Citrullinated peptide presentation by DCs and macrophages is constitutive, whereas B cells present modified peptides only with autophagy induced by serum starvation or BCR ligation.
Abstract
Antibody responses to citrullinated self-proteins are found in autoimmunities, particularly in rheumatoid arthritis, where they serve as a diagnostic indicator. We show here that processing of the protein hen egg-white lysozyme (HEL) resulted in citrullination of peptides presented on class II MHC molecules by antigen-presenting cells. The presentation of the citrullinated peptides but not of the unmodified peptides was associated with autophagy. Dendritic cells (DCs), macrophages, and thymic DCs presented citrullinated peptides constitutively. Their treatment with 3-methyladenine (3MA) blocked presentation of citrullinated HEL peptides, but presentation of unmodified peptides was not affected. Presentation of citrullinated peptides was not detected on B cells or B lymphoma cells under normal culture conditions. In B cells, engagement of the B cell antigen receptor was required for presentation of the citrullinated peptides, also inhibited by 3MA. B lymphoma–expressing HEL cells presented citrullinated peptides only after brief serum starvation. This presentation was reduced by 3MA or by reduction in Atg5 expression. Presentation of the unmodified peptides was not changed. The findings indicate a linkage between autophagy and autoreactivity through the generation of this neo-epitope.
Presentation by MHC molecules of peptides bearing posttranslational modifications can elicit highly specific T cell responses, speculated to be a component of autoimmunities. The T cells usually recognize the modified residue in the framework of the unmodified TCR contact residues of the peptide (Anderton, 2004; Engelhard et al., 2006; Petersen et al., 2009). One of the most compelling findings linking posttranslationally modified self-proteins to autoimmunity is the antibody response to citrullinated proteins in rheumatoid arthritis (RA). The presence of citrulline in proteins is caused by the enzymatic deimination of arginine residues; citrulline is not a natural amino acid encoded in DNA. The conversion from peptidylarginine to peptidylcitrulline is catalyzed by the peptidylarginine deiminase (PAD) family of enzymes, which differ in tissue expression. PAD2 and PAD4 are both expressed by cells of the immune system. In RA, the antibodies are found early in disease, at levels that correlate with severity (Klareskog et al., 2008; Wegner et al., 2010). Moreover, there is a strong correlation between antibody reactivity and the presence of RA-susceptible HLA alleles (de Vries et al., 2006; van der Helm-van Mil and Huizinga, 2008). Citrullinated peptides were shown to bind with higher affinity to the RA-susceptible HLA allele DR4 as a result of the more favorable interaction of the citrulline side chain with the positively charged P4 pocket (Hill et al., 2003).
We previously reported that immunization with the protein hen egg-white lysozyme (HEL) gave rise to T cells that recognized different sets of HEL peptides presented by class II MHC molecules (Ireland et al., 2006). In this study, we correlate the response of a T cell that exclusively recognizes the chemically dominant HEL peptide, 48–62 (DGSTDYGILQINSRW), with a citrulline to arginine change at residue 61 (48–62-Cit61) to autophagy.
RESULTS AND DISCUSSION
Primary APCs present citrullinated peptides in vivo
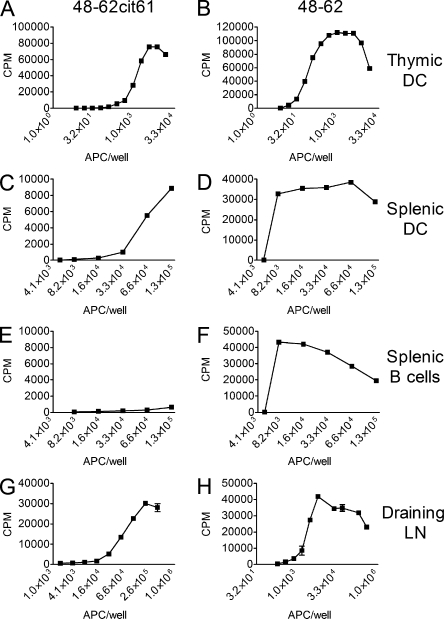
To determine whether presentation of citrullinated peptides by class II MHC molecules occurred normally from in vivo APCs presenting a self-protein, membrane HEL (mHEL) transgenic mice were examined. All APCs from these mice expressed HEL linked to the transmembrane region of Ld under the I-E promoter. Presentation of HEL 48–62 by APCs isolated from these mice was strong, found to be 3,400–20,000 peptide–MHC complexes per cell (Peterson et al., 1999). DCs identified as CD11c+ cells from the thymi and spleens of mHEL mice elicited a robust response to the dominant HEL peptide 48–62 (using as indicator cell the CD4 T cell hybridoma 3A9, which recognized the unmodified peptide; Fig. 1, B and D). The APCs also presented 48–62-Cit61 (using Granny as the indicator T cell that only recognized the citrullinated peptide; Fig. 1, A and C). In contrast, splenic CD19+ cells did not elicit a response from the 48–62-Cit61–reactive hybridoma, though the response of 3A9 was strong (Fig. 1, E and F). Thus, although DCs and macrophages presented the citrullinated derivative from HEL, B cells only presented the unmodified epitope. This finding is in concordance with our previously published results demonstrating that a B cell lymphoma did not present citrullinated peptides after processing HEL (Ireland et al., 2006). Lastly, we verified the presentation of citrullinated peptides by APCs in the draining lymph nodes of B10.BR mice immunized with 10 nmol HEL in adjuvant (Fig. 1, G and H).
Figure 1.
Presentation of citrullinated peptides by freshly harvested APCs. (A–F) IL-2 production from CD11c+ cells isolated from thymi (A and B), spleen (C and D), and splenic CD19 + cells (E and F). A, C, and E show response to the 48–62-Cit61 peptide by Granny, whereas B, D, and F show response to the unmodified 48–62 by 3A9. (G and H) The response to CD11c+, CD11b+ cells purified from the popliteal lymph nodes of mice 24 h after immunization with 10 nmol HEL and tested against (G) Granny or (H) 3A9 is shown. Data from each panel are representative of at least two independent experiments, and each data point represents duplicate wells. At least three mice were used in each experiment. Error bars indicate SEM.
Serum starvation induces presentation of citrullinated peptides by B lymphoma cells
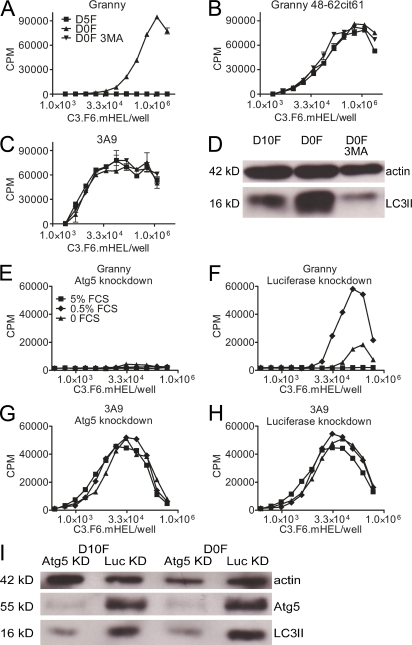
To determine the reasons for the lack of citrullination by B cells and explore conditions that may induce it, we examined C3.F6, a B lymphoma line which when cultured with HEL does not present the 48–62-Cit61 peptide (Ireland et al., 2006). We examined in particular C3.F6.mHEL that expresses HEL with a transmembrane linker and constitutively presents high levels of HEL peptides on I-Ak molecules. There was striking presentation of HEL 48–62-Cit61 if the cells were cultured without FCS for a limited period of time, as little as 3–4 h (Fig. 2 A). The response to the unmodified 48–62 was not changed by C3.F6.mHEL cultured in reduced FCS (Fig. 2 C). Moreover, total citrullinated peptides were detected by biochemical approaches in eluates from I-Ak molecules isolated from C3.F6.mHEL. The mean percent increase in citrullinated peptides was 580% in cells cultured in 1% FCS compared with those cultured in 5% (n = 4 experiments; Table S1).
Figure 2.
C3.F6.mHEL cells present citrullinated peptides after serum starvation–induced autophagy. (A–C) T cell responses to C3.F6.mHEL cultured in DME supplemented with 5% FCS (D5F), no serum (DOF), or no serum in the presence of 10 mM 3MA (D0F 3MA) for 4 h; the culture medium was then changed to 5% FCS, and the T cell hybridomas Granny (A and B) or 3A9 (C) were added. (D) Western blot of actin and LC3II in C3.F6.mHEL cultured in D10F, D0F, and D0F with 3MA. Data are representative of three independent experiments; each data point represents duplicate wells. Targeted knockdown of Atg5 expression inhibited citrullination of HEL peptides. (E–H) Responses of T cells to C3.F6.mHEL cells expressing either shRNA targeting Atg5 expression (E and G) or luciferase expression (F and H). Cells were cultured in media supplemented with 5% FCS, 0.5% FCS, or no FCS for 4 h and then replaced with media supplemented with 5% FCS. (I) Western blot of actin, Atg5 levels, and LC3II in the treated C3.F6.mHEL cells shows that Atg5-deficient cells had reduced levels of LC3II conversion after serum starvation. Data are representative of three independent experiments. Error bars indicate SEM.
We considered whether the B cell line was undergoing autophagy as a result of the stress imposed by lowering the FCS concentration. Several studies indicated that APCs constitutively underwent autophagy and that this process contributed to the repertoire of peptides presented on MHC class II molecules (Nimmerjahn et al., 2003; Dengjel et al., 2005; Dörfel et al., 2005; Paludan et al., 2005; Schmid et al., 2007). The serum-starved cells were treated with 3-methyladenine (3MA), a class III PI3 kinase inhibitor, during the period of serum starvation. 3MA inhibits autophagy (Seglen and Gordon, 1982; Petiot et al., 2000), although effects on other components of intracellular protein processing and or catabolism were not ruled out. After culture with HEL, the presentation of HEL 48–62-Cit61 by the serum-starved cell line was entirely reduced by treatment with 3MA (Fig. 2 A). In striking contrast, the T cell response to the unmodified 48–62 peptide was not affected at all (Fig. 2 C). There was no difference in responses among untreated, serum-starved cells, or serum-starved cells treated with 3MA when cultured just with the HEL 48–62-Cit61 peptide, where processing is not required, indicating that their levels of MHC molecules were not altered (Fig. 2 B). C3.F6.mHEL cultured without FCS had an increase in the levels of LC3II, which was reduced by 3MA treatment, confirming induction of autophagy by serum starvation and its inhibition by 3MA (Fig. 2 D).
Low expression of Atg5 inhibits presentation after serum starvation of B lymphoma cells
We next assessed an alternative method for inhibiting autophagy. We designed short hairpin RNA (shRNA) constructs that targeted expression of Atg5, a protein essential for autophagy, and cloned them into a lentiviral delivery system in which the infected cells expressing YFP were purified by flow cytometry. As a negative control, cells were infected with virus containing shRNA that targeted Luciferase expression. The shRNA system effectively reduced Atg5 expression by Western blot (Fig. 2 I). RT-PCR indicated a 72% decrease in Atg5 message relative to Luciferase knockdown controls (not depicted). The cells exhibited reduced levels of LC3II conversion (Fig. 2 I). There was no difference in the levels of messenger RNA (mRNA) to PAD2 or PAD4 (Fig. S1 C). Inhibition of Atg5 expression in serum-starved cells resulted in no presentation of citrullinated peptides from HEL (Fig. 2 E). In contrast, there was presentation of citrullinated peptides in the control cells in which luciferase expression was targeted (Fig. 2 F). Neither serum starvation nor expression of Atg5 had any bearing on presentation of the unmodified epitope, HEL 48–62 (Fig. 2 G).
PAD2 and PAD4 expression alone is not sufficient for citrullination of antigen, and PAD activity can be detected in isolated autophagosomes
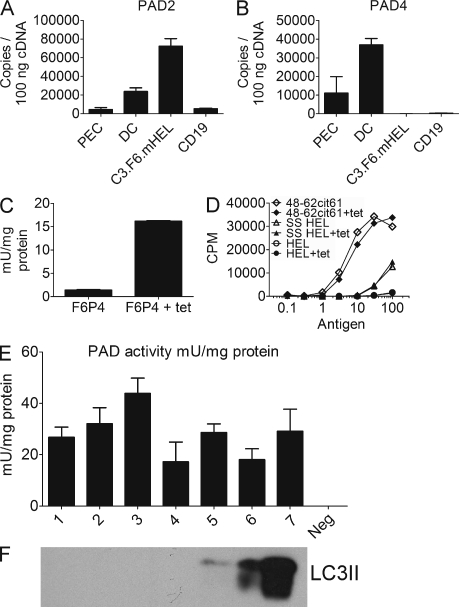
Whether the poor presentation by B cells and B lymphoma cells could be attributed to an absence or poor expression of the enzymes that convert arginine to citrulline was a concern. PAD2 and PAD4 are expressed in APCs, but PAD4 was shown to be expressed at lower levels in C3.F6 and B cells (Vossenaar et al., 2004; Ireland et al., 2006). Quantitation of PAD2 and PAD4 mRNA in the various APCs examined in this study confirmed these findings (Fig. 3, A and B). Although peritoneal exudate macrophages (PECs) and DCs expressed both enzymes, PAD4 expression was weakly detected in C3.F6. We generated a C3.F6 line in which expression of PAD4 was induced (C3.F6.P4). PAD4 was cloned from splenic APCs into a vector under control of the CMV promoter with tetracycline operon regulatory elements; such a vector was cotransfected with the tetracycline repressor. We verified that treatment of C3.F6.P4 with tetracycline led to expression of biologically active PAD4 by measuring PAD activity in cell lysates (Fig. 3 C). However, cells expressing PAD4 did not present 48–62-Cit61 peptide from HEL unless they were cultured in serum-free conditions (Fig. 3 D), indicating that PAD4 expression alone was not sufficient to allow citrullination of antigen and that other factors were playing a role. In support of this conclusion, we found that conditions that led to induced citrullination of HEL were not associated with changes in expression of either PAD2 or PAD4 (Fig. S1).
Figure 3.
PAD expression in APCs alone is not sufficient for citrullination of HEL; PAD activity can be detected in autophagosomes. (A and B) PAD2 (A) and PAD4 (B) mRNA was measured by RT-PCR and quantified using DNA standards. Data are presented as the mean of quadruplicate reaction replicates and are representative of at least two independent experiments. (C) PAD activity was measured biochemically in lysates of C3.F6.P4 cells after the addition of tetracycline. Data are the mean of triplicate reactions and are representative of three independent experiments. (D) Presentation of HEL or HEL peptide to Granny by C3.F6.P4 with or without tetracycline to induce PAD4 expression in normal culture conditions or after culture in DME without FCS (SS in the panel). Data are representative of three independent experiments. (E) PAD activity as measured by conversion of an artificial substrate was assessed in fractions of elicited PECs. The fractions are represented with numbers as follows: 1, whole cell lysate; 2, postnuclear supernatant; 3, nuclei; 4, second pellet enriched in mitochondria and peroxisomes; 5, light membrane fraction; 6, complex heavy fraction enriched in the ER; and 7, final autophagosome pellet. The negative control (Neg) contained substrate without protein. The data presented are pooled from three independent experiments. 20–60 mice were used in each experiment. (F) The fractions were analyzed by Western blot for LC3II enrichment. The data are representative three independent experiments. Error bars indicate SEM.
The results indicated that autophagy was involved, i.e., induced presentation after serum starvation and its inhibition by 3MA or by reduced Atg5 expression. However, the data also indicated that PAD expression alone was not the sole limiting factor for citrullination of antigen and raised the question of whether PAD enzymes gained access to antigen-loading compartments through the autophagic pathway. That the contents of autophagosomes contributed to antigen-processing compartments has been well documented. In addition, it has been shown that calcium, which was required for PAD activity, accumulated in autophagic vesicles (Fader et al., 2008). To address this question, we examined PECs that presented well the Cit61 epitope from HEL (Fig. 4, B and F). The PECs were fractionated, and the purified autophagic vesicles were tested for PAD enzymatic activity and LC3II (Seglen and Brinchmann, 2010). All the various fractions showed PAD activity, as expected from its known distribution in various cell compartments. Importantly the autophagosomes contained PAD activity and were enriched for LC3II (Fig. 3, E and F).
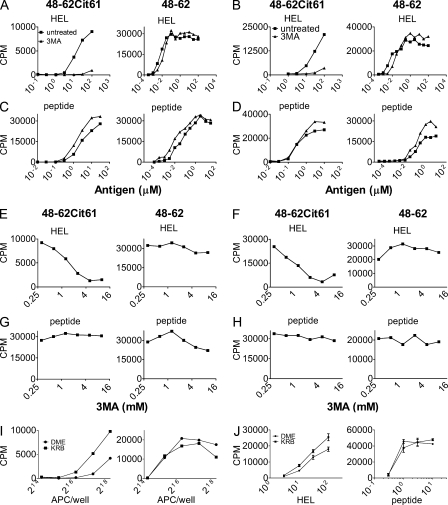
Figure 4.
Treatment with 3MA inhibits presentation of citrullinated peptides by DCs or PECs. (A and B) Presentation of HEL to Granny (left panel labeled 48-62Cit61) or 3A9 (right panel labeled 48–62) with and without 10 mM 3MA to DCs (A) or PECs (B). (C and D) Presentation of the synthetic peptide with and without 3MA to DCs (C) or PECs (D). (E–H) Presentation of 30 µM HEL or 3 µM of the synthetic peptide to DCs (E and G) or PECs (F and H) examining different concentrations of 3MA. (I and J) Presentation of citrullinated peptide was examined after a 1-h culture in Krebs-Ringer bicarbonate buffer (KRB) after a short 2-h pulse with HEL in DCs (I) or PECs (J). Data are representative of two to three experiments. Error bars indicate SEM.
Presentation of citrullinated peptide by primary APCs is inhibited by 3MA and enhanced by amino acid starvation
These findings relating autophagy to the presentation of the citrullinated peptides were then applied to presentation by DCs, PECs, and primary B cells. The citrullinated peptides were presented by either DCs (Fig. 4, A and E) or PECs (Fig. 4, B and F) after processing HEL. The addition of 3MA inhibited presentation in a dose-dependent manner. In contrast, presentation of HEL to 3A9 or presentation of synthetic peptide was not affected at all over a range of 3MA concentrations in either cell type (Fig. 4, C, D, G, and H). We did observe enhanced presentation in DCs (Fig. 4 I) and to a lesser extent in PECs (Fig. 4 J) when pulsed for 2 h with HEL and then cultured for 1 h in either DME or Krebs-Ringer bicarbonate buffer.
B cells present citrullinated peptides after engagement of their antigen receptor
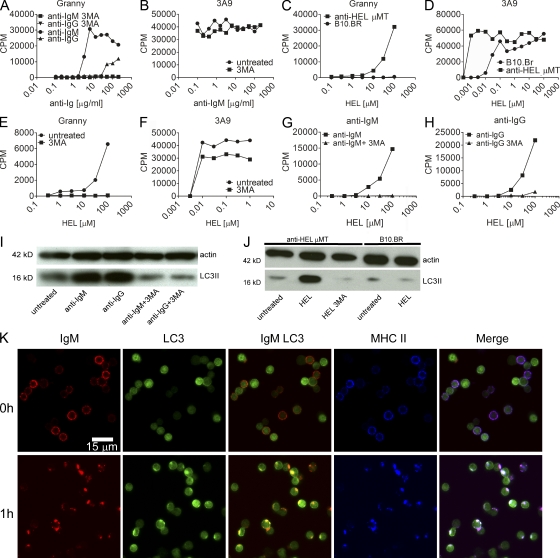
The conditions that might induce citrullination by primary B cells were evaluated using B cells from mHEL mice or monoclonal B cells from anti-HEL transgenic mice. LPS did not result in presentation of the 48–62-Cit61 epitope; culturing in starvation media resulted in poor survival and uninterpretable results. However, addition of anti-IgM antibodies induced strong presentation of the 48–62-Cit61 epitope from B cells of mHEL mice (Fig. 5 A). Identical results were found adding F(ab′)2 fragments of the antibody. Treatment of B cells with anti-IgG also induced a level of presentation of citrullinated peptides. Fig. 5 A also shows that presentation of citrullinated peptides by both anti-IgM and anti-IgG was inhibited by treatment with 3MA, again pointing to an autophagy response. 3MA did not inhibit the presentation of the unmodified 48–62 peptide to 3A9 by the B cells from mHEL mice (Fig. 5 B). There was an increase in LC3II levels in B cells treated with either anti-IgM or anti-IgG (Fig. 5 I), consistent with a report that BCR engagement induces autophagy in B cells (Watanabe and Tsubata, 2009).
Figure 5.
BCR engagement induces presentation of citrullinated peptides by primary B cells. (A and B) Presentation by CD19+ cells isolated from spleens of mHEL mice. (A) Effects of various concentrations of anti-IgM or anti-Ig on Granny ± 3MA. (B) The same as in A but on 3A9 ± 3MA. (C) Presentation of HEL by CD19+ cells from anti-HEL transgenic mice or B10.BR mice to Granny. (D) The same as in C but to 3A9. (E) The effect of 3MA on presentation by CD19+ cells from anti-HEL transgenic mice after processing HEL to Granny. (F) The same as in E but to 3A9. (G) Presentation of HEL by B cells from B10.BR mice to Granny after treatment with 40 µg/ml anti-IgM ± 3MA. (H) The same as in G but treating the cells with anti-IgG ± 3MA. (I and J) Western blots of actin and LC3II from mHEL B cells cultured overnight with anti-Ig (I) or anti-HEL B cells cultured with 30 µM HEL ± 3MA (J). (K) B cells from GFP-LC3 mice were labeled on ice with anti-IgM and fixed immediately or after 1 h at 37°C and stained to label MHC class II. All data are representative of at least two independent experiments.
Likewise, B cells from anti-HEL B cell transgenic mice presented the HEL 48–62-Cit61 epitope after taking up and processing HEL (Fig. 5 C). Such presentation was inhibited by 3MA (Fig. 5 E). In contrast, B cells from B10.BR mice, even with high concentrations of HEL, up to 100 µM HEL, did not present it. However, we observed enhanced presentation of citrullinated peptides from processed HEL in the presence of anti-IgM (Fig. 5 G) or anti-IgG (Fig. 5 H) by B cells from B10.BR mice, which was similarly inhibited by 3MA. In contrast to DCs and macrophages, in which 3MA has no effect on presentation of 48–62, we observed some effect in B cells presenting HEL to 3A9 (Fig. 5 F). It appears that autophagy, although not required for presentation of unmodified peptide, may make some contribution to processing or presentation of HEL by B cells. We found that anti-HEL B cells but not B10.BR B cells cultured with HEL had increased LC3II levels, which was inhibited by 3MA (Fig. 5 J).
To examine the intracellular events occurring after BCR engagement, we examined B cells from GFP-LC3 mice that express a GFP-LC3 fusion protein as a marker of autophagosomes (Mizushima and Kuma, 2008). As expected, treatment with anti-IgM led to capping and internalization of surface IgM (Fig. 5 K). After treatment with anti-IgM, GFP-positive vesicles were observed that colocalized with MHC II vesicles and internalized IgM, thereby demonstrating an association of autophagic vesicles with BCR-mediated uptake and processing for presentation on MHC II.
In summary, we presented evidence that autophagy was a key cellular event involved in the generation of citrullinated peptides by APCs. Our findings posit that presentation of citrullinated peptide is a result and a biochemical marker of an autophagy response. The inhibition by 3MA of presentation of HEL peptides in DCs and PECs was highly selective, only affecting that of the 48–62-Cit61 epitope. The findings argue for distinct sites in the APCs where each processing event takes place, i.e., the one that generates the 48–62-Cit61 peptide and the one that gives rise to the unmodified 48–62 peptides. The effective presentation of the citrullinated epitopes by DCs and macrophages compared with the very weak presentation by primary B cells or B cell lines could have several explanations. The levels of PAD4 were very low in B cells. A second component became evident in the C3.F6 line made to overexpress PAD4 in which again presentation was poor, requiring the stress signal of low serum to induce citrullination. In primary B cells, the effectiveness of capping surface Ig in inducing presentation of the citrullinated epitope was striking. These findings point to the intracellular events that bring the PAD molecule into the autophagic vesicles of the APCs as fundamental to whether or not citrullination of antigen takes place.
The connection between citrullination and autophagy in APCs raises several issues. One is the teleological explanation for the citrullination of proteins. Citrullination alters intramolecular interactions and can lead to increased accessibility to proteolysis (Tarcsa et al., 1996; Pritzker et al., 2000). In addition, the change from arginine to citrulline may expand the repertoire of presented peptides (Hill et al., 2003; de Haan et al., 2005; Ireland et al., 2006; Beltrami et al., 2008). Finally, the finding that an antibody to the B cell antigen receptor can drive the presentation of citrullinated epitopes is particularly of note in the context of RA, with its component of autoantibodies to Ig, such as rheumatoid factor. The combination of anti-Ig autoantibodies, autophagy-inducing events, and the appropriate class II HLA alleles may contribute to presentation of citrullinated self-peptides and a breach of immunological tolerance, precipitating events that lead to the initiation and/or progression of an autoimmune process.
MATERIALS AND METHODS
Cell culture.
CD4 T cell hybridomas were made by fusing cells obtained from popliteal lymph nodes 7 d after immunization with 10 nmol HEL in complete Freund’s adjuvant and fusing to the BW5147α-β-thymoma cells line, as previously described (Peterson et al., 1999). C3.F6.mHEL B lymphoma cells were used as APCs for examining the response to the various peptides. PECs were obtained by intraperitoneal injection with 1 ml thioglycollate or 100 mg concanavalin A. After 4 d, PECs were harvested. DCs were derived from bone marrow cultured for 8 d in GM-CSF–containing medium. Bone marrow was plated at 3.3 × 105 cells per well in a 6-well plate in DME supplemented with conditioned culture supernatant. The media was changed on day 4. Hybridoma activation was measured by IL-2 secretion as assayed by proliferation of the IL-2–dependent line CTLL. Where indicated, cells were purified using the Miltenyi magnetic cell separation reagents and LS columns (Miltenyi Biotec).
Mice and reagents.
B10.BR mice usually 6–12 wk old of both sexes were obtained from the Jackson Laboratory and maintained in facilities under specific pathogen–free conditions at Washington University in St. Louis (St. Louis, MO) in accordance with institutional animal care guidelines. Animal protocols were approved by the Washington University Animal Studies Committee. B10.BR mice expressing mHEL under the class II Eα promoter were generated and maintained by our laboratory (Peterson et al., 1999). GFP-LC3 mice were a gift from K. Boehle in the laboratory of K. Moley at Washington University in St. Louis. HEL was obtained from Sigma-Aldrich and purified to eliminate contaminant proteins and LPS. Dylight 567–labeled rabbit anti-IgM, rabbit anti-IgG, rabbit anti-IgM, and rabbit anti-IgM F(ab′)2 were obtained from Jackson ImmunoResearch Laboratories, Inc. Biotinylated anti–I-Ab was obtained from BD. Streptavidin-conjugated Alexa Fluor 647 was purchased from Invitrogen. Peptides were synthesized using Fmoc techniques. 3MA was obtained from Sigma-Aldrich and suspended at 100 mM in saline and before use heated at 56°C until dissolved and then used immediately.
Western blot.
100 µM chloroquine was added to the media during the final 15 min of culture. Cells were washed in serum-free media and lysed for 10 min on ice in lysis buffer (50 µM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.2% deoxycholic acid, and 0.1% SDS, with Complete Protease Inhibitor Cocktail [Roche]). Lysates were cleared by centrifugation in a tabletop centrifuge at maximum speed at 4°C for 10 min. Protein was quantified using a bicinchoninic acid protein assay (Thermo Fisher Scientific). Lysates were mixed with Laemmli buffer and resolved on a 15% polyacrylamide gel and transferred to a nitrocellulose membrane. Mouse anti-LC3 was purchased from Cosmo Bio Co., Ltd. Rabbit antiactin and goat anti–rabbit-peroxidase were purchased from Sigma-Aldrich. Goat anti–mouse horseradish peroxidase was purchased from Jackson ImmunoResearch Laboratories, Inc.
RT-PCR.
RNA was isolated using the RNeasy kit (QIAGEN) according to the manufacturer’s instructions. The RNA was treated with DNA-free (Invitrogen) to ensure the removal of any contaminating DNA that might be present, reverse transcribed using the high-capacity cDNA Reverse Transcription kit (Applied Biosystems), and amplified using Fast SYBR Green Master Mix (Applied Biosystems) and the primer sequences actin forward, 5′-GGCTGTATTCCCCTCCATCG-3′; and reverse, 5′-CCAGTTGGTAACAATATGT-3′; and ATG5 forward, 5′-TGTGCTTCGAGATGTGTGGTT-3′; and reverse, 5′-GTCAAATAGCTGACTCTTGGCAA-3′ in a Step One Plus Real time PCR System (Applied Biosystems). PAD2 and PAD4 primer and probe sets for TaqMan Gene Expression Assays were obtained from Applied Biosystems.
PAD4 expression.
Total RNA was extracted from CD11c+ cells isolated from B10.BR spleens using the RNeasy kit. The RNA was treated with DNA-free and reverse transcribed using the high-capacity cDNA Reverse Transcription kit. PAD4 was cloned into the pcDNA4/TO and cotransfected with pcDNA6/TR into C3.F6 (T-REx system; Invitrogen). Cells were maintained under 250 µg/ml Zeocin (Invitrogen) and 5 µg/ml Blasticidin (Invitrogen) selection. PAD4 expression was induced with 1 µg/ml tetracycline (Sigma-Aldrich).
Purification of autophagosomes.
Autophagosomes were isolated from PECs taken from at least 20 mice in a protocol described by Seglen and Brinchmann (2010). In brief, cells were cultured for 2 h in saline with 200 µM vinblastine (Sigma-Aldrich) in a glass tube in a shaking water bath at 37°C, then cooled, washed in cold saline, and swollen in 10% sucrose. The cells were disrupted with a single 200-kV pulse, the volume was increased with homogenization buffer (0.25 M sucrose, 10 mM Hepes, and 1 mM EDTA, pH 7.3), and six passes were made in a cell homogenizer (Isobiotec) with a 16-µm clearance. The lysates were incubated for 7 min in a shaking water bath at 37°C with glycyl-l-phenylalanine (Bachem) at 0.5 mM to eliminate lysosomes. The lysates (fraction 1) were spun at 4,000 rpm in an SS34 rotor (Beckman Coulter) for 2 min to remove the nuclei and intact cells (fraction 3). The pellet was washed once, and the supernatant (fraction 2) was fractionated over a two-step (9.5%/22.5%) Nycodenz (Axis-Shield) gradient for 1 h at 28,000 rpm in an SW28 rotor (Beckman Coulter). The pellet (fraction 4), upper interface (fraction 5), and bottom interface were collected. The bottom interface was then fractionated over a two-step gradient (33% Percoll/22.5% Nycodenz) for 30 min at 20,000 rpm in an SW28 rotor. The top band (fraction 6) and bottom band (fraction 7) were collected. Fractions 5, 6, and 7 were washed with homogenization buffer and pelleted at 10,800 rpm in an SW28 rotor. These fractions were disrupted with several quick passes through a 30-gauge needle, and protein concentration in each fraction was measured by bicinchoninic acid protein assay, and equivalent protein concentrations were used in the PAD assay.
PAD activity assay.
PAD activity was measured as previously described (Watanabe et al., 1988). In brief, cells were suspended at 108/ml in 0.1 M Tris HCl, pH 7.6, 10 µM 2-mercaptoethanol, and 1 mM EDTA and disrupted with a Dounce homogenizer. Lysates were cleared by centrifugation at 10,000 g for 10 min at 4°C. Lysates were mixed 1:1 with PAD assay buffer (200 mM Tris HCl, pH 7.5, 20 mM CaCl2, and 10 mM DTT) ± 20 mM BAEE and incubated for 1 h at 56°C. The reaction was stopped by adding 5 M HClO4 to one fifth of the total volume. Citrulline was measured using methods described previously using free citrulline as a standard (Holm et al., 2006).
Microscopy.
Fixed cells were permeabilized with Perm/Wash buffer (BD) and treated with 10% 24G2 supernatant to block Fc receptors. Cells were stained in solution on ice and cytospun onto glass slides for imaging. Cells were mounted with ProLong gold (Invitrogen). Cells were imaged with a laser-scanning confocal microscope (510; Carl Zeiss).
Quantitation of citrulline in naturally processed peptides.
C3.F6 mHEL were cultured in 2L roller bottles in DME supplemented with either 5 or 1% FCS and cultured at 37°C until they reached confluence. Naturally processed peptides were purified as previously described using 40F anti–I-Ak antibody–conjugated Sepharose (Suri et al., 2002). Citrulline was measured using methods described previously using free citrulline as a standard (Holm et al., 2006).
shRNA targeting.
Stable cell lines expressing shRNA designed to target expression of Luciferase as a control or Atg5 were generated with the previously described pFLRU lentiviral vector system (Langer et al., 2008). The pFLRU:shRNA:Luciferase and packaging vectors were a gift from J. Bednarski in the laboratory of B. Sleckman at Washington University in St. Louis. The primers used to make the Atg5-targeting vectors were 5′-GTGGAAAGGACGAAACACCGGAAGAACTTAGCCTATATTTCAAGAGAATATAGG-3′ and 5′-TCCAGCTCGAGAAAAAGGAAGAACTTAGCCTATATTCTCTTGAAATATAGG-3′ (bold portions indicate the specific targeting sequence for ATG5 knockdown, and the remainder of the sequences is for construction of the hairpin loop and the expression cassette). 2 µg of the pFLRU vectors was cotransfected with 1.6 µg pHR′Δ8.2R packaging vector and 0.4 µg pCMV-VSVg envelope plasmid into HEK293T cells with Lipofectamine 2000 (Invitrogen). Supernatants were harvested 24 h later, and after centrifuging three times for 5 min at 500 g to remove any HEK293T cells, supernatants were transferred to C3.F6.mHEL cells with Polybrene added to 5 µg/ml. After 24 h, cells were placed under puromycin selection, and after two passages, cells expressing high YFP were purified by flow cytometric cell sorting.
Online supplemental material.
Fig. S1 shows PAD2 and PAD4 expression in APCs under the experimental conditions included in this study. Table S1 includes a summary of four independent experiments that examined the level of citrulline in naturally processed peptides from cells cultured under normal conditions compared with those cultured in serum-starved conditions. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110640/DC1.
Acknowledgments
We would like to acknowledge Shirley Petzold, Katherine Frederick, and Jeremy Herzog for technical assistance.
This work was supported by National Institutes of Health grants AI022033 and AI024742.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- 3MA
- 3-methyladenine
- HEL
- hen egg-white lysozyme
- mHEL
- membrane HEL
- mRNA
- messenger RNA
- PAD
- peptidylarginine deiminase
- PEC
- peritoneal exudate macrophage
- RA
- rheumatoid arthritis
- shRNA
- short hairpin RNA
References
- Anderton S.M. 2004. Post-translational modifications of self antigens: implications for autoimmunity. Curr. Opin. Immunol. 16:753–758 10.1016/j.coi.2004.09.001 [DOI] [PubMed] [Google Scholar]
- Beltrami A., Rossmann M., Fiorillo M.T., Paladini F., Sorrentino R., Saenger W., Kumar P., Ziegler A., Uchanska-Ziegler B. 2008. Citrullination-dependent differential presentation of a self-peptide by HLA-B27 subtypes. J. Biol. Chem. 283:27189–27199 10.1074/jbc.M802818200 [DOI] [PubMed] [Google Scholar]
- de Haan E.C., Wagenaar-Hilbers J.P., Liskamp R.M., Moret E.E., Wauben M.H. 2005. Limited plasticity in T cell recognition of modified T cell receptor contact residues in MHC class II bound peptides. Mol. Immunol. 42:355–364 10.1016/j.molimm.2004.07.044 [DOI] [PubMed] [Google Scholar]
- de Vries R.R., Huizinga T.W., Toes R.E. 2006. HLA and RA revisited: citrullinated food for the SE hypothesis, the DR6 effect, and NIMA. Hum. Immunol. 67:454–459 10.1016/j.humimm.2006.03.016 [DOI] [PubMed] [Google Scholar]
- Dengjel J., Schoor O., Fischer R., Reich M., Kraus M., Müller M., Kreymborg K., Altenberend F., Brandenburg J., Kalbacher H., et al. 2005. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA. 102:7922–7927 10.1073/pnas.0501190102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörfel D., Appel S., Grünebach F., Weck M.M., Müller M.R., Heine A., Brossart P. 2005. Processing and presentation of HLA class I and II epitopes by dendritic cells after transfection with in vitro-transcribed MUC1 RNA. Blood. 105:3199–3205 10.1182/blood-2004-09-3556 [DOI] [PubMed] [Google Scholar]
- Engelhard V.H., Altrich-Vanlith M., Ostankovitch M., Zarling A.L. 2006. Post-translational modifications of naturally processed MHC-binding epitopes. Curr. Opin. Immunol. 18:92–97 10.1016/j.coi.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Fader C.M., Sánchez D., Furlán M., Colombo M.I. 2008. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 9:230–250 10.1111/j.1600-0854.2007.00677.x [DOI] [PubMed] [Google Scholar]
- Hill J.A., Southwood S., Sette A., Jevnikar A.M., Bell D.A., Cairns E. 2003. Cutting edge: the conversion of arginine to citrulline allows for a high-affinity peptide interaction with the rheumatoid arthritis-associated HLA-DRB1*0401 MHC class II molecule. J. Immunol. 171:538–541 [DOI] [PubMed] [Google Scholar]
- Holm A., Rise F., Sessler N., Sollid L.M., Undheim K., Fleckenstein B. 2006. Specific modification of peptide-bound citrulline residues. Anal. Biochem. 352:68–76 10.1016/j.ab.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Ireland J., Herzog J., Unanue E.R. 2006. Cutting edge: unique T cells that recognize citrullinated peptides are a feature of protein immunization. J. Immunol. 177:1421–1425 [DOI] [PubMed] [Google Scholar]
- Klareskog L., Rönnelid J., Lundberg K., Padyukov L., Alfredsson L. 2008. Immunity to citrullinated proteins in rheumatoid arthritis. Annu. Rev. Immunol. 26:651–675 10.1146/annurev.immunol.26.021607.090244 [DOI] [PubMed] [Google Scholar]
- Langer E.M., Feng Y., Zhaoyuan H., Rauscher F.J., III, Kroll K.L., Longmore G.D. 2008. Ajuba LIM proteins are snail/slug corepressors required for neural crest development in Xenopus. Dev. Cell. 14:424–436 10.1016/j.devcel.2008.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Kuma A. 2008. Autophagosomes in GFP-LC3 transgenic mice. Methods Mol. Biol. 445:119–124 10.1007/978-1-59745-157-4_7 [DOI] [PubMed] [Google Scholar]
- Nimmerjahn F., Milosevic S., Behrends U., Jaffee E.M., Pardoll D.M., Bornkamm G.W., Mautner J. 2003. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur. J. Immunol. 33:1250–1259 10.1002/eji.200323730 [DOI] [PubMed] [Google Scholar]
- Paludan C., Schmid D., Landthaler M., Vockerodt M., Kube D., Tuschl T., Münz C. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 307:593–596 10.1126/science.1104904 [DOI] [PubMed] [Google Scholar]
- Peterson D.A., DiPaolo R.J., Kanagawa O., Unanue E.R. 1999. Quantitative analysis of the T cell repertoire that escapes negative selection. Immunity. 11:453–462 10.1016/S1074-7613(00)80120-X [DOI] [PubMed] [Google Scholar]
- Petersen J., Purcell A.W., Rossjohn J. 2009. Post-translationally modified T cell epitopes: Immune recognition and immunotherapy. J. Mol. Med. 87:1045–1051 10.1007/s00109-009-0526-4 [DOI] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis E., Blommaart E.F., Meijer A.J., Codogno P. 2000. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275:992–998 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- Pritzker L.B., Joshi S., Gowan J.J., Harauz G., Moscarello M.A. 2000. Deimination of myelin basic protein. 1. Effect of deimination of arginyl residues of myelin basic protein on its structure and susceptibility to digestion by cathepsin D. Biochemistry. 39:5374–5381 10.1021/bi9925569 [DOI] [PubMed] [Google Scholar]
- Schmid D., Pypaert M., Münz C. 2007. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 26:79–92 10.1016/j.immuni.2006.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P.O., Brinchmann M.F. 2010. Purification of autophagosomes from rat hepatocytes. Autophagy. 6:542–547 10.4161/auto.6.4.11272 [DOI] [PubMed] [Google Scholar]
- Seglen P.O., Gordon P.B. 1982. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA. 79:1889–1892 10.1073/pnas.79.6.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri A., Vidavsky I., van der Drift K., Kanagawa O., Gross M.L., Unanue E.R. 2002. In APCs, the autologous peptides selected by the diabetogenic I-Ag7 molecule are unique and determined by the amino acid changes in the P9 pocket. J. Immunol. 168:1235–1243 [DOI] [PubMed] [Google Scholar]
- Tarcsa E., Marekov L.N., Mei G., Melino G., Lee S.C., Steinert P.M. 1996. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J. Biol. Chem. 271:30709–30716 10.1074/jbc.271.48.30709 [DOI] [PubMed] [Google Scholar]
- van der Helm-van Mil A.H., Huizinga T.W. 2008. Advances in the genetics of rheumatoid arthritis point to subclassification into distinct disease subsets. Arthritis Res. Ther. 10:205 10.1186/ar2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossenaar E.R., Radstake T.R., van der Heijden A., van Mansum M.A., Dieteren C., de Rooij D.J., Barrera P., Zendman A.J., van Venrooij W.J. 2004. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann. Rheum. Dis. 63:373–381 10.1136/ard.2003.012211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Tsubata T. 2009. Autophagy connects antigen receptor signaling to costimulatory signaling in B lymphocytes. Autophagy. 5:108–110 10.4161/auto.5.1.7278 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Akiyama K., Hikichi K., Ohtsuka R., Okuyama A., Senshu T. 1988. Combined biochemical and immunochemical comparison of peptidylarginine deiminases present in various tissues. Biochim. Biophys. Acta. 966:375–383 10.1016/0304-4165(88)90088-8 [DOI] [PubMed] [Google Scholar]
- Wegner N., Lundberg K., Kinloch A., Fisher B., Malmström V., Feldmann M., Venables P.J. 2010. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunol. Rev. 233:34–54 10.1111/j.0105-2896.2009.00850.x [DOI] [PubMed] [Google Scholar]