nakshatri H. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37:4850–4861. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]


