Critical injury in humans induces a genomic storm with simultaneous changes in expression of innate and adaptive immunity genes.
Abstract
Human survival from injury requires an appropriate inflammatory and immune response. We describe the circulating leukocyte transcriptome after severe trauma and burn injury, as well as in healthy subjects receiving low-dose bacterial endotoxin, and show that these severe stresses produce a global reprioritization affecting >80% of the cellular functions and pathways, a truly unexpected “genomic storm.” In severe blunt trauma, the early leukocyte genomic response is consistent with simultaneously increased expression of genes involved in the systemic inflammatory, innate immune, and compensatory antiinflammatory responses, as well as in the suppression of genes involved in adaptive immunity. Furthermore, complications like nosocomial infections and organ failure are not associated with any genomic evidence of a second hit and differ only in the magnitude and duration of this genomic reprioritization. The similarities in gene expression patterns between different injuries reveal an apparently fundamental human response to severe inflammatory stress, with genomic signatures that are surprisingly far more common than different. Based on these transcriptional data, we propose a new paradigm for the human immunological response to severe injury.
Traumatic injury with its potential for infection was likely a common cause of death for our human ancestors. Even today, massive injury remains the most common cause of death for those under the age of 45 yr in developed countries (Sasser et al., 2006; Probst et al., 2009). Only recently has the human injury response been studied systematically at the genomic level and only now is it beginning to become better understood. Unfortunately, billions of dollars worldwide have been invested on new biological therapeutics for severe injury, as well as for its sequelae, sepsis and septic shock, with disappointing, if not harmful, results. The current immune, inflammatory paradigm, based on an incomplete understanding of the functional integration of the complex host response, remains a major impediment to the development of effective innovative therapies.
Prior work has focused on the role of individual mediators (e.g., TNF or IL-1; Giannoudis, 2003; DeLong and Born, 2004; Giannoudis et al., 2004; Keel and Trentz, 2005) or processes such as apoptosis and cellular death in nosocomial infections and organ injury after trauma (Hotchkiss et al., 2009). Rather than using a reductionist approach, we examined the genome-wide expression patterns of blood leukocytes in the immediate postinjury period to better understand the overall priorities and patterns of gene expression underlying not only the initial injury response, but also the development of complications and delayed clinical recovery (Flohé et al., 2008). We have compared the genomic response by blood leukocytes to trauma with the changes in gene expression produced by major burns (>20% of body surface area), as well as the response by healthy subjects to the administration of low-dose bacterial endotoxin (Calvano et al., 2005). The results of this systems-wide approach to the study of severe human injury challenge several current clinical dogmas regarding the nature of the host response to severe injury. In addition, the datasets described in this report of our large clinical study are an important resource that will enable important future analyses like mathematical modeling and predicting patient outcomes.
Circulating blood leukocytes have the capacity to seek out, recognize, and mount an appropriate inflammatory response at the earliest sign of injury. Innate immune cells initially recognize and are activated by pathogen-associated molecular patterns (PAMPs) or endogenous alarmins and danger signals (Xu et al., 2009; Puneet et al., 2010; Zhang et al., 2010). Blood neutrophils, monocytes, and NK cells are implicated as primary effectors during the initial inflammation and activation of innate immunity. Severe trauma has also been characterized by immunosuppression, primarily seen on the adaptive immune system with T lymphocyte populations being the most markedly affected cell population (Hotchkiss and Karl, 2003; Keel and Trentz, 2005). Although antiinflammatory processes and reduced effector T cell function are necessary to limit or localize the response to severe trauma, a prolonged or exaggerated period of immune suppression or defective immune response leads to increased susceptibility to secondary infections (Hotchkiss and Karl, 2003).
We isolated whole blood leukocytes and performed genome-wide expression analysis from the Affymetrix U133 GeneChip using a cohort of 167 patients between the ages of 18 and 55 yr who consented to blood sampling from 1,637 adult severe blunt trauma patients who developed hypotension or acidosis and required resuscitation with blood products from seven US hospitals. Blood was sampled within 12 h and at 1, 4, 7, 14, 21, and 28 d after the injury. Genome-wide expression from patients with trauma was compared with age-, sex-, and ethnicity-matched healthy subjects and with 133 adult patients after severe burn injury (>20% of the body surface area) or 4 healthy adult subjects administered low-dose bacterial endotoxin.
There were multiple objectives of the present study. The first objective was to determine whether the multiple-organ dysfunction syndrome (MODS) phenotypes observed agreed with the current paradigm that explains whether the MODS seen after injury is the result of excessive proinflammatory responses (systemic inflammatory response syndrome [SIRS]) followed temporally by compensatory antiinflammatory response syndrome (CARS) and suppression of adaptive immunity. In those with MODS, this period of recovery from organ failure varied from a few days, nonrecovery at 28 d, or death. Unexpectedly, there were no clinical outcomes consistent with MODS followed by recovery and subsequently severe MODS that might be predicted as a second hit (Sauaia et al., 1994; Keel and Trentz, 2005).
The second objective was to determine whether there were recognizable gene expression changes in the blood leukocytes after severe blunt trauma. Our data indicate that severe trauma altered the expression of >80% of the leukocyte transcriptome during the first 28 d after injury, and these changes were highly reproducible within at least 30 discernable gene expression patterns. Of the most significantly regulated pathways, injury produced early activation of those involving innate and simultaneous suppression of those involving adaptive immunity. Interestingly, severity of injury, magnitude of physiological derangement, and volume of transfused blood minimally affected these patterns.
The third objective was to determine whether there were patterns of gene expression associated with two extremes of clinical recovery (uncomplicated versus complicated). Surprisingly, gene expression patterns were highly comparable between these two recovery extremes with selective differences in only magnitude and duration. Our data support a new paradigm for the host immunological response to injury.
RESULTS AND DISCUSSION
Trauma patients versus healthy subjects
The characteristics of the trauma patients and healthy subjects and the patient clinical outcomes are shown in Table I and Fig. 1 A. As seen in Fig. 1 A, the majority of trauma patients presented with mild to severe MODS but recovered before 28 d. There was only a small fraction of patients who either did not develop MODS or developed severe MODS and did not recover before 28 d. There were no patients who developed MODS initially, partially recovered, but went on to develop severe MODS.
Table I.
Characteristics and outcomes in the 167 trauma patients and 37 healthy control subjects
| Parameter | Controls (n = 37) | Total cohort (n = 167) | Uncomplicated recovery patient (<5 d; n = 55) | Complicated recovery patient (14 d, no recovery by 28 d, or death; n = 41) | Probability |
| Demographics | |||||
| Age (yr) | 30 ± 8 | 34 ± 1 (33, 25–44) | 33 ± 2 (32, 21–43) | 34 ± 2 (34, 26–42) | P = 0.466a |
| Sex (male/female) | 22/15 | 106/61 | 30/25 | 30/11 | P = 0.090c |
| APACHE II | ND | 27.3 ± 0.5 (28, 24–32) | 24.4 ± 0.8 (25, 21–29) | 29.4 ± 0.8 (29, 26–33) | P < 0.001a |
| Maximum abbreviated injury scale (AIS) | ND | 4.0 ± 0.1 (4, 3–5) | 3.8 ± 0.1 (4, 3–5) | 4.2 ± 0.1 (4, 4–5) | P = 0.050b |
| Head AIS | ND | 3.0 ± 0.1 (3, 2–4) | 2.9 ± 0.3 (3, 2–4) | 3.1 ± 0.3 (3, 2–4) | P = 0.659b |
| Face/neck AIS | ND | 1.7 ± 0.1 (2, 1–2) | 1.6 ± 0.2 (2, 1–2) | 1.6 ± 0.2 (1, 1–2) | P = 0.586b |
| Thorax AIS | ND | 3.4 ± 0.1 (3, 3–4) | 3.1 ± 0.2 (3, 3–4) | 3.3 ± 0.2 (3, 3–4) | P = 0.497b |
| Abdomen AIS | ND | 3.2 ± 0.1 (3, 2–4) | 3.4 ± 0.2 (4, 2–4) | 3.6 ± 0.2 (4, 3–4) | P = 0.561b |
| Spine AIS | ND | 2.1 ± 0.1 (2) | 2.0 ± 0.0 (2) | 2.0 ± 0.0 (2) | P = 1.000b |
| Upper extremity/lower extremity AIS | ND | 3.3 ± 0.1 (3, 3–4) | 3.1 ± 0.2 (3) | 3.4 ± 0.2 (3, 3–5) | P = 0.233b |
| ISS | ND | 31.3 ± 1.0 (33, 22–41) | 26.2 ± 1.8 (24, 17–35) | 35.7 ± 2.0 (38, 27–42) | P < 0.001a |
| New ISS | ND | 36.3 ± 1.0 (34, 27–43) | 32.6 ± 1.8 (29, 22–40) | 39.8 ± 1.9 (41, 29–44) | P = 0.004b |
| Total transfusion (ml) administered within the fist 24 h | 0 | 2,425 ± 158 (1,900, 1,050–3,000) | 1,705 ± 172 (1,400, 700–2,229) | 2,952 ± 423 (2,150, 1,050–3,500) | P = 0.005b |
| Total crystalloid (ml) administered within the fist 24 h | 0 | 12,891 ± 557 (10,800, 8,276–15,800) | 10,544 ± 765 (9,070, 7,409–12,163) | 15,226 ± 1,530 (12,935, 8,728–18,683) | P = 0.003b |
| Worst base deficit | ND | −9.8 ± 0.4 (−9.1, −12.0 to −6.4) | −9.2 ± 0.4 (−8.9, −11.6 to −6.4) | −10.6 ± 0.8 (−10.3, −13.8 to −6.0) | P = 0.133a |
| Lowest systolic blood pressure (mm Hg) | ND | 89.4 ± 1.5 (86, 78–103) | 92.3 ± 3.0 (88, 80–108) | 86.6 ± 3.0 (84, 77–97) | P = 0.121b |
| Outcomes | |||||
| Survival | ND | 96% (160/7) | 100% | 83% (34/7) | NA |
| Maximum modified Marshall score | ND | 5.5 ± 0.2 (5, 3–7) | 3.0 ± 0.1 (3, 2–4) | 8.8 ± 0.4 (8, 7–10) | NA |
| Hospital length of stay (d) | 0 | 24.8 ± 1.4 (21, 12–32) | 15.2 ± 1.7 (12, 9–18) | 35.8 ± 3.6 (30, 23–42) | NA |
| Intensive care unit length of stay (d) | 0 | 13.0 ± 0.9 (9, 5–18) | 4.8 ± 0.4 (5, 3–6) | 25.1 ± 2.3 (21, 18–30) | NA |
| Time to recovery (d) | 0 | 10.2 ± 0.6 (7, 4–15) | 2.9 ± 0.1 (3, 2–4) | 22.0 ± 0.9 (20, 18–28) | NA |
| Integral of MOF over days | 0 | 46.8 ± 3.4 (32, 14–69) | 10.8 ± 0.9 (11, 6–15) | 97.0 ± 6.6 (87, 67–113) | NA |
| Complications | |||||
| Noninfectious complications | 0 | 51.5% (86/81) | 5.5% (3/52) | 90.2% (37/4) | P < 0.001c |
| Nosocomial infections | 0 | 54.5% (91/76) | 20.0% (11/44) | 85.4% (35/6) | P < 0.001c |
| Surgical site infections | 0 | 22.2% (37/130) | 7.3% (4/51) | 41.5% (17/24) | P < 0.001c |
| Ventilator-associated pneumonia (cases/1,000 ICU days) | 0 | 24.0 | 3.8 | 25.3 | P = 0.030d |
MOF, multiple organ failure; NA, not applicable. Values represent the mean ± SEM, with median and middle quartiles indicated in parentheses. Significance was designated at the P < 0.05 level of confidence.
Data were analyzed by the Student’s t test.
Data were analyzed by the Mann-Whitney signed rank test.
Data were analyzed by the Fisher’s exact text.
Data were analyzed by the exact binomial test.
Figure 1.
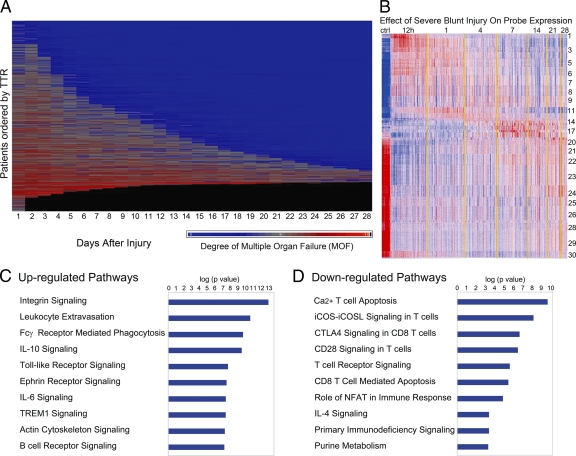
Organ injury and genomic changes associated with severe blunt trauma. (A) Whole blood was taken from severe blunt trauma patients, leukocytes were isolated, and total cellular RNA was extracted and hybridized onto an HU133 Plus 2.0 GeneChip. The continuum of clinical responses to severe blunt trauma in the 1,637 total patients from which the 167 sampling trauma patients were drawn is shown graphically. Each row represents an individual patient ordered by time to recovery (TTR), and the x axis represents time from injury in days. Patients are sorted from least to most severe organ injury and mortality. The presence and severity of organ injury is represented by colors from blue (least severe) to red (most severe). Black indicates death. (B) K-means clustering of the genes into 30 clusters based on patterns of expression over time. Red indicates increased and blue indicates decreased expression relative to the mean (white). 5,136 genes were differentially expressed between patients and controls (ctrl; FDR <0.001 and at least twofold change). (C and D) Summary of the canonical pathways most affected by trauma. The graph shows the −log10 (p value) of the enrichment of the pathway.
Our data showed that severe blunt trauma produced significant changes in the leukocyte messenger RNA abundance of 16,820 out of 20,720 Entrez genes on the microarrays, representing >80% of the human genome over the first 28 d (using a false discovery rate [FDR] adjusted probability <0.001; Fig. 1 B; Storey et al., 2005). The term “genomic storm” has been coined to capture the magnitude and rapidity with which the leukocyte transcriptome reorganized and reprioritized its expression patterns. To our knowledge, the magnitude and the extent of the change in expression represent the only examples of such a severe perturbation of the human genome after extreme stress. However, given the multitude of stressors to the body after injury, this observation is not entirely unexpected. In model systems, nearly all of the organism’s genes have been shown to be involved in the response to diverse internal or external stimuli (Arbeitman et al., 2002; Hillenmeyer et al., 2008).
Of the significant genes, 5,136 genes exhibited at least a twofold change in expression over the time course compared with healthy subjects (Fig. 1 B). The greatest changes in circulating leukocyte gene expression were seen at the earliest time point (<12 h from injury), and the number of genes whose expression decreased (n = 3,051; clusters 17–30) was greater than the number of genes whose expression increased (n = 2,085; clusters 1–16). At the first sampling time period (within 12 h after injury), which represented 167 samples (Fig. 1 B), 37 samples were drawn within 4 h after injury, 55 between 5 and 8 h and 75 between 9 and 12 h (Fig. S1). Sampling densely in the first 12 h allowed substantial precision to characterize the early genomic response. Additional plots for all 30 clusters are shown in Fig. S2 (A–C).
Not surprisingly, the genes whose expression increased the most were those involved in innate immunity and the inflammatory response, including NB1 (CD177), MMP8 (neutrophil collagenase), LTF (lactotransferrin), and HP (haptoglobin), which all reached maximum expression within 12 h. As shown in Fig. 1 C, 8 of the 10 gene families most increased after injury in leukocytes were directly involved in innate immunity, pathogen recognition, or inflammation. For example, the expression of all of the Toll-like receptor (TLR) genes (TLR1, TLR2, TLR4, TLR5, TLR8, TLR9, and TLR10), with the exception of TLR3 and TLR7, was significantly increased after injury, as was the expression of other pattern recognition receptors, NOD1 (nucleotide-binding oligomerization domain containing 1), NOD2 , NALP1 (NLR family, pyrin domain containing 1), and NALP3.
Also not surprising, 9 of the 10 gene families most suppressed after injury were involved in antigen presentation and T cell activation (Fig. 1 D). The genes whose expression decreased the greatest included S100A8 (calgranulin), MYBL1 (myeloblastosis viral homologue, v-myb), KLRF1 (Killer cell lectin like receptor subfamily F, member 1), TGFBR3 (TGF-β receptor III subunit), and TCRA (T cell receptor α subunit).
The pattern of gene expression points to the simultaneous initiation of a myriad of innate and adaptive immunological processes. The vast majority of the gene ontologies, whose expression was predominantly increased, were those involved in innate immunity, microbial recognition, inflammation, or, much later, B cell proliferation and immunoglobulin synthesis. Conversely, the gene ontologies whose expression was most decreased were those involved in T cell function and antigen presentation.
Because the cytokine milieu might be of interest in the human response to injury, quantitation of 17 immunity-related cytokines was performed (Fig. S3). Although there was substantial patient to patient variation within a sampling time point, four cytokines (IL-6, IL-1ra, IL-8, and MCP1) showed significant temporal variation over the 28-d period. Not surprisingly, these cytokines have been associated with the injury response in many other studies (DeLong and Born, 2004; Giannoudis et al., 2004).
Contribution of clinical parameters
We show that clinical parameters associated with poor clinical recovery, increased injury severity score (ISS), massive volumes of blood transfused, and increased degree of shock (base deficit; Sauaia et al., 1994), have a surprisingly limited effect on gene expression. A univariate analysis demonstrated minimal contributions with the expression of only ∼200 genes altered by blood transfusion in the first day after injury; no gene expression changes were associated with ISS and only 8 with base deficit within the first 12 h (Fig. S4 A). To control for several cofactors, a propensity analysis identified only ∼400 genes whose expression was dependent on the volume of transfused blood (Fig. S4 B). Similarly, the early genomic changes were not caused by differences in the patterns of blood leukocytes between the complicated and uncomplicated recovery patients (Fig. S5).
Severe trauma compared with burns or endotoxemia
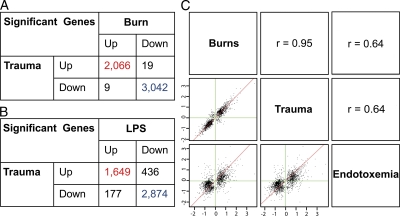
We show that the genomic response to trauma is remarkably similar to the changes in gene expression caused by severe burn injury or infusion of low-dose bacterial endotoxin (producing endotoxemia), differing primarily in the duration of the response. Of the 5,136 genes shown in Fig. 1 A that change twofold after severe trauma, 98% (all but 83) had the same direction of change after burns (Fig. 2 A), and 88% (4,533) changed similarly in endotoxemia (Fig. 2 B). Scatter plots of the fold changes of significant genes in trauma, burns, or endotoxemia showed that the Pearson correlation coefficient (r) between trauma and burns was 0.95 and between trauma and endotoxemia was 0.64 (Fig. 2 C). On a genome-wide scale, these findings demonstrate a common response pattern reflective of the large overlap in upstream receptors and signaling intermediates activated by each condition (i.e., TLR4; Baccala et al., 2009).
Figure 2.
Validation of the genomic response to trauma in burn patients and healthy adults challenged with low-dose bacterial endotoxin. (A and B) Comparison of direction of changes among the genes identified in Fig. 1 B, between trauma and burns (A) and trauma and endotoxemia (B). (C) Scatter plots of log2 fold changes (x and y axes) of 5,855 genes (FDR <0.001 and at least twofold change) in trauma, burns, or endotoxemia (bottom left) and corresponding Pearson correlation coefficient (r). The axes in C represent fold changes.
Despite markedly different clinical presentation of severe blunt trauma, burn injury, and endotoxemia, the early genomic changes were highly comparable particularly between the stressors, burns, or blunt trauma. These common response patterns likely resulted from their in vivo activation either through primarily a single TLR4 agonist in the case of endotoxemia or through the release of PAMPs, DAMPs (danger-associated molecular patterns), or alarmins in the case of burns or trauma. As previously mentioned, mitochondrial DNA (Zhang et al., 2010), histones (Xu et al., 2009), and pathogens (Calvano et al., 2005; Puneet et al., 2010) are stimuli that are available in the plasma to trigger multiple receptors like TLRs, PAMPs, and alarmins in burns and injury. Given the massive cellular necrosis and apoptosis associated with injury and trauma, intracellular proteins and cellular debris can be readily seen in the plasma. In another study (Liu et al., 2006), we also showed that the largest number (up to 70%) of unique proteins measured in blood plasma from humans after trauma were intracellular components. Certainly, these molecules also are readily available to serve as alarmins and trigger these pattern recognition receptors.
Our findings also suggest that not only is the genomic response to severe trauma dramatic, but the changes in gene expression are also long-lived. In more than half of the genes, messenger RNA abundance levels had not returned to baseline after 28 d in blunt trauma and 90 d in severe burns, reflecting prolonged aberrations in the leukocyte transcriptome. Perhaps the persistence of cellular debris in the plasma, in part, drives this prolonged genomic response, which is consistent with the concept of nonresolving inflammation (Nathan and Ding, 2010).
Complicated versus uncomplicated clinical recovery
The development of organ failures and infections are well known complications after injury that contribute to prolonged intensive care unit stays and higher costs for acute hospitalization and rehabilitation. To explore in greater detail the transcriptomic response to critical illness, we compared genomic patterns of the extremes in clinical recovery after trauma, those patients who were responding to the injury itself (uncomplicated: recovery in <5 d), and those who were, in addition, responding to ensuing complications (complicated: recovery >14 d, no recovery, or death; Fig. 1 A).
We were interested to identify whether the genomic patterns were different between patients with complicated and uncomplicated outcomes. That is, are there genes or pathways that behave differently in these extremes in clinical recovery? There were 2,391 genes in the circulating leukocytes whose expression was significantly different (FDR <0.001) at any one point over the time course when comparing patients with complicated versus uncomplicated outcomes. Of these genes, 1,201 had at least twofold changes in expression at any time point when compared with controls over the entire time course of the complicated or uncomplicated recovery (Fig. 3 A). Among the top up-regulated pathways associated with complicated recovery were IL-10, IL-6, and p38 MAPK signaling, whereas among the most down-regulated pathways were antigen presentation and T cell regulation (Fig. S6, A and B).
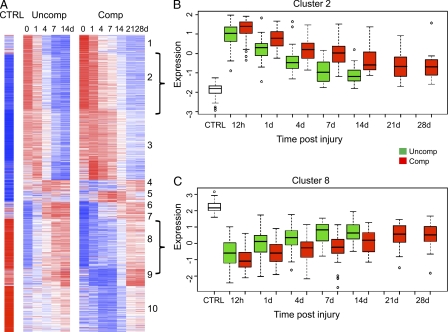
Figure 3.
Differences in gene expression patterns between patients with a complicated and uncomplicated clinical recovery. Heat map of 1,201 genes whose expression was at least twofold different at any time point when compared with controls (CTRL) for patients with a complicated (Comp) or uncomplicated recovery (Uncomp). (A) Cluster analysis of the two cohorts. The brackets to the right of the cluster indicate cluster 2 and 8 shown in B and C, respectively. (B) One cluster of genes whose expression was up-regulated in patients with a complicated recovery. (C) One cluster of genes whose expression was down-regulated in patients with a complicated recovery.
The expression patterns for all 30 clusters are shown in Fig. S2 A subcategorized by complicated or uncomplicated outcome; 28/30 box plots showed early up- or down-regulation of the gene clusters. In the 5,136 genes whose expression changed at least twofold after severe injury, the direction and magnitude of peak perturbations did not differ between clinical outcomes in this set (Fig. S2 B). In addition, a plot of the resolution time (half-time of recovery) from peak perturbation indicated that in complicated outcomes, genomic recovery was prolonged (Fig. S2 C). For example, cluster 2 showed a greater increase in early gene expression and a delayed return to baseline, whereas cluster 8 showed an exaggerated decrease in expression and a delayed return (Fig. 3, B and C). All 10 clusters are shown in Fig. S6 C. The direction of the responses for each of the 10 clusters was identical between a complicated and uncomplicated clinical recovery.
Unexpectedly, we show that the difference in gene expression between the two clinical recovery groups is not qualitative, but is only quantitative, the magnitude of the early response and the time required for expression to return to control values. In patients with an uncomplicated recovery, expression was returning or had returned to baseline within 7–14 d for both up- and down-regulated genes, but in patients with a complicated and prolonged recovery, the early changes in expression were greater, and the later changes had not returned to baseline, for the most part, by 28 d. There were no new genes or pathways that were recruited or any that dropped out in trauma patients with a complicated recovery versus those with an uncomplicated recovery.
Despite this clinical outcome, dichotomization and the modest, but significant differences in severity of injury, the volume of blood transfused, and the degree of base deficit between the two outcome groups (as shown in Table I), the genomic findings are unexpected. The overall changes in gene expression are remarkably similar, and the number of genes that are differentially expressed is small when compared with the changes in the transcriptome produced by severe injury.
Conclusion and perspectives
These experiments challenge the current paradigm regarding how the adult human responds to severe injury. Severe injury, whether a result of blunt trauma or burn injury produces a genomic storm in which up to 80% of the leukocyte transcriptome is altered. The changes occur rapidly in trauma within 4–12 h and are prolonged for days and weeks. We are not aware of any tumor or other clinical condition associated with such diversity and magnitude of genomic changes. Furthermore, delayed clinical recovery with organ injury is not associated with dramatic qualitative differences in the leukocyte transcriptome. In both clinical cohorts (complicated and uncomplicated recovery), all genes moved in the same direction despite the patient’s clinical course. We could find no evidence of a single gene or cluster of genes whose expression changed uniquely, associated with different clinical outcomes. Rather, the genomic changes in the dichotomous clinical recoveries in trauma represent variations in a common inflammatory stress response, which is unlikely to be distinguishable by a single or small set of biomarkers.
Our findings demonstrate that the genomic response to trauma not only induces the activation of a large number of inflammatory mediators, genes involved in pattern recognition, and antimicrobial functions, but also suppresses genes involved in antigen presentation, T cell proliferation and apoptosis, T cell receptor function, and NK cell function. Furthermore, the differences between complicated and uncomplicated outcomes are seen in all of these pathways. Such findings suggest that identifying single agents for the treatment of the multitude of immunological complications after trauma might be unlikely.
Furthermore, given the commonality of gene expression between burn injury and blunt trauma and the considerable overlap with a single, low-dose bolus of bacterial endotoxin, the genomic patterns in the human leukocyte transcriptome represent a fundamental response to severe inflammatory stress. The findings are consistent with a genomic storm that is neither chaotic nor erratic, but rather highly coordinated and reproducible. This storm likely represents a common transcriptional response to severe stress in humans regardless of its origin, with far more similarities than differences.
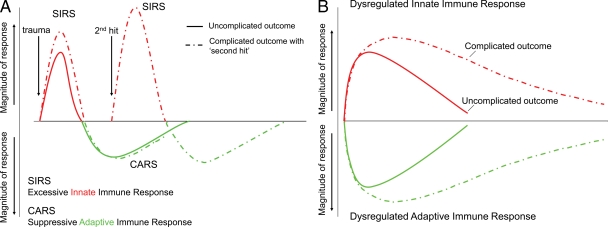
The current paradigm of the host response to severe trauma has been traditionally viewed as an early SIRS followed temporally by a compensatory antiinflammatory or immune-suppressive response syndrome (CARS; Fig. 4 A; Bone, 1996a; Hotchkiss and Karl, 2003). Current dogma argues that exaggerated inflammation contributes to adverse outcome (Giannoudis, 2003; Keel and Trentz, 2005; Xu et al., 2009; Zhang et al., 2010), and complicated outcomes are commonly associated with second hits or multiple inflammatory events induced by clinical episodes of infection or surgical stress, causing a secondary major genomic response (Nast-Kolb et al., 2001; Keel and Trentz, 2005). Much of this work is based on mouse models of trauma, burns, and sepsis, but we have accumulating evidence that the human genomic response to severe trauma can only be partially recapitulated by mouse models (unpublished data). However, the findings are unequivocal that the temporal nature of the current SIRS/CARS paradigm is not supported at the level of the leukocyte transcriptome.
Figure 4.
A genomic storm: Refining the immune, inflammatory paradigm in trauma. (A) The current paradigm explains complications of severe injury as a result of excessive proinflammatory responses (SIRS) followed temporally by compensatory antiinflammatory responses (CARS) and suppression of adaptive immunity. A second-hit phenomenon results from sequential insults, which leads to more severe, recurrent SIRS and organ dysfunction. (B) The proposed new paradigm involves simultaneous and rapid induction of innate (both pro- and antiinflammatory genes) and suppression of adaptive immunity genes. Complicated recoveries are delayed, resulting in a prolonged, dysregulated immune–inflammatory state.
The question of whether these prolonged changes in gene expression reflect an ongoing or repeated inflammatory stimulus or simply a response to the primary injury event cannot be resolved. The findings are most consistent with the nonresolving inflammation hypothesis (Nathan and Ding, 2010) that severely injured patients who are destined to die from their injuries have the same response as patients who subsequently recover. The difference is in the degree and the duration of the dysregulated acute inflammatory response.
The early peak and continuous genomic recovery over 28 d in the circulating blood leukocytes are also not consistent with a second-hit phenomenon causing recurrent major systemic inflammatory responses (Dewar et al., 2009). Nor are these data consistent with the current paradigm that the early transcriptional activation of innate immunity and microbial recognition precedes or induces a secondary or subsequent transcriptional activation of antiinflammatory or immune suppressive genes, or suppression of antigen presentation, or T cell response genes (Bone, 1996a,b). Collectively, these data demonstrate a potentially sustained genomic response implicating the potential acute onset of a chronic inflammatory process that may be associated with increased risk for late mortality. In fact, these unique findings may be partly responsible for the recent findings by Davidson et al. (2011) demonstrating increased 1-yr mortality in patients sustaining severe injury compared with noninjured matched controls.
We propose a new paradigm. At the level of the leukocyte transcriptome, alterations in the expression of classical inflammatory and antiinflammatory as well as adaptive immunity genes occur simultaneously, not sequentially after severe injury (Fig. 4 B). Our data show that the transcriptomic changes in the adaptive immune response occur very early and in fact are simultaneous with the proinflammatory reactions found in innate immunity. Given these findings, however, it can still be true that the phenotype of the immunosuppression of trauma may not be fully manifested until days after the injury, but initiation of these processes at the level of the leukocyte transcriptome occurs early and simultaneously with the activation of inflammation and innate immunity.
Severe blunt trauma and burn injury produce a global reprioritization of the leukocyte transcriptome affecting multiple cellular functions and pathways, a true genomic storm, the first hours of which are mimicked by endotoxemia. In the context of the host immune response, these changes are represented by simultaneous up-regulation of innate immune-related genes and the suppression of adaptive immune-related genes, regardless of clinical outcomes.
There are several caveats, however. We have looked initially at blood leukocyte populations, and compartmentalization of the inflammatory response is well known. Whether these changes in innate and adaptive immune responses are recapitulated in secondary lymphoid organs and the reticuloendothelial system is unknown. Second, our sampling intervals were extended as time from the injury progressed. There may well have been brief secondary responses that we could not detect. However, the absence of late episodes of new organ injury (Fig. 1 A) in this patient population argues strongly against evidence of any clinically relevant second inflammatory hit.
Although the mechanisms responsible for complicated clinical recovery after severe injury remain incompletely elucidated, we show in this study that the initial magnitude and duration of these genomic changes may discriminate complicated and uncomplicated recoveries. The changes in both innate and adaptive immunity are established soon after injury, so that early, targeted therapy to either or both immune pathways may be the approach that has the best possibility of improving patient outcomes.
MATERIALS AND METHODS
Study design.
Blood was sampled from 167 severe blunt trauma patients under the age of 55 yr who consented to blood sampling. The first blood sample was taken within 12 h of the injury and 1, 4, 7, 14, 21, and 28 d after injury. Study subjects were treated under the guidance of standard operating procedures developed, implemented, and audited across all participating centers to minimize treatment variation (Nathens et al., 2005; Minei et al., 2006; Moore et al., 2006; West et al., 2006; Harbrecht et al., 2007; Cuschieri et al., 2008; O’Keefe et al., 2008; West et al., 2008; Evans et al., 2009). Clinical outcomes and complications within 28 d after injury were recorded. Total blood leukocytes were isolated according to protocols published previously (Feezor et al., 2004; Laudanski et al., 2006). Total cellular RNA was extracted and hybridized onto an HU133 Plus 2.0 GeneChip (Affymetrix) according to the manufacturer’s recommendations. The study was approved by the Institutional Review Board of each institution. In addition to local institutional oversight, Massachusetts General Hospital reviewed and approved the program’s data center and databases.
Statistical analysis.
Trauma patients were divided into two groups to evaluate the impact of complications and expression differences associated with clinical recovery: (1) uncomplicated recovery in <5 d (n = 55) and (2) complicated recovery after 14 d, no recovery by 28 d, or death (n = 41; Fig. 1 B). Univariate analysis was performed to compare characteristics between groups using either the Student’s t test or Mann-Whitney signed rank test and Fisher’s exact test. Univariate analyses were conducted to examine the effect of blood transfusion, ISS, and base deficit on expression using a linear regression model. A propensity score for the effect of blood transfusion was developed from the confounding variables using logistic regression (D’Agostino, 1998; Hayes and Groner, 2008).
Statistical analysis was performed to identify genes differentially expressed between injured patients and healthy subjects and between trauma patients with different clinical outcomes using the software program EDGE (Storey et al., 2005). A k-means clustering was then applied to visualize major temporal patterns of the resulting significant genes with at least a twofold change in expression over time (Tavazoie et al., 1999; Calvano et al., 2005). Genes differentially expressed between patients and controls and between different clinical outcomes were then subjected to pathway analysis using Ingenuity Pathway Knowledge Base as previously described (Calvano et al., 2005; Laudanski et al., 2006). The results were independently validated with blood leukocyte genomics obtained from 133 adult patients with burns >20% of the total body surface area and from 4 healthy humans after administration of low-dose bacterial endotoxin (Calvano et al., 2005).
Additional information.
A supplemental web-based portal (Massachusetts General Hospital, 2011) is available to explore in greater detail the largest clinical and genomic database to date from severely injured humans. Data in this study have been deposited in the GEO DataSets site under accession number GSE11375.
Online supplemental material.
Fig. S1 shows the distribution of 167 samples assayed within the first 12 h of injury. Fig. S2 shows the comparison of gene expression patterns of the 5,136 genes with a greater than twofold change from control subjects for patients with a complicated (n = 55) and uncomplicated (n = 41) clinical recovery. Fig. S3 presents data on the plasma cytokine/chemokine levels after severe blunt injury. Fig. S4 shows the effect of transfusion, ISS, and base deficit on early patterns of gene expression. Fig. S5 presents data on the concentrations of blood leukocyte populations in the trauma patients. Fig. S6 depicts the differences in gene expression patterns between patients with a complicated and uncomplicated recovery. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111354/DC1.
Acknowledgments
This work was supported by grant 5U54 GM-062119-10, entitled Inflammation and the Host Response to Injury, a Large-Scale Collaborative Research Program (Glue Grant) funded by the National Institute of General Medical Sciences. The clinical trial number (ClinicalTrials.gov) is NCT00257231.
The authors have no conflicting financial interests.
Author contributions: W. Xiao, M. Mindrinos, J. Seok, and J. Cuschieri contributed equally to this paper. R. Tompkins, R. Davis, W. Xiao, M. Mindrinos, R. Maier, L. Moldawer, J. Cuschieri, L. Hennessy, E. Moore, J. Minei, P. Bankey, J. Johnson, J. Sperry, A. Nathens, T. Billiar, M. West, B. Brownstein, D. Herndon, H. Baker, C. Finnerty, M. Jeschke, M. Lopez, M. Klein, R. Gamelli, N. Gibran, B. Arnoldo, G. McDonald-Smith, D. Schoenfeld, J. Cobb, S. Warren, A. Cuenca, S. Lowry, and S. Calvano were responsible for study conception, design, and/or supervision. R. Tompkins, R. Davis, W. Xiao, M. Mindrinos, R. Maier, L. Moldawer, J. Cuschieri, L. Hennessy, E. Moore, J. Minei, P. Bankey, J. Johnson, J. Sperry, A. Nathens, T. Billiar, M. West, B. Brownstein, D. Herndon, H. Baker, C. Finnerty, M. Jeschke, M. Lopez, M. Klein, R. Gamelli, N. Gibran, B. Arnoldo, G. McDonald-Smith, D. Schoenfeld, J. Cobb, S. Warren, A. Cuenca, S. Lowry, S. Calvano, D. Hayden, and P. Mason were responsible for data acquisition. R. Tompkins, R. Davis, W. Xiao, J. Seok, M. Mindrinos, H. Gao, R. Maier, L. Moldawer, J. Cuschieri, L. Hennessy, E. Moore, J. Minei, P. Bankey, J. Johnson, J. Sperry, A. Nathens, T. Billiar, M. West, B. Brownstein, D. Herndon, H. Baker, C. Finnerty, M. Jeschke, M. Lopez, M. Klein, R. Gamelli, N. Gibran, B. Arnoldo, G. McDonald-Smith, D. Schoenfeld, J. Cobb, S. Warren, A. Cuenca, S. Lowry, S. Calvano, D. Hayden, P. Mason, and J. Storey were responsible for data analysis and/or interpretation. R. Tompkins, R. Davis, W. Xiao, J. Seok, M. Mindrinos, H. Gao, R. Maier, L. Moldawer, J. Cuschieri, G. McDonald-Smith, D. Schoenfeld, D. Hayden, S. Warren, and A. Cuenca were responsible for drafting or critical revision of the manuscript. W. Xiao, J. Seok, M. Mindrinos, H. Gao, D. Hayden, and D. Schoenfeld were responsible for statistical analysis. The Inflammation and Host Response to Injury Large-Scale Collaborative Research Program is also comprised of Lily Altstein, Ulysses G.J. Balis, David G. Camp II, Asit K. De, Brian G. Harbrecht, Shari E. Honari, Bruce A. McKinley, Carol L. Miller-Graziano, Frederick A. Moore, Grant E. O’Keefe, Laurence G. Rahme, Daniel G. Remick, Michael B. Shapiro, Richard D. Smith, Robert Tibshirani, Mehmet Toner, Bram Wispelwey, and Wing H. Wong.
Footnotes
Abbreviations used:
- CARS
- compensatory antiinflammatory response syndrome
- FDR
- false discovery rate
- ISS
- injury severity score
- MODS
- multiple-organ dysfunction syndrome
- PAMP
- pathogen-associated molecular pattern
- SIRS
- systemic inflammatory response syndrome
References
- Arbeitman M.N., Furlong E.E., Imam F., Johnson E., Null B.H., Baker B.S., Krasnow M.A., Scott M.P., Davis R.W., White K.P. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science. 297:2270–2275 10.1126/science.1072152 [DOI] [PubMed] [Google Scholar]
- Baccala R., Gonzalez-Quintial R., Lawson B.R., Stern M.E., Kono D.H., Beutler B., Theofilopoulos A.N. 2009. Sensors of the innate immune system: their mode of action. Nat Rev Rheumatol. 5:448–456 10.1038/nrrheum.2009.136 [DOI] [PubMed] [Google Scholar]
- Bone R.C. 1996a. A piece of my mind. Maumee: my Walden pond. JAMA. 276:1931 10.1001/jama.276.24.1931 [DOI] [PubMed] [Google Scholar]
- Bone R.C. 1996b. Sir Isaac Newton, sepsis, SIRS, and CARS. Crit. Care Med. 24:1125–1128 10.1097/00003246-199607000-00010 [DOI] [PubMed] [Google Scholar]
- Calvano S.E., Xiao W., Richards D.R., Felciano R.M., Baker H.V., Cho R.J., Chen R.O., Brownstein B.H., Cobb J.P., Tschoeke S.K., et al. ; Inflamm and Host Response to Injury Large Scale Collaborative Research Program 2005. A network-based analysis of systemic inflammation in humans. Nature. 437:1032–1037 10.1038/nature03985 [DOI] [PubMed] [Google Scholar]
- Cuschieri J., Freeman B., O’Keefe G., Harbrecht B.G., Bankey P., Johnson J.L., Minei J.P., Sperry J., West M., Nathens A., et al. ; Inflammation and the Host Response to Injury Collaborative Research Program 2008. Inflammation and the host response to injury a large-scale collaborative project: patient-oriented research core standard operating procedure for clinical care X. Guidelines for venous thromboembolism prophylaxis in the trauma patient. J. Trauma. 65:944–950 10.1097/TA.0b013e3181826df7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino R.B., Jr 1998. Propensity score methods for bias reduction in the comparison of a treatment to a non-randomized control group. Stat. Med. 17:2265–2281 [DOI] [PubMed] [Google Scholar]
- Davidson G.H., Hamlat C.A., Rivara F.P., Koepsell T.D., Jurkovich G.J., Arbabi S. 2011. Long-term survival of adult trauma patients. JAMA. 305:1001–1007 10.1001/jama.2011.259 [DOI] [PubMed] [Google Scholar]
- DeLong W.G., Jr, Born C.T. 2004. Cytokines in patients with polytrauma. Clin. Orthop. Relat. Res. (422):57–65 10.1097/01.blo.0000130840.64528.1e [DOI] [PubMed] [Google Scholar]
- Dewar D., Moore F.A., Moore E.E., Balogh Z. 2009. Postinjury multiple organ failure. Injury. 40:912–918 10.1016/j.injury.2009.05.024 [DOI] [PubMed] [Google Scholar]
- Evans H.L., Cuschieri J., Moore E.E., Shapiro M.B., Nathens A.B., Johnson J.L., Harbrecht B.G., Minei J.P., Bankey P.E., Maier R.V., West M.A.; Inflammation and Host Response to Injury Investigators 2009. Inflammation and the host response to injury, a Large-Scale Collaborative Project: patient-oriented research core standard operating procedures for clinical care IX. Definitions for complications of clinical care of critically injured patients. J. Trauma. 67:384–388 10.1097/TA.0b013e3181ad66a7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feezor R.J., Baker H.V., Mindrinos M., Hayden D., Tannahill C.L., Brownstein B.H., Fay A., MacMillan S., Laramie J., Xiao W., et al. ; Inflammation and Host Response to Injury, Large-Scale Collaborative Research Program 2004. Whole blood and leukocyte RNA isolation for gene expression analyses. Physiol. Genomics. 19:247–254 10.1152/physiolgenomics.00020.2004 [DOI] [PubMed] [Google Scholar]
- Flohé S.B., Flohé S., Schade F.U. 2008. Invited review: Deterioration of the immune system after trauma: signals and cellular mechanisms. Innate Immun. 14:333–344 10.1177/1753425908100016 [DOI] [PubMed] [Google Scholar]
- Giannoudis P.V. 2003. Current concepts of the inflammatory response after major trauma: An update. Injury. 34:397–404 10.1016/S0020-1383(02)00416-3 [DOI] [PubMed] [Google Scholar]
- Giannoudis P.V., Hildebrand F., Pape H.C. 2004. Inflammatory serum markers in patients with multiple trauma. Can they predict outcome? J. Bone Joint Surg. Br. 86:313–323 10.1302/0301-620X.86B3.15035 [DOI] [PubMed] [Google Scholar]
- Harbrecht B.G., Minei J.P., Shapiro M.B., Nathens A.B., Moore E.E., West M.A., Bankey P.E., Cuschieri J., Johnson J.L., Maier R.V.; Inflammation and the Host Response to Injury Scale Collaborative Research Project 2007. Inflammation and the host response to injury, a large-scale collaborative project: patient-oriented research core-standard operating procedures for clinical care: VI. Blood glucose control in the critically ill trauma patient. J. Trauma. 63:703–708 10.1097/TA.0b013e31811eadea [DOI] [PubMed] [Google Scholar]
- Hayes J.R., Groner J.I. 2008. Using multiple imputation and propensity scores to test the effect of car seats and seat belt usage on injury severity from trauma registry data. J. Pediatr. Surg. 43:924–927 10.1016/j.jpedsurg.2007.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E., Fung E., Wildenhain J., Pierce S.E., Hoon S., Lee W., Proctor M., St Onge R.P., Tyers M., Koller D., et al. 2008. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science. 320:362–365 10.1126/science.1150021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss R.S., Karl I.E. 2003. The pathophysiology and treatment of sepsis. N. Engl. J. Med. 348:138–150 10.1056/NEJMra021333 [DOI] [PubMed] [Google Scholar]
- Hotchkiss R.S., Strasser A., McDunn J.E., Swanson P.E. 2009. Cell death. N. Engl. J. Med. 361:1570–1583 10.1056/NEJMra0901217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keel M., Trentz O. 2005. Pathophysiology of polytrauma. Injury. 36:691–709 10.1016/j.injury.2004.12.037 [DOI] [PubMed] [Google Scholar]
- Laudanski K., Miller-Graziano C., Xiao W., Mindrinos M.N., Richards D.R., De A., Moldawer L.L., Maier R.V., Bankey P., Baker H.V., et al. 2006. Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways. Proc. Natl. Acad. Sci. USA. 103:15564–15569 10.1073/pnas.0607028103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Qian W.J., Gritsenko M.A., Xiao W., Moldawer L.L., Kaushal A., Monroe M.E., Varnum S.M., Moore R.J., Purvine S.O., et al. ; Inflammation and the Host Response to Injury Large Scale Collaborative Research Programm 2006. High dynamic range characterization of the trauma patient plasma proteome. Mol. Cell. Proteomics. 5:1899–1913 10.1074/mcp.M600068-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massachusetts General Hospital 2011. The Human Genomic Response to Severe Traumatic Injury: An Interactive Website for Exploring Gene Expression and Clinical Outcomes. Available at: http://web.mgh.harvard.edu/TRT/
- Minei J.P., Nathens A.B., West M., Harbrecht B.G., Moore E.E., Shapiro M.B., Bankey P.E., Johnson J.L., Freeman B., McKinley B.A., et al. ; Inflammation and the Host Response to Injury Large Scale Collaborative Research Program Investigators 2006. Inflammation and the Host Response to Injury, a Large-Scale Collaborative Project: patient-oriented research core—standard operating procedures for clinical care. II. Guidelines for prevention, diagnosis and treatment of ventilator-associated pneumonia (VAP) in the trauma patient. J. Trauma. 60:1106–1113 10.1097/01.ta.0000220424.34835.f1 [DOI] [PubMed] [Google Scholar]
- Moore F.A., McKinley B.A., Moore E.E., Nathens A.B., West M., Shapiro M.B., Bankey P., Freeman B., Harbrecht B.G., Johnson J.L., et al. ; Inflammation and the Host Response to Injury Collaborative Research Program 2006. Inflammation and the Host Response to Injury, a large-scale collaborative project: patient-oriented research core—standard operating procedures for clinical care. III. Guidelines for shock resuscitation. J. Trauma. 61:82–89 10.1097/01.ta.0000225933.08478.65 [DOI] [PubMed] [Google Scholar]
- Nast-Kolb D., Aufmkolk M., Rucholtz S., Obertacke U., Waydhas C. 2001. Multiple organ failure still a major cause of morbidity but not mortality in blunt multiple trauma. J. Trauma. 51:835–841 10.1097/00005373-200111000-00003 [DOI] [PubMed] [Google Scholar]
- Nathan C., Ding A. 2010. Nonresolving inflammation. Cell. 140:871–882 10.1016/j.cell.2010.02.029 [DOI] [PubMed] [Google Scholar]
- Nathens A.B., Johnson J.L., Minei J.P., Moore E.E., Shapiro M., Bankey P., Freeman B., Harbrecht B.G., Lowry S.F., McKinley B., et al. ; Inflammation and the Host Response to Injury Investigators 2005. Inflammation and the Host Response to Injury, a large-scale collaborative project: Patient-Oriented Research Core—standard operating procedures for clinical care. I. Guidelines for mechanical ventilation of the trauma patient. J. Trauma. 59:764–769 [PubMed] [Google Scholar]
- O’Keefe G.E., Shelton M., Cuschieri J., Moore E.E., Lowry S.F., Harbrecht B.G., Maier R.V.; Inflammation and the Host Response to Injury Collaborative Research Program 2008. Inflammation and the host response to injury, a large-scale collaborative project: patient-oriented research core—standard operating procedures for clinical care VIII—Nutritional support of the trauma patient. J. Trauma. 65:1520–1528 10.1097/TA.0b013e3181904b0c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst C., Pape H.C., Hildebrand F., Regel G., Mahlke L., Giannoudis P., Krettek C., Grotz M.R. 2009. 30 years of polytrauma care: An analysis of the change in strategies and results of 4849 cases treated at a single institution. Injury. 40:77–83 10.1016/j.injury.2008.10.004 [DOI] [PubMed] [Google Scholar]
- Puneet P., Yap C.T., Wong L., Lam Y., Koh D.R., Moochhala S., Pfeilschifter J., Huwiler A., Melendez A.J. 2010. SphK1 regulates proinflammatory responses associated with endotoxin and polymicrobial sepsis. Science. 328:1290–1294 10.1126/science.1188635 [DOI] [PubMed] [Google Scholar]
- Sasser S.M., Varghese M., Joshipura M., Kellermann A. 2006. Preventing death and disability through the timely provision of prehospital trauma care. Bull. World Health Organ. 84:507 10.2471/BLT.06.033605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauaia A., Moore F.A., Moore E.E., Haenel J.B., Read R.A., Lezotte D.C. 1994. Early predictors of postinjury multiple organ failure. Arch. Surg. 129:39–45 [DOI] [PubMed] [Google Scholar]
- Storey J.D., Xiao W., Leek J.T., Tompkins R.G., Davis R.W. 2005. Significance analysis of time course microarray experiments. Proc. Natl. Acad. Sci. USA. 102:12837–12842 10.1073/pnas.0504609102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S., Hughes J.D., Campbell M.J., Cho R.J., Church G.M. 1999. Systematic determination of genetic network architecture. Nat. Genet. 22:281–285 10.1038/10343 [DOI] [PubMed] [Google Scholar]
- West M.A., Shapiro M.B., Nathens A.B., Johnson J.L., Moore E.E., Minei J.P., Bankey P.E., Freeman B., Harbrecht B.G., McKinley B.A., et al. ; Inflammation and the Host Response to Injury Collaborative Research Program 2006. Inflammation and the host response to injury, a large-scale collaborative project: Patient-oriented research core-standard operating procedures for clinical care. IV. Guidelines for transfusion in the trauma patient. J. Trauma. 61:436–439 10.1097/01.ta.0000232517.83039.c4 [DOI] [PubMed] [Google Scholar]
- West M.A., Moore E.E., Shapiro M.B., Nathens A.B., Cuschieri J., Johnson J.L., Harbrecht B.G., Minei J.P., Bankey P.E., Maier R.V.; Inflammation and the Host Response to Injury Collaborative Research Program 2008. Inflammation and the host response to injury, a large-scale collaborative project: patient-oriented research core—standard operating procedures for clinical care VII—Guidelines for antibiotic administration in severely injured patients. J. Trauma. 65:1511–1519 10.1097/TA.0b013e318184ee35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhang X., Pelayo R., Monestier M., Ammollo C.T., Semeraro F., Taylor F.B., Esmon N.L., Lupu F., Esmon C.T. 2009. Extracellular histones are major mediators of death in sepsis. Nat. Med. 15:1318–1321 10.1038/nm.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Raoof M., Chen Y., Sumi Y., Sursal T., Junger W., Brohi K., Itagaki K., Hauser C.J. 2010. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature. 464:104–107 10.1038/nature08780 [DOI] [PMC free article] [PubMed] [Google Scholar]